

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0545.2
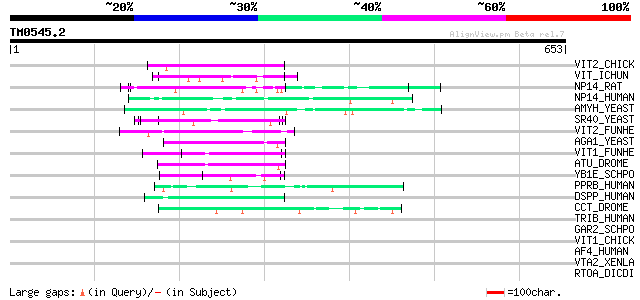
(653 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIT2_CHICK (P02845) Vitellogenin II precursor (Major vitellogeni... 56 3e-07
VIT_ICHUN (Q91062) Vitellogenin precursor (VTG) [Contains: Lipov... 54 1e-06
NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 k... 54 1e-06
NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130... 52 4e-06
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 52 6e-06
SR40_YEAST (P32583) Suppressor protein SRP40 50 2e-05
VIT2_FUNHE (Q98893) Vitellogenin II precursor (VTG II) [Contains... 49 3e-05
AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor 49 4e-05
VIT1_FUNHE (Q90508) Vitellogenin I precursor (VTG I) [Contains: ... 49 5e-05
ATU_DROME (Q94546) Another transcription unit protein 47 1e-04
YB1E_SCHPO (P87179) Serine-rich protein C30B4.01c precursor 47 2e-04
PPRB_HUMAN (Q15648) Peroxisome proliferator-activated receptor b... 47 2e-04
DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contai... 46 4e-04
CCT_DROME (O96433) Cyclin T 45 6e-04
TRIB_HUMAN (Q14669) Thyroid receptor interacting protein 12 (TRI... 44 0.001
GAR2_SCHPO (P41891) Protein gar2 44 0.001
VIT1_CHICK (P87498) Vitellogenin I precursor (Minor vitellogenin... 44 0.002
AF4_HUMAN (P51825) AF-4 protein (Proto-oncogene AF4) (FEL protein) 43 0.002
VTA2_XENLA (P18709) Vitellogenin A2 precursor (VTG A2) [Contains... 43 0.003
RTOA_DICDI (P54681) Protein rtoA (Ratio-A) 43 0.003
>VIT2_CHICK (P02845) Vitellogenin II precursor (Major vitellogenin)
[Contains: Lipovitellin I (LVI); Phosvitin (PV);
Lipovitellin II (LVII); YGP40]
Length = 1850
Score = 56.2 bits (134), Expect = 3e-07
Identities = 47/171 (27%), Positives = 77/171 (44%), Gaps = 10/171 (5%)
Query: 163 DDDVALADKLKIPKTR--KRGAT--------PTTSASQKRAKGAGGSTVGNPSPAKSASQ 212
D +A+K + PK R K+G T P S + + +T + S A S ++
Sbjct: 1087 DSMFKVANKTRHPKNRPSKKGNTVLAEFGTEPDAKTSSSSSSASSTATSSSSSSASSPNR 1146
Query: 213 REEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSS 272
++ M + + V Q + ++ +K++S K S+ SSSS S SS+S S
Sbjct: 1147 KKPMDEEENDQVKQARNKDASSSSRSSKSSNSSKRSSSKSSNSSKRSSSSSSSSSSSSRS 1206
Query: 273 PLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
+S S + +++ SS+ + SSSSS SSSSS S+ S
Sbjct: 1207 SSSSSSSSSNSKSSSSSSKSSSSSSRSRSSSKSSSSSSSSSSSSSSKSSSS 1257
Score = 43.1 bits (100), Expect = 0.002
Identities = 44/180 (24%), Positives = 77/180 (42%), Gaps = 6/180 (3%)
Query: 158 MEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQ 217
M+++++D V A + K + R + + S+ + +K + S + S + S+S
Sbjct: 1150 MDEEENDQVKQA-RNKDASSSSRSSKSSNSSKRSSSKSSNSSKRSSSSSSSSSSSSRSSS 1208
Query: 218 QGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKAS 277
+ ++SS ++S + K+S SS S SSSS S S +SSS K+S
Sbjct: 1209 SSSSSSSNSKSSSSSSKSSSSSSRSRSSSKSSSSSSS-SSSSSSSKSSSSRSSSSSSKSS 1267
Query: 278 GPKRVGMSKMAVPRATWFSSKPPPIALSS---SSSDLE-SSSSSLDSTESDPVWDKLTFY 333
+ ++ SS ++ S S LE SSSS S+ +W + Y
Sbjct: 1268 SHHSHSHHSGHLNGSSSSSSSSRSVSHHSHEHHSGHLEDDSSSSSSSSVLSKIWGRHEIY 1327
>VIT_ICHUN (Q91062) Vitellogenin precursor (VTG) [Contains:
Lipovitellin LV-1N; Lipovitellin LV-1C; Lipovitellin
LV-2]
Length = 1823
Score = 54.3 bits (129), Expect = 1e-06
Identities = 46/161 (28%), Positives = 71/161 (43%), Gaps = 13/161 (8%)
Query: 169 ADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAK------SASQREEMQQGGDQ 222
+DK K K ++ ++S+S + + G +P + +A + + Q+G D
Sbjct: 1119 SDKDKDAKKPPGSSSSSSSSSSSSSSSSSSDKSGKKTPRQGSTVNLAAKRASKKQRGKDS 1178
Query: 223 GVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRV 282
+SS D++ P + AK+ + G S H S SS+SSS +S K
Sbjct: 1179 SSSSSSSSSSSDSSKSPHKHGGAKRQHAGHGAPHLGPQS-HSSSSSSSSSSSSSSASKSF 1237
Query: 283 GMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
K + R KP P SSSSS +SSSSS S+ S
Sbjct: 1238 STVKPPMTR------KPRPARSSSSSSSSDSSSSSSSSSSS 1272
Score = 50.4 bits (119), Expect = 1e-05
Identities = 55/205 (26%), Positives = 85/205 (40%), Gaps = 42/205 (20%)
Query: 176 KTRKRGATPTTSASQ--KRAKGAGGSTVGNPSPAKSASQREEMQQGGDQ----------- 222
KT ++G+T +A + K+ +G S+ + S + S S + + GG +
Sbjct: 1154 KTPRQGSTVNLAAKRASKKQRGKDSSSSSSSSSSSSDSSKSPHKHGGAKRQHAGHGAPHL 1213
Query: 223 GVDQQASSPLQDTTSIPVEMAQAKKT--------------SGKRSSRKSGSSSSHHSKSS 268
G +SS ++S +++ T S SS S SSSS S SS
Sbjct: 1214 GPQSHSSSSSSSSSSSSSSASKSFSTVKPPMTRKPRPARSSSSSSSSDSSSSSSSSSSSS 1273
Query: 269 TSSSPLKASGPKRVGMSKMAV---------------PRATWFSSKPPPIALSSSSSDLES 313
+SSS +S + + +AV R S + P + SSSSS S
Sbjct: 1274 SSSSSSSSSSSESKSLEWLAVKDVNQSAFYNFKYVPQRKPQTSRRHTPASSSSSSSSSSS 1333
Query: 314 SSSSLDSTESDPVWDKLTFYFNSKP 338
SSSS S++SD +F +SKP
Sbjct: 1334 SSSSSSSSDSDMTVSAESFEKHSKP 1358
Score = 35.4 bits (80), Expect = 0.47
Identities = 38/117 (32%), Positives = 54/117 (45%), Gaps = 10/117 (8%)
Query: 207 AKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSK 266
AK++ + ++ ++GV + S L+ S + K SS S SSSS
Sbjct: 1088 AKASKKSKKNSTITEEGVGETIISQLKKILSSDKDKDAKKPPGSSSSSSSSSSSSS---- 1143
Query: 267 SSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
SS+SS P++ +A RA S K SSSSS SSSSS DS++S
Sbjct: 1144 SSSSSDKSGKKTPRQGSTVNLAAKRA---SKKQRGKDSSSSSS---SSSSSSDSSKS 1194
>NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 kDa
protein) (140 kDa nucleolar phosphoprotein) (Nopp140)
(Nucleolar and coiled-body phosphoprotein 1)
Length = 704
Score = 53.9 bits (128), Expect = 1e-06
Identities = 72/368 (19%), Positives = 132/368 (35%), Gaps = 24/368 (6%)
Query: 143 PTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVG 202
P + + + S++ ED +++ DK ++P ++ A P AS + G +
Sbjct: 73 PVAKKAKKETSSSDSSEDSSEEE----DKAQVPT--QKAAAPAKRASLPQHAGKAAAKAS 126
Query: 203 NPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSS 262
S ++ +S+ EE + + V Q+A P P + A+ SS SSS
Sbjct: 127 ESSSSEESSEEEEEKDKKKKPVQQKAVKPQAKAVRPPPKKAE--------SSESESDSSS 178
Query: 263 HHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSL---D 319
T A+ K ++ P +K P A + + SSSSS D
Sbjct: 179 EDEAPQTQKPKAAATAAKAPTKAQTKAPAKPGPPAKAQPKAANGKAGSSSSSSSSSSSDD 238
Query: 320 STESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNRPSPAREENIP 379
S E L K + + P + + + ++ + + P +++ P
Sbjct: 239 SEEEKKAAAPLKKTAPKKQVVAKAPVKVTAAPTQKSSSSEDSSSEEEEEQKKPMKKKAGP 298
Query: 380 HDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSEPSPRQFQTSPQQPRLSPQRLQQDAR 439
+ V + + +G P K A+A ++P+ +S + S + + A+
Sbjct: 299 YSSVP--PPSVSLSKKSVGAQSPKK----AAAQTQPADSSADSSEESDSSSEEEKKTPAK 352
Query: 440 AALLSPHAKGGSASSKTAASSHTS-SERLSALLVEDPLSIFQSFFDGTLDLESPPRQAET 498
+ AK K +SS +S S+ P+S +S PP
Sbjct: 353 TVVSKTPAKPAPVKKKAESSSDSSDSDSSEDEAPAKPVSATKSPLSKPAVTPKPPAAKAV 412
Query: 499 AETAQSGG 506
A Q G
Sbjct: 413 ATPKQPAG 420
Score = 53.5 bits (127), Expect = 2e-06
Identities = 85/379 (22%), Positives = 145/379 (37%), Gaps = 81/379 (21%)
Query: 140 IPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGS 199
+P+ +A+ + + ++ E ++++ DK K P +++ P A + K A S
Sbjct: 114 LPQHAGKAAAKASESSSSEESSEEEEEK--DKKKKP-VQQKAVKPQAKAVRPPPKKAESS 170
Query: 200 TVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGS 259
S + S+S+ E Q + A +P + T P + K K ++ K+GS
Sbjct: 171 E----SESDSSSEDEAPQTQKPKAAATAAKAPTKAQTKAPAKPGPPAKAQPKAANGKAGS 226
Query: 260 SSSHHSKSSTSSS--------PLKASGPKRVGMSKMAV---------------------- 289
SSS S SS+ S PLK + PK+ ++K V
Sbjct: 227 SSSSSSSSSSDDSEEEKKAAAPLKKTAPKKQVVAKAPVKVTAAPTQKSSSSEDSSSEEEE 286
Query: 290 -------PRATWFSSKPPP-IALSSSSSDLES----------SSSSLDSTESDPVWDKLT 331
+A +SS PPP ++LS S +S + SS DS+E +
Sbjct: 287 EQKKPMKKKAGPYSSVPPPSVSLSKKSVGAQSPKKAAAQTQPADSSADSSEESDSSSEEE 346
Query: 332 FYFNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNRPSPAREENIPHDEVQGGEDPII 391
+K + + P +P K+ ++ S+ + S + E+ P V + P+
Sbjct: 347 KKTPAKTVVSKTP-AKPAPVKKKAESSSD-------SSDSDSSEDEAPAKPVSATKSPLS 398
Query: 392 QPDQVIGVMP-PPKADVLASAHSEPSPRQFQTSPQQPRLSPQRLQQDARAALLSPHAKGG 450
