

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
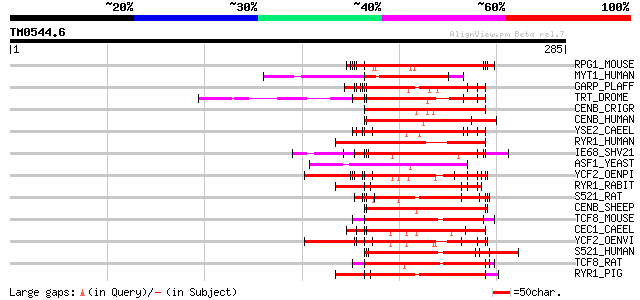
Query= TM0544.6
(285 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase regulat... 70 8e-12
MYT1_HUMAN (Q01538) Myelin transcription factor 1 (MYT1) (MYTI) ... 68 3e-11
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 66 1e-10
TRT_DROME (P19351) Troponin T, skeletal muscle (Upheld protein) ... 62 2e-09
CENB_CRIGR (P48988) Major centromere autoantigen B (Centromere p... 61 3e-09
CENB_HUMAN (P07199) Major centromere autoantigen B (Centromere p... 60 8e-09
YSE2_CAEEL (Q09936) Hypothetical protein C53C9.2 in chromosome X 59 1e-08
RYR1_HUMAN (P21817) Ryanodine receptor 1 (Skeletal muscle-type r... 59 1e-08
IE68_SHV21 (Q01042) Immediate-early protein 59 1e-08
ASF1_YEAST (P32447) Anti-silencing protein 1 59 1e-08
YCF2_OENPI (P31568) Protein ycf2 (Fragment) 59 1e-08
RYR1_RABIT (P11716) Ryanodine receptor 1 (Skeletal muscle-type r... 58 2e-08
S521_RAT (Q9QY02) Putative splicing factor YT521 (RA301-binding ... 58 3e-08
CENB_SHEEP (P49451) Major centromere autoantigen B (Centromere p... 57 4e-08
TCF8_MOUSE (Q64318) Transcription factor 8 (Zinc finger homeobox... 57 5e-08
CEC1_CAEEL (P34618) Chromo domain protein cec-1 57 5e-08
YCF2_OENVI (P31569) Protein ycf2 (Fragment) 57 7e-08
S521_HUMAN (Q96MU7) Putative splicing factor YT521 57 7e-08
TCF8_RAT (Q62947) Transcription factor 8 (Zinc finger homeodomai... 56 9e-08
RYR1_PIG (P16960) Ryanodine receptor 1 (Skeletal muscle-type rya... 56 9e-08
>RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase
regulator-interacting protein 1 (RPGR-interacting
protein 1)
Length = 1331
Score = 69.7 bits (169), Expect = 8e-12
Identities = 35/67 (52%), Positives = 50/67 (74%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
++V EE EEE EEE++++E++E++E+EEEE EEEEE EEE +++E E+EEEE E E
Sbjct: 919 EEVKEEEVEEEEEEEEEEEEVKEEKEEEEEEEREEEEEKEEEKEEEEEEDEKEEEEEEEE 978
Query: 238 REERVAE 244
EE E
Sbjct: 979 EEEEEEE 985
Score = 64.3 bits (155), Expect = 3e-10
Identities = 34/74 (45%), Positives = 53/74 (70%), Gaps = 1/74 (1%)
Query: 176 IPDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIE 235
+ ++ EEEEEE++EE +++E EE +E+EE+E +EEEE E+E +++E+EEEEEE E
Sbjct: 926 VEEEEEEEEEEEEVKEEKEEEEEEEREEEEEKEEEKEEEEEEDEKEEEEEEEEEEEEEEE 985
Query: 236 CEREERVAEWGFHE 249
E ++ V E F E
Sbjct: 986 DENKD-VLEASFTE 998
Score = 62.0 bits (149), Expect = 2e-09
Identities = 33/75 (44%), Positives = 50/75 (66%), Gaps = 4/75 (5%)
Query: 174 HLIPDQVSLEEEE----EEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEE 229
+L P+ + E E EE EEE ++E++E++ +EEEE EEEEE++EE +++E+ E
Sbjct: 893 YLAPEGFQMSEAEKPEGEEKEEEGGEEEVKEEEVEEEEEEEEEEEEVKEEKEEEEEEERE 952
Query: 230 EEEEIECEREERVAE 244
EEEE E E+EE E
Sbjct: 953 EEEEKEEEKEEEEEE 967
Score = 59.7 bits (143), Expect = 8e-09
Identities = 33/64 (51%), Positives = 47/64 (72%), Gaps = 2/64 (3%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQ--EEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
E+EEE EEE ++E+EE++E+ EEEE+ EE+EE EEE +++EKEEE+EE E E E+
Sbjct: 911 EKEEEGGEEEVKEEEVEEEEEEEEEEEEVKEEKEEEEEEEREEEEEKEEEKEEEEEEDEK 970
Query: 241 RVAE 244
E
Sbjct: 971 EEEE 974
Score = 57.8 bits (138), Expect = 3e-08
Identities = 30/68 (44%), Positives = 46/68 (67%), Gaps = 4/68 (5%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
++V E+EEEE EE ++++E EE++E+EEEE +EEEE EEE ++E+EEE+E +
Sbjct: 937 EEVKEEKEEEEEEEREEEEEKEEEKEEEEEEDEKEEEEEEEE----EEEEEEEDENKDVL 992
Query: 238 REERVAEW 245
EW
Sbjct: 993 EASFTEEW 1000
Score = 57.8 bits (138), Expect = 3e-08
Identities = 37/73 (50%), Positives = 48/73 (65%), Gaps = 6/73 (8%)
Query: 177 PDQVSLEEE--EEEIEEEDDQDEIEEDQEQEE---EELVEEEEELEEEVALDDDEKEEEE 231
P+ EEE EEE++EE+ ++E EE++E+EE EE EEEEE EE ++EKEEEE
Sbjct: 907 PEGEEKEEEGGEEEVKEEEVEEE-EEEEEEEEEVKEEKEEEEEEEREEEEEKEEEKEEEE 965
Query: 232 EEIECEREERVAE 244
EE E E EE E
Sbjct: 966 EEDEKEEEEEEEE 978
Score = 50.1 bits (118), Expect = 6e-06
Identities = 27/61 (44%), Positives = 40/61 (65%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E EEEE +EE+ ++E EED+++EEEE EEEEE EE+ D E EE + ++++
Sbjct: 950 EREEEEEKEEEKEEEEEEDEKEEEEEEEEEEEEEEEDENKDVLEASFTEEWVPFFSQDQI 1009
Query: 243 A 243
A
Sbjct: 1010 A 1010
Score = 43.9 bits (102), Expect = 5e-04
Identities = 25/66 (37%), Positives = 39/66 (58%)
Query: 179 QVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECER 238
+V L+ + + E Q E E EE+E EEE++EE +++E+EEEEEE++ E+
Sbjct: 884 KVQLDWKSHYLAPEGFQMSEAEKPEGEEKEEEGGEEEVKEEEVEEEEEEEEEEEEVKEEK 943
Query: 239 EERVAE 244
EE E
Sbjct: 944 EEEEEE 949
Score = 37.4 bits (85), Expect = 0.042
Identities = 26/104 (25%), Positives = 47/104 (45%), Gaps = 1/104 (0%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIEC-EREER 241
EEE+E+ EEE++++E EE++E E ++++E E D+ E IE + E+
Sbjct: 965 EEEDEKEEEEEEEEEEEEEEEDENKDVLEASFTEEWVPFFSQDQIASTEIPIEAGQYPEK 1024
Query: 242 VAEWGFHEGYRRCHEMVNQHHHHHHNVHGNHHHNHIRMCGISIV 285
E R H++ + H G N + +I+
Sbjct: 1025 RKPPVIAEKKEREHQVASYSRRKHSKKPGVQDKNRMEYLSCNIL 1068
>MYT1_HUMAN (Q01538) Myelin transcription factor 1 (MYT1) (MYTI)
(Proteolipid protein binding protein) (PLPB1)
Length = 1121
Score = 67.8 bits (164), Expect = 3e-11
Identities = 39/103 (37%), Positives = 61/103 (58%), Gaps = 3/103 (2%)
Query: 131 NRNGKPSFFLRPVRKDGRLELTEVRIHRAEILHASRHDGRLTLHLIPDQVSLEEEEEEIE 190
+ G+ F++P + E+ EV R++ L + + + L EEE+ E
Sbjct: 207 SEEGEKGLFIQPEDAE---EVVEVTTERSQDLCPQSLEDAASEESSKQKGILSHEEEDEE 263
Query: 191 EEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEE 233
EE++++E EED+E+EEEE EEEEE EEE +++E+EEEEEE
Sbjct: 264 EEEEEEEEEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 306
Score = 48.5 bits (114), Expect = 2e-05
Identities = 24/43 (55%), Positives = 33/43 (75%), Gaps = 1/43 (2%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDD 225
EEEEEE EEE++++E EE++E+EEEE EEEEE +V +D
Sbjct: 275 EEEEEE-EEEEEEEEEEEEEEEEEEEEEEEEEEAAPDVIFQED 316
Score = 37.4 bits (85), Expect = 0.042
Identities = 27/88 (30%), Positives = 44/88 (49%), Gaps = 14/88 (15%)
Query: 164 ASRHDGRLTLHLIPDQVSLEEEEEEIEEEDDQD-------EIEEDQEQEEEELVEEEEEL 216
A+ +G L + P+ EE E+ E QD + ++ +++ ++ EEE
Sbjct: 205 AASEEGEKGLFIQPEDA---EEVVEVTTERSQDLCPQSLEDAASEESSKQKGILSHEEED 261
Query: 217 EEEVALDDDEKEEEEEEIECEREERVAE 244
EEE ++E+EEEEE+ E E EE E
Sbjct: 262 EEE----EEEEEEEEEDEEEEEEEEEEE 285
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 65.9 bits (159), Expect = 1e-10
Identities = 35/64 (54%), Positives = 48/64 (74%), Gaps = 2/64 (3%)
Query: 181 SLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
S E +EE E ++D++E+EED+E+EEEE EEEEE EEE +++E+EEEEEE E E +E
Sbjct: 555 SKEVQEESKEVQEDEEEVEEDEEEEEEE--EEEEEEEEEEEEEEEEEEEEEEEDEDEEDE 612
Query: 241 RVAE 244
AE
Sbjct: 613 DDAE 616
Score = 65.1 bits (157), Expect = 2e-10
Identities = 33/62 (53%), Positives = 46/62 (73%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
EEE EE EEE++++E EE++E+EEEE EEEEE EEE ++DE + EE+E + E +E
Sbjct: 569 EEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDD 628
Query: 243 AE 244
AE
Sbjct: 629 AE 630
Score = 62.8 bits (151), Expect = 9e-10
Identities = 31/64 (48%), Positives = 47/64 (73%), Gaps = 2/64 (3%)
Query: 183 EEEEEEIEEEDDQDEIEEDQE--QEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEEEEE E+E+D+D+ EED++ +E+E+ EE+++ E++ DDDE E+E+EE E E EE
Sbjct: 600 EEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEEEEEE 659
Query: 241 RVAE 244
E
Sbjct: 660 EEEE 663
Score = 62.8 bits (151), Expect = 9e-10
Identities = 28/58 (48%), Positives = 46/58 (79%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEEEEE EEE++++E EE++E+EEEE +E+EE E++ D+D+ EE+E++ E + +E
Sbjct: 578 EEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDE 635
Score = 62.4 bits (150), Expect = 1e-09
Identities = 28/58 (48%), Positives = 45/58 (77%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEEEEE EEE++++E EE++E+EEEE EE+E+ E+E ++DE + EE+E + E ++
Sbjct: 576 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDD 633
Score = 62.4 bits (150), Expect = 1e-09
Identities = 28/59 (47%), Positives = 48/59 (80%), Gaps = 2/59 (3%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
++E+EEE+EE+++++E EE++E+EEEE EEEEE EEE ++DE EE+E++ E + ++
Sbjct: 565 VQEDEEEVEEDEEEEEEEEEEEEEEEE--EEEEEEEEEEEEEEDEDEEDEDDAEEDEDD 621
Score = 61.6 bits (148), Expect = 2e-09
Identities = 31/61 (50%), Positives = 45/61 (72%)
Query: 184 EEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVA 243
+E +E EEE ++DE EE++E+EEEE EEEEE EEE +++E E+EE+E + E +E A
Sbjct: 563 KEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDA 622
Query: 244 E 244
E
Sbjct: 623 E 623
Score = 60.8 bits (146), Expect = 4e-09
Identities = 30/58 (51%), Positives = 44/58 (75%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
E+EEEE EEE++++E EE++E+EEEE EEEE+ +EE D +E E++ EE E + EE
Sbjct: 574 EDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEE 631
Score = 60.5 bits (145), Expect = 5e-09
