

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0544.13
(545 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
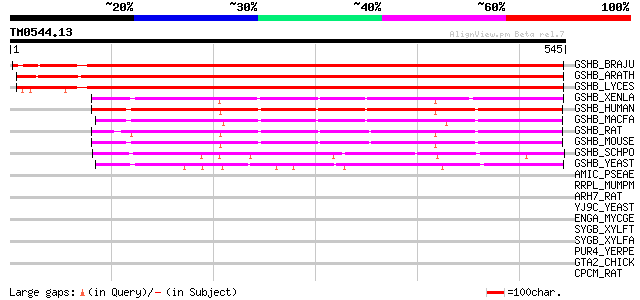
Score E
Sequences producing significant alignments: (bits) Value
GSHB_BRAJU (O23732) Glutathione synthetase, chloroplast precurso... 705 0.0
GSHB_ARATH (P46416) Glutathione synthetase, chloroplast precurso... 685 0.0
GSHB_LYCES (O22494) Glutathione synthetase, chloroplast precurso... 674 0.0
GSHB_XENLA (P35668) Glutathione synthetase (EC 6.3.2.3) (Glutath... 326 8e-89
GSHB_HUMAN (P48637) Glutathione synthetase (EC 6.3.2.3) (Glutath... 314 4e-85
GSHB_MACFA (Q8HXX5) Glutathione synthetase (EC 6.3.2.3) (Glutath... 307 5e-83
GSHB_RAT (P46413) Glutathione synthetase (EC 6.3.2.3) (Glutathio... 301 4e-81
GSHB_MOUSE (P51855) Glutathione synthetase (EC 6.3.2.3) (Glutath... 300 8e-81
GSHB_SCHPO (P35669) Glutathione synthetase large chain (EC 6.3.2... 241 3e-63
GSHB_YEAST (Q08220) Glutathione synthetase (EC 6.3.2.3) (Glutath... 234 4e-61
AMIC_PSEAE (P27017) Aliphatic amidase expression-regulating protein 32 4.2
RRPL_MUMPM (P30929) RNA polymerase beta subunit (EC 2.7.7.48) (L... 32 5.5
ARH7_RAT (O55043) Rho guanine nucleotide exchange factor 7 (PAK-... 32 5.5
YJ9C_YEAST (P47166) Hypothetical 81.2 kDa protein in NMD5-HOM6 i... 31 7.2
ENGA_MYCGE (P47571) GTP-binding protein engA 31 7.2
SYGB_XYLFT (Q87D45) Glycyl-tRNA synthetase beta chain (EC 6.1.1.... 31 9.4
SYGB_XYLFA (Q9PC26) Glycyl-tRNA synthetase beta chain (EC 6.1.1.... 31 9.4
PUR4_YERPE (Q8ZCQ2) Phosphoribosylformylglycinamidine synthase (... 31 9.4
GTA2_CHICK (Q08393) Glutathione S-transferase (EC 2.5.1.18) (Cla... 31 9.4
CPCM_RAT (P19225) Cytochrome P450 2C22 (EC 1.14.14.1) (CYPIIC22)... 31 9.4
>GSHB_BRAJU (O23732) Glutathione synthetase, chloroplast precursor
(EC 6.3.2.3) (Glutathione synthase) (GSH synthetase)
(GSH-S)
Length = 530
Score = 705 bits (1820), Expect = 0.0
Identities = 345/542 (63%), Positives = 438/542 (80%), Gaps = 14/542 (2%)
Query: 3 VGGCGSGCCFITSGTSHSFSHSTFPSFSSIQLHHQSLSLSLSFPKHHLKLIMSQHLSVSS 62
VGGC S ++ +S +F +T S SS++L+ QS L+ K +S S
Sbjct: 2 VGGCSS----LSYSSSSTFIATTTLS-SSLKLNPQSFIFHLNLRKRPPLRCLSSLTMESP 56
Query: 63 SPIVEEVNDSSSFDYHQIDPELLENVVYDALVYATLNGLLVGDKSVQRSGKVPGVGLVHL 122
PI+ D+ + D ++ +VYDALV+++L+GL+VGDK+ QRSG VPGVG++H
Sbjct: 57 KPIL---------DFEKFDDGFVQKLVYDALVWSSLHGLVVGDKTHQRSGTVPGVGMMHA 107
Query: 123 PFSLLPPPLPESYWKQACELAPLFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIHS 182
P +LLP P PESYW QACE+AP+FNELVDR+SLDGKF+Q+SLSRT+KAD FTSRLL+IHS
