

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0550.1
(1631 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
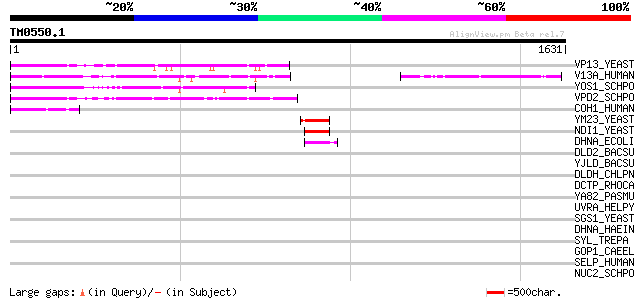
Score E
Sequences producing significant alignments: (bits) Value
VP13_YEAST (Q07878) Vacuolar protein sorting-associated protein ... 222 7e-57
V13A_HUMAN (Q96RL7) Vacuolar protein sorting 13A (Chorein) (Chor... 214 2e-54
YOS1_SCHPO (P87319) Hypothetical protein C21C3.01c in chromosome II 152 7e-36
VPD2_SCHPO (O42926) Putative vacuolar protein sorting-associated... 137 3e-31
COH1_HUMAN (Q7Z7G8) Cohen syndrome protein 1 76 7e-13
YM23_YEAST (P40215) Hypothetical 62.8 kDa protein in RPS16A-TIF3... 72 1e-11
NDI1_YEAST (P32340) Rotenone-insensitive NADH-ubiquinone oxidore... 59 1e-07
DHNA_ECOLI (P00393) NADH dehydrogenase (EC 1.6.99.3) 48 2e-04
DLD2_BACSU (P54533) Dihydrolipoyl dehydrogenase (EC 1.8.1.4) (E3... 42 0.011
YJLD_BACSU (P80861) NADH dehydrogenase-like protein yjlD (EC 1.6... 37 0.58
DLDH_CHLPN (Q9Z773) Dihydrolipoyl dehydrogenase (EC 1.8.1.4) (E3... 37 0.58
DCTP_RHOCA (P37735) C4-dicarboxylate-binding periplasmic protein... 37 0.58
YA82_PASMU (Q9CLW7) Hypothetical protein PM1082 36 0.99
UVRA_HELPY (P56474) UvrABC system protein A (UvrA protein) (Exci... 35 1.3
SGS1_YEAST (P35187) Helicase SGS1 (Helicase TPS1) 35 1.7
DHNA_HAEIN (P44856) NADH dehydrogenase (EC 1.6.99.3) 35 1.7
SYL_TREPA (O83595) Leucyl-tRNA synthetase (EC 6.1.1.4) (Leucine-... 35 2.2
GOP1_CAEEL (P46578) Hypothetical protein gop-1 35 2.2
SELP_HUMAN (P49908) Selenoprotein P precursor (SeP) 34 3.8
NUC2_SCHPO (P10505) Nuclear scaffold-like protein p76 34 3.8
>VP13_YEAST (Q07878) Vacuolar protein sorting-associated protein
VPS13
Length = 3144
Score = 222 bits (565), Expect = 7e-57
Identities = 217/902 (24%), Positives = 385/902 (42%), Gaps = 141/902 (15%)
Query: 1 MLEDQVAYLLQRYLGNYVRGLNKEALKISVWKGDVELKNMQLKPEALNALKLPVKVKAGF 60
MLE A LL R LG+YV + L + +W GDV+LKN++L+ + L++L LP+ VK+G
Sbjct: 1 MLESLAANLLNRLLGSYVENFDPNQLNVGIWSGDVKLKNLKLRKDCLDSLNLPIDVKSGI 60
Query: 61 LGSVKLKVPWSRLGQDPVLVYLDRIFLLAEPATQVEGCSEDAVQEA---KKIRIEEMELK 117
LG + L VPWS L PV + ++ +LL P ++ E+ ++ A K ++ E EL
Sbjct: 61 LGDLVLTVPWSSLKNKPVKIIIEDCYLLCSPRSEDHENDEEMIKRAFRLKMRKVSEWELT 120
Query: 118 LWEKSQQLKSEMKTINPSSQ----GSAKKLFSTIIGNLKLSISNIHIRYE--DGESNPGH 171
+ +SE KT + SS+ G + L + II NL+++I NIH+RYE DG G
Sbjct: 121 NQARILSTQSENKTSSSSSEKNNAGFMQSLTTKIIDNLQVTIKNIHLRYEDMDGIFTTG- 179
Query: 172 PFAAGVMLDKLSAVTVDDTGKETFITGGALDLIQKLSSMDLFAYQRITKNINSFLNCKMR 231
P + G+ L++LSAV+ D +FI IT+NI L
Sbjct: 180 PSSVGLTLNELSAVSTDSNWAPSFID--------------------ITQNITHKL----- 214
Query: 232 IVKQPLNPKSLSCWSVELDRLAVYLDSDIIPWHASKEWEDLLPSEWFQIFKFGTKDG-KP 290
+ L+ L +Y ++D P + + + L + F G KD
Sbjct: 215 ---------------LTLNSLCLYWNTDSPPLISDDDQDRSLEN-----FVRGFKDMIAS 254
Query: 291 ADRLLQKHSYVLEPVTGKGNYSKLLLNEVADS-KQPLQKAVVNLDDVTISLSKDGYRDIM 349
+ KH Y+L+PV+G G KL +N++ + +QP + D+ + L Y DI+
Sbjct: 255 KNSTAPKHQYILKPVSGLG---KLSINKLGSTEEQPHIDLQMFYDEFGLELDDTEYNDIL 311
Query: 350 KLADNFAAFNQRLKYAHFRPPVPVKADPRSWWKYAYRAVSDQMKKASGKMSWEQVLRYTS 409
+ + K+ RP V +P W+KY V +++ + + +WE +