Identities = 29/63 (46%), Positives = 48/63 (76%), Gaps = 4/63 (6%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
++V +EEEEE EEE++++E EE++E+EEEE EEE+E EE D+D+ EE+E++ E +
Sbjct: 570 EEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEE----DEDDAEEDEDDAEED 625
Query: 238 REE 240
++
Sbjct: 626 EDD 628
Score = 59.7 bits (143), Expect = 8e-09
Identities = 32/70 (45%), Positives = 46/70 (65%), Gaps = 8/70 (11%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEE--------EVALDDDEKEEEEEEI 234
EEEEEE EEED+ +E E+D E++E++ E+E++ EE E D+DE E+EE+E
Sbjct: 596 EEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEE 655
Query: 235 ECEREERVAE 244
E E EE +E
Sbjct: 656 EEEEEEEESE 665
Score = 59.3 bits (142), Expect = 1e-08
Identities = 29/58 (50%), Positives = 44/58 (75%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EE +E E+E++ +E EE++E+EEEE EEEEE EEE +++E+E+E+EE E + EE
Sbjct: 560 EESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEE 617
Score = 58.9 bits (141), Expect = 1e-08
Identities = 29/65 (44%), Positives = 49/65 (74%), Gaps = 3/65 (4%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEE---ELEEEVALDDDEKEEEEEEIECERE 239
EEEEEE EEE++++E EED+++E+E+ EE+E E +E+ A +DD++E+++EE + E E
Sbjct: 588 EEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDE 647
Query: 240 ERVAE 244
+ E
Sbjct: 648 DEDEE 652
Score = 57.4 bits (137), Expect = 4e-08
Identities = 25/58 (43%), Positives = 44/58 (75%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEEEEE EEE++++E EE++E E+EE ++ EE E++ D+D+ EE+++E + + E+
Sbjct: 585 EEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEED 642
Score = 55.8 bits (133), Expect = 1e-07
Identities = 25/58 (43%), Positives = 45/58 (77%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEEEEE EEE++++E EE++E++E+E E++ E +E+ A +D++ EE+++ E + EE
Sbjct: 584 EEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEE 641
Score = 55.5 bits (132), Expect = 1e-07
Identities = 28/66 (42%), Positives = 45/66 (67%)
Query: 179 QVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECER 238
Q +++EE++ EE + E ++ +++EEE+ E+EEE EEE +++E+EEEEEE E E
Sbjct: 542 QKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEE 601
Query: 239 EERVAE 244
EE E
Sbjct: 602 EEEEDE 607
Score = 53.9 bits (128), Expect = 4e-07
Identities = 25/58 (43%), Positives = 43/58 (74%), Gaps = 3/58 (5%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIE 235
D+ EE+E++ EE++D E ++D+E ++EE +E+E+ +EE D++E+EEEEEE E
Sbjct: 611 DEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEE---DEEEEEEEEEESE 665
Score = 53.9 bits (128), Expect = 4e-07
Identities = 32/72 (44%), Positives = 48/72 (66%), Gaps = 8/72 (11%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQE-----QEEEELVEEEEELEEEVALDDDEKEEEEE 232
++ L++++ +EED ++E +E QE QE+EE VEE+EE EEE +++E+EEEEE
Sbjct: 535 EEAELQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEE---EEEEEEEEEE 591
Query: 233 EIECEREERVAE 244
E E E EE E
Sbjct: 592 EEEEEEEEEEEE 603
Score = 48.9 bits (115), Expect = 1e-05
Identities = 23/58 (39%), Positives = 41/58 (70%), Gaps = 6/58 (10%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIE 235
D+ EE+E++ EE+DD+++ +E+ + E+E+ EE+EE EEE E+EE E++I+
Sbjct: 618 DEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEEEEEE------EEEESEKKIK 669
Score = 45.8 bits (107), Expect = 1e-04
Identities = 24/72 (33%), Positives = 47/72 (64%)
Query: 173 LHLIPDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEE 232
++++P + + +++ +IEE + Q + D+E++++E +E +E +EV D++E EE+EE
Sbjct: 518 VNVVPRRDNHKKKMAKIEEAELQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEE 577
Query: 233 EIECEREERVAE 244
E E E EE E
Sbjct: 578 EEEEEEEEEEEE 589
Score = 40.0 bits (92), Expect = 0.007
Identities = 15/67 (22%), Positives = 49/67 (72%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
+Q +E+++++ EE++ + + +E ++QE++E ++E+E++++ ++ + K++EE+E + +
Sbjct: 274 EQEKIEKKKKKQEEKEKKKQEKERKKQEKKERKQKEKEMKKQKKIEKERKKKEEKEKKKK 333
Query: 238 REERVAE 244
+ ++ E
Sbjct: 334 KHDKENE 340
Score = 36.2 bits (82), Expect = 0.094
Identities = 17/65 (26%), Positives = 46/65 (70%), Gaps = 1/65 (1%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKE-EEEEEIEC 236
D+ +++E+E++E++ ++ ++ QE++E++ E+E + +E+ EKE +++++IE
Sbjct: 261 DKKERKQKEKEMKEQEKIEKKKKKQEEKEKKKQEKERKKQEKKERKQKEKEMKKQKKIEK 320
Query: 237 EREER 241
ER+++
Sbjct: 321 ERKKK 325
Score = 35.8 bits (81), Expect = 0.12
Identities = 17/67 (25%), Positives = 43/67 (63%), Gaps = 1/67 (1%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
D+ ++EE ++ D ++ ++++E +E+E +E++++ +EE EKE +++E + E
Sbjct: 246 DKKEDHQQEEMLKTLDKKERKQKEKEMKEQEKIEKKKKKQEEKEKKKQEKERKKQE-KKE 304
Query: 238 REERVAE 244
R+++ E
Sbjct: 305 RKQKEKE 311
Score = 34.3 bits (77), Expect = 0.36
Identities = 16/64 (25%), Positives = 42/64 (65%), Gaps = 7/64 (10%)
Query: 185 EEEEIEEEDDQDEIEEDQEQEE-------EELVEEEEELEEEVALDDDEKEEEEEEIECE 237
+ ++ ++E+ ++ +ED +QEE +E ++E+E++E+ ++ +K++EE+E + +
Sbjct: 234 DTKDNDKENISEDKKEDHQQEEMLKTLDKKERKQKEKEMKEQEKIEKKKKKQEEKEKKKQ 293
Query: 238 REER 241
+ER
Sbjct: 294 EKER 297
Score = 34.3 bits (77), Expect = 0.36
Identities = 14/64 (21%), Positives = 43/64 (66%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
D+ ++ E+++E ++++ + + +E++++E +E+E E+ +EKE++++E E +
Sbjct: 239 DKENISEDKKEDHQQEEMLKTLDKKERKQKEKEMKEQEKIEKKKKKQEEKEKKKQEKERK 298
Query: 238 REER 241
++E+
Sbjct: 299 KQEK 302
Score = 32.3 bits (72), Expect = 1.4
Identities = 16/59 (27%), Positives = 40/59 (67%), Gaps = 1/59 (1%)
Query: 184 EEEEEIEEEDD-QDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREER 241
+E+E+IE++ Q+E E+ ++++E + E++E ++E + +K E+E + + E+E++
Sbjct: 273 KEQEKIEKKKKKQEEKEKKKQEKERKKQEKKERKQKEKEMKKQKKIEKERKKKEEKEKK 331
Score = 30.4 bits (67), Expect = 5.2
Identities = 15/50 (30%), Positives = 30/50 (60%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEE 232
E +E E +EE+ ++ ++ E +EEE EEE + EE + + K ++++
Sbjct: 373 EHKEGEHKEEEHKEGEHKEGEHKEEEHKEEEHKKEEHKSKEHKSKGKKDK 422
Score = 30.4 bits (67), Expect = 5.2
Identities = 16/59 (27%), Positives = 34/59 (57%)
Query: 184 EEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
+EEE E E + E +E++ +EEE EE + E + D+ ++++ + + ++E+V
Sbjct: 380 KEEEHKEGEHKEGEHKEEEHKEEEHKKEEHKSKEHKSKGKKDKGKKDKGKHKKAKKEKV 438
Score = 29.6 bits (65), Expect = 8.8
Identities = 10/51 (19%), Positives = 35/51 (68%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEE 233
+++E+E ++++ ++ ++++E ++++ +E+E + +EE + ++E EE
Sbjct: 291 KKQEKERKKQEKKERKQKEKEMKKQKKIEKERKKKEEKEKKKKKHDKENEE 341
>TRT_DROME (P19351) Troponin T, skeletal muscle (Upheld protein)
(Intended thorax protein)
Length = 396
Score = 61.6 bits (148), Expect = 2e-09
Identities = 47/136 (34%), Positives = 66/136 (47%), Gaps = 32/136 (23%)
Query: 98 DGGDEIWRRTMMRAEGRGNRRKLRSFPPPLSSLNRNGKPSFFLRPVRKDGRLELTEVRIH 157
D ++IW + GR + KL + F RP +K G E E
Sbjct: 293 DSNEKIWNEKKEQYTGR-QKSKLPKW--------------FGERPGKKAGEPETPE---- 333
Query: 158 RAEILHASRHDGRLTLHLIPDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELE 217
D + ++ D E EEE +EEED++DE +E++E+EEEE EEEEE E
Sbjct: 334 -------GEEDAKADEDIVEDDE--EVEEEVVEEEDEEDEEDEEEEEEEEEEEEEEEEEE 384
Query: 218 EEVALDDDEKEEEEEE 233
EE ++E+EEEEEE
Sbjct: 385 EE----EEEEEEEEEE 396
Score = 60.1 bits (144), Expect = 6e-09
Identities = 34/57 (59%), Positives = 41/57 (71%), Gaps = 6/57 (10%)
Query: 184 EEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
E++EE+EEE ++E EED+E EEEE EEEEE EEE E+EEEEEE E E EE
Sbjct: 346 EDDEEVEEEVVEEEDEEDEEDEEEEEEEEEEEEEEE------EEEEEEEEEEEEEEE 396
Score = 55.8 bits (133), Expect = 1e-07
Identities = 29/62 (46%), Positives = 42/62 (66%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E E E + + D+D +E+D+E EEE + EE+EE EE+ +++E+EEEEEE E E EE
Sbjct: 330 ETPEGEEDAKADEDIVEDDEEVEEEVVEEEDEEDEEDEEEEEEEEEEEEEEEEEEEEEEE 389
Query: 243 AE 244
E
Sbjct: 390 EE 391
Score = 53.5 bits (127), Expect = 6e-07
Identities = 36/73 (49%), Positives = 45/73 (61%), Gaps = 5/73 (6%)
Query: 177 PDQVSLEEEEEEIEEEDDQDE-IEEDQEQEEEELVEEE----EELEEEVALDDDEKEEEE 231
P + + E E E EE+ DE I ED E+ EEE+VEEE EE EEE +++E+EEEE
Sbjct: 322 PGKKAGEPETPEGEEDAKADEDIVEDDEEVEEEVVEEEDEEDEEDEEEEEEEEEEEEEEE 381
Query: 232 EEIECEREERVAE 244
EE E E EE E
Sbjct: 382 EEEEEEEEEEEEE 394
Score = 30.8 bits (68), Expect = 4.0
Identities = 25/117 (21%), Positives = 53/117 (44%), Gaps = 19/117 (16%)
Query: 125 PPLSSLNRNGKPSFFLRPVRKDGRLELTEVRIHRAEILHASRHDGRLTLHLIPDQVSLEE 184
PP + G P F R +K L D +L ++ + +
Sbjct: 24 PPQTPAEGEGDPEFIKRQDQKRSDL------------------DDQLKEYITEWRKQRSK 65
Query: 185 EEEEIEE-EDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EE+E+++ ++ Q + + + +EE+++ + ++E EE + +EK++ E E + R E
Sbjct: 66 EEDELKKLKEKQAKRKVTRAEEEQKMAQRKKEEEERRVREAEEKKQREIEEKAMRLE 122
Score = 30.0 bits (66), Expect = 6.7
Identities = 16/62 (25%), Positives = 33/62 (52%), Gaps = 6/62 (9%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E ++ +EED+ +++E Q + + EEE+++ + +KEEEE + E++
Sbjct: 58 EWRKQRSKEEDELKKLKEKQAKRKVTRAEEEQKMAQR------KKEEEERRVREAEEKKQ 111