Sbjct: 108 PIALLPTPFPESYWNQACEVAPIFNELVDRISLDGKFIQDSLSRTRKADVFTSRLLEIHS 167
Query: 183 KMLEINKKEEIRMGLHRSDYMLDEKTKRLLQIEMNTISTSFSGVGCVTTELHRNILSHYG 242
KMLE NK+EEIR+GLHRSDYMLDE+TK LLQIEMNTIS SF G G + TELH+++L +G
Sbjct: 168 KMLESNKREEIRLGLHRSDYMLDEETKSLLQIEMNTISCSFPGFGRLVTELHQSLLRSHG 227
Query: 243 KLLGLDSIRVPANSATNKFAETLAKAWLEYNNPRAVIMIMVQPEEHNMYDQHMISAVLRD 302
LGLDS RVP N +T++FA+ +AKAWLEYNNPRAV+M++VQP+E NMYDQH +S+VLR+
Sbjct: 228 DHLGLDSKRVPRNGSTSQFADAMAKAWLEYNNPRAVVMVVVQPDERNMYDQHWLSSVLRE 287
Query: 303 TYHIPTIRKTLAEVDQEGAVLPDGTLSVDGQAIAIVYFRAGYTPKDYPSESEWRARLLME 362
++I TIRK+LAEV+ EG V DGTL+V GQA+++VY+R+GYTP+DYPSESEW ARLL+E
Sbjct: 288 KHNIVTIRKSLAEVETEGRVHEDGTLTVGGQAVSVVYYRSGYTPRDYPSESEWNARLLIE 347
Query: 363 QSSAIKCPPISYHLVGTKKIQQELAKPNVLERFFENKDDVAKLRECFAGLWSLDDSDIVK 422
QSSA+KCP I+YHL GTKKIQQELAKP VLERF +NKDD+AKLR+CFAGLWSLDD +I+K
Sbjct: 348 QSSAVKCPSIAYHLAGTKKIQQELAKPGVLERFMDNKDDIAKLRKCFAGLWSLDDPEIIK 407
Query: 423 QAIEKPELFVMKPQREGGGNNIYGDDLRETLIKLQKSGSEEDAAYILMQRIFPTSTAAIL 482
+AIEKPELFVMKPQREGGGNNIYGDD+RE L++LQ+ G EE+AAYILMQRIFP + L
Sbjct: 408 KAIEKPELFVMKPQREGGGNNIYGDDVRENLLRLQREGEEENAAYILMQRIFPKVSNVFL 467
Query: 483 MRNGCWHKDHVLTEFGIFGTYLRNKDKVIINSESGYMVRTKMSSSDEGGVLPGFGVIDSV 542
+R+G +HKD ++E G++G YLRNK++VIIN SGY++RTK+SSSDEGGV G+ V+DS+
Sbjct: 468 LRDGVYHKDQAISELGVYGAYLRNKERVIINEHSGYLMRTKVSSSDEGGVAAGYAVLDSI 527
Query: 543 YL 544
YL
Sbjct: 528 YL 529
>GSHB_ARATH (P46416) Glutathione synthetase, chloroplast precursor
(EC 6.3.2.3) (Glutathione synthase) (GSH synthetase)
(GSH-S)
Length = 539
Score = 685 bits (1767), Expect = 0.0
Identities = 339/540 (62%), Positives = 433/540 (79%), Gaps = 5/540 (0%)
Query: 7 GSGCCFITSGTSHSFSHSTFPSFSSIQLHHQSLSLS-LSFPKHHLKLIMSQH-LSVSSSP 64
GSGC ++ +S + + + F S SS SL L+ SF + K + +Q L S
Sbjct: 2 GSGCSSLSYSSSSTCNATVF-SISSSPSSSSSLKLNPSSFLFQNPKTLRNQSPLRCGRSF 60
Query: 65 IVEEVNDSSSFDYHQIDPELLENVVYDALVYATLNGLLVGDKSVQRSGKVPGVGLVHLPF 124
+E + FD ++D E ++ +VYDALV+++L+GL+VGDKS Q+SG VPGVGL+H P
Sbjct: 61 KME--SQKPIFDLEKLDDEFVQKLVYDALVWSSLHGLVVGDKSYQKSGNVPGVGLMHAPI 118
Query: 125 SLLPPPLPESYWKQACELAPLFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIHSKM 184
+LLP PE+YWKQAC + PLFNEL+DRVSLDGKFLQ+SLSRTKK D FTSRLLDIHSKM
Sbjct: 119 ALLPTAFPEAYWKQACNVTPLFNELIDRVSLDGKFLQDSLSRTKKVDVFTSRLLDIHSKM 178
Query: 185 LEINKKEEIRMGLHRSDYMLDEKTKRLLQIEMNTISTSFSGVGCVTTELHRNILSHYGKL 244
LE NKKE+IR+GLHR DYMLDE+T LLQIEMNTIS SF G+ + ++LH+++L YG
Sbjct: 179 LERNKKEDIRLGLHRFDYMLDEETNSLLQIEMNTISCSFPGLSRLVSQLHQSLLRSYGDQ 238