Sbjct: 312 HVLSSIQLRQITKKFKKARPSFAVSENPTEWFKYIAACVINEIHEKNKMWTWESMKEKCE 371
Query: 410 LRKRYIYLYASLLK--------SDPSQVTISGNKEIEDLDHELDIELILQWRMLAHK--- 458
R+ Y L+ LK DP Q ++ +L +L + I+ +R +A +
Sbjct: 372 QRRLYTKLWVEKLKLKNLEAPLRDPIQ-----EAQLSELHKDLTYDEIILFRSVAKRQYA 426
Query: 459 -----FVEQSAEPNLSVRKQ---------KAGNSWWSFGWTGKSPKEDSEELSFSEEDWN 504
E S P S + K SW S W GK +E E+L +EE
Sbjct: 427 QYKLGMTEDSPTPTASSNIEPQTSNKSATKNNGSWLSSWWNGKPTEEVDEDLIMTEEQRQ 486
Query: 505 RLNKIIGYKEGDDGQSPVNSKADVMHTFLVVHMNHNASKLIGEAQDL-VAELSCEDLSCS 563
L I + E +D + + + + + + + + Q+L + + E+
Sbjct: 487 ELYDAIEFDENEDKGPVLQVPRERVELRVTSLLKKGSFTIRKKKQNLNLGSIIFENCKVD 546
Query: 564 VKLYPETKVFDIKLGSYQL--SSPKGLL---------AESAASFDS-------------L 599
P++ + +L + L SP L ++ +S D+ L
Sbjct: 547 FAQRPDSFLSSFQLNKFSLEDGSPNALYKHIISVRNSSKDQSSIDNHATGEEEEEDEPLL 606
Query: 600 VGVFKYKPFDDKVDWSMVAKASPCYMTYMKDSINQIVKFFETNATVSQTIALETAAAVQL 659
F+ P D D ++ K + Y I ++ KFF+ + +TI A +
Sbjct: 607 RASFELNPLDGLADSNLNIKLLGMTVFYHVHFITEVHKFFKASNQHMETIG-NIVNAAEA 665
Query: 660 KIDEVKRTAQQQMNRALKDHARFSLDLDIAAPKITIPTDFYPDNTHATKLLLDLGNLMIR 719
++ + + L+DH ++ LD+ AP I +P D P + ++D G++ I
Sbjct: 666 TVEGWTTQTRMGIESLLEDHKTVNVSLDLQAPLIILPLD--PHDWDTPCAIIDAGHMSIL 723
Query: 720 TQ-------DDSRQESAEDN-----------MYLRFDLVLSDVSAFLFDGDYHWSEISVN 761
+ + ++ S E+ M+ RF ++ D F+ + ++
Sbjct: 724 SDLVPKEKIKEIKELSPEEYDKIDGNEINRLMFDRFQILSQDTQIFVGPD----IQSTIG 779
Query: 762 KLTHSTNTSFFPIIDRCGVILQLQQILLETPY-YPSTRLAVRLPSLAFHFSPARYHRLMH 820
K+ +++T+ F I+D+ + L + +L Y P+ R+ LP L+ + +Y +M+
Sbjct: 780 KINTASSTNDFRILDKMKLELTVDLSILPKAYKLPTIRVFGHLPRLSLSINDIQYKTIMN 839
Query: 821 VI 822
+I
Sbjct: 840 LI 841
>V13A_HUMAN (Q96RL7) Vacuolar protein sorting 13A (Chorein)
(Chorea-acanthocytosis protein)
Length = 3174
Score = 214 bits (545), Expect = 2e-54
Identities = 217/870 (24%), Positives = 375/870 (42%), Gaps = 128/870 (14%)
Query: 1 MLEDQVAYLLQRYLGNYVRGLNKEALKISVWKGDVELKNMQLKPEALNALKLPVKVKAGF 60
+ E V +L R+LG+YV L+ L + +WKG V LKN+Q+K AL+ L +P KVK G
Sbjct: 2 VFESVVVDVLNRFLGDYVVDLDTSQLSLGIWKGAVALKNLQIKENALSQLDVPFKVKVGH 61
Query: 61 LGSVKLKVPWSRLGQDPVLVYLDRIFLLAEPATQVEGCSEDAVQEAKKI-RIEEMELKLW 119
+G++KL +PW L PV L+ I+LL P+++++ D ++E K++ ++ ELK
Sbjct: 62 IGNLKLIIPWKNLYTQPVEAVLEEIYLLIVPSSRIK---YDPLKEEKQLMEAKQQELKRI 118
Query: 120 EKSQQLKSEMKTINPSSQGS-AKKLFSTIIGNLKLSISNIHIRYEDGESNPGHPFAAGVM 178
E+++Q + + P Q + A+KL + II NL++ IS+IHIRYED +N P + G+
Sbjct: 119 EEAKQKVVDQEQHLPEKQDTFAEKLVTQIIKNLQVKISSIHIRYEDDITNRDKPLSFGIS 178
Query: 179 LDKLSAVTVDDTGKETFITGGALDLIQKLSSMD-LFAYQRITKNINSFLNCKMRIVKQPL 237
L LS T D + L++KL +D LFAY
Sbjct: 179 LQNLSMQTTDQYWVPC-LHDETEKLVRKLIRLDNLFAY---------------------W 216
Query: 238 NPKSLSCWSVELDRLAVYLDSDIIPWHASKEWEDLLPSEWFQIFKFGTKDGKPADRLLQK 297
N KS + + D L + I+ E+++P +
Sbjct: 217 NVKSQMFYLSDYDNSLDDLKNGIVN-------ENIVP---------------------EG 248
Query: 298 HSYVLEPVTGKGNYSKLLLNEVA--DSKQPLQKAVVNLDDVTISLSKDGYRDIMKLADNF 355
+ +V P++ +KL++N + D P + L ++ I +K Y IM+L ++
Sbjct: 249 YDFVFRPISAN---AKLVMNRRSDFDFSAPKINLEIELHNIAIEFNKPQYFSIMELLESV 305
Query: 356 AAFNQRLKYAHFRPPVPVKADPRSWWKYAYRAVSD-QMKKASGKMSWEQVLRYTSLRKRY 414