Query: 243 AE 244
E
Sbjct: 112 RE 113
>CENB_CRIGR (P48988) Major centromere autoantigen B (Centromere
protein B) (CENP-B)
Length = 606
Score = 61.2 bits (147), Expect = 3e-09
Identities = 37/68 (54%), Positives = 48/68 (70%), Gaps = 6/68 (8%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEE----EEL--EEEVALDDDEKEEEEEEIEC 236
EEEEEE EEE++++E E ++E+EEEE EEE EE+ EEEV + DE +EEEEE E
Sbjct: 408 EEEEEEEEEEEEEEEGEGEEEEEEEEEGEEEGGEGEEVGEEEEVEEEGDESDEEEEEEEE 467
Query: 237 EREERVAE 244
E EE +E
Sbjct: 468 EEEESSSE 475
Score = 58.9 bits (141), Expect = 1e-08
Identities = 37/71 (52%), Positives = 47/71 (66%), Gaps = 9/71 (12%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEE--------ELVEEEEELEEEV-ALDDDEKEEEEEE 233
EEEEEE EEE+++ E EE++E+EEE E V EEEE+EEE D++E+EEEEEE
Sbjct: 410 EEEEEEEEEEEEEGEGEEEEEEEEEGEEEGGEGEEVGEEEEVEEEGDESDEEEEEEEEEE 469
Query: 234 IECEREERVAE 244
E E AE
Sbjct: 470 EESSSEGLEAE 480
Score = 41.6 bits (96), Expect = 0.002
Identities = 24/52 (46%), Positives = 32/52 (61%)
Query: 195 QDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAEWG 246
+ E EE++E+EEEE EEE E EEE +++ +EE E E EE V E G
Sbjct: 404 EGEEEEEEEEEEEEEEEEEGEGEEEEEEEEEGEEEGGEGEEVGEEEEVEEEG 455
Score = 38.9 bits (89), Expect = 0.015
Identities = 27/91 (29%), Positives = 43/91 (46%), Gaps = 35/91 (38%)
Query: 185 EEEEIEEEDDQDEIEEDQEQEEEE--------------------------LVEEEEEL-- 216
EEEE+EEE D+ + EE++E+EEEE V+EE +
Sbjct: 447 EEEEVEEEGDESDEEEEEEEEEEEESSSEGLEAEDWAQGVVEASGGFGGYSVQEEAQCPT 506
Query: 217 -------EEEVALDDDEKEEEEEEIECEREE 240
E+ + D+E+E+EEE+ E E ++
Sbjct: 507 LHFLEGGEDSDSDSDEEEEDEEEDEEDEEDD 537
Score = 37.7 bits (86), Expect = 0.032
Identities = 28/94 (29%), Positives = 43/94 (44%), Gaps = 31/94 (32%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEE-----------------------LVEEEE 214
++V EEE EE +E D++E EE++E+EE V+EE
Sbjct: 443 EEVGEEEEVEEEGDESDEEEEEEEEEEEESSSEGLEAEDWAQGVVEASGGFGGYSVQEEA 502
Query: 215 ELE--------EEVALDDDEKEEEEEEIECEREE 240
+ E+ D DE+EE+EEE E + E+
Sbjct: 503 QCPTLHFLEGGEDSDSDSDEEEEDEEEDEEDEED 536
Score = 36.2 bits (82), Expect = 0.094
Identities = 23/40 (57%), Positives = 24/40 (59%), Gaps = 6/40 (15%)
Query: 211 EEEEELEEEVALDDDEKEEEEEEIECEREERVAEWGFHEG 250
EEEEE EEE E+EEEEEE E E EE E G EG
Sbjct: 406 EEEEEEEEE------EEEEEEEEGEGEEEEEEEEEGEEEG 439
Score = 35.8 bits (81), Expect = 0.12
Identities = 17/41 (41%), Positives = 28/41 (67%), Gaps = 3/41 (7%)
Query: 172 TLHLIP---DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEEL 209
TLH + D S +EEEE EEED++DE ++D +++ +E+
Sbjct: 506 TLHFLEGGEDSDSDSDEEEEDEEEDEEDEEDDDDDEDGDEV 546
Score = 32.0 bits (71), Expect = 1.8
Identities = 23/84 (27%), Positives = 34/84 (40%), Gaps = 35/84 (41%)
Query: 183 EEEEEEIEEEDDQDEIE-----------------------------------EDQEQEEE 207
EEEEEE EEE + +E ED + + +
Sbjct: 462 EEEEEEEEEESSSEGLEAEDWAQGVVEASGGFGGYSVQEEAQCPTLHFLEGGEDSDSDSD 521
Query: 208 ELVEEEEELEEEVALDDDEKEEEE 231
E E+EEE EE+ DDD+++ +E
Sbjct: 522 EEEEDEEEDEEDEEDDDDDEDGDE 545
Score = 30.8 bits (68), Expect = 4.0
Identities = 28/97 (28%), Positives = 43/97 (43%), Gaps = 16/97 (16%)
Query: 191 EEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAEWGFHEG 250
E+ D D EE++++EE+E E+EE DDD+ E+ +E E +A + +
Sbjct: 514 EDSDSDSDEEEEDEEEDE--EDEE--------DDDDDEDGDEVPVPSFGEAMAYFAMVKR 563
Query: 251 YRR---CHEMVNQH--HHHHHNVHGNHHHNHIRMCGI 282
Y + V H H H VH NH G+
Sbjct: 564 YLTSFPIDDRVQSHILHLEHDLVHVT-RKNHAWQAGV 599
>CENB_HUMAN (P07199) Major centromere autoantigen B (Centromere
protein B) (CENP-B)
Length = 599
Score = 59.7 bits (143), Expect = 8e-09
Identities = 34/60 (56%), Positives = 42/60 (69%), Gaps = 5/60 (8%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVE-----EEEELEEEVALDDDEKEEEEEEIECE 237
EEEEEE EEE + +E EE+ E+EEEE E EEEE+EEE +D DE+EEE+EE E
Sbjct: 410 EEEEEEEEEEGEGEEEEEEGEEEEEEGGEGEELGEEEEVEEEGDVDSDEEEEEDEESSSE 469
Score = 55.1 bits (131), Expect = 2e-07
Identities = 30/67 (44%), Positives = 43/67 (63%)
Query: 184 EEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVA 243
E EEE EEE++++E E + E+EEEE EEEEE E L ++E+ EEE +++ + EE
Sbjct: 404 EGEEEEEEEEEEEEEEGEGEEEEEEGEEEEEEGGEGEELGEEEEVEEEGDVDSDEEEEED 463
Query: 244 EWGFHEG 250
E EG
Sbjct: 464 EESSSEG 470
Score = 39.7 bits (91), Expect = 0.009
Identities = 22/53 (41%), Positives = 34/53 (63%), Gaps = 4/53 (7%)
Query: 183 EEEEEEIEEEDDQ----DEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEE 231
EEEEEE EEE+++ +E+ E++E EEE V+ +EE EE+ + E E+
Sbjct: 423 EEEEEEGEEEEEEGGEGEELGEEEEVEEEGDVDSDEEEEEDEESSSEGLEAED 475
Score = 35.8 bits (81), Expect = 0.12
Identities = 16/40 (40%), Positives = 27/40 (67%), Gaps = 2/40 (5%)
Query: 172 TLHLIP--DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEEL 209
TLH + + + EEE+ EEEDD+DE ++D E++ +E+
Sbjct: 500 TLHFLEGGEDSDSDSEEEDDEEEDDEDEDDDDDEEDGDEV 539
Score = 35.0 bits (79), Expect = 0.21
Identities = 24/91 (26%), Positives = 38/91 (41%), Gaps = 35/91 (38%)
Query: 185 EEEEIEEEDDQDEIEEDQEQEEEE-----------------------------------L 209
EEEE+EEE D D EE++E EE
Sbjct: 444 EEEEVEEEGDVDSDEEEEEDEESSSEGLEAEDWAQGVVEAGGSFGAYGAQEEAQCPTLHF 503
Query: 210 VEEEEELEEEVALDDDEKEEEEEEIECEREE 240
+E E+ + + +DDE+E++E+E + + EE
Sbjct: 504 LEGGEDSDSDSEEEDDEEEDDEDEDDDDDEE 534
Score = 34.3 bits (77), Expect = 0.36
Identities = 28/89 (31%), Positives = 42/89 (46%), Gaps = 10/89 (11%)
Query: 200 EDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECER-EERVAEWGFHEGYRR---CH 255
ED + + EE +EEE+ E+E DDD+ EE+ +E+ E +A + + Y
Sbjct: 508 EDSDSDSEEEDDEEEDDEDE---DDDDDEEDGDEVPVPSFGEAMAYFAMVKRYLTSFPID 564
Query: 256 EMVNQH--HHHHHNVHGNHHHNHIRMCGI 282
+ V H H H VH NH R G+
Sbjct: 565 DRVQSHILHLEHDLVHVT-RKNHARQAGV 592
>YSE2_CAEEL (Q09936) Hypothetical protein C53C9.2 in chromosome X
Length = 397
Score = 59.3 bits (142), Expect = 1e-08
Identities = 30/54 (55%), Positives = 41/54 (75%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIE 235
+E E EE EEE+++++IEE +EEEE EEEEE EEE +++E+EEEEEE E
Sbjct: 344 VEPEPEEEEEEEEEEKIEEPAAKEEEEEEEEEEEEEEEELEEEEEEEEEEEEDE 397
Score = 55.5 bits (132), Expect = 1e-07
Identities = 31/58 (53%), Positives = 43/58 (73%), Gaps = 4/58 (6%)
Query: 177 PDQVSLEEEEEEIEEED-DQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEE 233
P + EEEEEE EEE ++ +E++E+EEEE EEEEELEEE +++E+EEEE+E
Sbjct: 343 PVEPEPEEEEEEEEEEKIEEPAAKEEEEEEEEEEEEEEEELEEE---EEEEEEEEEDE 397
Score = 50.8 bits (120), Expect = 4e-06
Identities = 28/58 (48%), Positives = 38/58 (65%), Gaps = 6/58 (10%)
Query: 187 EEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAE 244
E +E E +++E EE++E+ EE +EEEE EEE E+EEEEEE+E E EE E
Sbjct: 342 EPVEPEPEEEEEEEEEEKIEEPAAKEEEEEEEE------EEEEEEEELEEEEEEEEEE 393
Score = 48.5 bits (114), Expect = 2e-05
Identities = 28/66 (42%), Positives = 38/66 (57%), Gaps = 4/66 (6%)
Query: 183 EEEEEEIEEEDDQDEIEEDQ----EQEEEELVEEEEELEEEVALDDDEKEEEEEEIECER 238
EE E + E D ++ + E EEEE EEEE++EE A +++E+EEEEEE E E
Sbjct: 323 EERESSRQSEIDSQSVKASEPVEPEPEEEEEEEEEEKIEEPAAKEEEEEEEEEEEEEEEE 382
Query: 239 EERVAE 244
E E
Sbjct: 383 LEEEEE 388
Score = 46.2 bits (108), Expect = 9e-05
Identities = 26/67 (38%), Positives = 41/67 (60%), Gaps = 5/67 (7%)
Query: 179 QVSLEEEEEEIEEEDDQDEIEEDQEQEEEEL-----VEEEEELEEEVALDDDEKEEEEEE 233
Q ++ + + E + + EE++E+EEE++ EEEEE EEE +++E EEEEEE
Sbjct: 330 QSEIDSQSVKASEPVEPEPEEEEEEEEEEKIEEPAAKEEEEEEEEEEEEEEEELEEEEEE 389
Query: 234 IECEREE 240
E E E+
Sbjct: 390 EEEEEED 396
Score = 42.0 bits (97), Expect = 0.002
Identities = 21/40 (52%), Positives = 31/40 (77%), Gaps = 2/40 (5%)
Query: 176 IPDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEE 215
I + + EEEEEE EEE++++E E++E+EEEE EEE+E
Sbjct: 360 IEEPAAKEEEEEEEEEEEEEEEELEEEEEEEEE--EEEDE 397
>RYR1_HUMAN (P21817) Ryanodine receptor 1 (Skeletal muscle-type
ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle
calcium release channel)
Length = 5038
Score = 59.3 bits (142), Expect = 1e-08
Identities = 33/77 (42%), Positives = 49/77 (62%), Gaps = 10/77 (12%)
Query: 168 DGRLTLHLIPDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEK 227
D + L +I +V EEEEEE EEE+ ++E EE++E++EEE +E+E DE+
Sbjct: 1858 DVKQILKMIEPEVFTEEEEEEDEEEEGEEEDEEEKEEDEEETAQEKE----------DEE 1907
Query: 228 EEEEEEIECEREERVAE 244
+EEEE E E+EE + E
Sbjct: 1908 KEEEEAAEGEKEEGLEE 1924
Score = 41.6 bits (96), Expect = 0.002
Identities = 26/59 (44%), Positives = 38/59 (64%), Gaps = 5/59 (8%)
Query: 186 EEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAE 244
+E++++ E E E+EEEE +EEEE EEE D++EKEE+EEE E+E+ E
Sbjct: 1856 DEDVKQILKMIEPEVFTEEEEEE--DEEEEGEEE---DEEEKEEDEEETAQEKEDEEKE 1909
Score = 39.7 bits (91), Expect = 0.009
Identities = 21/49 (42%), Positives = 31/49 (62%)
Query: 202 QEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAEWGFHEG 250
+E+EEE+ EE EE +EE +D+E+ +E+E E + EE AE EG
Sbjct: 1873 EEEEEEDEEEEGEEEDEEEKEEDEEETAQEKEDEEKEEEEAAEGEKEEG 1921
Score = 33.1 bits (74), Expect = 0.80
Identities = 22/50 (44%), Positives = 31/50 (62%), Gaps = 5/50 (10%)