Query: 245 LGLDSIRVPANSATNKFAETLAKAWLEYNNPRAVIMIMVQPEEHNMYDQHMISAVLRDTY 304
+G+DS RVP N++T +FA+ LAKAWLEY+NPRAV+M++VQPEE NMYDQH++S++LR+ +
Sbjct: 239 IGIDSERVPINTSTIQFADALAKAWLEYSNPRAVVMVIVQPEERNMYDQHLLSSILREKH 298
Query: 305 HIPTIRKTLAEVDQEGAVLPDGTLSVDGQAIAIVYFRAGYTPKDYPSESEWRARLLMEQS 364
+I IRKTLAEV++EG+V D TL V GQA+A+VYFR+GYTP D+PSESEW ARLL+E+S
Sbjct: 299 NIVVIRKTLAEVEKEGSVQEDETLIVGGQAVAVVYFRSGYTPNDHPSESEWNARLLIEES 358
Query: 365 SAIKCPPISYHLVGTKKIQQELAKPNVLERFFENKDDVAKLRECFAGLWSLDDSDIVKQA 424
SA+KCP I+YHL G+KKIQQELAKP VLERF +NK+D+AKLR+CFAGLWSLDDS+IVKQA
Sbjct: 359 SAVKCPSIAYHLTGSKKIQQELAKPGVLERFLDNKEDIAKLRKCFAGLWSLDDSEIVKQA 418
Query: 425 IEKPELFVMKPQREGGGNNIYGDDLRETLIKLQKSGSEEDAAYILMQRIFPTSTAAILMR 484
IEKP LFVMKPQREGGGNNIYGDD+RE L++LQK G E +AAYILMQRIFP + L+R
Sbjct: 419 IEKPGLFVMKPQREGGGNNIYGDDVRENLLRLQKEGEEGNAAYILMQRIFPKVSNMFLVR 478
Query: 485 NGCWHKDHVLTEFGIFGTYLRNKDKVIINSESGYMVRTKMSSSDEGGVLPGFGVIDSVYL 544
G +HK ++E G++G YLR+KD+VI+N +SGY++RTK++SSDEGGV GFGV+DS+YL
Sbjct: 479 EGVYHKHQAISELGVYGAYLRSKDEVIVNEQSGYLMRTKIASSDEGGVAAGFGVLDSIYL 538
>GSHB_LYCES (O22494) Glutathione synthetase, chloroplast precursor
(EC 6.3.2.3) (Glutathione synthase) (GSH synthetase)
(GSH-S)
Length = 546
Score = 674 bits (1740), Expect = 0.0
Identities = 345/553 (62%), Positives = 421/553 (75%), Gaps = 24/553 (4%)
Query: 7 GSGCC-----FITSGTSH-----SFSHS-TFPSFSSIQLHHQSLSLSLSFPKHHLKLI-- 53
GSGC T TSH S S+S F S + H S + PK LK
Sbjct: 2 GSGCSSPSISLTTIATSHFQSQESLSNSLNFYSPTRFLEPHLLKSSKIFIPKSPLKCAKV 61
Query: 54 --MSQHLSVSSSPIVEEVNDSSSFDYHQIDPELLENVVYDALVYATLNGLLVGDKSVQRS 111
M L S+ PIV D H ID +L++ + DALV+ L GLLVGD++ +RS
Sbjct: 62 PEMQTQLEDSAKPIV---------DPHDIDSKLVQKLANDALVWCPLRGLLVGDRNSERS 112
Query: 112 GKVPGVGLVHLPFSLLPPPLPESYWKQACELAPLFNELVDRVSLDGKFLQESLSRTKKAD 171
G +PGV +VH P +L+P PES+WKQACE+AP+FNELVDRVS DG+FLQ+SLSRT+KAD
Sbjct: 113 GTIPGVDMVHAPVALIPMSFPESHWKQACEVAPIFNELVDRVSQDGEFLQQSLSRTRKAD 172
Query: 172 EFTSRLLDIHSKMLEINKKEEIRMGLHRSDYMLDEKTKRLLQIEMNTISTSFSGVGCVTT 231
FTSRLL+IHSKMLEINK EEIR+GLHRSDYMLDE+TK LLQIE+NTIS+SF G+ C+ +
Sbjct: 173 PFTSRLLEIHSKMLEINKLEEIRLGLHRSDYMLDEQTKLLLQIELNTISSSFPGLSCLVS 232
Query: 232 ELHRNILSHYGKLLGLDSIRVPANSATNKFAETLAKAWLEYNNPRAVIMIMVQPEEHNMY 291
ELHR++L Y + + D R+PAN+A N+FAE LAKAW EY +PRAVI+ +VQ EE NMY
Sbjct: 233 ELHRSLLQQYREDIASDPNRIPANNAVNQFAEALAKAWNEYGDPRAVIIFVVQAEERNMY 292
Query: 292 DQHMISAVLRDTYHIPTIRKTLAEVDQEGAVLPDGTLSVDGQAIAIVYFRAGYTPKDYPS 351
DQH +SA LR+ + + TIRKTLAE+D G + DGTL VDGQA+A++YFRAGY P DY S
Sbjct: 293 DQHWLSASLRERHQVTTIRKTLAEIDALGELQQDGTLVVDGQAVAVIYFRAGYAPSDYHS 352
