

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0559.5
(263 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
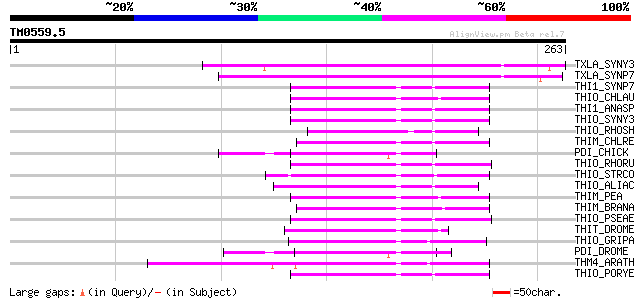
Score E
Sequences producing significant alignments: (bits) Value
TXLA_SYNY3 (P73920) Thiol:disulfide interchange protein txlA hom... 131 2e-30
TXLA_SYNP7 (P35088) Thiol:disulfide interchange protein txlA 121 2e-27
THI1_SYNP7 (P12243) Thioredoxin 1 (TRX-1) (Thioredoxin M) 62 1e-09
THIO_CHLAU (Q7M1B9) Thioredoxin (TRX) 59 9e-09
THI1_ANASP (P06544) Thioredoxin 1 (TRX-1) (Thioredoxin M) 59 1e-08
THIO_SYNY3 (P52231) Thioredoxin (TRX) 59 2e-08
THIO_RHOSH (P08058) Thioredoxin (TRX) 58 3e-08
THIM_CHLRE (P23400) Thioredoxin M-type, chloroplast precursor (T... 58 3e-08
PDI_CHICK (P09102) Protein disulfide-isomerase precursor (EC 5.3... 58 3e-08
THIO_RHORU (P10473) Thioredoxin (TRX) 57 3e-08
THIO_STRCO (P52230) Thioredoxin (TRX) 57 5e-08
THIO_ALIAC (P80579) Thioredoxin (TRX) 57 5e-08
THIM_PEA (P48384) Thioredoxin M-type, chloroplast precursor (TRX-M) 57 5e-08
THIM_BRANA (Q9XGS0) Thioredoxin M-type, chloroplast precursor (T... 57 6e-08
THIO_PSEAE (Q9X2T1) Thioredoxin (TRX) 56 1e-07
THIT_DROME (Q8IFW4) Thioredoxin T (ThioredoxinT) 55 1e-07
THIO_GRIPA (P50338) Thioredoxin 55 1e-07
PDI_DROME (P54399) Protein disulfide-isomerase precursor (EC 5.3... 55 1e-07
THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast precursor ... 55 2e-07
THIO_PORYE (P50254) Thioredoxin 55 2e-07
>TXLA_SYNY3 (P73920) Thiol:disulfide interchange protein txlA
homolog
Length = 180
Score = 131 bits (329), Expect = 2e-30
Identities = 68/174 (39%), Positives = 100/174 (57%), Gaps = 3/174 (1%)
Query: 92 PTKDINRKVAIASTLAALGLFLATRLDF-GVSLKDLTANALPYEQALSNGKPTVVEFYAD 150
P K N +A+ + + ++L + G+SL+ A+P AL NG+PT+VEFYAD
Sbjct: 3 PAKIRNALLAVVAIALSAAVYLGFQTQTQGISLEAQAQRAIPLATALDNGRPTLVEFYAD 62
Query: 151 WCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEE 210
WC C+ +APD+ ++++ Y VNF MLNVDN W E+ + V+GIPHF +LD G
Sbjct: 63 WCTSCQAMAPDLAELKKNYGGSVNFAMLNVDNNKWLPEVLRYRVDGIPHFVYLDDTGTAI 122
Query: 211 GNVVGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVH-QVVDPRSHG 263
+G P ++L +N+ AL E +P+A V G+ S E R + PRSHG
Sbjct: 123 AESIGEQPLRVLEQNITALVAHE-PIPYANVTGQTSVVENRTIEADPTSPRSHG 175
>TXLA_SYNP7 (P35088) Thiol:disulfide interchange protein txlA
Length = 191
Score = 121 bits (303), Expect = 2e-27
Identities = 62/166 (37%), Positives = 93/166 (55%), Gaps = 4/166 (2%)
Query: 100 VAIASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELA 159
+A A L L + + + SL L A PYE A++N +P ++EFYADWC C+ +A
Sbjct: 17 IAAALVLTILVVLGSRQPSAAASLASLAEQATPYEVAIANDRPMLLEFYADWCTSCQAMA 76
Query: 160 PDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPR 219
+ ++Q Y DR++FVMLN+DN W E+ ++ V+GIP F +L+ +G +G +G LPR