Q L Y F+P VP+ R WW YA V + + SW+ + ++ K+Y
Sbjct: 306 DMMAQNLPYRKFKPDVPLHHHAREWWAYAIHGVLEVNVCPRLWMWSWKHIRKHRQKVKQY 365
Query: 415 IYLYASLL--KSDPSQVTISGNKEIEDLDHELDIELILQWRMLAHKFVEQSAEP--NLSV 470
LY L K P ++ +S +E+L+ LD+ I R A V+++ V
Sbjct: 366 KELYKKKLTSKKPPGELLVS----LEELEKTLDVFNITIARQTAEVEVKKAGYKIYKEGV 421
Query: 471 RKQKAGNSW--WSFGWTGKSPKEDSEELS-------FSEEDWNRLNKIIGYKEGDDGQSP 521
+ + W W + W+ ++ E ++ + E+ L + IGY E P
Sbjct: 422 KDPEDNKGWFSWLWSWSEQNTNEQQPDVQPETLEEMLTPEEKALLYEAIGYSE--TAVDP 479
Query: 522 VNSKADVMHTF--------LVVHMNHNASKLIGEAQDLVAELSCEDLSCSVKLYPETKV- 572
K F +V+ NH +L+ ++ E+ S + P +
Sbjct: 480 TLLKTFEALKFFVHLKSMSIVLRENHQKPELV--------DIVIEEFSTLIVQRPGAQAI 531
Query: 573 -FDIKLGSYQL-----SSPKGLLAESAASFDSLVGV-FKYKPFDDKVDWSMVAKASPCYM 625
F+ K+ S+ + +S K L S SL + F+ P D+ V + +A P +
Sbjct: 532 KFETKIDSFHITGLPDNSEKPRLLSSLDDAMSLFQITFEINPLDETVSQRCIIEAEPLEI 591
Query: 626 TYMKDSINQIVKFFETNATVSQTIALETAAAVQLKIDEVKRTAQQQMNRALKDHARFSLD 685
Y ++N IV+FF V + AA K++E + + ++ L
Sbjct: 592 IYDARTVNSIVEFFRPPKEVH---LAQLTAATLTKLEEFRSKTATGLLYIIETQKVLDLK 648
Query: 686 LDIAAPKITIPTD--FYPDNTHATKLLLDLGNLMIRT----------QDDSRQESAEDNM 733
+++ A I +P D F P + LLLDLG+L + + Q ++ + D
Sbjct: 649 INLKASYIIVPQDGIFSPT---SNLLLLDLGHLKVTSKSRSELPDVKQGEANLKEIMDRA 705
Query: 734 YLRFDLVLSDVSAFLFDGDYHWSEISVNKLTHSTNTSFFPIIDRCGVILQLQQILLETPY 793
Y FD+ L+ V +W E KL+ ST P+ + L + ++
Sbjct: 706 YDSFDIQLTSVQLLYSRVGDNWRE--ARKLSVSTQHILVPM--HFNLELSKAMVFMDV-R 760
Query: 794 YPSTRLAVRLPSLAFHFSPARYHRLMHVIK 823
P ++ +LP ++ S + +M +I+
Sbjct: 761 MPKFKIYGKLPLISLRISDKKLQGIMELIE 790
Score = 53.5 bits (127), Expect = 5e-06
Identities = 94/493 (19%), Positives = 217/493 (43%), Gaps = 42/493 (8%)
Query: 1148 STKEHELNDTLE-SFVKAQIIIYDQNSTRYNNIDKQVIVTLATLTFFCRRPTILAIMEFI 1206
+T ++ + D L +VKA+ + D ST YNN+ + + V ++L +L + ++
Sbjct: 952 TTLDNTMEDLLTLEYVKAEKNVPDLKST-YNNVLQLIKVNFSSLDIHLHTEALLNTINYL 1010
Query: 1207 NSINIENGNLATSSDSSSTSMVKNDLSNDLDVLHVTTVEEHAVKGLLGKGKSRVMFSLTL 1266
+ N+ S+ S + + + DV +++ A+K L + + +
Sbjct: 1011 H-------NILPQSEEKSAPVSTTETEDKGDV-----IKKLALK--LSTNEDIITLQILA 1056
Query: 1267 KMAQAQILLMKENETKLACLSQESLLAEIKVFPSSFSIKAALGNLKISDDSLSSSHFYYW 1326
+++ QI + ++ + ++ + E L +E+ + PS I A L N+ + D +++ Y
Sbjct: 1057 ELSCLQIFI-QDQKCNISEIKIEGLDSEMIMRPSETEINAKLRNIIVLDSDITA--IYKK 1113
Query: 1327 ACDMRNPGGRSFVELEFTSFSCDDE--DYEGYDFSLFGELSEVRIVYLNRFLQEIVGYFM 1384
A + SF + + + D D + + + +V++ +FL I+ F+
Sbjct: 1114 AVYITGKEVFSFKMVSYMDATAGSAYTDMNVVDIQVNLIVGCIEVVFVTKFLYSILA-FI 1172
Query: 1385 GLVPNSPRSVVKVTDQVTNSEKWFSASDIEGSPAVKFDLSLRKPIILMPRNRDSLDFLRL 1444
+ +++ + T Q + S + D++++ P++++P++ S +
Sbjct: 1173 DNFQAAKQALAEATVQAAGMAATGVRELAQRSSRMALDINIKAPVVVIPQSPVSENVFVA 1232
Query: 1445 DIVHITVRNTFQWIGGSKSEINAVHLETLMVQVEDINLNVGTGTDLGESIIKDV---NGL 1501
D IT+ NTF I S+S V ++ + +++ ++ L + + D+ L
Sbjct: 1233 DFGLITMTNTFHMITESQSSPPPV-IDLITIKLSEMRLYRSRFINDAYQEVLDLLLPLNL 1291
Query: 1502 SVIIHRSL-RDLSHQFPSIEIIIKMEELKAAMSNKEYQIITECAVSN-FSEVPDIPSPL- 1558
V++ R+L + + P + +++ ++ +S ++ I + N + E SP
Sbjct: 1292 EVVVERNLCWEWYQEVPCFNVNAQLKPMEFILSQEDITTIFKTLHGNIWYEKDGSASPAV 1351