Query: 352 ESEWRARLLMEQSSAIKCPPISYHLVGTKKIQQELAKPNVLERFFENKDDVAKLRECFAG 411
ESEW+ARLLMEQS A+KCP ISYHL G+KKIQQELAKPNVLERF ENKDD+AKLR+CFAG
Sbjct: 353 ESEWKARLLMEQSRAVKCPSISYHLAGSKKIQQELAKPNVLERFLENKDDIAKLRKCFAG 412
Query: 412 LWSLDDSDIVKQAIEKPELFVMKPQREGGGNNIYGDDLRETLIKLQKSGSEEDAAYILMQ 471
LWSLD+SDIVK AIE+PEL+VMKPQREGGGNNIYG+D+R L+KLQK G+ DAAYILMQ
Sbjct: 413 LWSLDESDIVKDAIERPELYVMKPQREGGGNNIYGEDVRGALLKLQKEGTGSDAAYILMQ 472
Query: 472 RIFPTSTAAILMRNGCWHKDHVLTEFGIFGTYLRNKDKVIINSESGYMVRTKMSSSDEGG 531
RIFP + +ILMR G HK+ ++E GI+GTYLRNK +V+IN ++GY++RTK+SSSDEGG
Sbjct: 473 RIFPKISHSILMREGISHKEETISELGIYGTYLRNKTEVLINQQAGYLMRTKVSSSDEGG 532
Query: 532 VLPGFGVIDSVYL 544
V GF V+DS+YL
Sbjct: 533 VAAGFAVLDSIYL 545
>GSHB_XENLA (P35668) Glutathione synthetase (EC 6.3.2.3)
(Glutathione synthase) (GSH synthetase) (GSH-S)
Length = 474
Score = 326 bits (836), Expect = 8e-89
Identities = 189/474 (39%), Positives = 287/474 (59%), Gaps = 21/474 (4%)
Query: 81 DPELLENVVYDALVYATLNGLLVGDKSVQRSGKVPGVGLVHLPFSLLPPPLPESYWKQAC 140
D +LLE + A+ A L G+L+ K S V PF+LLP P+P++ ++QA
Sbjct: 11 DTKLLEELAPIAIDAALLQGVLMRTKESPNSSDVVSFA----PFALLPSPVPKALFEQAK 66
Query: 141 ELAPLFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIHSKMLEINKKEEIRMGLHRS 200
+ FN LVDR+S D FL++ LS T K D+F RL IH ++ + + +E+ +G++RS
Sbjct: 67 CVQEDFNTLVDRISQDTSFLEQVLSSTIKVDDFIRRLFAIHKQVQQEDCTQEVFLGINRS 126
Query: 201 DYML---DEKTKRLLQIEMNTISTSFSGVGCVTTELHRNILSHYGKLLGLDSIRVPANSA 257
DYM D+ T L QIE+NTI+ SF G+ T +H+++L K +S + N A
Sbjct: 127 DYMFDCRDDGTPALKQIEINTIAASFGGLASRTPAVHQHVLKFLRK--SEESSSILTNDA 184
Query: 258 TNKFAETLAKAWLEYNNPRAVIMIMVQPEEHNMYDQHMISAVLRDTYHIPTIRKTLAEVD 317
+A AW Y + A +M +V+ E+ N+ DQ I A L ++ IR+ LA+V
Sbjct: 185 VEGIGWGIAHAWALYGSVDATVMFLVENEQRNILDQRFIEAELCKR-NVRVIRRRLADVF 243
Query: 318 QEGAVLPDGTLSVDGQAIAIVYFRAGYTPKDYPSESEWRARLLMEQSSAIKCPPISYHLV 377
+ G + + L +DG +A+ YFR GY P+DY +E +W ARL++E+S A+KCP + LV
Sbjct: 244 ERGTLDEERHLFIDGYEVAVAYFRTGYVPQDY-TEQDWEARLMLERSRAVKCPDVPTQLV 302
Query: 378 GTKKIQQELAKPNVLERFFENKDD-VAKLRECFAGLWSLD----DSDIVKQAIEKPELFV 432
GTKK+QQEL++P +LE+F +K + VA++RE F GL+SLD + V+ A+ P+ FV
Sbjct: 303 GTKKVQQELSRPQILEKFLPDKPEAVARIRETFTGLYSLDIGEEGDEAVRVALANPDQFV 362
Query: 433 MKPQREGGGNNIYGDDLRETLIKLQK-SGSEEDAAYILMQRIFPTSTAAILMR-NGCWHK 490
+KPQREGGGNN+YG++L+E KLQ+ SEE +YILM +I P L+R G
Sbjct: 363 LKPQREGGGNNLYGEELKE---KLQECKDSEERTSYILMDKINPKPLKNCLLRAGGRVQI 419
Query: 491 DHVLTEFGIFGTYLRNKDKVIINSESGYMVRTKMSSSDEGGVLPGFGVIDSVYL 544
++E G+FG Y+R++D++I + G+++RTK +GGV G V+D+ YL