Sbjct: 77 GRIAALKQDYSDRLDFVMLNIDNDKWLPEVLDYNVDGIPQFVYLNGQGQPQGISIGELPR 136
Query: 220 QLLLENVDALARGEASVPHARVVGKYSSAEA---RKVHQVVDPRSH 262
+L N+DAL + +P+ G S A DPRSH
Sbjct: 137 SVLAANLDALVEAQ-PLPYTNARGNLSEFSADLQPSRSSQTDPRSH 181
>THI1_SYNP7 (P12243) Thioredoxin 1 (TRX-1) (Thioredoxin M)
Length = 106
Score = 62.4 bits (150), Expect = 1e-09
Identities = 32/94 (34%), Positives = 50/94 (53%), Gaps = 3/94 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
++ L + P +V+F+A WC CR +AP V +I QQY D+V V +N D ++G
Sbjct: 13 QEVLESSIPVLVDFWAPWCGPCRMVAPVVDEIAQQYSDQVKVVKVNTDEN--PSVASQYG 70
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
+ IP + K+G VVG +P+ L +D
Sbjct: 71 IRSIPTL-MIFKDGQRVDTVVGAVPKTTLANTLD 103
>THIO_CHLAU (Q7M1B9) Thioredoxin (TRX)
Length = 109
Score = 59.3 bits (142), Expect = 9e-09
Identities = 31/94 (32%), Positives = 49/94 (51%), Gaps = 3/94 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E+ L + P VV+F+A WC CR +AP + K+ +Y R+ +N D+ Q + G
Sbjct: 13 EKVLQSKTPVVVDFWAPWCGPCRVIAPILDKLAGEYAGRLTIAKVNTDDN--VQYASQLG 70
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
++G+P K+G E G +VG P + E D
Sbjct: 71 LKGLPTLVIF-KDGREVGRLVGARPEAMYREIFD 103
>THI1_ANASP (P06544) Thioredoxin 1 (TRX-1) (Thioredoxin M)
Length = 106
Score = 58.9 bits (141), Expect = 1e-08
Identities = 30/94 (31%), Positives = 52/94 (54%), Gaps = 3/94 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
++ L + P +V+F+A WC CR +AP V +I QQY+ ++ V +N D Q ++G
Sbjct: 13 QEVLDSDVPVLVDFWAPWCGPCRMVAPVVDEIAQQYEGKIKVVKVNTDEN--PQVASQYG 70
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
+ IP + K G + VVG +P+ L + ++
Sbjct: 71 IRSIPTL-MIFKGGQKVDMVVGAVPKTTLSQTLE 103
>THIO_SYNY3 (P52231) Thioredoxin (TRX)
Length = 106
Score = 58.5 bits (140), Expect = 2e-08
Identities = 32/94 (34%), Positives = 48/94 (51%), Gaps = 3/94 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E L + P +V+F+A WC CR +AP V +I QQY+ +V V LN D ++G
Sbjct: 13 EDVLDSELPVLVDFWAPWCGPCRMVAPVVDEISQQYEGKVKVVKLNTDEN--PNTASQYG 70
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
+ IP + K G VVG +P+ L ++
Sbjct: 71 IRSIPTL-MIFKGGQRVDMVVGAVPKTTLASTLE 103
>THIO_RHOSH (P08058) Thioredoxin (TRX)
Length = 105
Score = 57.8 bits (138), Expect = 3e-08
Identities = 30/81 (37%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query: 142 PTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFA 201
P VV+F+A+WC CR++ P + ++ ++Y +V V +NVD + GV GIP
Sbjct: 20 PVVVDFWAEWCGPCRQIGPALEELSKEYAGKVKIVKVNVDENPESPAM--LGVRGIPAL- 76
Query: 202 FLDKEGNEEGNVVGRLPRQLL 222
FL K G N VG P+ L
Sbjct: 77 FLFKNGQVVSNKVGAAPKAAL 97
>THIM_CHLRE (P23400) Thioredoxin M-type, chloroplast precursor
(TRX-M) (Thioredoxin CH2)
Length = 140
Score = 57.8 bits (138), Expect = 3e-08