Query: 178 DQVSLEEEEEEI--EEEDDQDEIEEDQEQEEEELVEE---EEELEEEVAL 222
D+ EE+EEE E+ED++ E EE E E+EE +EE + +L E V L
Sbjct: 1888 DEEEKEEDEEETAQEKEDEEKEEEEAAEGEKEEGLEEGLLQMKLPESVKL 1937
>IE68_SHV21 (Q01042) Immediate-early protein
Length = 407
Score = 59.3 bits (142), Expect = 1e-08
Identities = 31/62 (50%), Positives = 39/62 (62%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
EEE EE EEE +++E EE+ E+E EE E EEE EEE ++ +E EE E E E E
Sbjct: 148 EEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEEEAEEAEEE 207
Query: 243 AE 244
AE
Sbjct: 208 AE 209
Score = 58.2 bits (139), Expect = 2e-08
Identities = 31/67 (46%), Positives = 41/67 (60%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
++ EE EEE EEE+ ++ E++E EEEE EEE E EE ++ E+E EEEE E E
Sbjct: 107 EEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEE 166
Query: 238 REERVAE 244
EE E
Sbjct: 167 AEEEAEE 173
Score = 57.4 bits (137), Expect = 4e-08
Identities = 32/62 (51%), Positives = 39/62 (62%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E EEEE EE +++ E EE +E E EE EEEE EEE A + +E+E EE E E E EE
Sbjct: 105 EAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAE 164
Query: 243 AE 244
E
Sbjct: 165 EE 166
Score = 57.0 bits (136), Expect = 5e-08
Identities = 32/74 (43%), Positives = 42/74 (56%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E EE+E EEE+ ++ EE +E+E EE EEEE EEE A +++ +E EEEE E EE
Sbjct: 100 EAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAE 159
Query: 243 AEWGFHEGYRRCHE 256
E E E
Sbjct: 160 EEEAEEEAEEEAEE 173
Score = 56.6 bits (135), Expect = 7e-08
Identities = 32/59 (54%), Positives = 39/59 (65%), Gaps = 1/59 (1%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEE-EVALDDDEKEEEEEEIECEREE 240
EEE EE EE ++ +E EE+ E+ EEE E EEE EE E A + +E EEE EE E E EE
Sbjct: 182 EEEAEEAEEAEEAEEAEEEAEEAEEEAEEAEEEAEEAEEAEEAEEAEEEAEEAEEEEEE 240
Score = 56.2 bits (134), Expect = 9e-08
Identities = 40/100 (40%), Positives = 52/100 (52%), Gaps = 4/100 (4%)
Query: 146 DGRLELTEVRIHRAEILHASRHDGRLTLHLIPDQVSLEEEEEEIEEEDDQDEIEEDQEQE 205
+GR E E AE A + ++ + E E EE E E+++ E EE +E E
Sbjct: 91 EGREEAEE---EEAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAE 147
Query: 206 EEELVEEEEELEEEVALDDDEKE-EEEEEIECEREERVAE 244
EEE E EEE EEE A ++ E+E EE EE E E EE E
Sbjct: 148 EEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEE 187
Score = 55.5 bits (132), Expect = 1e-07
Identities = 32/79 (40%), Positives = 41/79 (51%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
++ EE EEE EE +++ EE+ E+E EE EE EE EEE + +E EE EE E E
Sbjct: 139 EEEEAEEAEEEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAE 198
Query: 238 REERVAEWGFHEGYRRCHE 256
E AE E E
Sbjct: 199 EEAEEAEEEAEEAEEEAEE 217
Score = 53.5 bits (127), Expect = 6e-07
Identities = 31/74 (41%), Positives = 39/74 (51%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E EEEE EE ++++ E ++E EEEE EE EE EE ++E EEE EE E E
Sbjct: 137 EAEEEEAEEAEEEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEE 196
Query: 243 AEWGFHEGYRRCHE 256
AE E E
Sbjct: 197 AEEEAEEAEEEAEE 210
Score = 52.8 bits (125), Expect = 1e-06
Identities = 30/67 (44%), Positives = 38/67 (55%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
++ EEE EE EE ++ E E ++E EE E EE EE EEE ++E EE EEE E
Sbjct: 159 EEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEEEAEEAEEEAEEAEEEAEEA 218
Query: 238 REERVAE 244
E AE
Sbjct: 219 EEAEEAE 225
Score = 52.4 bits (124), Expect = 1e-06
Identities = 28/61 (45%), Positives = 36/61 (58%)
Query: 184 EEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVA 243
EE EE EEE +++ E ++ +E EE EE EE EEE ++E EE EE E E E A
Sbjct: 172 EEAEEAEEEAEEEAEEAEEAEEAEEAEEEAEEAEEEAEEAEEEAEEAEEAEEAEEAEEEA 231
Query: 244 E 244
E
Sbjct: 232 E 232
Score = 51.6 bits (122), Expect = 2e-06
Identities = 30/56 (53%), Positives = 37/56 (65%), Gaps = 1/56 (1%)
Query: 185 EEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EE E EE +++E EE +E EEEE E EEE EEE A + + +EEE EE E E EE
Sbjct: 88 EEGEGREEAEEEEAEE-KEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEE 142
Score = 51.2 bits (121), Expect = 3e-06
Identities = 28/67 (41%), Positives = 39/67 (57%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
++ EEEE E EE ++ +E E ++ +EE E E EEE EEE ++ +EE EEE E
Sbjct: 129 EEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEA 188
Query: 238 REERVAE 244
E AE
Sbjct: 189 EEAEEAE 195
Score = 47.8 bits (112), Expect = 3e-05
Identities = 28/62 (45%), Positives = 36/62 (57%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
EE E E EE + E E++E EE+E EEE E EE A +++ +E E EE E E EE
Sbjct: 80 EEREGEGGEEGEGREEAEEEEAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAE 139
Query: 243 AE 244
E
Sbjct: 140 EE 141
Score = 47.8 bits (112), Expect = 3e-05
Identities = 31/65 (47%), Positives = 38/65 (57%), Gaps = 6/65 (9%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEE----EEEEIECER 238
EE EEE EE ++ +E EE +E+ EE EEE E EE A + +E EE EEE E E
Sbjct: 179 EEAEEEAEEAEEAEEAEEAEEEAEE--AEEEAEEAEEEAEEAEEAEEAEEAEEEAEEAEE 236
Query: 239 EERVA 243
EE A
Sbjct: 237 EEEEA 241
Score = 47.0 bits (110), Expect = 5e-05
Identities = 28/62 (45%), Positives = 37/62 (59%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
EE E E E+++ E +E +E+E EE EE EE E E A ++E+ EEEE E E EE
Sbjct: 88 EEGEGREEAEEEEAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAE 147
Query: 243 AE 244
E
Sbjct: 148 EE 149
Score = 46.6 bits (109), Expect = 7e-05
Identities = 30/87 (34%), Positives = 43/87 (48%), Gaps = 14/87 (16%)
Query: 172 TLHLIPDQVSLEEEEEEIEEEDDQ--------------DEIEEDQEQEEEELVEEEEELE 217
T H ++ E+ EE+EEE ++ +E E +E EEEE E+E E E
Sbjct: 50 TTHQQQAALTEEQRREEVEEEGEERERRGEEEREGEGGEEGEGREEAEEEEAEEKEAEEE 109
Query: 218 EEVALDDDEKEEEEEEIECEREERVAE 244
E +++ +EEE EE E E EE E
Sbjct: 110 EAEEAEEEAEEEEAEEAEAEEEEAEEE 136
>ASF1_YEAST (P32447) Anti-silencing protein 1
Length = 279
Score = 59.3 bits (142), Expect = 1e-08
Identities = 36/89 (40%), Positives = 52/89 (57%), Gaps = 11/89 (12%)
Query: 155 RIHRAEILHASRHDGRLTLHLIPDQVSLEEEEEEIEEEDDQDEIEEDQEQ--------EE 206
R+ R I+ + ++G L P+Q +++EEEE +EE+D DE +ED E E
Sbjct: 145 RVTRFNIVWDNENEGDL---YPPEQPGVDDEEEEDDEEEDDDEDDEDDEDDDQEDGEGEA 201
Query: 207 EELVEEEEELEEEVALDDDEKEEEEEEIE 235
EE EEEEE EE+ ++ EEEEE+IE
Sbjct: 202 EEAAEEEEEEEEKTEDNETNLEEEEEDIE 230
Score = 39.7 bits (91), Expect = 0.009
Identities = 20/60 (33%), Positives = 37/60 (61%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
++ + EEEEEE + ED++ +EE++E E +EEE EE ++D +E +++ + E
Sbjct: 202 EEAAEEEEEEEEKTEDNETNLEEEEEDIENSDGDEEEGEEEVGSVDKNEDGNDKKRRKIE 261
Score = 36.6 bits (83), Expect = 0.072
Identities = 22/58 (37%), Positives = 34/58 (57%), Gaps = 7/58 (12%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
E EE EEE+++++ E++ E L EEEE++E D DE+E EEE ++ E
Sbjct: 200 EAEEAAEEEEEEEEKTEDN----ETNLEEEEEDIENS---DGDEEEGEEEVGSVDKNE 250
Score = 33.9 bits (76), Expect = 0.47
Identities = 19/62 (30%), Positives = 34/62 (54%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E E + E + EE+++ EEE+ E++E+ E++ D + + EE E E E EE+
Sbjct: 156 ENEGDLYPPEQPGVDDEEEEDDEEEDDDEDDEDDEDDDQEDGEGEAEEAAEEEEEEEEKT 215
Query: 243 AE 244
+
Sbjct: 216 ED 217
>YCF2_OENPI (P31568) Protein ycf2 (Fragment)
Length = 721
Score = 58.9 bits (141), Expect = 1e-08
Identities = 36/85 (42%), Positives = 54/85 (63%), Gaps = 7/85 (8%)
Query: 152 TEVRIHRAEILHASRHDGRLTLHLIPDQVSLEEEEEEIEEEDDQ-DEIEEDQEQEEEELV 210
TEV + E + + + L + LEEE+EE++EE+D+ DE EE+ ++EE+EL
Sbjct: 509 TEVAVWGVEEEGEADDEEDVLLEAQQEDELLEEEDEELDEEEDELDEEEEEPKEEEDELH 568
Query: 211 EEEEELEEEVALDDDEKEEEEEEIE 235
EEEEE EEE E+EEEE+E++
Sbjct: 569 EEEEEEEEE------EEEEEEDELQ 587
Score = 58.2 bits (139), Expect = 2e-08
Identities = 31/64 (48%), Positives = 49/64 (76%), Gaps = 4/64 (6%)
Query: 183 EEEEEEIEEEDDQ-DEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREER 241
++E+E +EEED++ DE E++ ++EEEE EEE+EL EE +++E+EEEEEE E E +E
Sbjct: 533 QQEDELLEEEDEELDEEEDELDEEEEEPKEEEDELHEE---EEEEEEEEEEEEEDELQEN 589
Query: 242 VAEW 245
+E+
Sbjct: 590 DSEF 593
Score = 53.1 bits (126), Expect = 7e-07
Identities = 32/70 (45%), Positives = 48/70 (67%), Gaps = 5/70 (7%)
Query: 176 IPDQVSLEEEEEEIEEEDDQDEI--EEDQE--QEEEELVEEEEE-LEEEVALDDDEKEEE 230
+ ++ ++EE+ + E +DE+ EED+E +EE+EL EEEEE EEE L ++E+EEE
Sbjct: 516 VEEEGEADDEEDVLLEAQQEDELLEEEDEELDEEEDELDEEEEEPKEEEDELHEEEEEEE 575
Query: 231 EEEIECEREE 240
EEE E E +E
Sbjct: 576 EEEEEEEEDE 585
Score = 51.6 bits (122), Expect = 2e-06
Identities = 24/47 (51%), Positives = 36/47 (76%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKE 228
L+EEE+E++EE+++ + EED+ EEEE EEEEE EEE L +++ E
Sbjct: 546 LDEEEDELDEEEEEPKEEEDELHEEEEEEEEEEEEEEEDELQENDSE 592
Score = 51.2 bits (121), Expect = 3e-06
Identities = 28/59 (47%), Positives = 40/59 (67%), Gaps = 1/59 (1%)
Query: 187 EEIEEEDDQDEIEEDQEQEEEELVEEEEEL-EEEVALDDDEKEEEEEEIECEREERVAE 244
EE E DD++++ + +QE+E L EE+EEL EEE LD++E+E +EEE E EE E
Sbjct: 517 EEEGEADDEEDVLLEAQQEDELLEEEDEELDEEEDELDEEEEEPKEEEDELHEEEEEEE 575