Query: 1559 --NQYSSKTLNGATDDIVPEVTSGADSVTTDV----EASVLLKIWVSINL----VELSLY 1608
+QYS+ +G T + + GA VT V ++L+K ++I+ + + LY
Sbjct: 1352 TKDQYSA--TSGVTTN-ASHHSGGATVVTAAVVEVHSRALLVKTTLNISFKTDDLTMVLY 1408
Query: 1609 TGISRDASLATVQ 1621
+ + AS V+
Sbjct: 1409 SPGPKQASFTDVR 1421
>YOS1_SCHPO (P87319) Hypothetical protein C21C3.01c in chromosome II
Length = 3011
Score = 152 bits (384), Expect = 7e-36
Identities = 167/763 (21%), Positives = 325/763 (41%), Gaps = 102/763 (13%)
Query: 1 MLEDQVAYLLQRYLGNYVRGLNKEALKISVWKGDVELKNMQLKPEALNALKLPVKVKAGF 60
MLE VA LL + LG+YV L+ + L I VW G V L N+++KPEAL+ L +P+++ +G
Sbjct: 1 MLEGLVAGLLNKILGSYVDNLDTKQLNIGVWGGHVSLHNLRIKPEALDKLGIPIEITSGL 60
Query: 61 LGSVKLKVPWSRLGQDPVLVYLDRIFLLAEPATQVEGCSEDAVQEAKKIRIEEME-LKLW 119
+G+ L++PWS L + + ++ I+L P + ++ Q + ++ E+++ ++
Sbjct: 61 IGTFTLEIPWSNLRNKSLTINIEDIYLSIHPQAKNSLTRDELEQSQQALKQEQLDSFEIL 120
Query: 120 EKSQQLKSEMKTINPS---SQGSAKKLFSTIIGNLKLSISNIHIRYEDGESNPGHPFAAG 176
K+ + E + NP+ Q + L + + N+++ I IH+R+ED S+ P++ G
Sbjct: 121 RKNFRETLEESSSNPNISRKQSFIEYLIAKLTDNIQIYIERIHLRFEDNLSDLEKPYSLG 180
Query: 177 VMLDKLSAVTVDDTGKETFITGGAL--DLIQKLSSMDLFAYQRITKNINSFLNCKMRIVK 234
+ L L + D + E ++ + I K+ ++D F+ I+K C++
Sbjct: 181 LTLYSLRVTSTDASFTEYLLSTDPIPSSCIHKIITVDYFSIYWISK-------CEIS--- 230
Query: 235 QPLNPKSLSCWSVELDRLAVYLDSDIIPWHASKEWEDLLPSEWFQIFKFGTKDGKPADRL 294
C + E DI + ++L+PS + PA
Sbjct: 231 --------KCTTTE----------DIFSY-----LKNLIPS----------AEKSPA--- 254
Query: 295 LQKHSYVLEPVTGKGNYSKLLLNEVADSKQPLQKAVVNLDDVTISLSKDGYRDIMKLADN 354
++Y+L+P+ + +L D L + +++++++I+LS Y ++ + D
Sbjct: 255 ---YNYILKPLRATAHV--VLFRHPTDQIMQL-RGKLSVEEISITLSDHMYYSLLGVIDY 308
Query: 355 FAAFNQRLKYAHFRPPVPVKADPRSWWKYAYRAVSDQMKKASGKMSWEQVLRYTSLRKRY 414
F ++ Y +RP K P W+KYA V D + ++ +W+ R R Y
Sbjct: 309 FRVVMKQQYYLQYRPKSTPKEKPLEWFKYAILVVKDSVHESRYHWTWKYFKRRRDDRIAY 368
Query: 415 IYLYAS-LLKSDPSQVTISGNKEIEDLDHELDIELILQWRMLAH-KFVEQSAEPNLSVRK 472
+++ L S+ I K+IE + D ++++R H +E+ L +
Sbjct: 369 MHIIRKRYLNEQISKEEIDLQKKIEKRNSTYD---LIKYRSRVHTSLIEERNSIYLKPKT 425
Query: 473 QKAGNSW-WSFGWTGKSPKEDSEELS-----------------FSEEDWNRLNKIIGYKE 514
A + W G+ K +D + L+ FS +W+
Sbjct: 426 SAAHGLYDWFSGYIRKPQSQDEDTLASTDKTAADLTDQEQKEFFSAIEWSG-QLYPDTVN 484
Query: 515 GDDGQSPVNSKADVMHTFLVV--HMNHNASKLIGEAQDLVAELSCEDLSCSVKLYPETKV 572
D N + + V+ H+N LI + + A C S+KL K
Sbjct: 485 LDPDMCMANVEVSIAKGSFVIQSHINGRVIPLIKQRFESFA-TECFIRPQSLKLKVSLKD 543
Query: 573 FDIKLG--SYQLSSPKGLLA----ESAASFDSLVGVFKYKPFDDKVDWSMVAKASPCYMT 626
D+ G + +L + + A E + S + ++ F +D V KAS +
Sbjct: 544 LDMFDGITNPELEPARVIFAKPSVEESESLQKIPEAYRTHLFFLLLDTKPVYKASSTLIV 603
Query: 627 YMKD--------SINQIVKFFETNATVSQTIALETAAAVQLKIDEVKRTAQQQMNRALKD 678
+++ I ++ FF T + ++ E + K+ + R + ++ AL+
Sbjct: 604 HLRTLVIIYNRVCIESLLAFFVPPRTKIEHVS-EWGYSAAAKVMTLARQTRASLDYALEM 662
Query: 679 HARFSLDLDIAAPKITIPTDFYPDNTHATKLLLDLGNLMIRTQ 721
H + +D+ AP I + + + + L LD+G ++ TQ
Sbjct: 663 HKTSDMTIDLQAPLIVVREEC--TDLKSPTLFLDVGRALVHTQ 703
>VPD2_SCHPO (O42926) Putative vacuolar protein sorting-associated
protein vps1302
Length = 3131
Score = 137 bits (344), Expect = 3e-31
Identities = 197/875 (22%), Positives = 364/875 (41%), Gaps = 110/875 (12%)