+P V P PP A +A +P+QP S Q+ Q + S
Sbjct: 399 KP----AVTPKPPAAKAVA-------------TPKQPAGSGQKPQSRKADSSSSEEESSS 441
Query: 451 SASSKTAASSHTSSERLSA 469
S T S T R++A
Sbjct: 442 SEEEATKKSVTTPKARVTA 460
Score = 47.0 bits (110), Expect = 2e-04
Identities = 51/201 (25%), Positives = 83/201 (40%), Gaps = 24/201 (11%)
Query: 131 RTLIGQAEPIPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPKT--RKRGATPTTSA 188
+T++ + P P + + S++ + D +D A A + K+ K TP A
Sbjct: 352 KTVVSKTPAKPAPVKK---KAESSSDSSDSDSSEDEAPAKPVSATKSPLSKPAVTPKPPA 408
Query: 189 SQKRA---KGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQA 245
++ A + AG A S+S EE ++ + ++P T+ A
Sbjct: 409 AKAVATPKQPAGSGQKPQSRKADSSSSEEESSSSEEEATKKSVTTPKARVTAKAAPSLPA 468
Query: 246 KKT--SGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIA 303
K+ +G SS S SSSS K + P K + AVP KP P+
Sbjct: 469 KQAPRAGGDSSSDSESSSSEEEKKTPPKPPAKKKA------AGAAVP-------KPTPVK 515
Query: 304 LSSSSSDLESSSSSLDSTESD 324
+++ S SSSSS DS+E +
Sbjct: 516 KAAAESS-SSSSSSEDSSEEE 535
Score = 33.9 bits (76), Expect = 1.4
Identities = 39/192 (20%), Positives = 74/192 (38%), Gaps = 16/192 (8%)
Query: 139 PIPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGG 198
P+ +P P A A + A + P++RK ++ + S + A
Sbjct: 396 PLSKPAVTPKPPAAKAVATPKQP-------AGSGQKPQSRKADSSSSEEESSSSEEEATK 448
Query: 199 STVGNPSP------AKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKR 252
+V P A S ++ + GGD D ++SS ++ + P A+ KK +G
Sbjct: 449 KSVTTPKARVTAKAAPSLPAKQAPRAGGDSSSDSESSSSEEEKKTPPKPPAK-KKAAGAA 507
Query: 253 SSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLE 312
+ + + SS+SSS +S ++ A P+ + K + S + +
Sbjct: 508 VPKPTPVKKAAAESSSSSSSSEDSSEEEKKKPKSKATPKPQ--AGKANGVPASQNGKAGK 565
Query: 313 SSSSSLDSTESD 324
S + TE +
Sbjct: 566 ESEEEEEDTEQN 577
>NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130
kDa protein) (140 kDa nucleolar phosphoprotein)
(Nopp140) (Nucleolar and coiled-body phosphoprotein 1)
Length = 699
Score = 52.4 bits (124), Expect = 4e-06
Identities = 77/351 (21%), Positives = 129/351 (35%), Gaps = 44/351 (12%)
Query: 140 IPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGS 199
+P A +S++ E DDDD++ D+ K P ++G P A++ K A S
Sbjct: 114 LPPGKAAAKASESSSSEESRDDDDEE----DQKKQPV--QKGVKPQAKAAKAPPKKAKSS 167
Query: 200 TVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGS 259
+ S ++ + + + V Q +P + + P K ++ K+ S
Sbjct: 168 DSDSDSSSEDEPPKNQKPKITPVTVKAQTKAPPKPARAAP-----------KIANGKAAS 216
Query: 260 SSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLD 319
SSS SS+SSS + K K VP+ + P + ++++ SSSS D
Sbjct: 217 SSS---SSSSSSSSDDSEEEKAAATPKKTVPKKQVVAKAP----VKAATTPTRKSSSSED 269
Query: 320 STESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNRPSPAREENIP 379
S+ + K P ++ P P K +L PP+ + P
Sbjct: 270 SSSDEEEEQKKPMKNKPGPYSYAPPPSAPPPKK----SLGTQPPKKAVEKQQPVESSEDS 325
Query: 380 HDEVQGGEDPIIQPDQVIGV-----MPPP--KADVLASAHSEPSPRQFQTSPQQPRLSPQ 432
DE + +P V PPP KA +S S+ + +P +P + +
Sbjct: 326 SDESDSSSEEEKKPPTKAVVSKATTKPPPAKKAAESSSDSSDSDSSEDDEAPSKPAGTTK 385
Query: 433 RLQQDARAALLSPHAK---------GGSASSKTAASSHTSSERLSALLVED 474
SP K GG T + +SSE S+ E+
Sbjct: 386 NSSNKPAVTTKSPAVKPAAAPKQPVGGGQKLLTRKADSSSSEEESSSSEEE 436
Score = 41.6 bits (96), Expect = 0.007
Identities = 47/178 (26%), Positives = 68/178 (37%), Gaps = 42/178 (23%)
Query: 159 EDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQ 218
ED D+ + +++ K P T+ + TT P PAK A++
Sbjct: 323 EDSSDESDSSSEEEKKPPTKAVVSKATTK----------------PPPAKKAAESSSDSS 366
Query: 219 GGDQGVDQQASS------------PLQDTTSIPVEMAQAKKT----SGKRSSRKSGSSSS 262
D D +A S P T S V+ A A K K +RK+ SSSS
Sbjct: 367 DSDSSEDDEAPSKPAGTTKNSSNKPAVTTKSPAVKPAAAPKQPVGGGQKLLTRKADSSSS 426
Query: 263 HHSKSSTSSSPLK----ASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSS 316
SS+ K + PK + +++P +K P SSSD +SSSS
Sbjct: 427 EEESSSSEEEKTKKMVATTKPKATAKAALSLP------AKQAPQGSRDSSSDSDSSSS 478
Score = 38.9 bits (89), Expect = 0.043
Identities = 42/190 (22%), Positives = 68/190 (35%), Gaps = 18/190 (9%)
Query: 142 RPTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTV 201
+P P +S+ D +DD A + K TT + + A V
Sbjct: 351 KPPPAKKAAESSSDSSDSDSSEDDEAPSKPAGTTKNSSNKPAVTTKSPAVKPAAAPKQPV 410
Query: 202 GN-----PSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKT--SGKRSS 254
G A S+S EE ++ + ++ T+ AK+ + SS
Sbjct: 411 GGGQKLLTRKADSSSSEEESSSSEEEKTKKMVATTKPKATAKAALSLPAKQAPQGSRDSS 470
Query: 255 RKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESS 314
S SSSS + TS S +K K G ++ P + ++ +S
Sbjct: 471 SDSDSSSSEEEEEKTSKSAVKKKPQKVAG-----------GAAPSKPASAKKGKAESSNS 519
Query: 315 SSSLDSTESD 324
SSS DS+E +
Sbjct: 520 SSSDDSSEEE 529
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 51.6 bits (122), Expect = 6e-06
Identities = 74/383 (19%), Positives = 134/383 (34%), Gaps = 30/383 (7%)
Query: 136 QAEPIPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKG 195
++ P PTP +S +S+ + A + + TT +S
Sbjct: 389 ESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTS 448
Query: 196 AGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSR 255
+ + P P S+S E + +S+P+ +S E + A TS S
Sbjct: 449 STTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESS 508
Query: 256 KSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSS 315
+ + S + +SS+P S V +T SS P SSS+++ S+
Sbjct: 509 SAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSTP 568
Query: 316 SSLDSTESD----PVWDKLTFYFNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNRPS 371
+ +TES P T +S P+ P + ++ S+ P + +
Sbjct: 569 VTSSTTESSSAPVPTPSSSTTESSSAPV--------PTPSSSTTES-SSAPAPTPSSSTT 619
Query: 372 PAREENIPHDEVQGGEDPIIQP-----DQVIGVMPPPKADVLASAHSE-PSPRQFQTSPQ 425
+ + + P+ P + +P P + S+ + P+P T
Sbjct: 620 ESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESS 679
Query: 426 QPRLSPQRLQQDARAALLSPHAKGGSASSKTAASSHTSSERLSALLVEDPLSIFQSFFDG 485
++ + + A + S + SA T +SS T S S+ V P S
Sbjct: 680 SAPVTSSTTESSS-APVTSSTTESSSAPVPTPSSSTTES---SSAPVPTPSS-------S 728
Query: 486 TLDLESPPRQAETAETAQSGGVP 508
T + S P ++ T +S P
Sbjct: 729 TTESSSAPVPTPSSSTTESSSAP 751
Score = 49.7 bits (117), Expect = 2e-05
Identities = 84/385 (21%), Positives = 137/385 (34%), Gaps = 37/385 (9%)
Query: 136 QAEPIPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKG 195
++ P PTP +S +S+ + A T A T+S ++ +
Sbjct: 323 ESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAP 382
Query: 196 AGGSTVGN---PSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKR 252
ST + P P S+S E + +S+P+ TS E + A TS
Sbjct: 383 VTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPV---TSSTTESSSAPVTS--- 436
Query: 253 SSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLE 312
S+ +S S+ S + +SS+P+ S V +T SS P SSS+++
Sbjct: 437 STTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESS 496
Query: 313 SSSSSLDSTESD----PVWDKLTFYFNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQN 368
S+ + +TES P T +S P P + ++ S+ P
Sbjct: 497 SAPVTSSTTESSSAPVPTPSSSTTESSSAP--------APTPSSSTTES-SSAPVTSSTT 547
Query: 369 RPSPAREENIPHDEVQGGEDPIIQPDQVIGVMP---PPKADVLASAHSEPSPRQFQT-SP 424
S A + P+ P P + +S+ P+P T S
Sbjct: 548 ESSSAPVPTPSSSTTESSSTPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESS 607
Query: 425 QQPRLSP-QRLQQDARAALLSPHAKGGSASSKTAASSHTSSERLSALLVEDPLSIFQSFF 483
P +P + + A + S + SA T +SS T S S+ V P S
Sbjct: 608 SAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTES---SSAPVPTPSS------ 658
Query: 484 DGTLDLESPPRQAETAETAQSGGVP 508
T + S P ++ T +S P
Sbjct: 659 -STTESSSAPVPTPSSSTTESSSAP 682
Score = 43.1 bits (100), Expect = 0.002
Identities = 45/205 (21%), Positives = 79/205 (37%), Gaps = 10/205 (4%)
Query: 129 SIRTLIGQAEPIPRP----TPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATP 184
S T + P+P P T +S V S+T E T T
Sbjct: 657 SSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTE 716
Query: 185 TTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQ 244