Score = 43.9 bits (102), Expect = 5e-04
Identities = 25/61 (40%), Positives = 37/61 (59%), Gaps = 1/61 (1%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREER 241
+E EEE+E +D+ E+E +++E E EE E EEEV ++E E EEE+E +E
Sbjct: 236 VEGTEEEVEGTEDE-EVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEE 294
Query: 242 V 242
V
Sbjct: 295 V 295
Score = 43.9 bits (102), Expect = 5e-04
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 5/71 (7%)
Query: 177 PDQVSLEEEEEEIEEEDDQ-----DEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEE 231
P + +E EEE+E +++ +E+E +++E E EE E EEEV ++E E E
Sbjct: 181 PTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTE 240
Query: 232 EEIECEREERV 242
EE+E +E V
Sbjct: 241 EEVEGTEDEEV 251
Score = 43.1 bits (100), Expect = 8e-04
Identities = 27/66 (40%), Positives = 39/66 (58%), Gaps = 3/66 (4%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELE---EEVALDDDEKEEEEEEI 234
++V EEE E E+++ + EE+ E EEE+ EEE+E EEV ++E E EEE+
Sbjct: 322 EEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEV 381
Query: 235 ECEREE 240
E EE
Sbjct: 382 EGTEEE 387
Score = 42.7 bits (99), Expect = 0.001
Identities = 27/61 (44%), Positives = 37/61 (60%), Gaps = 3/61 (4%)
Query: 183 EEEEEEIEEEDD--QDEIEEDQEQEEEELVEEEEELEEEV-ALDDDEKEEEEEEIECERE 239
EEE E EEE + ++E+E +++E E EE E EEEV +D+E E EEE+E E
Sbjct: 270 EEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEE 329
Query: 240 E 240
E
Sbjct: 330 E 330
Score = 42.4 bits (98), Expect = 0.001
Identities = 27/62 (43%), Positives = 38/62 (60%), Gaps = 4/62 (6%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEEL---EEEV-ALDDDEKEEEEEEIECER 238
E+EE E E+++ + EE+ E EEE+ EEE+ EEEV +D+E E EEE+E
Sbjct: 247 EDEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTE 306
Query: 239 EE 240
EE
Sbjct: 307 EE 308
Score = 42.4 bits (98), Expect = 0.001
Identities = 28/67 (41%), Positives = 40/67 (58%), Gaps = 6/67 (8%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQE--EEELVEEEEELE----EEVALDDDEKEEEEEEIE 235
+E EEE+E +++ E ED+E E EEE+ EEE+E EEV ++E E EEE+E
Sbjct: 273 VEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVE 332
Query: 236 CEREERV 242
+E V
Sbjct: 333 GTEDEEV 339
Score = 42.4 bits (98), Expect = 0.001
Identities = 28/64 (43%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQE--EEELVEEEEEL---EEEVALDDDEKEEEEEEIEC 236
+E EEE+E +++ E ED+E E EEE+ EEE+ EEEV ++E E EEE+E
Sbjct: 317 VEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEG 376
Query: 237 EREE 240
EE
Sbjct: 377 TEEE 380
Score = 42.4 bits (98), Expect = 0.001
Identities = 26/62 (41%), Positives = 41/62 (65%), Gaps = 7/62 (11%)
Query: 179 QVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECER 238
+V++ EEE E +D++D + E Q+E+EL+EEE+E LD++E E +EEE E +
Sbjct: 510 EVAVWGVEEEGEADDEEDVLLE--AQQEDELLEEEDE-----ELDEEEDELDEEEEEPKE 562
Query: 239 EE 240
EE
Sbjct: 563 EE 564
Score = 42.4 bits (98), Expect = 0.001
Identities = 28/61 (45%), Positives = 35/61 (56%), Gaps = 1/61 (1%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEE-LEEEVALDDDEKEEEEEEIECEREER 241
EEE E EEE + E E + +EE E EEE E EEEV ++E E EEE+E +E
Sbjct: 343 EEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEE 402
Query: 242 V 242
V
Sbjct: 403 V 403
Score = 42.0 bits (97), Expect = 0.002
Identities = 25/60 (41%), Positives = 36/60 (59%), Gaps = 2/60 (3%)
Query: 183 EEEEEEIEEEDD--QDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
+EE E EEE + ++E+E +++E E EE E EEEV ++E E EEE+E EE
Sbjct: 314 DEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEE 373
Score = 42.0 bits (97), Expect = 0.002
Identities = 28/65 (43%), Positives = 39/65 (59%), Gaps = 6/65 (9%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQE--EEELVEEEEELE----EEVALDDDEKEEEEEEIE 235
+E EEE+E +++ E ED+E E EEE+ EEE+E EEV ++E E EEE+E
Sbjct: 295 VEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVE 354
Query: 236 CEREE 240
EE
Sbjct: 355 GTEEE 359
Score = 42.0 bits (97), Expect = 0.002
Identities = 28/68 (41%), Positives = 41/68 (60%), Gaps = 5/68 (7%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEEL---EEEVALDDDEKEE--EEE 232
++V EEE E E+++ + EE+ E EEE+ EEE+ EEEV +DE+ E E+E
Sbjct: 198 EEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEDE 257
Query: 233 EIECEREE 240
E+E EE
Sbjct: 258 EVEGTEEE 265
Score = 41.6 bits (96), Expect = 0.002
Identities = 25/59 (42%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEV-ALDDDEKEEEEEEIECEREE 240
E E E E E ++E+E +++E E EE E EEEV +D+E E EEE+E EE
Sbjct: 294 EVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEE 352
Score = 41.6 bits (96), Expect = 0.002
Identities = 27/67 (40%), Positives = 38/67 (56%), Gaps = 4/67 (5%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELV----EEEEELEEEVALDDDEKEEEEEE 233
++V EEE E E+++ + EE+ E EEE+ EE E EEEV ++E E EEE
Sbjct: 300 EEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEE 359
Query: 234 IECEREE 240
+E EE
Sbjct: 360 VEGTEEE 366
Score = 41.6 bits (96), Expect = 0.002
Identities = 22/43 (51%), Positives = 34/43 (78%), Gaps = 5/43 (11%)
Query: 178 DQVSLEEEEEEIEEEDDQ--DEIEEDQEQEEEELVEEEEELEE 218
++ L+EEEEE +EE+D+ +E EE++E+EEE EEE+EL+E
Sbjct: 549 EEDELDEEEEEPKEEEDELHEEEEEEEEEEEE---EEEDELQE 588
Score = 40.8 bits (94), Expect = 0.004
Identities = 27/59 (45%), Positives = 34/59 (56%), Gaps = 1/59 (1%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEE-EELEEEVALDDDEKEEEEEEIECEREE 240
+EE E EEE + E E + +EE E EEE E EEEV ++E E EEE+E EE
Sbjct: 336 DEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEE 394
Score = 40.8 bits (94), Expect = 0.004
Identities = 26/63 (41%), Positives = 38/63 (60%), Gaps = 5/63 (7%)
Query: 185 EEEEIE-EEDDQDEIEEDQEQEEEELVEEEEELE----EEVALDDDEKEEEEEEIECERE 239
E+EE+E E++ + EE+ E EEE+ EEE+E EEV ++E E EEE+E +
Sbjct: 255 EDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTED 314
Query: 240 ERV 242
E V
Sbjct: 315 EEV 317
Score = 40.4 bits (93), Expect = 0.005
Identities = 24/68 (35%), Positives = 41/68 (60%), Gaps = 5/68 (7%)
Query: 182 LEEEEEEIEEEDDQDEI-----EEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIEC 236
+ EEE+E+ E+ + E+ EE+ E ++EE V E + E+E+ ++DE+ +EEE+
Sbjct: 495 IPEEEDELPEDALETEVAVWGVEEEGEADDEEDVLLEAQQEDELLEEEDEELDEEEDELD 554
Query: 237 EREERVAE 244
E EE E
Sbjct: 555 EEEEEPKE 562
Score = 40.0 bits (92), Expect = 0.007
Identities = 27/62 (43%), Positives = 38/62 (60%), Gaps = 6/62 (9%)
Query: 185 EEEEIE-EEDDQDEIEEDQEQEEEELVEEEEELE----EEV-ALDDDEKEEEEEEIECER 238
E+EE+E E++ + EE+ E EEE+ EEE+E EEV +D+E E EEE+E
Sbjct: 211 EDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEDEEVEGTEEEVEGTE 270
Query: 239 EE 240
EE
Sbjct: 271 EE 272
Score = 39.7 bits (91), Expect = 0.009
Identities = 28/65 (43%), Positives = 40/65 (61%), Gaps = 6/65 (9%)
Query: 182 LEEEEEEIE-EEDDQDEIEEDQEQEEEELVE--EEEEL---EEEVALDDDEKEEEEEEIE 235
+E EEE+E E++ + EE+ E E+E VE E+EE+ EEEV ++E E EEE+E
Sbjct: 222 VEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVE 281
Query: 236 CEREE 240
EE
Sbjct: 282 GTEEE 286
Score = 39.7 bits (91), Expect = 0.009
Identities = 28/63 (44%), Positives = 36/63 (56%), Gaps = 3/63 (4%)
Query: 183 EEEEEEIEEEDD--QDEIEEDQEQEEEELVEEEEELEEEVALDDDEKE-EEEEEIECERE 239
EEE E EEE + +DE E E+E E EE E EEEV ++E E E+EE+E +
Sbjct: 197 EEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTED 256
Query: 240 ERV 242
E V
Sbjct: 257 EEV 259
Score = 38.5 bits (88), Expect = 0.019
Identities = 27/67 (40%), Positives = 37/67 (54%), Gaps = 4/67 (5%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELE----EEVALDDDEKEEEEEE 233
++V EEE E EE+ + EE + EEE E+EE+E EEV ++E E EEE
Sbjct: 213 EEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEDEEVEGTEEEVEGTEEE 272
Query: 234 IECEREE 240
+E EE
Sbjct: 273 VEGTEEE 279
Score = 38.5 bits (88), Expect = 0.019
Identities = 27/68 (39%), Positives = 39/68 (56%), Gaps = 5/68 (7%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELV----EEEEELEEEVALDDDEKE-EEEE 232
++V EEE E E+++ + EE+ E EEE+ EE E EEEV ++E E E+E
Sbjct: 278 EEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDE 337
Query: 233 EIECEREE 240
E+E EE
Sbjct: 338 EVEGTEEE 345
Score = 37.7 bits (86), Expect = 0.032
Identities = 26/61 (42%), Positives = 36/61 (58%), Gaps = 3/61 (4%)
Query: 183 EEEEEEIEEEDD--QDEIEEDQEQEEEELVEEEEELEEEVALDDDEKE-EEEEEIECERE 239
EEE E EEE + ++E+E +E+ E EE E EEEV ++E E E+EE+E E
Sbjct: 263 EEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEE 322
Query: 240 E 240
E
Sbjct: 323 E 323
Score = 35.0 bits (79), Expect = 0.21
Identities = 24/52 (46%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEE-LEEEVALDDDEKEEEEEE 233
EEE E EEE + E E + +EE E EEE E EEEV +DE+ E E+
Sbjct: 357 EEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEK 408
Score = 34.3 bits (77), Expect = 0.36
Identities = 24/60 (40%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEV-ALDDDEKEEEEEEIECEREE 240
LE E + ++E+E E+E E EE E EEEV +D+E E EEE+E EE
Sbjct: 170 LELEGALVGSSPTEEEVE-GTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEE 228