Query: 1 MLEDQVAYLLQRYLGNYVRGLNKEALKISVWKGDVELKNMQLKPEALNALKLPVKVKAGF 60
MLE +A L R LG Y+ + LK++VW GDV L+N+QLK A L+LP+ V+ G
Sbjct: 1 MLEGLLANFLNRLLGEYIENFDATQLKVAVWNGDVTLRNLQLKRSAFRKLELPISVQYGL 60
Query: 61 LGSVKLKVPWSRLGQDPVLVYLDRIFLLAEPATQVEGCSEDAVQEAKKIRIEEMELKLWE 120
+ + LK+PWS L PV +Y+ I LA V+ SE V + + + +++LWE
Sbjct: 61 VEELTLKIPWSSLKNKPVEIYIVGIRALASMEENVKSQSE--VDPHQVLESKRRQMQLWE 118
Query: 121 KSQQLKSEMKTINPSSQGSAKKLFSTIIGNLKLSISNIHIRYED---GESNPGHPFAAGV 177
SQ K+E +P +Q + L + +I N++++I +IHIR+E PG ++ G+
Sbjct: 119 ASQIGKAE-TAYDPKTQTFTESLITRMIDNIQINIRDIHIRFEHLPVSNVVPG-GYSFGL 176
Query: 178 MLDKLSAVTVDDTGKETFITGGALDLIQKLSSMDLFAYQRITKNINSFLNCKMRIVKQPL 237
+L + S + ++ TF+ T++ + C
Sbjct: 177 LLSEFSIESCNEEWTSTFVE---------------------TESQTIYKLC--------- 206
Query: 238 NPKSLSCWSVELDRLAVYLDSDIIPWHASKEWEDLLPSEWFQIFKFGTKDGKPADRLLQK 297
SL + + D A +D I +E F+ D + ++
Sbjct: 207 ---SLRGFGIYSDETAECIDKTDI-------------NELMNTFQALITDFQSREK---- 246
Query: 298 HSYVLEPVTGKGNYSKLLLNEVADSKQPLQKAVVNLDDVTISLSKDGYRDIMKLADNFAA 357
Y++ PVTG +K+ +N++ + P + ++ +SL+ M LA
Sbjct: 247 -DYIIAPVTG---MAKVTINKLPTPEIPRFLSQISFRGFDVSLNDSQISCGMSLAHELQD 302
Query: 358 FNQRLKYAHFRPPVPVKAD-PRSWWKYAYRAVSDQMKKASGKMSWEQVLRYTSLRKRYIY 416
+L + R V D P + ++ ++ +++ SW + + R+ YI
Sbjct: 303 VMSKLAFRK-RIVGNVSLDTPLDYLRFIFQKTLHDIQQKHYARSWPAIKAFCEKRRNYIK 361
Query: 417 LYASLLKSDPSQVTISGNKEIEDLDHELDIELILQWRMLAHKFVE-QSAEPNLSVRKQKA 475
LY K Q++ +KE++ L+ LDI + +R LA++ ++ + EP KQ+
Sbjct: 362 LYKK--KFLAVQLSADESKELDVLELNLDISQLKLFRSLAYQEIKNEGFEP--QPPKQQG 417
Query: 476 GNSWWSFGWTGKSPK--EDSEELSFSEEDWNRLN-KIIGYKEGDDGQSPVNSKADV---- 528
SW + G S + E +++ + D LN II D QS S DV
Sbjct: 418 WGSWMWNSFRGASDENGEVTDDQRKTVIDAIGLNDSIIESAHVGDLQSN-TSLFDVRLEV 476
Query: 529 -MHTFLVVHMNHNASKLIGEAQDLVAELSC--EDLSC-----SVKLYPETKVFDIKLGSY 580
F ++ + + E + ++ C +D S S+K+Y + + F Y
Sbjct: 477 PSGKFSLLKYPKKENVISLELLNFFSQFKCNKKDYSFVANLGSLKMYNDDRNF-----LY 531
Query: 581 QLSSPKGLLAESAASFDSLVGVFKYKPFDDKVDWSMVAKASPCYMTYMKDSINQIVKFFE 640
+ + ++ S +L F+ DD D + + ++ VK F+
Sbjct: 532 PRETSNKEIETTSPSIFTL--SFERSSSDDN-DTDTLGINLRALEFFYDPNLLLRVKGFQ 588
Query: 641 TNATVSQTIALETAAAVQLKIDEVKRTAQQQMNRALKDHARFSLDLDIAAPKITIPTDFY 700
++ + A D +T Q + L + + +++ P + P D
Sbjct: 589 SSLFANDNGRKLVRLANDAVTDFTSQTVQ-NLQEYLNEKRKLNVNFQFQTPLLIFPEDC- 646
Query: 701 PDNTHATKLLLDLGNLMIRTQ----DDSRQESAEDNMY-LRFDLV-LSDVSAFLFDGDYH 754
+N + L +D G + I +Q +D S + +Y L +D +S SA L G +
Sbjct: 647 -NNPESFSLFVDAGFISITSQNVETNDKESNSESETLYHLIYDRYRVSLESARLLLGPLN 705
Query: 755 WSEISVNKLTHSTNTSFFPIIDRCGVILQLQQILLETPYYPSTRLAVRLPSLAFHFSPAR 814
+ NK+ + +++ + +Q + TP YP ++ +P L+F S A+
Sbjct: 706 ELKNESNKINEE-----YYLMNELNTEIFIQAQRVPTPQYPRIKVEGNMPCLSFILSDAQ 760
Query: 815 YHRLMH----VIKIFEEGDDGSSEFLRPWNQADLE 845
+ L + ++K + DD + RP + L+
Sbjct: 761 FRILNNLASTMLKDGKASDDDDNGDWRPESSESLD 795
>COH1_HUMAN (Q7Z7G8) Cohen syndrome protein 1
Length = 4022
Score = 76.3 bits (186), Expect = 7e-13
Identities = 48/205 (23%), Positives = 95/205 (45%), Gaps = 8/205 (3%)
Query: 1 MLEDQVAYLLQRYLGNYVRGLNKEALKISVWKGDVELKNMQLKPEAL-NALKLPVKVKAG 59
MLE V +L Y+ Y++ L L++S+W GDV L ++LK + L LKLP +G