Sbjct: 420 SECISELGMFGVYVRHRDQMIYYDQVGHLLRTKAIEHSDGGVAAGVAVLDNPYL 473
>GSHB_HUMAN (P48637) Glutathione synthetase (EC 6.3.2.3)
(Glutathione synthase) (GSH synthetase) (GSH-S)
Length = 474
Score = 314 bits (804), Expect = 4e-85
Identities = 185/472 (39%), Positives = 285/472 (60%), Gaps = 19/472 (4%)
Query: 81 DPELLENVVYDALVYATLNGLLVGDKSVQRSGKVPGVGLVHLPFSLLPPPLPESYWKQAC 140
D + LE + A+ A G+L+ S +V + + PF+L P +P + +QA
Sbjct: 11 DKQQLEELARQAVDRALAEGVLLRTSQEPTSSEV----VSYAPFTLFPSLVPSALLEQAY 66
Query: 141 ELAPLFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIHSKMLEINKKEEIRMGLHRS 200
+ FN LVD VS + FL+++LS T K D+FT+RL DIH ++L+ + + +GL+RS
Sbjct: 67 AVQMDFNLLVDAVSQNAAFLEQTLSSTIKQDDFTARLFDIHKQVLKEGIAQTVFLGLNRS 126
Query: 201 DYMLD---EKTKRLLQIEMNTISTSFSGVGCVTTELHRNILSHYGKLLGLDSIRVPANSA 257
DYM + + L QIE+NTIS SF G+ T +HR++LS K ++ ++ +N+
Sbjct: 127 DYMFQRSADGSPALKQIEINTISASFGGLASRTPAVHRHVLSVLSKT--KEAGKILSNNP 184
Query: 258 TNKFAETLAKAWLEYNNPRAVIMIMVQPEEHNMYDQHMISAVLRDTYHIPTIRKTLAEVD 317
+ A +AKAW Y +P A+++++ Q +E N++DQ I L +I IR+T ++
Sbjct: 185 SKGLALGIAKAWELYGSPNALVLLIAQEKERNIFDQRAIENELL-ARNIHVIRRTFEDIS 243
Query: 318 QEGAVLPDGTLSVDGQAIAIVYFRAGYTPKDYPSESEWRARLLMEQSSAIKCPPISYHLV 377
++G++ D L VDGQ IA+VYFR GY P+ Y S W ARLL+E+S A KCP I+ L
Sbjct: 244 EKGSLDQDRRLFVDGQEIAVVYFRDGYMPRQY-SLQNWEARLLLERSHAAKCPDIATQLA 302
Query: 378 GTKKIQQELAKPNVLERFFENKDD-VAKLRECFAGLWSLD----DSDIVKQAIEKPELFV 432
GTKK+QQEL++P +LE + + VA+LR FAGL+SLD + +A+ P FV
Sbjct: 303 GTKKVQQELSRPGMLEMLLPGQPEAVARLRATFAGLYSLDVGEEGDQAIAEALAAPSRFV 362
Query: 433 MKPQREGGGNNIYGDDLRETLIKLQKSGSEEDAAYILMQRIFPTSTAAILMRNGCWHK-D 491
+KPQREGGGNN+YG+++ + L +L+ SEE A+YILM++I P L+R G +
Sbjct: 363 LKPQREGGGNNLYGEEMVQALKQLK--DSEERASYILMEKIEPEPFENCLLRPGSPARVV 420
Query: 492 HVLTEFGIFGTYLRNKDKVIINSESGYMVRTKMSSSDEGGVLPGFGVIDSVY 543
++E GIFG Y+R + +++N G+++RTK +GGV G V+D+ Y
Sbjct: 421 QCISELGIFGVYVRQEKTLVMNKHVGHLLRTKAIEHADGGVAAGVAVLDNPY 472
>GSHB_MACFA (Q8HXX5) Glutathione synthetase (EC 6.3.2.3)
(Glutathione synthase) (GSH synthetase) (GSH-S)
(QccE-14611)
Length = 474
Score = 307 bits (786), Expect = 5e-83
Identities = 184/468 (39%), Positives = 281/468 (59%), Gaps = 19/468 (4%)
Query: 85 LENVVYDALVYATLNGLLVGDKSVQRSGKVPGVGLVHLPFSLLPPPLPESYWKQACELAP 144
LE + A+ A G+L+ S +V + + PF+L P +P + +QA +
Sbjct: 15 LEELARQAVDRALAEGVLLRTSQEPTSSEV----VSYAPFTLFPSLVPSALLEQAYAVQM 70
Query: 145 LFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIHSKMLEINKKEEIRMGLHRSDYML 204
FN LVD V+ + FL+++LS T + D+FT+RL DIH ++L+ + + +GL+RSDYM