Score = 33.5 bits (75), Expect = 0.61
Identities = 25/56 (44%), Positives = 34/56 (60%), Gaps = 4/56 (7%)
Query: 183 EEEEEEIEEEDD--QDEIEEDQEQEEEELVEEEEELEEEVALDDDEKE-EEEEEIE 235
EEE E EEE + ++E+E E+E E EE E EEEV ++E E E+EE+E
Sbjct: 350 EEEVEGTEEEVEGTEEEVEGT-EEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVE 404
Score = 33.1 bits (74), Expect = 0.80
Identities = 25/79 (31%), Positives = 38/79 (47%), Gaps = 12/79 (15%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEE-----------EEELEEEVALDDDEKEEEE 231
++ E E+EE+DD DE ++ E+L E + L+ E + +E EEE
Sbjct: 440 QKYESELEEDDDDDEDVFAPQKMLEDLFSELVWSPRIWHPWDFLLDCEAEIPAEEIPEEE 499
Query: 232 EEI-ECEREERVAEWGFHE 249
+E+ E E VA WG E
Sbjct: 500 DELPEDALETEVAVWGVEE 518
Score = 32.7 bits (73), Expect = 1.0
Identities = 19/49 (38%), Positives = 27/49 (54%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEE 231
EEE E EEE + E E + +EE E EEE E E+ ++ EK+ +
Sbjct: 364 EEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEKDSSQ 412
Score = 32.3 bits (72), Expect = 1.4
Identities = 22/62 (35%), Positives = 35/62 (55%), Gaps = 4/62 (6%)
Query: 182 LEEEEEEIE-EEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
+E EEE+E E++ + EE+ E EEE+ EEE+E +D+E E E++ +
Sbjct: 360 VEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVE---GTEDEEVEGTEKDSSQFDND 416
Query: 241 RV 242
RV
Sbjct: 417 RV 418
Score = 32.3 bits (72), Expect = 1.4
Identities = 14/30 (46%), Positives = 22/30 (72%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEE 207
++ L EEEEE EEE++++E +E QE + E
Sbjct: 563 EEDELHEEEEEEEEEEEEEEEDELQENDSE 592
>RYR1_RABIT (P11716) Ryanodine receptor 1 (Skeletal muscle-type
ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle
calcium release channel)
Length = 5037
Score = 58.2 bits (139), Expect = 2e-08
Identities = 30/68 (44%), Positives = 48/68 (70%), Gaps = 2/68 (2%)
Query: 168 DGRLTLHLIPDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEK 227
D + L +I +V EEEEEE EEE++++E EED+E++EE+ EEEEE E+ +++
Sbjct: 1858 DVKQILKMIEPEVFTEEEEEEEEEEEEEEEEEEDEEEKEED--EEEEEKEDAEKEEEEAP 1915
Query: 228 EEEEEEIE 235
E E+E++E
Sbjct: 1916 EGEKEDLE 1923
Score = 53.1 bits (126), Expect = 7e-07
Identities = 26/60 (43%), Positives = 44/60 (73%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
EEEEEE EEE++++E EE++E++EEE +E+ E EEE A + ++++ EE ++ + E V
Sbjct: 1876 EEEEEEEEEEEEEEEDEEEKEEDEEEEEKEDAEKEEEEAPEGEKEDLEEGLLQMKLPESV 1935
Score = 51.6 bits (122), Expect = 2e-06
Identities = 25/55 (45%), Positives = 40/55 (72%)
Query: 186 EEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
E E+ E++++E EE++E+EEEE EEE+E +EE +D ++EEEE E E+E+
Sbjct: 1867 EPEVFTEEEEEEEEEEEEEEEEEEDEEEKEEDEEEEEKEDAEKEEEEAPEGEKED 1921
Score = 51.6 bits (122), Expect = 2e-06
Identities = 25/58 (43%), Positives = 41/58 (70%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
+E+ ++I + + + E++E+EEEE EEEEE E+E ++DE+EEE+E+ E E EE
Sbjct: 1856 DEDVKQILKMIEPEVFTEEEEEEEEEEEEEEEEEEDEEEKEEDEEEEEKEDAEKEEEE 1913
Score = 50.1 bits (118), Expect = 6e-06
Identities = 23/50 (46%), Positives = 38/50 (76%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEE 232
EEEEEE EEE++++E +E++++E+EE E+E+ +EE + EKE+ EE
Sbjct: 1875 EEEEEEEEEEEEEEEEDEEEKEEDEEEEEKEDAEKEEEEAPEGEKEDLEE 1924
Score = 34.3 bits (77), Expect = 0.36
Identities = 21/47 (44%), Positives = 29/47 (61%), Gaps = 2/47 (4%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEE--EEELEEEVAL 222
D+ EE+EEE E+ED + E EE E E+E+L E + +L E V L
Sbjct: 1891 DEEEKEEDEEEEEKEDAEKEEEEAPEGEKEDLEEGLLQMKLPESVKL 1937
>S521_RAT (Q9QY02) Putative splicing factor YT521 (RA301-binding
protein)
Length = 738
Score = 57.8 bits (138), Expect = 3e-08
Identities = 30/60 (50%), Positives = 42/60 (70%), Gaps = 9/60 (15%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREER 241
+EE+EE E+ DD +E++ED E+EE+E +EEEE DE+EEEEEE E E++ER
Sbjct: 208 VEEDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEE---------DEEEEEEEEEEYEQDER 258
Score = 56.6 bits (135), Expect = 7e-08
Identities = 28/64 (43%), Positives = 45/64 (69%), Gaps = 2/64 (3%)
Query: 183 EEE--EEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEE EE++EE+++ DE +D E+ +E+ EEE+E E+E D++E+EEEEEE E + +
Sbjct: 200 EEEGGEEDVEEDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEEDEEEEEEEEEEYEQDERD 259
Query: 241 RVAE 244
+ E
Sbjct: 260 QKEE 263
Score = 51.2 bits (121), Expect = 3e-06
Identities = 28/70 (40%), Positives = 44/70 (62%), Gaps = 8/70 (11%)
Query: 184 EEEEEIEEEDDQDEI-------EEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIEC 236
EE+EE++E+ D DE EED+E++EEE EEEEE EEE + DE++++EE +
Sbjct: 209 EEDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEEDEEEEE-EEEEEYEQDERDQKEEGNDY 267
Query: 237 EREERVAEWG 246
+ ++ G
Sbjct: 268 DTRSEASDSG 277
Score = 51.2 bits (121), Expect = 3e-06
Identities = 24/58 (41%), Positives = 42/58 (72%), Gaps = 2/58 (3%)
Query: 188 EIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAEW 245
E EEE ++++EED+E +E+ +++EE++E+ ++DE+E+EEEE E E EE E+
Sbjct: 198 ENEEEGGEEDVEEDEEVDEDG--DDDEEVDEDAEEEEDEEEDEEEEDEEEEEEEEEEY 253
Score = 50.8 bits (120), Expect = 4e-06
Identities = 26/64 (40%), Positives = 44/64 (68%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECE 237
+Q + E EEE EED +++ E D++ +++E V+E+ E EE+ D++E++EEEEE E E
Sbjct: 192 EQGNNTENEEEGGEEDVEEDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEEDEEEEEEEEE 251
Query: 238 REER 241
E+
Sbjct: 252 EYEQ 255
Score = 47.4 bits (111), Expect = 4e-05
Identities = 23/55 (41%), Positives = 38/55 (68%), Gaps = 1/55 (1%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEE 232
++V + EEEE EEED+++E EE++E+EEEE ++E + +EE D D + E +
Sbjct: 222 EEVDEDAEEEEDEEEDEEEEDEEEEEEEEEEYEQDERDQKEE-GNDYDTRSEASD 275
Score = 40.0 bits (92), Expect = 0.007
Identities = 17/56 (30%), Positives = 36/56 (63%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEE 233
D ++E+ EE E+E++ +E E+++E+EEEE E++E +++ +D + E +
Sbjct: 220 DDEEVDEDAEEEEDEEEDEEEEDEEEEEEEEEEYEQDERDQKEEGNDYDTRSEASD 275
Score = 40.0 bits (92), Expect = 0.007
Identities = 24/74 (32%), Positives = 40/74 (53%), Gaps = 4/74 (5%)
Query: 184 EEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEE----EVALDDDEKEEEEEEIECERE 239
+ E +Q E++E+ EE VEE+EE++E + +D+D +EEE+EE + E E
Sbjct: 182 DHETGSSASSEQGNNTENEEEGGEEDVEEDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEE 241
Query: 240 ERVAEWGFHEGYRR 253
+ E E Y +
Sbjct: 242 DEEEEEEEEEEYEQ 255
Score = 38.1 bits (87), Expect = 0.025
Identities = 17/46 (36%), Positives = 28/46 (59%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKE 228
EEE+EE E+E++++E EE+ EQ+E + EE + + D E
Sbjct: 234 EEEDEEEEDEEEEEEEEEEYEQDERDQKEEGNDYDTRSEASDSGSE 279
Score = 32.7 bits (73), Expect = 1.0
Identities = 30/100 (30%), Positives = 40/100 (40%), Gaps = 17/100 (17%)
Query: 179 QVSLEE---EEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDD---------- 225
Q S EE EE + E E E E EE++EE+ +D+D
Sbjct: 168 QSSKEEGNSEEYGSDHETGSSASSEQGNNTENEEEGGEEDVEEDEEVDEDGDDDEEVDED 227
Query: 226 ----EKEEEEEEIECEREERVAEWGFHEGYRRCHEMVNQH 261
E EEE+EE E E EE E + + R E N +
Sbjct: 228 AEEEEDEEEDEEEEDEEEEEEEEEEYEQDERDQKEEGNDY 267
>CENB_SHEEP (P49451) Major centromere autoantigen B (Centromere
protein B) (CENP-B) (Fragment)
Length = 239
Score = 57.4 bits (137), Expect = 4e-08
Identities = 33/67 (49%), Positives = 42/67 (62%), Gaps = 4/67 (5%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEE----ELVEEEEELEEEVALDDDEKEEEEEEIECER 238
EEEEEE EE + ++E EED E+EEE E + EEEE+EEE +D E+EEEEEE
Sbjct: 52 EEEEEEEEEGEGEEEEEEDGEEEEEAGEGEELGEEEEVEEEGDVDTVEEEEEEEEESSSE 111
Query: 239 EERVAEW 245
+W
Sbjct: 112 GLEAEDW 118
Score = 52.8 bits (125), Expect = 1e-06
Identities = 28/61 (45%), Positives = 40/61 (64%)
Query: 184 EEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVA 243
E EEE EEE++++E E + E+EEEE EEEEE E L ++E+ EEE +++ EE
Sbjct: 45 EGEEEEEEEEEEEEEEGEGEEEEEEDGEEEEEAGEGEELGEEEEVEEEGDVDTVEEEEEE 104
Query: 244 E 244
E
Sbjct: 105 E 105
Score = 42.4 bits (98), Expect = 0.001
Identities = 17/38 (44%), Positives = 27/38 (70%)
Query: 172 TLHLIPDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEEL 209
TLH + + E + EE EE+DD+DE +ED E+E++E+
Sbjct: 142 TLHFLEGEEDSESDSEEEEEDDDEDEDDEDDEEEDDEV 179
Score = 42.0 bits (97), Expect = 0.002
Identities = 24/47 (51%), Positives = 29/47 (61%)
Query: 200 EDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAEWG 246
E++E+EEEE EEE E EEE D +E+EE E E EE V E G
Sbjct: 47 EEEEEEEEEEEEEEGEGEEEEEEDGEEEEEAGEGEELGEEEEVEEEG 93
Score = 37.0 bits (84), Expect = 0.055
Identities = 43/151 (28%), Positives = 56/151 (36%), Gaps = 53/151 (35%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQEEE---------------------------------- 207
L EEEE EEE D D +EE++E+EEE
Sbjct: 83 LGEEEEV-EEEGDVDTVEEEEEEEEESSSEGLEAEDWAQGVVEAGGSFGGYGAQEEAQCP 141
Query: 208 --ELVEEEEELE---EEVALDDDEKEEEEEEIECEREERVAEWGFHEGY----------R 252
+E EE+ E EE DDDE E++E++ E + E V +G Y
Sbjct: 142 TLHFLEGEEDSESDSEEEEEDDDEDEDDEDDEEEDDEVPVPSFGEAMAYFAMVKRYLTSS 201