++SA + + P P S+S E + +S+P+ +S E +
Sbjct: 717 SSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSS 776
Query: 245 AKKTSGKRSSRKSGSS-----SSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKP 299
A + S+ +S S+ SS ++SS + P +S + + P ++ S
Sbjct: 777 APVPTPSSSTTESSSAPVPTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSS 836
Query: 300 PPIALSSSSSDLESSSSSLDSTESD 324
P+ + SSS ESSS+ + S+ ++
Sbjct: 837 VPVP-TPSSSTTESSSAPVSSSTTE 860
Score = 42.0 bits (97), Expect = 0.005
Identities = 48/201 (23%), Positives = 76/201 (36%), Gaps = 21/201 (10%)
Query: 136 QAEPIPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGA-TPTTSASQKRAK 194
++ P PTP +S +S+ + A T A PT S+S +
Sbjct: 605 ESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESS 664
Query: 195 GAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTT----SIPVEMAQAKKTSG 250
A P P S+S E + +S+P+ +T S PV + T
Sbjct: 665 SA-------PVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTES 717
Query: 251 KRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSD 310
+ + SSS+ + +SS+P+ S V +T SS P SSS+++
Sbjct: 718 SSAPVPTPSSST----TESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTE 773
Query: 311 LESS-----SSSLDSTESDPV 326
S+ SSS + S PV
Sbjct: 774 SSSAPVPTPSSSTTESSSAPV 794
Score = 41.6 bits (96), Expect = 0.007
Identities = 71/367 (19%), Positives = 125/367 (33%), Gaps = 49/367 (13%)
Query: 144 TPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGN 203
T +S +S T + + + + ATPTT++ K
Sbjct: 222 TTTSSTSESSTTTSSTSESSTTTSSTSESSTSSSTTAPATPTTTSCTKE----------K 271
Query: 204 PSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSH 263
P+P + S +E + + P DTT + KT K+++ + SS
Sbjct: 272 PTPPTTTSCTKE-----------KPTPPHHDTTPCTKKKTTTSKTCTKKTTTPVPTPSS- 319
Query: 264 HSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
S + +SS+P+ S V +T SS P SSS+++ S+ + +TES
Sbjct: 320 -STTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTES 378
Query: 324 D--PVWDKLTFYFNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNRPSPAREENIPHD 381
PV T P T S+ P + + + + +
Sbjct: 379 SSAPVTSSTT-----------ESSSAPVPTPSSSTTESSSAP--VTSSTTESSSAPVTSS 425
Query: 382 EVQGGEDPIIQPDQVIGVMPPPKADVLASAHSEPSPRQFQTSPQQPRLSPQRLQQDARAA 441
+ P+ P + +S+ P+P T ++ + + A
Sbjct: 426 TTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSS-AP 484
Query: 442 LLSPHAKGGSASSKTAASSHTSSERLSALLVEDPLSIFQSFFDGTLDLESPPRQAETAET 501
+ +P + +SS SS T S S+ V P S T + S P ++ T
Sbjct: 485 VPTPSSSTTESSSAPVTSSTTES---SSAPVPTPSS-------STTESSSAPAPTPSSST 534
Query: 502 AQSGGVP 508
+S P
Sbjct: 535 TESSSAP 541
Score = 36.6 bits (83), Expect = 0.21
Identities = 41/187 (21%), Positives = 75/187 (39%), Gaps = 11/187 (5%)
Query: 136 QAEPIPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPK--TRKRGATPTTSASQKRA 193
++ P PTP +S +S+ + + + P T + + P TS++ +
Sbjct: 701 ESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTE-- 758
Query: 194 KGAGGSTVGNPSPAKSASQREEMQ-QGGDQGVDQQASSPLQDTTSIPVEMAQAK-KTSGK 251
S+ P+P+ S ++ + +S+P+ +S E + A T
Sbjct: 759 ----SSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSVAPVPTPSS 814
Query: 252 RSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVP-RATWFSSKPPPIALSSSSSD 310
S+ S + SS SST SS + P + P ++ S P+ SSSS+
Sbjct: 815 SSNITSSAPSSTPFSSSTESSSVPVPTPSSSTTESSSAPVSSSTTESSVAPVPTPSSSSN 874
Query: 311 LESSSSS 317
+ SS+ S
Sbjct: 875 ITSSAPS 881
Score = 31.6 bits (70), Expect = 6.8
Identities = 37/188 (19%), Positives = 67/188 (34%), Gaps = 6/188 (3%)
Query: 136 QAEPIPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGA-TPTTSASQKRAK 194
++ P PTP +S +S+ + A T A PT S+S +
Sbjct: 731 ESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESS 790
Query: 195 GAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTT---SIPVEMAQAKKTSGK 251
A T + + S + + +S+P +T S+PV + T
Sbjct: 791 SAPVPTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSVPVPTPSSSTTESS 850
Query: 252 RSSRKSGSSSSHHSKSST--SSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSS 309
+ S ++ S + T SSS + +S P + S +T + P S +
Sbjct: 851 SAPVSSSTTESSVAPVPTPSSSSNITSSAPSSIPFSSTTESFSTGTTVTPSSSKYPGSQT 910
Query: 310 DLESSSSS 317
+ SS++
Sbjct: 911 ETSVSSTT 918
>SR40_YEAST (P32583) Suppressor protein SRP40
Length = 406
Score = 49.7 bits (117), Expect = 2e-05
Identities = 43/178 (24%), Positives = 79/178 (44%), Gaps = 3/178 (1%)
Query: 147 ASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSP 206
+S +S++ + D D + + ++ + S+S+ + +G S+ + S
Sbjct: 54 SSSSSSSSSSSSDSSDSSDSESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSS 113
Query: 207 AKSASQRE---EMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSH 263
+S+S+ E E ++ + ++ A + T + +SG SS +S S S
Sbjct: 114 DESSSESESEDETKKRARESDNEDAKETKKAKTEPESSSSSESSSSGSSSSSESESGSES 173
Query: 264 HSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDST 321
S SS+SSS S S+ + ++ SS + S SSSD +SSSSS S+
Sbjct: 174 DSDSSSSSSSSSDSESDSESDSQSSSSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSS 231
Score = 48.5 bits (114), Expect = 5e-05
Identities = 44/174 (25%), Positives = 80/174 (45%), Gaps = 10/174 (5%)
Query: 154 ATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQR 213
A+ +++ D+ +++ +K K+ ++ ++S+S + + S+ G S + S+S
Sbjct: 2 ASKKIKVDEVPKLSVKEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSS 61
Query: 214 EEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSP 273
D + +SS ++S + +S SS +S SSSS S SS+SSS
Sbjct: 62 SSSSDSSDSSDSESSSSSSSSSSS-------SSSSSDSESSSESDSSSSGSSSSSSSSSD 114
Query: 274 LKASGPKRVGMSKMAVPRATWFSSKPPPIALS---SSSSDLESSSSSLDSTESD 324
+S + +K + +K A + SSSS SSS S S+ES+
Sbjct: 115 ESSSESESEDETKKRARESDNEDAKETKKAKTEPESSSSSESSSSGSSSSSESE 168
Score = 45.8 bits (107), Expect = 4e-04
Identities = 44/150 (29%), Positives = 63/150 (41%), Gaps = 5/150 (3%)
Query: 176 KTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDT 235
K R R + + K+AK S+ + S + +S E + G + D +SS +
Sbjct: 127 KKRARESDNEDAKETKKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSS----S 182
Query: 236 TSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWF 295
+S E + SS S SSS S SS SSS +S S +
Sbjct: 183 SSSDSESDSESDSQSSSSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSS 242
Query: 296 SSKPPPIALSSSSSDLESS-SSSLDSTESD 324
S + SSSSSD S S+S DS++SD
Sbjct: 243 DSDSSGSSDSSSSSDSSSDESTSSDSSDSD 272
Score = 45.4 bits (106), Expect = 5e-04
Identities = 40/166 (24%), Positives = 64/166 (38%)
Query: 152 ASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSAS 211
+S+ E ED+ +D +T+K P +S+S + + S+ + S ++S S
Sbjct: 116 SSSESESEDETKKRARESDNEDAKETKKAKTEPESSSSSESSSSGSSSSSESESGSESDS 175
Query: 212 QREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSS 271
S ++S + + +S SS S SSSS S SS S
Sbjct: 176 DSSSSSSSSSDSESDSESDSQSSSSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSD 235
Query: 272 SPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSS 317
S +S S + + S + S S SD +S SSS
Sbjct: 236 SDSDSSSDSDSSGSSDSSSSSDSSSDESTSSDSSDSDSDSDSGSSS 281
>VIT2_FUNHE (Q98893) Vitellogenin II precursor (VTG II) [Contains:
Lipovitellin 1 (LV1); Phosvitin (PV); Lipovitellin 2
(LV2)]
Length = 1687
Score = 49.3 bits (116), Expect = 3e-05
Identities = 51/219 (23%), Positives = 89/219 (40%), Gaps = 28/219 (12%)
Query: 130 IRTLIGQAEPIPRPTPQASPRVASATVEMEDDD-------------DDDVALADKLKIPK 176
I +IG + TP A P + +E++ + +++ L DK + K
Sbjct: 1013 IYAIIGNHSLLVNVTPAAGPSIERIEIEVQFGEQAAEKILKEVYLNEEEEVLEDKNVLMK 1072
Query: 177 TRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTT 236
+K +P S K + + GS+ + S + S+S SS
Sbjct: 1073 LKKI-LSPGLKNSTKASSSSSGSSRSSRSRSSSSSSSSSSSSSSRSSSSSSRSSSSLRRN 1131
Query: 237 SIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFS 296
S +++A + KRSS S SSSS S SS+SSS K +K + +
Sbjct: 1132 SKMLDLADPLNITSKRSSSSSSSSSSSSSSSSSSSSSSK---------TKWQLHERNFTK 1182
Query: 297 SKPPPIALSSSSSDLESSSSSLDSTESDPVWDKLTFYFN 335
++S + +SS+SS +S +++K+T+ N