Sbjct: 71 DFNLLVDAVNQNAAFLEQTLSSTIEQDDFTARLFDIHKQVLKEGIAQTVFLGLNRSDYMF 130
Query: 205 DEKT---KRLLQIEMNTISTSFSGVGCVTTELHRNILSHYGKLLGLDSIRVPANSATNKF 261
T L QIE+NTIS SF G+ T +HR++LS K ++ ++ +N+ +
Sbjct: 131 QRSTDGSPALKQIEINTISASFGGLASRTPAVHRHVLSVLSKT--KEAGKILSNNPSKGL 188
Query: 262 AETLAKAWLEYNNPRAVIMIMVQPEEHNMYDQHMISAVLRDTYHIPTIRKTLAEVDQEGA 321
A +AKAW Y + A+++++ Q +E N++DQ I L +I IR+T ++ ++G+
Sbjct: 189 ALGIAKAWELYGSTNALVLLIAQEKERNIFDQRAIENELL-ARNIHVIRRTFEDISEKGS 247
Query: 322 VLPDGTLSVDGQAIAIVYFRAGYTPKDYPSESEWRARLLMEQSSAIKCPPISYHLVGTKK 381
+ D L VDGQ IA+VYFR GY P+ Y S W ARLL+E+S A KCP I+ L GTKK
Sbjct: 248 LDQDRRLFVDGQEIAVVYFRDGYMPRQY-SLQNWEARLLLERSCAAKCPDIATQLAGTKK 306
Query: 382 IQQELAKPNVLERFFENKDD-VAKLRECFAGLWSLDDSDIVKQAIEK----PELFVMKPQ 436
+QQEL++P +LE + + VA+LR FAGL+SLD + QAI K P FV+KPQ
Sbjct: 307 VQQELSRPGMLEMLLPGQPEAVARLRATFAGLYSLDMGEEGDQAIAKALAAPSRFVLKPQ 366
Query: 437 REGGGNNIYGDDLRETLIKLQKSGSEEDAAYILMQRIFPTSTAAILMRNGC-WHKDHVLT 495
REGGGNN+YG+++ + L +L+ SEE A+YILM++ P L+R G ++
Sbjct: 367 REGGGNNLYGEEMVQALKQLK--DSEERASYILMEKTEPEPFENCLLRPGSPAQVVQCIS 424
Query: 496 EFGIFGTYLRNKDKVIINSESGYMVRTKMSSSDEGGVLPGFGVIDSVY 543
E GIFG Y+R++ +++N G+++RTK +GGV G V+D+ Y
Sbjct: 425 ELGIFGVYVRHEKTLVMNKHVGHLLRTKAIEHADGGVAAGVAVLDNPY 472
>GSHB_RAT (P46413) Glutathione synthetase (EC 6.3.2.3) (Glutathione
synthase) (GSH synthetase) (GSH-S)
Length = 474
Score = 301 bits (770), Expect = 4e-81
Identities = 182/474 (38%), Positives = 280/474 (58%), Gaps = 23/474 (4%)
Query: 81 DPELLENVVYDALVYATLNGLLVGDKSVQRSGKVPGVG--LVHLPFSLLPPPLPESYWKQ 138
D + LE + A+ A G+L+ RS K P + + PF+L P P+P + +Q
Sbjct: 11 DEKQLEELAQQAIDRALAEGVLL------RSAKNPSSSDVVTYAPFTLFPSPVPSTLLEQ 64
Query: 139 ACELAPLFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIHSKMLEINKKEEIRMGLH 198
A + FN LVD VS + FL+++LS T K DE+T+RL DI+ ++L+ + + +GL+
Sbjct: 65 AYAVQMDFNILVDAVSQNSAFLEQTLSSTIKKDEYTARLFDIYKQVLKEGIAQTVFLGLN 124
Query: 199 RSDYMLD---EKTKRLLQIEMNTISTSFSGVGCVTTELHRNILSHYGKLLGLDSIRVPAN 255
RSDYM + +K L QIE+NTIS SF G+ T +HR++L+ K ++ ++ +N
Sbjct: 125 RSDYMFQCSADGSKALKQIEINTISASFGGLASRTPAVHRHVLNVLNKT--NEASKILSN 182
Query: 256 SATNKFAETLAKAWLEYNNPRAVIMIMVQPEEHNMYDQHMISAVLRDTYHIPTIRKTLAE 315
+ + A +AKAW Y + AV++++ Q +E N++DQ I L D I IR+ +
Sbjct: 183 NPSKGLALGIAKAWELYGSANAVVLLIAQEKERNIFDQRAIENELLDR-KIHVIRRRFED 241
Query: 316 VDQEGAVLPDGTLSVDGQAIAIVYFRAGYTPKDYPSESEWRARLLMEQSSAIKCPPISYH 375
V + G++ + L ++ Q +A+VYFR GY P Y +++ W ARLL+E+S A KCP I+
Sbjct: 242 VSERGSLDQNRRLFMEDQEVAVVYFRDGYMPSQYNAQN-WEARLLLERSCAAKCPDIATQ 300