Sbjct: 1 MLESYVTPILMSYVNRYIKNLKPSDLQLSLWGGDVVLSKLELKLDVLEQELKLPFTFLSG 60
Query: 60 FLGSVKLKVPWSRLGQDPVLVYLDRIFLLAEPATQVEGCSEDAVQEAKKIRIEEMELKLW 119
+ +++ VPW++LG +PV++ ++ + + + ++ E + R K
Sbjct: 61 HIHELRIHVPWTKLGSEPVVITINTMECILKLKDGIQDDHESCGSNSTN-RSTAESTKSS 119
Query: 120 EKSQQLKSEMKTINPSSQGSAKKLFSTIIGNLKLSISNIHIRYEDGESNPGHPFAAGVML 179
K ++++ T G + L ++ N+ + I+N+ ++Y + + V +
Sbjct: 120 IKPRRMQQAAPTDPDLPPGYVQSLIRRVVNNVNIVINNLILKYVEDD------IVLSVNI 173
Query: 180 DKLSAVTVDDTGKETFITGGALDLI 204
TV + F+ A DL+
Sbjct: 174 TSAECYTVGELWDRAFMDISATDLV 198
>YM23_YEAST (P40215) Hypothetical 62.8 kDa protein in RPS16A-TIF34
intergenic region
Length = 560
Score = 72.0 bits (175), Expect = 1e-11
Identities = 36/84 (42%), Positives = 57/84 (67%), Gaps = 3/84 (3%)
Query: 856 MVSFICASLTIGGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKR 915
++SF+ GGPTGVEFAA L D+V++DL + P + +K+TL+EA +IL+MFDK
Sbjct: 274 LLSFVVVG---GGPTGVEFAAELRDYVDQDLRKWMPELSKEIKVTLVEALPNILNMFDKY 330
Query: 916 ITAFADDKFQRDGIDVKTGSVVVK 939
+ +A D F+ + ID++ ++V K
Sbjct: 331 LVDYAQDLFKEEKIDLRLKTMVKK 354
>NDI1_YEAST (P32340) Rotenone-insensitive NADH-ubiquinone
oxidoreductase, mitochondrial precursor (EC 1.6.5.3)
(Internal NADH dehydrogenase)
Length = 513
Score = 58.9 bits (141), Expect = 1e-07
Identities = 28/73 (38%), Positives = 46/73 (62%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQR 926
GGPTGVE A L D+V++DL + P + + V+I L+EA +L+MF+K+++++A +
Sbjct: 236 GGPTGVEAAGELQDYVHQDLRKFLPALAEEVQIHLVEALPIVLNMFEKKLSSYAQSHLEN 295
Query: 927 DGIDVKTGSVVVK 939
I V + V K
Sbjct: 296 TSIKVHLRTAVAK 308
>DHNA_ECOLI (P00393) NADH dehydrogenase (EC 1.6.99.3)
Length = 433
Score = 48.1 bits (113), Expect = 2e-04
Identities = 33/96 (34%), Positives = 55/96 (56%), Gaps = 5/96 (5%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGI-KDIVKITLLEAGDHILSMFDKRITAFADDKFQ 925
GG TGVE +A LH+ V + Y G+ + + +TL+EAG+ IL RI+A A ++
Sbjct: 177 GGATGVELSAELHNAVKQLHSYGYKGLTNEALNVTLVEAGERILPALPPRISAAAHNELT 236
Query: 926 RDGIDVKTGSVVVKSTDPISGLSETSSDHDDIESEL 961
+ G+ V T ++V + + GL + D + IE++L
Sbjct: 237 KLGVRVLTQTMVTSADE--GGLH--TKDGEYIEADL 268
>DLD2_BACSU (P54533) Dihydrolipoyl dehydrogenase (EC 1.8.1.4) (E3
component of branched-chain alpha-keto acid
dehydrogenase complex) (LPD-Val) (Dihydrolipoamide
dehydrogenase)
Length = 474
Score = 42.4 bits (98), Expect = 0.011
Identities = 32/112 (28%), Positives = 53/112 (46%), Gaps = 22/112 (19%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQR 926
GG G+E+A+ LHDF VK+T++E D IL D I+ + ++
Sbjct: 189 GGVIGIEWASMLHDFG--------------VKVTVIEYADRILPTEDLEISKEMESLLKK 234
Query: 927 DGIDVKTGSVVVKSTDPISGLSETSSDHDDIESELDNQGVIDVAISERLFVT 978
GI TG+ V+ T +++TS DDI + + G +E++ V+
Sbjct: 235 KGIQFITGAKVLPDT-----MTKTS---DDISIQAEKDGETVTYSAEKMLVS 278
>YJLD_BACSU (P80861) NADH dehydrogenase-like protein yjlD (EC
1.6.99.-) (Glucose starvation-inducible protein 5)
(GSI5)
Length = 391
Score = 36.6 bits (83), Expect = 0.58
Identities = 33/131 (25%), Positives = 55/131 (41%), Gaps = 21/131 (16%)
Query: 823 KIFEEGDDGSSEFLRPWNQADLEGWLSLLTWKVMVSFICASLTIGGP--TGVEFAASLHD 880
K+F+ +D E+ + N+AD A++ IGG TGVE L D