Sbjct: 1183 DHIHQHSVSKERLNSKSSASSFES-----IYNKITYLSN 1216
Score = 41.6 bits (96), Expect = 0.007
Identities = 35/103 (33%), Positives = 44/103 (41%), Gaps = 6/103 (5%)
Query: 247 KTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSK------PP 300
K S K SS SGSS S S+SS+SSS +S R S + +SK P
Sbjct: 1082 KNSTKASSSSSGSSRSSRSRSSSSSSSSSSSSSSRSSSSSSRSSSSLRRNSKMLDLADPL 1141
Query: 301 PIALSSSSSDLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQH 343
I SSS SSSSS S+ S K + + + H
Sbjct: 1142 NITSKRSSSSSSSSSSSSSSSSSSSSSSKTKWQLHERNFTKDH 1184
>AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor
Length = 725
Score = 48.9 bits (115), Expect = 4e-05
Identities = 40/147 (27%), Positives = 68/147 (46%), Gaps = 6/147 (4%)
Query: 182 ATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVE 241
+ PTT++ + ++ S + S+S ++SP +TS +
Sbjct: 179 SNPTTTSLSSTSTSPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLT 238
Query: 242 MAQAKKTSGKRSSRKSGSSSSHHSKSSTS-SSPLKASGPKRVGMSKMAVPRATWFSSKPP 300
+ TS +SS + SSS+ S SSTS SS ++ P S + +++ +S P
Sbjct: 239 STSSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSPSSKSTSASSTSTSSYSTSTSP 298
Query: 301 PIALSSSSSDLES---SSSSLDSTESD 324
+L+SSS L S SS+S+ ST +D
Sbjct: 299 --SLTSSSPTLASTSPSSTSISSTFTD 323
Score = 40.4 bits (93), Expect = 0.015
Identities = 40/150 (26%), Positives = 61/150 (40%), Gaps = 4/150 (2%)
Query: 175 PKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQD 234
P T +T +TS S + ST + + S+S SS L
Sbjct: 181 PTTTSLSST-STSPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTS 239
Query: 235 TTSIPVEMAQAK-KTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRAT 293
T+S +Q+ TS +S S+S+ S +STS S K++ S + +
Sbjct: 240 TSSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSPSS-KSTSASSTSTSSYSTSTSP 298
Query: 294 WFSSKPPPIALSSSSSDLESSSSSLDSTES 323
+S P +A S+S S SS+ DST S
Sbjct: 299 SLTSSSPTLA-STSPSSTSISSTFTDSTSS 327
Score = 40.0 bits (92), Expect = 0.019
Identities = 38/140 (27%), Positives = 56/140 (39%), Gaps = 4/140 (2%)
Query: 185 TTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQ 244
TT S + + S + SP S + ++SP +TS
Sbjct: 150 TTVVSSSAIEPSSASII---SPVTSTLSSTTSSNPTTTSLSSTSTSPSSTSTSPSSTSTS 206
Query: 245 AKKTSGKRSSRKSGSSSSHHSKSSTS-SSPLKASGPKRVGMSKMAVPRATWFSSKPPPIA 303
+ TS SS + SSS+ S SSTS SS L ++ S+ + ++ +S P
Sbjct: 207 SSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSST 266
Query: 304 LSSSSSDLESSSSSLDSTES 323
+SSSS S SS S S
Sbjct: 267 STSSSSTSTSPSSKSTSASS 286
Score = 36.2 bits (82), Expect = 0.28
Identities = 29/98 (29%), Positives = 44/98 (44%), Gaps = 12/98 (12%)
Query: 228 ASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKM 287
+S+P + S + TS +S S S+S+ S +STSSS S
Sbjct: 178 SSNPTTTSLSSTSTSPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTS---------- 227
Query: 288 AVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESDP 325
P +T SS + SS+S+ S+S+S ST + P
Sbjct: 228 --PSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSP 263
Score = 33.1 bits (74), Expect = 2.4
Identities = 32/143 (22%), Positives = 58/143 (40%), Gaps = 14/143 (9%)
Query: 182 ATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVE 241
++ +TS SQ + ST +PS ++S ++SP +TS
Sbjct: 241 SSSSTSTSQSSTSTSSSSTSTSPSSTSTSSS-------------STSTSPSSKSTSASST 287
Query: 242 MAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMS-KMAVPRATWFSSKPP 300
+ TS S S + + S SSTS S +G S + + +S P
Sbjct: 288 STSSYSTSTSPSLTSSSPTLASTSPSSTSISSTFTDSTSSLGSSIASSSTSVSLYSPSTP 347
Query: 301 PIALSSSSSDLESSSSSLDSTES 323
++ S+SS++ + S + + E+
Sbjct: 348 VYSVPSTSSNVATPSMTSSTVET 370
Score = 32.3 bits (72), Expect = 4.0
Identities = 29/107 (27%), Positives = 41/107 (38%), Gaps = 7/107 (6%)
Query: 409 ASAHSEPSPRQFQTSPQQPRLSPQRLQQDARAALLSPHAKGGSASSKTAAS-------SH 461
+S+ + S TS SP + + SP +K SASS + +S S
Sbjct: 241 SSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSPSSKSTSASSTSTSSYSTSTSPSL 300
Query: 462 TSSERLSALLVEDPLSIFQSFFDGTLDLESPPRQAETAETAQSGGVP 508
TSS A SI +F D T L S + T+ + S P
Sbjct: 301 TSSSPTLASTSPSSTSISSTFTDSTSSLGSSIASSSTSVSLYSPSTP 347
>VIT1_FUNHE (Q90508) Vitellogenin I precursor (VTG I) [Contains:
Lipovitellin 1 (LV1); Phosvitin (PV); Lipovitellin 2
(LV2)]
Length = 1704
Score = 48.5 bits (114), Expect = 5e-05
Identities = 41/122 (33%), Positives = 60/122 (48%), Gaps = 4/122 (3%)
Query: 203 NPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSS 262
N S + S+S + + +SS +S +++A +S SSR+S SSSS
Sbjct: 1080 NSSSSSSSSSSSSSESRSSRSSSSSSSS---SRSSRKIDLAARTNSSSSSSSRRSRSSSS 1136
Query: 263 HHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTE 322
S SS+SSS +S +R S + ++ SS+ SSSSS SS+SSL S
Sbjct: 1137 SSSSSSSSSSS-SSSSSRRSSSSSSSSSSSSSRSSRRVNSTRSSSSSSRTSSASSLASFF 1195
Query: 323 SD 324
SD
Sbjct: 1196 SD 1197
Score = 44.7 bits (104), Expect = 8e-04
Identities = 41/164 (25%), Positives = 76/164 (46%), Gaps = 15/164 (9%)
Query: 157 EMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEM 216
E E+ ++ L KI +R+ ++ ++S+S ++ + + S + +S++
Sbjct: 1057 EDEETEEGGPVLVKLNKILSSRRNSSSSSSSSSSSSSESRSSRSSSSSSSSSRSSRK--- 1113
Query: 217 QQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKA 276
+D A + ++S + + +S SS S SSSS S SS+SSS +
Sbjct: 1114 -------IDLAARTNSSSSSSSRRSRSSSSSSSSSSSSSSSSSSSSRRSSSSSSSSSSSS 1166
Query: 277 S-GPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLD 319
S +RV ++ + + S+ +L+S SD SSSSS D
Sbjct: 1167 SRSSRRVNSTRSSSSSSRTSSAS----SLASFFSDSSSSSSSSD 1206
Score = 38.1 bits (87), Expect = 0.073
Identities = 31/85 (36%), Positives = 38/85 (44%)
Query: 239 PVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSK 298
PV + K S +R+S S SSSS S S SS +S K+ + T SS
Sbjct: 1066 PVLVKLNKILSSRRNSSSSSSSSSSSSSESRSSRSSSSSSSSSRSSRKIDLAARTNSSSS 1125
Query: 299 PPPIALSSSSSDLESSSSSLDSTES 323
SSSS SSSSS S+ S
Sbjct: 1126 SSSRRSRSSSSSSSSSSSSSSSSSS 1150
Score = 38.1 bits (87), Expect = 0.073
Identities = 25/83 (30%), Positives = 41/83 (49%)
Query: 241 EMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPP 300
++ +++ S SS S SSS S S+SSS + +++ ++ ++ S +
Sbjct: 1073 KILSSRRNSSSSSSSSSSSSSESRSSRSSSSSSSSSRSSRKIDLAARTNSSSSSSSRRSR 1132
Query: 301 PIALSSSSSDLESSSSSLDSTES 323
+ SSSSS SSSSS S S
Sbjct: 1133 SSSSSSSSSSSSSSSSSSSSRRS 1155
>ATU_DROME (Q94546) Another transcription unit protein
Length = 725
Score = 47.4 bits (111), Expect = 1e-04
Identities = 46/160 (28%), Positives = 69/160 (42%), Gaps = 12/160 (7%)
Query: 175 PKTRKRGATPT--TSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPL 232
P++RK G+T + + ++ R G+ S + + + R G +G ++ S+
Sbjct: 132 PQSRKSGSTHSRRSGSAHSRRSGSARSRKSGSAQSDRSESRSRSHSGSLKGNEESRSN-- 189
Query: 233 QDTTSIPVEMAQAKKTSGKRSSRKSGSSSSH-HSKSSTSSSPLKASGPKRVGMSKMAVP- 290
I VE A +K S RS +SGS +S SK+ T S SG S + VP
Sbjct: 190 SPNLQIDVERANSKSGSRSRSRSRSGSRTSRSRSKTGTPSPNRSRSGSASGSGSDVGVPK 249
Query: 291 -RATWFSSKPPPIALSSSSSDLESS-----SSSLDSTESD 324
+A S S S SD+E S S L T+SD
Sbjct: 250 KKARKASGSDQEKKKSGSDSDIEESPTKAKKSRLIDTDSD 289
Score = 31.2 bits (69), Expect = 8.9
Identities = 29/122 (23%), Positives = 47/122 (37%), Gaps = 4/122 (3%)
Query: 160 DDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSP-AKSASQREEMQQ 218
DDD + T + G+ P + S++ G+ S G+ S ++S S +
Sbjct: 7 DDDSGSSGSSRSGSRSVTPQGGSAPGSQRSRRSGSGSDRSRSGSRSSRSRSGSGSPRSAR 66
Query: 219 GGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASG 278
G + S L + + + SG SRKSG+ S S S S K+
Sbjct: 67 SGSA---ESRHSQLSGSARSKRSRSAHSRRSGSARSRKSGTPESPQSHRSGSLQSRKSGS 123
Query: 279 PK 280
P+
Sbjct: 124 PQ 125
Score = 31.2 bits (69), Expect = 8.9
Identities = 26/100 (26%), Positives = 41/100 (41%), Gaps = 5/100 (5%)
Query: 187 SASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAK 246
SA KR++ A G+ KS + +++ SP + P Q++
Sbjct: 80 SARSKRSRSAHSRRSGSARSRKSGTPESPQSHRSGSLQSRKSGSPQSRRSGSP----QSR 135
Query: 247 KTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSK 286
K SG SR+SGS+ S S S+ S A + S+