Identities = 29/91 (31%), Positives = 51/91 (55%), Gaps = 3/91 (3%)
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L + P +V+F+A WC CR +AP V +I +YKD++ V LN D + E+G+
Sbjct: 49 LESSVPVLVDFWAPWCGPCRIIAPVVDEIAGEYKDKLKCVKLNTDES--PNVASEYGIRS 106
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
IP + K G + ++G +P+ +++ V+
Sbjct: 107 IPTI-MVFKGGKKCETIIGAVPKATIVQTVE 136
>PDI_CHICK (P09102) Protein disulfide-isomerase precursor (EC
5.3.4.1) (PDI) (Prolyl 4-hydroxylase beta subunit)
(Cellular thyroid hormone binding protein) (Retina
cognin) (R-cognin)
Length = 515
Score = 57.8 bits (138), Expect = 3e-08
Identities = 39/104 (37%), Positives = 52/104 (49%), Gaps = 5/104 (4%)
Query: 100 VAIASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELA 159
VA+ +AALGL + GV L A +EQAL+ + +VEFYA WC C+ LA
Sbjct: 10 VALLCLVAALGLAEPLEEEDGV----LVLRAANFEQALAAHRHLLVEFYAPWCGHCKALA 65
Query: 160 PDVYKIEQQYKDRVNFVML-NVDNTNWEQELDEFGVEGIPHFAF 202
P+ K Q K + + L VD T + +FGV G P F
Sbjct: 66 PEYAKAAAQLKAEGSEIRLAKVDATEEAELAQQFGVRGYPTIKF 109
Score = 48.1 bits (113), Expect = 2e-05
Identities = 24/69 (34%), Positives = 35/69 (49%), Gaps = 2/69 (2%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E A K VEFYA WC C++LAP K+ + Y+D N V+ +D+T E++
Sbjct: 384 EVAFDENKNVFVEFYAPWCGHCKQLAPIWDKLGETYRDHENIVIAKMDST--ANEVEAVK 441
Query: 194 VEGIPHFAF 202
+ P F
Sbjct: 442 IHSFPTLKF 450
>THIO_RHORU (P10473) Thioredoxin (TRX)
Length = 104
Score = 57.4 bits (137), Expect = 3e-08
Identities = 31/95 (32%), Positives = 48/95 (49%), Gaps = 3/95 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E L P V+F+A+WC CR+ AP + ++ D+V +N+D Q ++G
Sbjct: 11 EDVLKADGPNXVDFWAEWCGPCRQXAPALEELATALGDKVTVAKINIDEN--PQTPSKYG 68
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDA 228
V GIP + K+G +G LP+ L E V+A
Sbjct: 69 VRGIPTL-MIFKDGQVAATKIGALPKTKLFEWVEA 102
>THIO_STRCO (P52230) Thioredoxin (TRX)
Length = 110
Score = 57.0 bits (136), Expect = 5e-08
Identities = 31/106 (29%), Positives = 57/106 (53%), Gaps = 4/106 (3%)
Query: 122 SLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVD 181
+LK +T ++ + L N KP +V+F+A WC CR++AP + I +Y D++ V LN+D
Sbjct: 4 TLKHVTDDSFE-QDVLKNDKPVLVDFWAAWCGPCRQIAPSLEAIAAEYGDKIEIVKLNID 62
Query: 182 NTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
++GV IP + + G +VG P+ ++ +++
Sbjct: 63 EN--PGTAAKYGVMSIPTLN-VYQGGEVAKTIVGAKPKAAIVRDLE 105
>THIO_ALIAC (P80579) Thioredoxin (TRX)
Length = 105
Score = 57.0 bits (136), Expect = 5e-08
Identities = 30/97 (30%), Positives = 50/97 (50%), Gaps = 3/97 (3%)
Query: 126 LTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNW 185
+T ++QA+ KP +V+F+A WC CR +AP + + + + D+V LNVD
Sbjct: 3 MTLTDANFQQAIQGDKPVLVDFWAAWCGPCRMMAPVLEEFAEAHADKVTVAKLNVDEN-- 60