Query: 253 RCHEMVNQH--HHHHHNVHGNHHHNHIRMCG 281
+ V H H H VH NH R G
Sbjct: 202 PIDDRVQSHILHLEHDLVHVT-RKNHARQAG 231
Score = 30.8 bits (68), Expect = 4.0
Identities = 20/42 (47%), Positives = 26/42 (61%), Gaps = 5/42 (11%)
Query: 203 EQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAE 244
+ E EE EEEEE EEE +++ + EEEEE + E EE E
Sbjct: 43 KSEGEE--EEEEEEEEE---EEEGEGEEEEEEDGEEEEEAGE 79
>TCF8_MOUSE (Q64318) Transcription factor 8 (Zinc finger homeobox
protein 1a) (MEB1) (Delta EF1)
Length = 1117
Score = 57.0 bits (136), Expect = 5e-08
Identities = 30/73 (41%), Positives = 43/73 (58%)
Query: 177 PDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIEC 236
P Q +E E EED+ E EE++E +E E ++E +E E +++E+EEEEEE E
Sbjct: 999 PSQADSDERESLTREEDEDSEKEEEEEDKEMEELQEGKECENPQGEEEEEEEEEEEEEEE 1058
Query: 237 EREERVAEWGFHE 249
E EE A+ HE
Sbjct: 1059 EEEEVEADEAEHE 1071
Score = 53.1 bits (126), Expect = 7e-07
Identities = 29/61 (47%), Positives = 42/61 (68%), Gaps = 2/61 (3%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E+E+ E EEE++ E+EE QE +E E + EEE EEE +++E+EEEEEE+E + E
Sbjct: 1014 EDEDSEKEEEEEDKEMEELQEGKECENPQGEEEEEEEE--EEEEEEEEEEEVEADEAEHE 1071
Query: 243 A 243
A
Sbjct: 1072 A 1072
Score = 40.0 bits (92), Expect = 0.007
Identities = 34/107 (31%), Positives = 52/107 (47%), Gaps = 30/107 (28%)
Query: 168 DGRLTLHLIPDQVSLEEEEEEIEE----------------EDDQDEIEEDQEQEEEELVE 211
D R +L D+ S +EEEEE +E E++++E EE++E+EEEE VE
Sbjct: 1005 DERESLTREEDEDSEKEEEEEDKEMEELQEGKECENPQGEEEEEEEEEEEEEEEEEEEVE 1064
Query: 212 EEEELEEEVALDDD-------------EKEEEEEEIECERE-ERVAE 244
+E E A D E++ E E+E E E E+++E
Sbjct: 1065 ADEAEHEAAAKTDGTVEVGAAQQAGSLEQKASESEMESESESEQLSE 1111
>CEC1_CAEEL (P34618) Chromo domain protein cec-1
Length = 304
Score = 57.0 bits (136), Expect = 5e-08
Identities = 28/63 (44%), Positives = 46/63 (72%), Gaps = 2/63 (3%)
Query: 184 EEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDD--EKEEEEEEIECEREER 241
EEEE +E ED+++ E + +E+E++ E+E+E EE+V L+ + EKEEEEE++E ++EE
Sbjct: 166 EEEESVEGEDEEESQEVEDLKEDEKMEEDEKEEEEDVQLESEKNEKEEEEEKVEEKKEEE 225
Query: 242 VAE 244
E
Sbjct: 226 EEE 228
Score = 54.3 bits (129), Expect = 3e-07
Identities = 31/83 (37%), Positives = 54/83 (64%), Gaps = 12/83 (14%)
Query: 174 HLIPDQVSLEEEEEEIEEEDD---------QDEIEEDQ--EQEEEELVEEEEELEEEVAL 222
H D+ S E+EEE+E++++ Q+E EE++ E E+EE +E E+L+E+ +
Sbjct: 132 HSSADKKSKAEDEEEVEDDEEPVPKKKKEVQEEPEEEESVEGEDEEESQEVEDLKEDEKM 191
Query: 223 DDDEKEEEEE-EIECEREERVAE 244
++DEKEEEE+ ++E E+ E+ E
Sbjct: 192 EEDEKEEEEDVQLESEKNEKEEE 214
Score = 51.2 bits (121), Expect = 3e-06
Identities = 27/64 (42%), Positives = 44/64 (68%), Gaps = 2/64 (3%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEE--ELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEE +E+E+ + +++EED+++EEE +L E+ E EEE +++KEEEEEE E E +
Sbjct: 176 EEESQEVEDLKEDEKMEEDEKEEEEDVQLESEKNEKEEEEEKVEEKKEEEEEEEEEEIQL 235
Query: 241 RVAE 244
+ E
Sbjct: 236 VIVE 239
Score = 50.8 bits (120), Expect = 4e-06
Identities = 27/66 (40%), Positives = 43/66 (64%)
Query: 179 QVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECER 238
+V E EEEE E +D++E +E ++ +E+E +EE+E+ EEE + EK E+EEE E
Sbjct: 160 EVQEEPEEEESVEGEDEEESQEVEDLKEDEKMEEDEKEEEEDVQLESEKNEKEEEEEKVE 219
Query: 239 EERVAE 244
E++ E
Sbjct: 220 EKKEEE 225
Score = 45.4 bits (106), Expect = 2e-04
Identities = 24/52 (46%), Positives = 36/52 (69%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEI 234
+EEEE+++ E +++E EE++E+ EE+ EEEEE EEE+ L EK E I
Sbjct: 196 KEEEEDVQLESEKNEKEEEEEKVEEKKEEEEEEEEEEIQLVIVEKTVIETTI 247
Score = 38.9 bits (89), Expect = 0.015
Identities = 22/72 (30%), Positives = 47/72 (64%), Gaps = 6/72 (8%)
Query: 177 PDQVSLEEEE--EEIEEEDDQDEIEEDQEQ--EEEELVEEEEELEEEVALDDDEKEEEEE 232
PD + EE ++ + +D++E+E+D+E ++++ V+EE E EE V +D+E+ +E E
Sbjct: 124 PDSDTDEEHSSADKKSKAEDEEEVEDDEEPVPKKKKEVQEEPEEEESVEGEDEEESQEVE 183
Query: 233 EIECEREERVAE 244
++ + +E++ E
Sbjct: 184 DL--KEDEKMEE 193
Score = 35.4 bits (80), Expect = 0.16
Identities = 21/61 (34%), Positives = 33/61 (53%), Gaps = 4/61 (6%)
Query: 188 EIEEEDDQDE----IEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVA 243
+I + D DE ++ + E+EE VE++EE + + E+ EEEE +E E EE
Sbjct: 121 DIRPDSDTDEEHSSADKKSKAEDEEEVEDDEEPVPKKKKEVQEEPEEEESVEGEDEEESQ 180
Query: 244 E 244
E
Sbjct: 181 E 181
Score = 32.7 bits (73), Expect = 1.0
Identities = 22/64 (34%), Positives = 36/64 (55%), Gaps = 4/64 (6%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
EEEEE++EE+ ++ EE++E+EE +LV E+ + E ++ E E E+ V
Sbjct: 212 EEEEEKVEEKKEE---EEEEEEEEIQLVIVEKTVIETTIVEPAVATPEPSE-PSSSEKAV 267
Query: 243 AEWG 246
E G
Sbjct: 268 VENG 271
>YCF2_OENVI (P31569) Protein ycf2 (Fragment)
Length = 630
Score = 56.6 bits (135), Expect = 7e-08
Identities = 33/82 (40%), Positives = 51/82 (61%), Gaps = 1/82 (1%)
Query: 152 TEVRIHRAEILHASRHDGRLTLHLIPDQVSLEEEEEEIEEEDDQ-DEIEEDQEQEEEELV 210
TEV + E + + + L + LEEE+EE++EE+D+ DE EE+ ++EE+EL
Sbjct: 420 TEVAVWGVEEEGEADDEEDVLLEAQQEDELLEEEDEELDEEEDELDEEEEEPKEEEDELH 479
Query: 211 EEEEELEEEVALDDDEKEEEEE 232
EEEEE EEE +D+ +E + E
Sbjct: 480 EEEEEEEEEEEEEDELQENDSE 501
Score = 55.8 bits (133), Expect = 1e-07
Identities = 31/64 (48%), Positives = 47/64 (73%), Gaps = 6/64 (9%)
Query: 183 EEEEEEIEEEDDQ-DEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREER 241
++E+E +EEED++ DE E++ ++EEEE EEE+EL EE +E+EEEEEE E E +E
Sbjct: 444 QQEDELLEEEDEELDEEEDELDEEEEEPKEEEDELHEE-----EEEEEEEEEEEDELQEN 498
Query: 242 VAEW 245
+E+
Sbjct: 499 DSEF 502
Score = 51.2 bits (121), Expect = 3e-06
Identities = 28/59 (47%), Positives = 40/59 (67%), Gaps = 1/59 (1%)
Query: 187 EEIEEEDDQDEIEEDQEQEEEELVEEEEEL-EEEVALDDDEKEEEEEEIECEREERVAE 244
EE E DD++++ + +QE+E L EE+EEL EEE LD++E+E +EEE E EE E
Sbjct: 428 EEEGEADDEEDVLLEAQQEDELLEEEDEELDEEEDELDEEEEEPKEEEDELHEEEEEEE 486
Score = 50.4 bits (119), Expect = 5e-06
Identities = 33/71 (46%), Positives = 45/71 (62%), Gaps = 9/71 (12%)
Query: 179 QVSLEEEEEEIEEEDDQDEIEEDQ------EQEEEELVEEEEEL---EEEVALDDDEKEE 229
+V++ EEE E +D++D + E Q E+E+EEL EEE+EL EEE ++DE E
Sbjct: 421 EVAVWGVEEEGEADDEEDVLLEAQQEDELLEEEDEELDEEEDELDEEEEEPKEEEDELHE 480
Query: 230 EEEEIECEREE 240
EEEE E E EE
Sbjct: 481 EEEEEEEEEEE 491
Score = 43.1 bits (100), Expect = 8e-04
Identities = 26/60 (43%), Positives = 36/60 (59%), Gaps = 2/60 (3%)
Query: 183 EEEEEEIEEEDD--QDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEE E EEE + ++E+E +++E E EE E EEEV ++E E EEE+E EE
Sbjct: 197 EEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEE 256
Score = 43.1 bits (100), Expect = 8e-04
Identities = 26/60 (43%), Positives = 36/60 (59%), Gaps = 2/60 (3%)
Query: 183 EEEEEEIEEEDD--QDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEE E EEE + ++E+E +++E E EE E EEEV ++E E EEE+E EE
Sbjct: 240 EEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEE 299
Score = 43.1 bits (100), Expect = 8e-04
Identities = 27/66 (40%), Positives = 39/66 (58%), Gaps = 3/66 (4%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELE---EEVALDDDEKEEEEEEI 234
++V EEE E E+++ + EE+ E EEE+ EEE+E EEV ++E E EEE+
Sbjct: 248 EEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEV 307
Query: 235 ECEREE 240
E EE
Sbjct: 308 EGTEEE 313
Score = 42.7 bits (99), Expect = 0.001
Identities = 28/66 (42%), Positives = 40/66 (60%), Gaps = 5/66 (7%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQE--EEELVEEEEEL---EEEVALDDDEKEEEEEEIEC 236
+E EEE+E +++ E ED+E E EEE+ EEE+ EEEV ++E E EEE+E
Sbjct: 200 VEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEG 259
Query: 237 EREERV 242
+E V
Sbjct: 260 TEDEEV 265
Score = 42.4 bits (98), Expect = 0.001
Identities = 28/64 (43%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQE--EEELVEEEEEL---EEEVALDDDEKEEEEEEIEC 236
+E EEE+E +++ E ED+E E EEE+ EEE+ EEEV ++E E EEE+E
Sbjct: 243 VEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEG 302
Query: 237 EREE 240
EE
Sbjct: 303 TEEE 306
Score = 42.0 bits (97), Expect = 0.002
Identities = 28/69 (40%), Positives = 39/69 (55%), Gaps = 5/69 (7%)
Query: 177 PDQVSLEEEEEEIE-EEDDQDEIEEDQEQEEEELV----EEEEELEEEVALDDDEKEEEE 231
P + +E EEE+E E++ + EE+ E EEE+ EE E EEEV ++E E E
Sbjct: 181 PTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTE 240
Query: 232 EEIECEREE 240
EE+E EE
Sbjct: 241 EEVEGTEEE 249
Score = 40.8 bits (94), Expect = 0.004
Identities = 26/60 (43%), Positives = 35/60 (58%), Gaps = 2/60 (3%)
Query: 183 EEEEEEIEEEDD--QDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREE 240
EEE E EEE + ++E+E +E+ E EE E EEEV ++E E EEE+E EE
Sbjct: 233 EEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEE 292
Score = 40.4 bits (93), Expect = 0.005
Identities = 28/60 (46%), Positives = 35/60 (57%), Gaps = 2/60 (3%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEE-EELEEEV-ALDDDEKEEEEEEIECEREE 240
+EE E EEE + E E + +EE E EEE E EEEV +D+E E EEE+E EE
Sbjct: 219 DEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEE 278
Score = 40.4 bits (93), Expect = 0.005
Identities = 24/68 (35%), Positives = 41/68 (60%), Gaps = 5/68 (7%)