Sbjct: 136 K-SGSTHSRRSGSAHSRRSGSARSRKSGSAQSDRSESRSR 174
>YB1E_SCHPO (P87179) Serine-rich protein C30B4.01c precursor
Length = 374
Score = 47.0 bits (110), Expect = 2e-04
Identities = 35/96 (36%), Positives = 46/96 (47%), Gaps = 5/96 (5%)
Query: 228 ASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKM 287
+SSP +T+ + + +S SS S SSSS S SS+SSS +S S
Sbjct: 141 SSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 200
Query: 288 AVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
+ SS PI S+SSS SSSSS S+ S
Sbjct: 201 SSS-----SSSSVPITSSTSSSHSSSSSSSSSSSSS 231
Score = 47.0 bits (110), Expect = 2e-04
Identities = 38/150 (25%), Positives = 65/150 (43%), Gaps = 8/150 (5%)
Query: 177 TRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTT 236
T + +T++S + + ST SP+ S+S +SS ++
Sbjct: 126 TVSSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 185
Query: 237 SIPVEMAQAKKTSGKRSSRKSG-----SSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPR 291
S + + +S SS S S+SS HS SS+SSS +S S +
Sbjct: 186 SSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSSSSSSSRPSSSSSFITTMS 245
Query: 292 ATWFSSK---PPPIALSSSSSDLESSSSSL 318
++ F S P + SS+SS++ SS+++L
Sbjct: 246 SSTFISTVTVTPSSSSSSTSSEVPSSTAAL 275
Score = 43.9 bits (102), Expect = 0.001
Identities = 34/95 (35%), Positives = 43/95 (44%)
Query: 229 SSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMA 288
SS TT+ P + + +S SS S SSSS S SS+SSS +S S +
Sbjct: 145 SSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 204
Query: 289 VPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
SS + SSSSS SSSS S+ S
Sbjct: 205 SSSVPITSSTSSSHSSSSSSSSSSSSSSRPSSSSS 239
Score = 42.4 bits (98), Expect = 0.004
Identities = 42/143 (29%), Positives = 60/143 (41%), Gaps = 25/143 (17%)
Query: 181 GATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPV 240
G TT +S + S+ +PS + + + +SP ++S
Sbjct: 121 GVLQTTVSSSSVSSTTSSSSSSSPSSSSTTT----------------TTSPSSSSSS--- 161
Query: 241 EMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPP 300
+ + +S SS S SSSS S SS+SSS +S S +VP + SS
Sbjct: 162 --SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHS 219
Query: 301 PIALSSSSSDLESSSSSLDSTES 323
SSSSS SSSSS S+ S
Sbjct: 220 ----SSSSSSSSSSSSSRPSSSS 238
Score = 39.3 bits (90), Expect = 0.033
Identities = 33/99 (33%), Positives = 45/99 (45%), Gaps = 16/99 (16%)
Query: 228 ASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKM 287
+SS + TTS + + ++ +S S SSSS S SS+SSS +S
Sbjct: 128 SSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSS-------- 179
Query: 288 AVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESDPV 326
SS + SSSSS SSSSS S+ S P+
Sbjct: 180 --------SSSSSSSSSSSSSSSSSSSSSSSSSSSSVPI 210
Score = 36.2 bits (82), Expect = 0.28
Identities = 30/91 (32%), Positives = 42/91 (45%)
Query: 249 SGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSS 308
+G + S SS S + SS+SSSP +S S + ++ SS + SSSS
Sbjct: 120 NGVLQTTVSSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSS 179
Query: 309 SDLESSSSSLDSTESDPVWDKLTFYFNSKPI 339
S SSSSS S+ S + +S PI
Sbjct: 180 SSSSSSSSSSSSSSSSSSSSSSSSSSSSVPI 210
>PPRB_HUMAN (Q15648) Peroxisome proliferator-activated receptor
binding protein (PBP) (PPAR binding protein) (Thyroid
hormone receptor-associated protein complex 220 kDa
component) (Trap220) (Thyroid receptor interacting
protein 2) (TRIP2) (p53 regulatory
Length = 1581
Score = 47.0 bits (110), Expect = 2e-04
Identities = 83/310 (26%), Positives = 114/310 (36%), Gaps = 53/310 (17%)
Query: 171 KLKIPKTRK---RGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQ 227
K K PK +K G +P+ S+S R ST G+ SP + G Q
Sbjct: 1006 KDKPPKRKKADTEGKSPSHSSSN-RPFTPPTSTGGSKSPGSA---------GRSQTPPGV 1055
Query: 228 ASSPLQDTT-SIPVEMAQAKKTSGKRSSRKSGS-----SSSHHSKSSTSSSPLKASGPKR 281
A+ P+ T IP K S SGS S SHHS SS+SSS SG +
Sbjct: 1056 ATPPIPKITIQIPKGTVMVGKPSSHSQYTSSGSVSSSGSKSHHSHSSSSSSSASTSGKMK 1115
Query: 282 VGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESDPVWDKLTFYFNSKPIAW 341
SSK + S SS + SS S S++S NS
Sbjct: 1116 --------------SSKSEGSSSSKLSSSMYSSQGSSGSSQSK----------NSSQSGG 1151
Query: 342 QHPGCEPYYTKECRDNLSNFPPQDMQNRPSPAREENI-------PHDEVQGGEDPIIQP- 393
+ PG P TK + S+ Q +PS ++ H GG D + P
Sbjct: 1152 K-PGSSP-ITKHGLSSGSSSTKMKPQGKPSSLMNPSLSKPNISPSHSRPPGGSDKLASPM 1209
Query: 394 DQVIGVMPPPKADVLASAHSEPSPRQFQTSPQQPRLSPQRLQQDARAALLSPHAKGGSAS 453
V G P KA S+ S S +S + S + + P + +AS
Sbjct: 1210 KPVPGTPPSSKAKSPISSGSGGSHMSGTSSSSGMKSSSGLGSSGSLSQKTPPSSNSCTAS 1269
Query: 454 SKTAASSHTS 463
S + +SS +S
Sbjct: 1270 SSSFSSSGSS 1279
Score = 39.7 bits (91), Expect = 0.025
Identities = 75/300 (25%), Positives = 122/300 (40%), Gaps = 71/300 (23%)
Query: 179 KRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSI 238
K G + +S+++ + +G S + NPS +K + G G D+ AS +
Sbjct: 1160 KHGLSSGSSSTKMKPQGKPSSLM-NPSLSKPNISPSHSRPPG--GSDKLASPMKPVPGTP 1216
Query: 239 PVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSK 298
P A++ +SG S SG+SSS K SSS L +SG S K
Sbjct: 1217 PSSKAKSPISSGSGGSHMSGTSSSSGMK---SSSGLGSSGS---------------LSQK 1258
Query: 299 PPPIALS--SSSSDLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKECRD 356
PP + S +SSS SS SS+ S+++ + +SK G P K
Sbjct: 1259 TPPSSNSCTASSSSFSSSGSSMSSSQNQ--------HGSSK-------GKSPSRNK---- 1299
Query: 357 NLSNFPPQDMQNRPS-PAREENIPHDEV---QGGEDPIIQPDQVIGVMPPPKADVLASAH 412
+PS A + + H V GGEDP+ D +GV + ++S H
Sbjct: 1300 ------------KPSLTAVIDKLKHGVVTSGPGGEDPL---DGQMGVSTNSSSHPMSSKH 1344
Query: 413 SEPSPRQFQTSPQQPRLSPQRLQQDARAALLSPHAKGGSASSKTAASSHTSSERLSALLV 472
+ S +FQ +R + D + +S +S KT+ S + S ++ +++
Sbjct: 1345 N-MSGGEFQ---------GKREKSDKDKSKVSTSGSSVDSSKKTSESKNVGSTGVAKIII 1394
Score = 33.1 bits (74), Expect = 2.4
Identities = 30/111 (27%), Positives = 45/111 (40%), Gaps = 11/111 (9%)
Query: 169 ADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQA 228
+DKL P G P++ A + G+GGS + S S+S + G G Q
Sbjct: 1202 SDKLASPMKPVPGTPPSSKAKSPISSGSGGSHMSGTS---SSSGMKSSSGLGSSGSLSQK 1258
Query: 229 SSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGP 279
+ P ++ + +S SS S SSS + SS SP + P
Sbjct: 1259 TPPSSNSCT--------ASSSSFSSSGSSMSSSQNQHGSSKGKSPSRNKKP 1301
>DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contains:
Dentin phosphoprotein (Dentin phosphophoryn) (DPP);
Dentin sialoprotein (DSP)]
Length = 1253
Score = 45.8 bits (107), Expect = 4e-04
Identities = 39/165 (23%), Positives = 64/165 (38%), Gaps = 4/165 (2%)
Query: 159 EDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQ 218
EDDD D + + G + K G G S + + S++ +
Sbjct: 511 EDDDSDSTSDTNNSDSNGNGNNG----NDDNDKSDSGKGKSDSSDSDSSDSSNSSDSSDS 566
Query: 219 GGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASG 278
D +SS + S + + + + SS S SS S S S+ SS K+
Sbjct: 567 SDSDSSDSNSSSDSDSSDSDSSDSSDSDSSDSSNSSDSSDSSDSSDSSDSSDSSDSKSDS 626
Query: 279 PKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
K S + ++ S + +S SSD +SS+S DS++S
Sbjct: 627 SKSESDSSDSDSKSDSSDSNSSDSSDNSDSSDSSNSSNSSDSSDS 671
Score = 39.7 bits (91), Expect = 0.025
Identities = 37/165 (22%), Positives = 59/165 (35%), Gaps = 4/165 (2%)
Query: 159 EDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQ 218
+ D D + +D R + + S+ + + S+ + S + S
Sbjct: 843 DSSDSSDSSDSDS----SNRSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSNESSNSSDSS 898
Query: 219 GGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASG 278
D +S + S + S S + S SS+ S SS SS +S
Sbjct: 899 DSSNSSDSDSSDSSNSSDSSDSSNSSDSSESSNSSDNSNSSDSSNSSDSSDSSDSSNSSD 958
Query: 279 PKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
G S + + SS + SS SSD SS S DS++S
Sbjct: 959 SSNSGDSSNSSDSSDSNSSDSSDSSNSSDSSDSSDSSDSSDSSDS 1003
Score = 39.3 bits (90), Expect = 0.033
Identities = 38/163 (23%), Positives = 59/163 (35%), Gaps = 1/163 (0%)
Query: 161 DDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGG 220
D D +D + + +S S + + S N S + +S +