Query: 186 EQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLL 222
+ +FG+ IP L K G ++G P++ L
Sbjct: 61 PETTSQFGIMSIPTL-ILFKGGRPVKQLIGYQPKEQL 96
>THIM_PEA (P48384) Thioredoxin M-type, chloroplast precursor (TRX-M)
Length = 172
Score = 57.0 bits (136), Expect = 5e-08
Identities = 27/94 (28%), Positives = 50/94 (52%), Gaps = 3/94 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E + + P +V+F+A WC CR +AP + ++ ++Y ++ LN D + ++G
Sbjct: 79 ELVIGSETPVLVDFWAPWCGPCRMIAPIIDELAKEYAGKIKCYKLNTDES--PNTATKYG 136
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
+ IP F K G + +V+G +P+ L E V+
Sbjct: 137 IRSIPTVLFF-KNGERKDSVIGAVPKATLSEKVE 169
>THIM_BRANA (Q9XGS0) Thioredoxin M-type, chloroplast precursor
(TRX-M)
Length = 177
Score = 56.6 bits (135), Expect = 6e-08
Identities = 28/91 (30%), Positives = 49/91 (53%), Gaps = 3/91 (3%)
Query: 137 LSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEG 196
L +P VV+F+A WC C+ + P V ++ QQY ++ F LN D++ ++GV
Sbjct: 86 LKADEPVVVDFWAPWCGPCKMIDPIVNELAQQYTGKIKFFKLNTDDS--PATPGKYGVRS 143
Query: 197 IPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
IP K G ++ ++G +P+ L ++D
Sbjct: 144 IPTIMIFVK-GEKKDTIIGAVPKTTLATSID 173
>THIO_PSEAE (Q9X2T1) Thioredoxin (TRX)
Length = 108
Score = 55.8 bits (133), Expect = 1e-07
Identities = 29/95 (30%), Positives = 51/95 (53%), Gaps = 3/95 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
+ L P +V+++A+WC C+ +AP + ++ + Y+ ++ LN+D + ++G
Sbjct: 15 QDVLKADGPVLVDYWAEWCGPCKMIAPVLDEVARDYQGKLKVCKLNIDEN--QDTPPKYG 72
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDA 228
V GIP L K+GN E VG L + L +DA
Sbjct: 73 VRGIPTL-MLFKDGNVEATKVGALSKSQLAAFLDA 106
>THIT_DROME (Q8IFW4) Thioredoxin T (ThioredoxinT)
Length = 157
Score = 55.5 bits (132), Expect = 1e-07
Identities = 29/78 (37%), Positives = 44/78 (56%), Gaps = 3/78 (3%)
Query: 131 LPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELD 190
L + L+ K V++FYADWC C+ +AP + ++ +Y DRV + +NVD E
Sbjct: 11 LDQQLILAEDKLVVIDFYADWCGPCKIIAPKLDELAHEYSDRVVVLKVNVDEN--EDITV 68
Query: 191 EFGVEGIPHFAFLDKEGN 208
E+ V +P F F+ K GN
Sbjct: 69 EYNVNSMPTFVFI-KGGN 85
>THIO_GRIPA (P50338) Thioredoxin
Length = 109
Score = 55.5 bits (132), Expect = 1e-07
Identities = 28/94 (29%), Positives = 50/94 (52%), Gaps = 3/94 (3%)
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
+E+ +++ +P +V+F+A WC CR +A + +I YKD++ V +N D E+
Sbjct: 12 HEEVINSRQPVLVDFWAPWCGPCRMIASTIDEIAHDYKDKLKVVKVNTDQN--PTIATEY 69
Query: 193 GVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENV 226
G+ IP + G + VVG +P+ LL +
Sbjct: 70 GIRSIP-TVMIFINGKKVDTVVGAVPKLTLLNTL 102
>PDI_DROME (P54399) Protein disulfide-isomerase precursor (EC
5.3.4.1) (PDI)
Length = 496
Score = 55.5 bits (132), Expect = 1e-07
Identities = 26/74 (35%), Positives = 41/74 (55%), Gaps = 2/74 (2%)