Query: 376 LVGTKKIQQELAKPNVLERFFENKDD-VAKLRECFAGLWSLD----DSDIVKQAIEKPEL 430
L GTKK+QQEL++ +LE + + VA+LR FAGL+SLD V +A+ P
Sbjct: 301 LAGTKKVQQELSRVGLLEALLPGQPEAVARLRATFAGLYSLDMGEEGDQAVAEALAAPSH 360
Query: 431 FVMKPQREGGGNNIYGDDLRETLIKLQKSGSEEDAAYILMQRIFPTSTAAILMRNGC-WH 489
FV+KPQREGGGNN YG+++ L +L+ SEE A+YILM++I P L+R G
Sbjct: 361 FVLKPQREGGGNNFYGEEMVHALEQLK--DSEERASYILMEKIEPEPFRNCLLRPGSPAQ 418
Query: 490 KDHVLTEFGIFGTYLRNKDKVIINSESGYMVRTKMSSSDEGGVLPGFGVIDSVY 543
++E GIFG Y+R +++N G+++RTK +GGV G V+D+ Y
Sbjct: 419 VVQCISELGIFGVYVRQGTTLVMNKHVGHLLRTKAIEHADGGVAAGVAVLDNPY 472
>GSHB_MOUSE (P51855) Glutathione synthetase (EC 6.3.2.3)
(Glutathione synthase) (GSH synthetase) (GSH-S)
Length = 474
Score = 300 bits (767), Expect = 8e-81
Identities = 178/472 (37%), Positives = 279/472 (58%), Gaps = 19/472 (4%)
Query: 81 DPELLENVVYDALVYATLNGLLVGDKSVQRSGKVPGVGLVHLPFSLLPPPLPESYWKQAC 140
D + LE + A+ A G+L+ S V + + PF+L P P+P + +QA
Sbjct: 11 DEKQLEELAKQAIDRALAEGVLLRSAQHPSSSDV----VTYAPFTLFPSPVPSALLEQAY 66
Query: 141 ELAPLFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIHSKMLEINKKEEIRMGLHRS 200
+ FN LVD VS + FL+++LS T K D++T+RL DI+ ++L+ + + +GL+RS
Sbjct: 67 AVQMDFNILVDAVSQNPAFLEQTLSSTIKKDDYTARLFDIYKQVLKEGIAQTVFLGLNRS 126
Query: 201 DYMLD---EKTKRLLQIEMNTISTSFSGVGCVTTELHRNILSHYGKLLGLDSIRVPANSA 257
DYM + +K L QIE+NTIS SF G+ T +HR++L+ K ++ ++ +N+
Sbjct: 127 DYMFQCGADGSKALKQIEINTISASFGGLASRTPAVHRHVLNVLNKT--KEASKILSNNP 184
Query: 258 TNKFAETLAKAWLEYNNPRAVIMIMVQPEEHNMYDQHMISAVLRDTYHIPTIRKTLAEVD 317
+ A +AKAW Y + AV++++ Q +E N++DQ + L D I IR +V
Sbjct: 185 SKGLALGIAKAWELYGSANAVVLLIAQEKERNIFDQRAVENELLDR-KIHVIRGRFEDVS 243
Query: 318 QEGAVLPDGTLSVDGQAIAIVYFRAGYTPKDYPSESEWRARLLMEQSSAIKCPPISYHLV 377
+ G++ + L +D Q +A+VYFR GY P Y S++ W ARL++E+S A KCP I+ L
Sbjct: 244 ERGSLDQNRRLFMDDQEVAVVYFRDGYMPSQYNSQN-WEARLMLERSRAAKCPDIAIQLA 302
Query: 378 GTKKIQQELAKPNVLERFFENKDD-VAKLRECFAGLWSLD----DSDIVKQAIEKPELFV 432
GTKK+QQEL++ +LE + + VA+LR FAGL+SLD + +A+ P FV
Sbjct: 303 GTKKVQQELSRVGLLEALLPGQPEAVARLRATFAGLYSLDMGEEGDQAIAEALAAPSHFV 362
Query: 433 MKPQREGGGNNIYGDDLRETLIKLQKSGSEEDAAYILMQRIFPTSTAAILMRNGC-WHKD 491
+KPQREGGGNN+YG+++ + L +L+ SEE A+YILM++I P L+R G
Sbjct: 363 LKPQREGGGNNLYGEEMVQALEQLK--DSEERASYILMEKIEPEPFRNCLLRPGSPAQVV 420
Query: 492 HVLTEFGIFGTYLRNKDKVIINSESGYMVRTKMSSSDEGGVLPGFGVIDSVY 543
++E GIFG Y+R +++N G+++RTK +GGV G V+D+ Y
Sbjct: 421 QCISELGIFGVYVRQGTTLVMNKHVGHLLRTKAVEHADGGVAAGVAVLDNPY 472
>GSHB_SCHPO (P35669) Glutathione synthetase large chain (EC 6.3.2.3)
(Glutathione synthase large chain) (GSH synthetase large