Sbjct: 739 DSSDSSDSSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSSDSSNSSDSNDSSNSSDSSDSS 798
Query: 221 DQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPK 280
+ D SS D++ + S S S+SS S SS SS +
Sbjct: 799 NSS-DSSNSSDSSDSSDSSDSDSSNSSDSSNSSDSSDSSNSSDSSDSSDSSDSSDSDSSN 857
Query: 281 RVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
R S + + SS + SS SSD SS+S DS++S
Sbjct: 858 RSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSNESSNSSDSSDS 900
Score = 38.9 bits (89), Expect = 0.043
Identities = 35/143 (24%), Positives = 57/143 (39%), Gaps = 6/143 (4%)
Query: 182 ATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVE 241
++ ++ +S + + S + S + +S D D SS D++
Sbjct: 718 SSDSSDSSNSNSSDSDSSNSSDSSDSSDSSDSSNSSDSSDSS-DSSNSSDSSDSSDSSDS 776
Query: 242 MAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPP 301
+ + SS S SS S +S S++SS S S + SS
Sbjct: 777 SDSSNSSDSNDSSNSSDSSDSSNSSDSSNSSDSSDSSDSSDSDSSNSSD-----SSNSSD 831
Query: 302 IALSSSSSDLESSSSSLDSTESD 324
+ SS+SSD SS S DS++SD
Sbjct: 832 SSDSSNSSDSSDSSDSSDSSDSD 854
Score = 38.1 bits (87), Expect = 0.073
Identities = 38/165 (23%), Positives = 66/165 (39%), Gaps = 2/165 (1%)
Query: 160 DDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQG 219
D D +D + ++ ++ +S + + S+ + S + S+ +
Sbjct: 538 DKSDSGKGKSDSSDSDSSDSSNSSDSSDSSDSDSSDSNSSSDSDSSDSDSSDSSDSDSSD 597
Query: 220 GDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSS-SHHSKSSTSSSPLKASG 278
D SS D++ + + +K S K S S S S S S S++S S +
Sbjct: 598 SSNSSDSSDSSDSSDSSDSS-DSSDSKSDSSKSESDSSDSDSKSDSSDSNSSDSSDNSDS 656
Query: 279 PKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
S + + SS + SSSSSD +SS S DS++S
Sbjct: 657 SDSSNSSNSSDSSDSSDSSDSSSSSDSSSSSDSSNSSDSSDSSDS 701
Score = 38.1 bits (87), Expect = 0.073
Identities = 33/140 (23%), Positives = 58/140 (40%), Gaps = 4/140 (2%)
Query: 185 TTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQ 244
++ + + + S N S + +S + D + SS + +S +
Sbjct: 582 SSDSDSSDSSDSDSSDSSNSSDSSDSSDSSDSSDSSDSSDSKSDSSKSESDSSDSDSKSD 641
Query: 245 AKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIAL 304
+ ++ SS S SS S +S +S+ SS S S + ++ SS +
Sbjct: 642 SSDSNSSDSSDNSDSSDSSNSSNSSDSSDSSDSSDS----SSSSDSSSSSDSSNSSDSSD 697
Query: 305 SSSSSDLESSSSSLDSTESD 324
SS SS+ SS S DS++SD
Sbjct: 698 SSDSSNSSESSDSSDSSDSD 717
Score = 37.0 bits (84), Expect = 0.16
Identities = 36/141 (25%), Positives = 55/141 (38%), Gaps = 3/141 (2%)
Query: 185 TTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQG--VDQQASSPLQDTTSIPVEM 242
+ S+ + + S N S + +S + D D SS D+++
Sbjct: 966 SNSSDSSDSNSSDSSDSSNSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSNSSDSS 1025
Query: 243 AQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPI 302
+ + SS S SS S +S S+ SS S G S + + SS
Sbjct: 1026 NSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSS-GSSDSSDSSDSSDSSDSSDS 1084
Query: 303 ALSSSSSDLESSSSSLDSTES 323
+ SS SSD SS S DS++S
Sbjct: 1085 SDSSDSSDSSESSDSSDSSDS 1105
Score = 36.6 bits (83), Expect = 0.21
Identities = 38/164 (23%), Positives = 58/164 (35%), Gaps = 8/164 (4%)
Query: 161 DDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGG 220
D D +D + + +S S + +G S + S + +S +
Sbjct: 1029 DSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSSGSSDSSDSSDSSDSSDSSDSSDSS 1088
Query: 221 DQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPK 280
D D SS D++ + + SS S SS S +S S+ SS S
Sbjct: 1089 DSS-DSSESSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDS 1147
Query: 281 RVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESD 324
SS + SS SSD SS S DS++S+
Sbjct: 1148 SDSSDSSD-------SSDSSDSSDSSDSSDSSDSSDSSDSSDSN 1184
Score = 36.6 bits (83), Expect = 0.21
Identities = 35/163 (21%), Positives = 61/163 (36%), Gaps = 2/163 (1%)
Query: 161 DDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGG 220
D D +D + + +S S + + S N S + ++S +
Sbjct: 978 DSSDSSNSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSNSSDSSNSSDSSDSSDSS 1037
Query: 221 DQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPK 280
D +S+ + S + + + +SG S S SS S +S S + +
Sbjct: 1038 DSSDSSDSSNSSDSSDSS--DSSDSSDSSGSSDSSDSSDSSDSSDSSDSSDSSDSSDSSE 1095
Query: 281 RVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
S + + SS + SS SSD SS+S DS++S
Sbjct: 1096 SSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSNSSDSSDS 1138
Score = 36.2 bits (82), Expect = 0.28
Identities = 38/162 (23%), Positives = 57/162 (34%), Gaps = 11/162 (6%)
Query: 162 DDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGD 221
D D +D + + +S S + + S + S + +S + D
Sbjct: 970 DSSDSNSSDSSDSSNSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSNSSDSSNSSD 1029
Query: 222 QGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKR 281
D SS D++ + + SS SGSS S S S+ SS S
Sbjct: 1030 SS-DSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSSGSSDSSDSSDSSDSSDSSDSSDSS 1088
Query: 282 VGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
SS+ + SS SSD SS S DS++S
Sbjct: 1089 DSSD----------SSESSDSSDSSDSSDSSDSSDSSDSSDS 1120
Score = 35.8 bits (81), Expect = 0.36
Identities = 35/138 (25%), Positives = 52/138 (37%), Gaps = 1/138 (0%)
Query: 187 SASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAK 246
S+ + + S N + + S S D SS D++ +
Sbjct: 710 SSDSSDSDSSDSSDSSNSNSSDSDSSNSSDSSDSSDSSDSSNSSDSSDSSDSSNSSDSSD 769
Query: 247 KTSGKRSSRKSGSSSSHHSK-SSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALS 305
+ SS S SS S+ S SS SS +S S + + SS + S
Sbjct: 770 SSDSSDSSDSSNSSDSNDSSNSSDSSDSSNSSDSSNSSDSSDSSDSSDSDSSNSSDSSNS 829
Query: 306 SSSSDLESSSSSLDSTES 323
S SSD +SS S DS++S
Sbjct: 830 SDSSDSSNSSDSSDSSDS 847
Score = 33.1 bits (74), Expect = 2.4
Identities = 35/136 (25%), Positives = 52/136 (37%), Gaps = 2/136 (1%)
Query: 186 TSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQA 245
+S S + + S + S + +S + D D SS D+++ +
Sbjct: 1081 SSDSSDSSDSSDSSESSDSSDSSDSSDSSDSSDSSDSS-DSSDSSDSSDSSNSSDSSDSS 1139
Query: 246 KKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALS 305
+ SS S SS S S S+ SS S S + + SS + S
Sbjct: 1140 DSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSS-DSSDSSDSNESSDSSDSSDSSDS 1198
Query: 306 SSSSDLESSSSSLDST 321
S+SSD SS S DST
Sbjct: 1199 SNSSDSSDSSDSSDST 1214
Score = 32.7 bits (73), Expect = 3.1
Identities = 35/138 (25%), Positives = 53/138 (38%), Gaps = 2/138 (1%)
Query: 186 TSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQA 245
+S S + + S + S + +S + D D SS D++ +
Sbjct: 1072 SSDSSDSSDSSDSSDSSDSSDSSESSDSSDSSDSSDSS-DSSDSSDSSDSSDSSDSSDSS 1130
Query: 246 KKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALS 305
+ SS S SS S S S+ SS S S + + SS + S
Sbjct: 1131 NSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSS-DSSDSSDSSDSSDSSDSNESSDS 1189
Query: 306 SSSSDLESSSSSLDSTES 323
S SSD SS+S DS++S
Sbjct: 1190 SDSSDSSDSSNSSDSSDS 1207
Score = 32.3 bits (72), Expect = 4.0
Identities = 34/138 (24%), Positives = 54/138 (38%), Gaps = 2/138 (1%)
Query: 186 TSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQA 245
+S S + + S + S + +S + D D SS D++ +
Sbjct: 1075 SSDSSDSSDSSDSSDSSDSSESSDSSDSSDSSDSSDSS-DSSDSSDSSDSSDSSDSSNSS 1133
Query: 246 KKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALS 305
+ SS S SS S S S+ SS S S + + S++ + S
Sbjct: 1134 DSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSS-DSSDSSDSSDSSDSNESSDSSDS 1192
Query: 306 SSSSDLESSSSSLDSTES 323
S SSD +SS S DS++S
Sbjct: 1193 SDSSDSSNSSDSSDSSDS 1210
>CCT_DROME (O96433) Cyclin T
Length = 1097
Score = 45.1 bits (105), Expect = 6e-04
Identities = 69/321 (21%), Positives = 117/321 (35%), Gaps = 59/321 (18%)
Query: 176 KTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDT 235
K+++ G +S +R+ +G + G+ S S+S QQ G + +
Sbjct: 409 KSKQPGYNNRMPSSHQRSSSSGLGSSGSGSQRSSSSSSSSSQQPGRPSMPVDYHKSSRGM 468
Query: 236 TSIPVEM---AQAKKTSGKRSSRKSGSSSSHHSKSSTSSS-------------------P 273
+ V M K TSG + + H S S++S+S P