Query: 182 LEEEEEEIEEEDDQDEI-----EEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIEC 236
+ EEE+E+ E+ + E+ EE+ E ++EE V E + E+E+ ++DE+ +EEE+
Sbjct: 406 IPEEEDELPEDALETEVAVWGVEEEGEADDEEDVLLEAQQEDELLEEEDEELDEEEDELD 465
Query: 237 EREERVAE 244
E EE E
Sbjct: 466 EEEEEPKE 473
Score = 40.0 bits (92), Expect = 0.007
Identities = 27/67 (40%), Positives = 40/67 (59%), Gaps = 4/67 (5%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEEL---EEEVALDDDEKE-EEEEE 233
++V EEE E E+++ + EE+ E EEE+ EEE+ EEEV ++E E E+EE
Sbjct: 205 EEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEE 264
Query: 234 IECEREE 240
+E EE
Sbjct: 265 VEGTEEE 271
Score = 39.7 bits (91), Expect = 0.009
Identities = 27/60 (45%), Positives = 34/60 (56%), Gaps = 2/60 (3%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEE--LEEEVALDDDEKEEEEEEIECEREE 240
EEE E EEE + E E + +EE E EEE E +EEV ++E E EEE+E EE
Sbjct: 226 EEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEE 285
Score = 38.5 bits (88), Expect = 0.019
Identities = 24/55 (43%), Positives = 35/55 (63%), Gaps = 4/55 (7%)
Query: 185 EEEEIE-EEDDQDEIEEDQEQEEEELVEEEEELE---EEVALDDDEKEEEEEEIE 235
E+EE+E E++ + EE+ E EEE+ EEE+E EEV ++E E EEE+E
Sbjct: 261 EDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVE 315
Score = 33.1 bits (74), Expect = 0.80
Identities = 25/79 (31%), Positives = 38/79 (47%), Gaps = 12/79 (15%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEE-----------EEELEEEVALDDDEKEEEE 231
++ E E+EE+DD DE ++ E+L E + L+ E + +E EEE
Sbjct: 351 QKYESELEEDDDDDEDVFAPQKMLEDLFSELVWSPRIWHPWDFLLDCEAEIPAEEIPEEE 410
Query: 232 EEI-ECEREERVAEWGFHE 249
+E+ E E VA WG E
Sbjct: 411 DELPEDALETEVAVWGVEE 429
Score = 32.3 bits (72), Expect = 1.4
Identities = 18/40 (45%), Positives = 23/40 (57%)
Query: 203 EQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E+E E EE E EEEV ++E E EEE+E +E V
Sbjct: 183 EEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEV 222
Score = 31.6 bits (70), Expect = 2.3
Identities = 23/60 (38%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 182 LEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKE-EEEEEIECEREE 240
LE E + ++E+E E+E E EE E EEEV ++E E E+EE+E EE
Sbjct: 170 LELEGALVGSSPTEEEVE-GTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEE 228
>S521_HUMAN (Q96MU7) Putative splicing factor YT521
Length = 727
Score = 56.6 bits (135), Expect = 7e-08
Identities = 29/59 (49%), Positives = 44/59 (74%), Gaps = 2/59 (3%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREER 241
E EEE +EE+ ++DE E+ +E+EE+ E+ EE EEE +++E+EEEEEE E E++ER
Sbjct: 199 ENEEEGVEEDVEEDEEVEEDAEEDEEVDEDGEEEEEEE--EEEEEEEEEEEEEYEQDER 255
Score = 55.1 bits (131), Expect = 2e-07
Identities = 28/65 (43%), Positives = 46/65 (70%), Gaps = 2/65 (3%)
Query: 184 EEEEEIEEEDDQDE-IEED-QEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREER 241
EE+EE+EE+ ++DE ++ED +E+EEEE EEEEE EEE + DE++++EE + +
Sbjct: 210 EEDEEVEEDAEEDEEVDEDGEEEEEEEEEEEEEEEEEEEEYEQDERDQKEEGNDYDTRSE 269
Query: 242 VAEWG 246
++ G
Sbjct: 270 ASDSG 274
Score = 53.5 bits (127), Expect = 6e-07
Identities = 26/57 (45%), Positives = 39/57 (67%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECERE 239
EE EE+ EE+++ DE E++E+EEEE EEEEE EEE D+ +++EE + + E
Sbjct: 213 EEVEEDAEEDEEVDEDGEEEEEEEEEEEEEEEEEEEEYEQDERDQKEEGNDYDTRSE 269
Score = 50.1 bits (118), Expect = 6e-06
Identities = 31/77 (40%), Positives = 47/77 (60%), Gaps = 4/77 (5%)
Query: 185 EEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAE 244
+E+ E++++ +EED E++EE VEE+ E +EEV D+D +EEEEEE E E EE E
Sbjct: 192 DEQGNNTENEEEGVEEDVEEDEE--VEEDAEEDEEV--DEDGEEEEEEEEEEEEEEEEEE 247
Query: 245 WGFHEGYRRCHEMVNQH 261
+ + R E N +
Sbjct: 248 EEYEQDERDQKEEGNDY 264
Score = 41.2 bits (95), Expect = 0.003
Identities = 20/51 (39%), Positives = 30/51 (58%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKE 228
D+ EEEEEE EEE++++E EE+ EQ+E + EE + + D E
Sbjct: 226 DEDGEEEEEEEEEEEEEEEEEEEEYEQDERDQKEEGNDYDTRSEASDSGSE 276
Score = 37.7 bits (86), Expect = 0.032
Identities = 28/81 (34%), Positives = 40/81 (48%), Gaps = 10/81 (12%)
Query: 178 DQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEE-----EE 232
++ + E D+Q E++E+ EE VEE+EE+EE D E++EE EE
Sbjct: 177 EEYGSDHETGSSGSSDEQGNNTENEEEGVEEDVEEDEEVEE-----DAEEDEEVDEDGEE 231
Query: 233 EIECEREERVAEWGFHEGYRR 253
E E E EE E E Y +
Sbjct: 232 EEEEEEEEEEEEEEEEEEYEQ 252
>TCF8_RAT (Q62947) Transcription factor 8 (Zinc finger homeodomain
enhancer-binding protein) (Zfhep)
Length = 1109
Score = 56.2 bits (134), Expect = 9e-08
Identities = 31/73 (42%), Positives = 44/73 (59%), Gaps = 2/73 (2%)
Query: 177 PDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIEC 236
P Q +E E EED+ E EE++E +E E ++E++E E +++E+EEEEEE E
Sbjct: 998 PSQADSDERESLTREEDEDSEKEEEEEDKEMEELQEDKECENPQEEEEEEEEEEEEEEEE 1057
Query: 237 EREERVAEWGFHE 249
E EE AE HE
Sbjct: 1058 EEEE--AEEAEHE 1068
Score = 55.5 bits (132), Expect = 1e-07
Identities = 31/64 (48%), Positives = 43/64 (66%), Gaps = 1/64 (1%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
E+E+ E EEE++ E+EE QE +E E +EEEE EEE +++E+EEEEE E E E
Sbjct: 1013 EDEDSEKEEEEEDKEMEELQEDKECENPQEEEEEEEEEE-EEEEEEEEEEAEEAEHEAAA 1071
Query: 243 AEWG 246
A+ G
Sbjct: 1072 AKTG 1075
Score = 44.3 bits (103), Expect = 3e-04
Identities = 26/69 (37%), Positives = 39/69 (55%), Gaps = 7/69 (10%)
Query: 183 EEEEEEIEEEDDQDEIEED-------QEQEEEELVEEEEELEEEVALDDDEKEEEEEEIE 235
+ E+EE EE+ + +E++ED +E+EEEE EEEEE EEE ++ E E +
Sbjct: 1016 DSEKEEEEEDKEMEELQEDKECENPQEEEEEEEEEEEEEEEEEEEEAEEAEHEAAAAKTG 1075
Query: 236 CEREERVAE 244
EE A+
Sbjct: 1076 GAVEEEAAQ 1084
Score = 42.0 bits (97), Expect = 0.002
Identities = 29/78 (37%), Positives = 41/78 (52%), Gaps = 13/78 (16%)
Query: 168 DGRLTLHLIPDQVSLEEEEEEIEE-------------EDDQDEIEEDQEQEEEELVEEEE 214
D R +L D+ S +EEEEE +E +++++E EE++E+EEEE EE E
Sbjct: 1004 DERESLTREEDEDSEKEEEEEDKEMEELQEDKECENPQEEEEEEEEEEEEEEEEEEEEAE 1063
Query: 215 ELEEEVALDDDEKEEEEE 232
E E E A EEE
Sbjct: 1064 EAEHEAAAAKTGGAVEEE 1081
Score = 36.2 bits (82), Expect = 0.094
Identities = 21/64 (32%), Positives = 35/64 (53%), Gaps = 2/64 (3%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECER--EE 240
EEEEEE EEE++++E E ++ + E + +EEE A +++ E +R EE
Sbjct: 1045 EEEEEEEEEEEEEEEEEAEEAEHEAAAAKTGGAVEEEAAQQAGSFQQKASGSESKRLSEE 1104
Query: 241 RVAE 244
+ E
Sbjct: 1105 KTNE 1108
Score = 35.4 bits (80), Expect = 0.16
Identities = 15/45 (33%), Positives = 30/45 (66%)
Query: 185 EEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEE 229
+EEE EEE++++E EE++E+E EE E + A++++ ++
Sbjct: 1041 QEEEEEEEEEEEEEEEEEEEEAEEAEHEAAAAKTGGAVEEEAAQQ 1085
>RYR1_PIG (P16960) Ryanodine receptor 1 (Skeletal muscle-type
ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle
calcium release channel)
Length = 5035
Score = 56.2 bits (134), Expect = 9e-08
Identities = 33/77 (42%), Positives = 47/77 (60%), Gaps = 10/77 (12%)
Query: 168 DGRLTLHLIPDQVSLEEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEK 227
D + L +I +V EEEEEE EE++E+EEE+ E+EE+ EEE +DE+
Sbjct: 1859 DVKQILKMIEPEVFTEEEEEE----------EEEEEEEEEDEEEKEEDEEEEAREKEDEE 1908
Query: 228 EEEEEEIECEREERVAE 244
+EEEE E E+EE + E
Sbjct: 1909 KEEEETAEGEKEEYLEE 1925
Score = 45.8 bits (107), Expect = 1e-04
Identities = 25/62 (40%), Positives = 41/62 (65%), Gaps = 2/62 (3%)
Query: 183 EEEEEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERV 242
+E+ ++I + + + E++E+EEEE EEEEE +EE +D+E+E E+E E + EE
Sbjct: 1857 DEDVKQILKMIEPEVFTEEEEEEEEE--EEEEEEDEEEKEEDEEEEAREKEDEEKEEEET 1914
Query: 243 AE 244
AE
Sbjct: 1915 AE 1916
Score = 45.1 bits (105), Expect = 2e-04
Identities = 26/66 (39%), Positives = 39/66 (58%)
Query: 186 EEEIEEEDDQDEIEEDQEQEEEELVEEEEELEEEVALDDDEKEEEEEEIECEREERVAEW 245
+E++++ E E E+EEEE EEEEE E+E ++DE+EE E+ + E+EE
Sbjct: 1857 DEDVKQILKMIEPEVFTEEEEEEEEEEEEEEEDEEEKEEDEEEEAREKEDEEKEEEETAE 1916
Query: 246 GFHEGY 251
G E Y
Sbjct: 1917 GEKEEY 1922
Score = 33.9 bits (76), Expect = 0.47
Identities = 22/50 (44%), Positives = 31/50 (62%), Gaps = 5/50 (10%)
Query: 178 DQVSLEEEEEE--IEEEDDQDEIEEDQEQEEEELVEE---EEELEEEVAL 222
D+ EE+EEE E+ED++ E EE E E+EE +EE + +L E V L
Sbjct: 1889 DEEEKEEDEEEEAREKEDEEKEEEETAEGEKEEYLEEGLLQMKLPESVKL 1938
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.132 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,563,070
Number of Sequences: 164201
Number of extensions: 2048759
Number of successful extensions: 82444
Number of sequences better than 10.0: 2250
Number of HSP's better than 10.0 without gapping: 1498
Number of HSP's successfully gapped in prelim test: 813
Number of HSP's that attempted gapping in prelim test: 26257
Number of HSP's gapped (non-prelim): 21106
length of query: 285
length of database: 59,974,054
effective HSP length: 109
effective length of query: 176
effective length of database: 42,076,145
effective search space: 7405401520
effective search space used: 7405401520
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0544.6