Query: 136 ALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVE 195
AL K +VEFYA WC C++LAP ++ ++YKD + V+ +D+T EL+ +
Sbjct: 381 ALDKSKSVLVEFYAPWCGHCKQLAPIYDQLAEKYKDNEDIVIAKMDST--ANELESIKIS 438
Query: 196 GIPHFAFLDKEGNE 209
P + KE N+
Sbjct: 439 SFPTIKYFRKEDNK 452
Score = 47.0 bits (110), Expect = 5e-05
Identities = 29/102 (28%), Positives = 52/102 (50%), Gaps = 5/102 (4%)
Query: 102 IASTLAALGLFLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPD 161
+A++ A +++ GV L A ++Q +++ + +VEFYA WC C+ LAP+
Sbjct: 10 LAASYVAASAEAEVKVEEGV----LVATVDNFKQLIADNEFVLVEFYAPWCGHCKALAPE 65
Query: 162 VYKIEQQYKDRVNFVML-NVDNTNWEQELDEFGVEGIPHFAF 202
K QQ ++ + + L VD T + +++ V G P F
Sbjct: 66 YAKAAQQLAEKESPIKLAKVDATVEGELAEQYAVRGYPTLKF 107
>THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast precursor
(TRX-M4)
Length = 193
Score = 55.1 bits (131), Expect = 2e-07
Identities = 39/175 (22%), Positives = 79/175 (44%), Gaps = 16/175 (9%)
Query: 66 PSTVNGESETCKPTSSTADASGLPQLPTKDINRKVAIASTLAALGLFLATRLDFGVSL-- 123
P + S + P+ S S L + + ++ + A+LG TR+ G +
Sbjct: 17 PIAASVSSSSAAPSVSRRRISPARFLEFRGLKSSRSLVTQSASLGANRRTRIARGGRIAC 76
Query: 124 --KDLTANALPYE---------QALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDR 172
+D TA A+ + L + P +VEF+A WC CR + P V ++ + + +
Sbjct: 77 EAQDTTAAAVEVPNLSDSEWQTKVLESDVPVLVEFWAPWCGPCRMIHPIVDQLAKDFAGK 136
Query: 173 VNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
F +N D + + +G+ +P + K G ++ +++G +PR+ L + ++
Sbjct: 137 FKFYKINTDES--PNTANRYGIRSVP-TVIIFKGGEKKDSIIGAVPRETLEKTIE 188
>THIO_PORYE (P50254) Thioredoxin
Length = 107
Score = 55.1 bits (131), Expect = 2e-07
Identities = 26/94 (27%), Positives = 50/94 (52%), Gaps = 3/94 (3%)
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
++ ++N P +V+F+A WC CR ++P V +I ++Y+ + V +N D+ E+G
Sbjct: 13 QEVINNNLPVLVDFWAPWCGPCRMVSPVVDEIAEEYESSIKVVKINTDDN--PTIAAEYG 70
Query: 194 VEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
+ IP + K G V+G +P+ L ++
Sbjct: 71 IRSIPTL-MIFKAGERVDTVIGAVPKSTLASTLN 103
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,136,944
Number of Sequences: 164201
Number of extensions: 1310392
Number of successful extensions: 3638
Number of sequences better than 10.0: 243
Number of HSP's better than 10.0 without gapping: 188
Number of HSP's successfully gapped in prelim test: 55
Number of HSP's that attempted gapping in prelim test: 3339
Number of HSP's gapped (non-prelim): 322
length of query: 263
length of database: 59,974,054
effective HSP length: 108
effective length of query: 155
effective length of database: 42,240,346
effective search space: 6547253630
effective search space used: 6547253630
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0559.5