Sbjct: 469 PPVGVGMPPHGSHKMTSGSKPQQPQQQPVPHPSASNSSASGMSSKDKSQSNKMYPNAPPP 528
Query: 274 LKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESDPVWDKLTFY 333
S P+ MS+ P A+ S PPP S S+ + E + + Y
Sbjct: 529 YSNSAPQNPLMSRGGYPGASNGSQPPPPAGYGGHRSKSGSTVHGMPPFEQQLPYSQSQSY 588
Query: 334 --FNSKPI----AWQHPGCEPYYTKECRDNLSNFPPQDMQNRPSPAREENIPHDEVQGGE 387
+P+ Q P ++ + +++L F P+ + P +
Sbjct: 589 GHMQQQPVPQSQQQQMPPEASQHSLQSKNSL--FSPEWPDIKKEPMSQS----------- 635
Query: 388 DPIIQPDQVIGVMPPPKA---DVLASAHSEPSPRQFQTSPQQPRLSPQRLQQDARAALLS 444
QP G++PPP D ++H PR + SP++ RL+P + +D ++
Sbjct: 636 ----QPQPFNGLLPPPAPPGHDYKLNSH----PRD-KESPKKERLTPTK--KDKHRPVMP 684
Query: 445 PHAKG----GSASSKTAASSH 461
P G GS SSK H
Sbjct: 685 PVGSGNSSSGSGSSKPMLPPH 705
>TRIB_HUMAN (Q14669) Thyroid receptor interacting protein 12
(TRIP12)
Length = 1992
Score = 43.9 bits (102), Expect = 0.001
Identities = 43/187 (22%), Positives = 74/187 (38%), Gaps = 14/187 (7%)
Query: 143 PTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAK-------- 194
P+ P S ++ + A + TRK GAT S SQKR +
Sbjct: 101 PSETNKPHSKSKKRHLDQEQQLKSAQSPSTSKAHTRKSGATGG-SRSQKRKRTESSCVKS 159
Query: 195 GAGGSTVG----NPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSG 250
G+G + G + P K AS+ + G + +S+ ++S V A + G
Sbjct: 160 GSGSESTGAEERSAKPTKLASKSATSAKAGCSTITDSSSAASTSSSSSAVASASSTVPPG 219
Query: 251 KRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSD 310
R + + + S+S++S SP ++S K + + W + P ++L +
Sbjct: 220 ARVKQGKDQNKARRSRSASSPSPRRSSREKEQSKTG-GSSKFDWAARFSPKVSLPKTKLS 278
Query: 311 LESSSSS 317
L SS S
Sbjct: 279 LPGSSKS 285
Score = 40.8 bits (94), Expect = 0.011
Identities = 76/334 (22%), Positives = 116/334 (33%), Gaps = 44/334 (13%)
Query: 152 ASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSAS 211
+SA + + +D D +++ K + K+ S KR+ + +PS AK
Sbjct: 38 SSAVIVPQPEDPDRANTSERQKTGQVPKKD----NSRGVKRSASPDYNRTNSPSSAKKPK 93
Query: 212 QREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGS---SSSHHSKSS 268
+ + + S ++ AQ+ TS K +RKSG+ S S K +
Sbjct: 94 ALQHTESPSETNKPHSKSKKRHLDQEQQLKSAQSPSTS-KAHTRKSGATGGSRSQKRKRT 152
Query: 269 TSSSPLKASGPKRVGM-------SKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDST 321
SS SG + G +K+A AT + I SSS++ SSSS++ S
Sbjct: 153 ESSCVKSGSGSESTGAEERSAKPTKLASKSATSAKAGCSTITDSSSAASTSSSSSAVASA 212
Query: 322 ESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNRPSPAREENIPHD 381
S P A G + + R S PSP R
Sbjct: 213 SS-----------TVPPGARVKQGKDQNKARRSRSASS----------PSPRRSSREKEQ 251
Query: 382 EVQGGEDPIIQPDQVIGVMPPPKADV-----LASAHSEPSPRQFQTSPQQPRLSPQRLQQ 436
GG + + PK + S S+P P Q R S ++ +
Sbjct: 252 SKTGGSSKFDWAARFSPKVSLPKTKLSLPGSSKSETSKPGPSGLQAKLASLRKSTKKRSE 311
Query: 437 DARAALLSPHAKGGSASSKTAASSHTSSERLSAL 470
A L S S KT S ++S R S L
Sbjct: 312 SPPAELPSLRR---STRQKTTGSCASTSRRGSGL 342
>GAR2_SCHPO (P41891) Protein gar2
Length = 500
Score = 43.9 bits (102), Expect = 0.001
Identities = 49/193 (25%), Positives = 74/193 (37%), Gaps = 29/193 (15%)
Query: 166 VALADKLKIPKT-----RKRGATPTTSASQKRAKGAG-------GSTVGNPSPAKSASQR 213
+A DK + K+ K+GA S S+K K A T +P +K ++R
Sbjct: 1 MAKKDKTSVKKSVKETASKKGAIEKPSKSKKITKEAAKEIAKQSSKTDVSPKKSKKEAKR 60
Query: 214 EEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSP 273
+ + V +Q S ++ +S E + S SS SSS S SS SSS
Sbjct: 61 ASSPEPSKKSVKKQKKSKKKEESSSESESESSSSESESSSSESESSSSESESSSSESSSS 120
Query: 274 -----------------LKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSS 316
++S + AV + + SSSS+ ES SS
Sbjct: 121 ESEEEVIVKTEEKKESSSESSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESESESS 180
Query: 317 SLDSTESDPVWDK 329
S +S E + V +K
Sbjct: 181 SSESEEEEEVVEK 193
Score = 39.7 bits (91), Expect = 0.025
Identities = 34/151 (22%), Positives = 67/151 (43%), Gaps = 8/151 (5%)
Query: 176 KTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEM----QQGGDQGVDQQASSP 231
++ ++ + +S ++ + + + S + S+ EE+ ++ + + +SS
Sbjct: 86 ESESESSSSESESSSSESESSSSESESSSSESSSSESEEEVIVKTEEKKESSSESSSSSE 145
Query: 232 LQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSP---LKASGPKRVGMSKMA 288
++ V++ + K++S SS S S S S SS S ++ + K+ G S+ +
Sbjct: 146 SEEEEEAVVKIEEKKESSSDSSSESSSSESESESSSSESEEEEEVVEKTEEKKEGSSESS 205
Query: 289 VPRATWFSSKPPPIALSSSSSDLESSSSSLD 319
+ S S SSSD ES SSS D
Sbjct: 206 SDSES-SSDSSSESGDSDSSSDSESESSSED 235
>VIT1_CHICK (P87498) Vitellogenin I precursor (Minor vitellogenin)
[Contains: Lipovitellin I (LVI); Phosvitin (PV);
Lipovitellin II (LVII); YGP42]
Length = 1912
Score = 43.5 bits (101), Expect = 0.002
Identities = 42/136 (30%), Positives = 63/136 (45%), Gaps = 15/136 (11%)
Query: 182 ATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVE 241
+TP++S S RA + G N +S + ++ GD +SS ++S
Sbjct: 1112 STPSSSDSDNRA--SQGDPQINLKSRQSKANEKKFYPFGDSSSSGSSSSSSSSSSS---- 1165
Query: 242 MAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPP 301
+ +S RSS S SSSS S SS+SSS K+S S+ + + SS
Sbjct: 1166 --SSDSSSSSRSSSSSDSSSSSSSSSSSSSSKSKSS-------SRSSKSNRSSSSSNSKD 1216
Query: 302 IALSSSSSDLESSSSS 317
+ SSS S+ + SSSS
Sbjct: 1217 SSSSSSKSNSKGSSSS 1232
Score = 40.0 bits (92), Expect = 0.019
Identities = 54/225 (24%), Positives = 85/225 (37%), Gaps = 32/225 (14%)
Query: 122 VEIFNHISIRTLIGQAEPIPRPTP--------QASPRVASATVEMEDDDDDDVALADKLK 173
V ++N I L +P+ P QA + + V + +D + + + +
Sbjct: 1014 VPLYNAIGEHALRMSFKPVYSDVPIEKIQVTIQAGDQAPTKMVRLVTFEDPERQESSRKE 1073
Query: 174 IPKTRKR-------GATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQ---- 222
+ K K+ AT + +S A S+ S S+ QG Q
Sbjct: 1074 VMKRVKKILDDTDNQATRNSRSSSSSASSISESSESTTSTPSSSDSDNRASQGDPQINLK 1133
Query: 223 ----GVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASG 278
+++ P D++S + +S SS SSSS S SS SSS +S
Sbjct: 1134 SRQSKANEKKFYPFGDSSS----SGSSSSSSSSSSSSSDSSSSSRSSSSSDSSSSSSSSS 1189
Query: 279 PKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
SK ++ SSK + SS+S D SSSS +S S
Sbjct: 1190 SSSSSKSK-----SSSRSSKSNRSSSSSNSKDSSSSSSKSNSKGS 1229
Score = 39.7 bits (91), Expect = 0.025
Identities = 37/176 (21%), Positives = 70/176 (39%), Gaps = 3/176 (1%)
Query: 152 ASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSAS 211
++ + D D+ + D K+R+ A + +G S+ + S + S+
Sbjct: 1109 STTSTPSSSDSDNRASQGDPQINLKSRQSKANEKKFYPFGDSSSSGSSSSSSSSSSSSSD 1168
Query: 212 QREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSS--- 268
+ +SS ++S +++ K++ SS S SSS SKS+
Sbjct: 1169 SSSSSRSSSSSDSSSSSSSSSSSSSSKSKSSSRSSKSNRSSSSSNSKDSSSSSSKSNSKG 1228
Query: 269 TSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESD 324
+SSS KASG ++ + S+ ++ + +SSSSS S+
Sbjct: 1229 SSSSSSKASGTRQKAKKQSKTTSFPHASAAEGERSVHEQKQETQSSSSSSSRASSN 1284
Score = 35.4 bits (80), Expect = 0.47
Identities = 31/119 (26%), Positives = 51/119 (42%), Gaps = 2/119 (1%)
Query: 206 PAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHS 265
P + S R+E+ + + +D + +++ S + ++S +S S S S + +
Sbjct: 1064 PERQESSRKEVMKRVKKILDDTDNQATRNSRSSSSSASSISESSESTTSTPSSSDSDNRA 1123
Query: 266 KSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESD 324
LK+ K K P SS + SSSSS +SSSSS S+ SD
Sbjct: 1124 SQGDPQINLKSRQSK--ANEKKFYPFGDSSSSGSSSSSSSSSSSSSDSSSSSRSSSSSD 1180
Score = 34.7 bits (78), Expect = 0.81
Identities = 25/108 (23%), Positives = 49/108 (45%), Gaps = 2/108 (1%)
Query: 180 RGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIP 239
+ + + S+ R+ + S + S +KS S+ G Q+A + T+
Sbjct: 1195 KSKSSSRSSKSNRSSSSSNSKDSSSSSSKSNSKGSSSSSSKASGTRQKAKKQSKTTSFPH 1254
Query: 240 VEMAQAKKT--SGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMS 285
A+ +++ K+ ++ S SSSS S +S S+S +S + G+S
Sbjct: 1255 ASAAEGERSVHEQKQETQSSSSSSSRASSNSRSTSSSTSSSSESSGVS 1302
>AF4_HUMAN (P51825) AF-4 protein (Proto-oncogene AF4) (FEL protein)
Length = 1210
Score = 43.1 bits (100), Expect = 0.002
Identities = 65/264 (24%), Positives = 88/264 (32%), Gaps = 40/264 (15%)
Query: 222 QGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKR 281