Sbjct: 133 KVFQHVEDRVREYSKTKNEAD------------------ATILIGGGGLTGVELVGELAD 174
Query: 881 FVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQRDGIDVKTGSVVVKS 940
+ +L + Y +K+ L+EAG IL + + A ++ G++ TG V
Sbjct: 175 IM-PNLAKKYGVDHKEIKLKLVEAGPKILPVLPDDLIERATASLEKRGVEFLTGLPVTNV 233
Query: 941 TDPISGLSETS 951
+ L + S
Sbjct: 234 EGNVIDLKDGS 244
>DLDH_CHLPN (Q9Z773) Dihydrolipoyl dehydrogenase (EC 1.8.1.4) (E3
component of 2-oxoglutarate dehydrogenase complex)
(Dihydrolipoamide dehydrogenase)
Length = 461
Score = 36.6 bits (83), Expect = 0.58
Identities = 22/67 (32%), Positives = 34/67 (49%), Gaps = 14/67 (20%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFDKRITAFADDKFQR 926
GG G EFA+ H V+IT++EA DHIL++ +K ++ +KF +
Sbjct: 178 GGVIGCEFASLFHTLG--------------VEITVIEALDHILAVNNKEVSQTVTNKFTK 223
Query: 927 DGIDVKT 933
GI + T
Sbjct: 224 QGIRILT 230
>DCTP_RHOCA (P37735) C4-dicarboxylate-binding periplasmic protein
precursor
Length = 333
Score = 36.6 bits (83), Expect = 0.58
Identities = 37/131 (28%), Positives = 59/131 (44%), Gaps = 14/131 (10%)
Query: 857 VSFICASLTIGGPTGVEFAASLHDFVNEDLVRLYPGIKDIVKITLLEAGDHILSMFD-KR 915
V + SL GP GV+ DF DL ++ + + K+T EAG +LS + K
Sbjct: 87 VQMLAPSLAKFGPLGVQ------DFEVFDLPYIFKDYEALHKVTQGEAGKMLLSKLEAKG 140
Query: 916 ITAFADDKFQRDGIDVKTGSVVVKSTDPISGLSETSSDHDDIESELDNQGVID--VAISE 973
IT A F +G + + + + D GL +E+E++ G + +A SE
Sbjct: 141 ITGLA---FWDNGFKIMSANTPLTMPDDFLGLKMRIQSSKVLEAEMNALGAVPQVMAFSE 197
Query: 974 --RLFVTGVLD 982
+ TGV+D
Sbjct: 198 VYQALQTGVVD 208
>YA82_PASMU (Q9CLW7) Hypothetical protein PM1082
Length = 343
Score = 35.8 bits (81), Expect = 0.99
Identities = 24/80 (30%), Positives = 41/80 (51%), Gaps = 10/80 (12%)
Query: 831 GSSEFLRPWNQADLEGWLSLLTWKVMVSFICASLTIGGPTGVEFAASLHDFV-------N 883
G+S+ L P +AD + +L LL + V F+C L I P ++ +LHDF
Sbjct: 150 GNSQLLDPNCEAD-QAYLQLL--EEAVDFMCEKLDIDAPEPIDITEALHDFTTLLNIHDK 206
Query: 884 EDLVRLYPGIKDIVKITLLE 903
E+ ++ Y I++ + LL+
Sbjct: 207 EEFIKRYQQIQETPEKCLLD 226
>UVRA_HELPY (P56474) UvrABC system protein A (UvrA protein)
(Excinuclease ABC subunit A)
Length = 935
Score = 35.4 bits (80), Expect = 1.3
Identities = 27/92 (29%), Positives = 40/92 (43%), Gaps = 6/92 (6%)
Query: 939 KSTDPISGLSETSSDHDDIESELDNQGVIDVAISERLFVTGVLDELKVCFSYSYQPDQSL 998
K T ++G+ +H D LD + S TGV+DE+++ F+ +Q
Sbjct: 657 KKTQSLNGVEIVGLEHLDKVIYLDQAPIGKTPRSNPATYTGVMDEIRILFA-----EQKE 711
Query: 999 MKVLLNQEKRLFEFRAIGGQVEVSMKDNDIFI 1030
K+L R F F GG+ E D DI I
Sbjct: 712 AKILGYSASR-FSFNVKGGRCEKCQGDGDIKI 742
>SGS1_YEAST (P35187) Helicase SGS1 (Helicase TPS1)
Length = 1447
Score = 35.0 bits (79), Expect = 1.7
Identities = 43/189 (22%), Positives = 78/189 (40%), Gaps = 12/189 (6%)
Query: 1131 PKFSRIIGLLPTDAPS-TSTKEHELNDTLESFVKAQIIIYDQNSTRYNNIDKQVIVTLAT 1189
P + I +P P+ T+TK+HE+ TL + + + Y S +Y ++ I +
Sbjct: 58 PGTTNFITSIPASGPTNTATKQHEVMQTLSN--DTEWLSYTATSNQYADVPMVDIPASTS 115
Query: 1190 LTFFCRRPTILAIMEFINSINIENGNLATSSDSSSTSMVKNDLSNDLDVLHVTTVEEHAV 1249
+ R P F N+ + +DSS +N+ + +D + E+ +
Sbjct: 116 VVSNPRTPNGSKTHNF-NTFRPHMASSLVENDSSRNLGSRNNNKSVIDNSSIGKQLENDI 174
Query: 1250 KGLLGKGKSRVMFSLTLKMAQAQILLMKENETKLACLSQESLLAEIKVFPSSFSIKAALG 1309