Q V + Q TS + TS S S T P +S P
Sbjct: 393 QHVSSVTQNQKQYDTSSKTHSNSQQGTSSMLEDDLQLSDSEDSDSEQTPEKPPSSSAPPS 452
Query: 282 VGMS-KMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESDPVWDKLTFYFNSKPIA 340
S V A S++ + S SSSD ES SSS DS E++P+
Sbjct: 453 APQSLPEPVASAHSSSAESESTSDSDSSSDSESESSSSDSEENEPL-------------- 498
Query: 341 WQHPGCEPYYTKECRDNLSNF---------PPQDMQNRPSPAREENIPHDEVQGGEDPII 391
+ P EP + L N+ PP+ ++ P R H E +G D
Sbjct: 499 -ETPAPEPEPPTTNKWQLDNWLTKVSQPAAPPEGPRSTEPPRR-----HPESKGSSDSAT 552
Query: 392 QPDQVIGVMPPPKADVLA----SAHSEPSPRQFQTSPQQPRLSPQRLQQDARAALLSPHA 447
+ PPPK+ A P R Q SP Q PQR +
Sbjct: 553 SQEHSESKDPPPKSSSKAPRAPPEAPHPGKRSCQKSPAQQE-PPQRQTVGTKQP-----K 606
Query: 448 KGGSASSKTAASSHTSSERLSALL 471
K AS++ + + ER LL
Sbjct: 607 KPVKASARAGSRTSLQGEREPGLL 630
Score = 31.2 bits (69), Expect = 8.9
Identities = 34/136 (25%), Positives = 52/136 (38%), Gaps = 32/136 (23%)
Query: 206 PAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRS------------ 253
P K + QR+ + G + D++S ++A+ +K +R
Sbjct: 777 PGKGSRQRKAEDKQPPAGKKHSSEKRSSDSSS---KLAKKRKGEAERDCDNKKIRLEKEI 833
Query: 254 -SRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSS---SSS 309
S+ S SSSSH S T K S P K +P PPP++ SS +
Sbjct: 834 KSQSSSSSSSHKESSKT-----KPSRPSSQSSKKEMLP--------PPPVSSSSQKPAKP 880
Query: 310 DLESSSSSLDSTESDP 325
L+ S D+ DP
Sbjct: 881 ALKRSRREADTCGQDP 896
>VTA2_XENLA (P18709) Vitellogenin A2 precursor (VTG A2) [Contains:
Lipovitellin I; Lipovitellin II; Phosvitin]
Length = 1807
Score = 42.7 bits (99), Expect = 0.003
Identities = 42/191 (21%), Positives = 75/191 (38%), Gaps = 10/191 (5%)
Query: 165 DVALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGV 224
D + + K + ++ ++S+S + + S+ +PS + S+S + ++
Sbjct: 1115 DAEVVEARKQQSSLSSSSSSSSSSSSSSSSSSSSSSSSSPSSSSSSSYSKRSKRREHNPH 1174
Query: 225 DQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASG------ 278
Q+ SS Q + G++ S SSSS S SS+SSS +S
Sbjct: 1175 HQRESSSSSSQEQNKKRNLQENRKHGQKGMSSSSSSSSSSSSSSSSSSSSSSSSSSSSEE 1234
Query: 279 ----PKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESDPVWDKLTFYF 334
R +K A ++ K + SSSSS SSS + + + L F
Sbjct: 1235 NRPHKNRQHDNKQAKMQSNQHQQKKNKFSESSSSSSSSSSSEMWNKKKHHRNFYDLNFRR 1294
Query: 335 NSKPIAWQHPG 345
++ +H G
Sbjct: 1295 TARTKGTEHRG 1305
Score = 42.0 bits (97), Expect = 0.005
Identities = 45/189 (23%), Positives = 82/189 (42%), Gaps = 13/189 (6%)
Query: 147 ASPRVAS---ATVEMEDDDD---DDVALADKLK----IPKTRKRGATPTTSASQKRAKGA 196
A P+ AS VE+E + D+ A+ +LK I ++RK S+++ K
Sbjct: 1049 AGPKAASKIMGLVEVEGTEGEPMDETAVTKRLKMILGIDESRKDTNETALYRSKQKKKNK 1108
Query: 197 GGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTS-GKRSSR 255
+ + ++ Q+ + +SS ++S + + +S KRS R
Sbjct: 1109 IHNRRLDAEVVEARKQQSSLSSSSSSSSSSSSSSSSSSSSSSSSSPSSSSSSSYSKRSKR 1168
Query: 256 KSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSS 315
+ + HH + S+SSS + + + + ++ + SS + SSSSS SSS
Sbjct: 1169 R--EHNPHHQRESSSSSSQEQNKKRNLQENRKHGQKGMSSSSSSSSSSSSSSSSSSSSSS 1226
Query: 316 SSLDSTESD 324
SS S+E +
Sbjct: 1227 SSSSSSEEN 1235
Score = 32.7 bits (73), Expect = 3.1
Identities = 61/285 (21%), Positives = 96/285 (33%), Gaps = 28/285 (9%)
Query: 200 TVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTS----------IPVEMAQAKKTS 249
T G P + ++R +M G D+ + L + + E+ +A+K
Sbjct: 1066 TEGEPMDETAVTKRLKMILGIDESRKDTNETALYRSKQKKKNKIHNRRLDAEVVEARKQQ 1125
Query: 250 GKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSS 309
SS S SSSS S SS+SSS +S P S + R+ P SSSSS
Sbjct: 1126 SSLSSSSSSSSSSSSSSSSSSSSS-SSSSPSSSSSSSYS-KRSKRREHNPHHQRESSSSS 1183
Query: 310 DLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKECRDNLSNFPPQDMQNR 369
E + K N K G + + S+ +
Sbjct: 1184 SQEQNK-------------KRNLQENRK---HGQKGMSSSSSSSSSSSSSSSSSSSSSSS 1227
Query: 370 PSPAREENIPHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSEPSPRQFQTSPQQPRL 429
S + EEN PH Q ++ +S+ S S +
Sbjct: 1228 SSSSSEENRPHKNRQHDNKQAKMQSNQHQQKKNKFSESSSSSSSSSSSEMWNKKKHHRNF 1287
Query: 430 SPQRLQQDARAALLSPHAKGGSASSKTAASSHTSSERLSALLVED 474
++ AR S+SS++++SS S+ R A + D
Sbjct: 1288 YDLNFRRTARTKGTEHRGSRLSSSSESSSSSSESAYRHKAKFLGD 1332
>RTOA_DICDI (P54681) Protein rtoA (Ratio-A)
Length = 400
Score = 42.7 bits (99), Expect = 0.003
Identities = 37/142 (26%), Positives = 58/142 (40%), Gaps = 11/142 (7%)
Query: 181 GATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPV 240
G+ +T +S ++ +G S G+ S S S E + G + + S +
Sbjct: 175 GSQGSTGSSNSGSESSGSSNSGSESSGSSNSGSESSSGSSNSGSESSSGSSNSGS----- 229
Query: 241 EMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPP 300
E + SG SS S +S S S S++S +SG G + SS
Sbjct: 230 ESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSG------SSNSG 283
Query: 301 PIALSSSSSDLESSSSSLDSTE 322
+ SS+S ESSS S S++
Sbjct: 284 SESSGSSNSGSESSSDSGSSSD 305
Score = 42.7 bits (99), Expect = 0.003
Identities = 38/136 (27%), Positives = 58/136 (41%), Gaps = 8/136 (5%)
Query: 187 SASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAK 246
S S ++ +G S G+ S + S S+ G Q ++S Q +T +Q+
Sbjct: 98 STSNSGSEASGSSNSGSQSTSNSGSEASGSSNSGSQSSTDSSNSGSQGSTGSSNSGSQSS 157
Query: 247 KTSGKRSSRKSGSSSSHHSKSST--SSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIAL 304
S S+ S SS+ S+ ST S+S ++SG G + SS
Sbjct: 158 TDSSNSGSQSSTDSSNSGSQGSTGSSNSGSESSGSSNSGSESSGSSNSGSESSS------ 211
Query: 305 SSSSSDLESSSSSLDS 320
SS+S ESSS S +S
Sbjct: 212 GSSNSGSESSSGSSNS 227
Score = 41.2 bits (95), Expect = 0.009
Identities = 37/138 (26%), Positives = 59/138 (41%), Gaps = 8/138 (5%)
Query: 187 SASQKRAKGAGGSTVGNPSPAKSASQREEMQQG----GDQGVDQQASSPLQDTTSIPVEM 242
S S ++ +G S G+ S S++ + G G Q ++S Q +T
Sbjct: 116 STSNSGSEASGSSNSGSQSSTDSSNSGSQGSTGSSNSGSQSSTDSSNSGSQSSTDSSNSG 175
Query: 243 AQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPI 302
+Q S S SGSS+S S +S+S ++S G S ++ S+
Sbjct: 176 SQGSTGSSNSGSESSGSSNSGSESSGSSNSGSESSS----GSSNSGSESSSGSSNSGSES 231
Query: 303 ALSSSSSDLESSSSSLDS 320
+ SS+S ESSS S +S
Sbjct: 232 SSGSSNSGSESSSGSSNS 249
Score = 38.9 bits (89), Expect = 0.043
Identities = 42/144 (29%), Positives = 63/144 (43%), Gaps = 12/144 (8%)
Query: 181 GATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPV 240
G+ +T +S ++G+ GS+ N S S + G ++SS ++ S
Sbjct: 164 GSQSSTDSSNSGSQGSTGSS--NSGSESSGSSNSGSESSGSSNSGSESSSGSSNSGS--- 218
Query: 241 EMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPK-RVGMSKMAVPRATWFSSKP 299
E + SG SS SGSS+S S SSS SG + G S ++ S+
Sbjct: 219 ESSSGSSNSGSESS--SGSSNS----GSESSSGSSNSGSESSSGSSNSGSESSSGSSNSG 272
Query: 300 PPIALSSSSSDLESSSSSLDSTES 323
+ SS+S ESS SS +ES
Sbjct: 273 SESSSGSSNSGSESSGSSNSGSES 296
Score = 38.5 bits (88), Expect = 0.056
Identities = 32/141 (22%), Positives = 57/141 (39%), Gaps = 1/141 (0%)
Query: 181 GATPTTSASQKRAKGA-GGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIP 239
G+ +T +S ++G+ G S G+ S S++ + Q ++ +
Sbjct: 131 GSQSSTDSSNSGSQGSTGSSNSGSQSSTDSSNSGSQSSTDSSNSGSQGSTGSSNSGSESS 190
Query: 240 VEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKP 299
++SG +S SS S +S S +SS + G S ++ S+
Sbjct: 191 GSSNSGSESSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSG 250
Query: 300 PPIALSSSSSDLESSSSSLDS 320
+ SS+S ESSS S +S
Sbjct: 251 SESSSGSSNSGSESSSGSSNS 271
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 77,708,397
Number of Sequences: 164201
Number of extensions: 3454671
Number of successful extensions: 15862
Number of sequences better than 10.0: 487
Number of HSP's better than 10.0 without gapping: 81
Number of HSP's successfully gapped in prelim test: 417
Number of HSP's that attempted gapping in prelim test: 12771
Number of HSP's gapped (non-prelim): 1618
length of query: 653
length of database: 59,974,054
effective HSP length: 117
effective length of query: 536
effective length of database: 40,762,537
effective search space: 21848719832
effective search space used: 21848719832
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0545.2