K R+ SL + + + LL++ K + + SL + K S I+ L
Sbjct: 175 K----LEVIRLQGSLIMALKEQSKLLLQ----KCSIIESTSLSEDAKRLQLSRDIRPQLS 226
Query: 1310 NLKISDDSL 1318
N+ I DSL
Sbjct: 227 NMSIRIDSL 235
>DHNA_HAEIN (P44856) NADH dehydrogenase (EC 1.6.99.3)
Length = 444
Score = 35.0 bits (79), Expect = 1.7
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 3/76 (3%)
Query: 867 GGPTGVEFAASLHDFVNEDLVRLYPGIKD--IVKITLLEAGDHILSMFDKRITAFADDKF 924
GG TGVE A L+ EDL G D +++TL+EAG +L + ++A D+
Sbjct: 188 GGATGVELTAELYHAA-EDLSSYGYGKIDSSCLQVTLVEAGTRLLPALPENLSAAVLDEL 246
Query: 925 QRDGIDVKTGSVVVKS 940
+ G +V+ +++ ++
Sbjct: 247 KEMGTNVQLNTMITEA 262
>SYL_TREPA (O83595) Leucyl-tRNA synthetase (EC 6.1.1.4)
(Leucine--tRNA ligase) (LeuRS)
Length = 878
Score = 34.7 bits (78), Expect = 2.2
Identities = 25/92 (27%), Positives = 41/92 (44%), Gaps = 1/92 (1%)
Query: 15 GNYVRGLNKEALKISVWKGDVELKNMQLKPEALNALKLPV-KVKAGFLGSVKLKVPWSRL 73
G YV E L+ V K LKN+ + + A +V F+G ++ PW+
Sbjct: 617 GTYVHARTGEKLECVVAKMSKALKNVVNPDDMIAAYGADACRVYEMFMGPLEASKPWNTQ 676
Query: 74 GQDPVLVYLDRIFLLAEPATQVEGCSEDAVQE 105
G V +L++I++LA G +D+ E
Sbjct: 677 GLVGVFRFLEKIWVLAGRVAAANGIPQDSRAE 708
>GOP1_CAEEL (P46578) Hypothetical protein gop-1
Length = 892
Score = 34.7 bits (78), Expect = 2.2
Identities = 40/151 (26%), Positives = 61/151 (39%), Gaps = 29/151 (19%)
Query: 1080 GLIPTESDDKFYEAPE--TLAD--SDVYMQSPGGTSEYPSSSSNEIKFNYSSLEPPKFSR 1135
G+ TE D+ F++ PE TL D DV + + P N S +F
Sbjct: 450 GVEATEEDEIFHDVPEEQTLEDLVDDVLVDTENSAISDPEPK------NVESESRSRFQS 503
Query: 1136 IIGLLPTDAPSTSTKEHELNDTLESFVKAQIIIYDQNSTRYNNI---------------D 1180
+ LP PSTS + L D L S +KA + D N R + D
Sbjct: 504 AVDELPP--PSTSGCDGRLFDALSSIIKA--VGTDDNRIRPITLELACLVIRQILMTVDD 559
Query: 1181 KQVIVTLATLTFFCRRPTILAIMEFINSINI 1211
++V +L L F R + +I +++N N+
Sbjct: 560 EKVHTSLTKLCFEVRLKLLSSIGQYVNGENL 590
>SELP_HUMAN (P49908) Selenoprotein P precursor (SeP)
Length = 381
Score = 33.9 bits (76), Expect = 3.8
Identities = 14/39 (35%), Positives = 22/39 (55%)
Query: 333 LDDVTISLSKDGYRDIMKLADNFAAFNQRLKYAHFRPPV 371
L+D+ + L K+GY +I + N + RLKY H + V
Sbjct: 69 LEDLRVKLKKEGYSNISYIVVNHQGISSRLKYTHLKNKV 107
>NUC2_SCHPO (P10505) Nuclear scaffold-like protein p76
Length = 665
Score = 33.9 bits (76), Expect = 3.8
Identities = 19/65 (29%), Positives = 40/65 (61%), Gaps = 1/65 (1%)
Query: 108 KIRIEEMELKLWEKSQQLKSEMKTINPSSQGSAKKLFSTIIGNLKLSISNIHIRYEDGES 167
K+ I EL +EKS+++ +++ ++PS + ++FST + +L+ S+ ++ +E E+
Sbjct: 368 KLGITYFELVDYEKSEEVFQKLRDLSPS-RVKDMEVFSTALWHLQKSVPLSYLAHETLET 426
Query: 168 NPGHP 172
NP P
Sbjct: 427 NPYSP 431
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 186,194,502
Number of Sequences: 164201
Number of extensions: 8042604
Number of successful extensions: 21529
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 21472
Number of HSP's gapped (non-prelim): 57
length of query: 1631
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1507
effective length of database: 39,613,130
effective search space: 59696986910
effective search space used: 59696986910
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0550.1