chain) (GSH-S) (Phytochelatin synthetase)
Length = 498
Score = 241 bits (616), Expect = 3e-63
Identities = 165/498 (33%), Positives = 265/498 (53%), Gaps = 42/498 (8%)
Query: 82 PELLENVVYDALVYATLNGLLVGDKSVQRSGKVPGVGLVHLPFSLLPPPLPESYWKQACE 141
PE +E + A +A +G++ + SV + G+ +P +L P +P + +A
Sbjct: 8 PEQIEELGKGARDFAFAHGVVFTELSVSKEGRNIAT---QIPITLFPSVIPHGAFVEAVS 64
Query: 142 LAPLFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIHSKMLEI------NKKEEIRM 195
+ +N+L +++ D +FL+ L K DEF ++L +++ K E N+ + + +
Sbjct: 65 VQKAYNKLYAKIANDYEFLRLHLQSITKYDEFMNKLWNLYQKHREAVAHLKENQFQPLSL 124
Query: 196 GLHRSDYML--DEKTKRLLQIEMNTISTSFSGVGCVTTELHR--------------NILS 239
G+ RSDYM+ D+ Q+E NTIS SF GV + LH N L+
Sbjct: 125 GVFRSDYMVHQDDSFIGCKQVEFNTISVSFGGVSKAVSNLHAYCSQSGLYRKPLTTNYLT 184
Query: 240 HYGKLLGLDSIRVPANSATNKFAETL-AKAWLEYNNPRAVIMIMVQPEEHNMYDQHMISA 298
+ G+ + A A + + + +K + +N + +++ +V+ E N+ DQ +
Sbjct: 185 VNTSVSGICTGISNAVDAYRDYVKNITSKMNIASDNTKPIVLFVVKGGERNITDQRTLEY 244
Query: 299 VLRDTYHIPTIRKTLAEV-----DQEGAVLPDGTLSVDGQAIAIVYFRAGYTPKDYPSES 353
L + +H+ + R +AE+ D+ L T S +A+VY+R GY DYPS+
Sbjct: 245 ELLNRFHVISKRIDIAELNSLIHDKSSNKLYMKT-SFTTYEVAVVYYRVGYALDDYPSQE 303
Query: 354 EWRARLLMEQSSAIKCPPISYHLVGTKKIQQELAKPNVLERFFENKDDVAKLRECFAGLW 413
W RL +E + AIKCP IS HL G+KKIQQ LA+ N LERF E D++ +R FA ++
Sbjct: 304 AWDMRLTIENTLAIKCPSISTHLAGSKKIQQVLAESNALERFLEG-DELQAVRSTFADMY 362
Query: 414 SLDDS----DIVKQAIEKPELFVMKPQREGGGNNIYGDDLRETLIKLQKSGSEEDAAYIL 469
LDD+ + +K A EKPE FV+KPQREGGGNN YG D+ L K+ + EE +YIL
Sbjct: 363 PLDDTPRGKEGIKLAFEKPEDFVLKPQREGGGNNTYGKDIPGLLSKMPQ---EEWDSYIL 419
Query: 470 MQRIFPTSTAAILMRNGCWHKDHVLTEFGIFGTYLRN--KDKVIINSESGYMVRTKMSSS 527
M+ I + +++ K V+ E GI GT + N D+V+ N +SG++ RTK +
Sbjct: 420 MRYINAVPSQNYILKGERPEKFDVVDEIGILGTIVWNIKTDEVVQNGQSGFICRTKPKKT 479
Query: 528 DEGGVLPGFGVIDSVYLT 545
+EGGV G+ + S+ L+
Sbjct: 480 NEGGVATGYASLSSIELS 497
>GSHB_YEAST (Q08220) Glutathione synthetase (EC 6.3.2.3)
(Glutathione synthase) (GSH synthetase) (GSH-S)
Length = 491
Score = 234 bits (597), Expect = 4e-61
Identities = 153/489 (31%), Positives = 266/489 (54%), Gaps = 38/489 (7%)
Query: 85 LENVVYDALVYATLNGLLVGDKSVQRSGKVPGVGLVHLPFSLLPPPLPESYWKQACELAP 144
L ++ + +A NGL + + + V P ++ P P+P + +A ++ P
Sbjct: 11 LNELIQEVNQWAITNGLSMYPPKFEENPSNASVS----PVTIYPTPIPRKCFDEAVQIQP 66
Query: 145 LFNELVDRVSLDGKFLQESLSRTKKA-----DEFTSRLLDIHSKMLEIN--KKEEIRMGL 197
+FNEL R++ D L +T +A EFT +L ++ L+ KK+ R+G+
Sbjct: 67 VFNELYARITQDMAQPDSYLHKTTEALALSDSEFTGKLWSLYLATLKSAQYKKQNFRLGI 126
Query: 198 HRSDYMLDEK--TKRLLQIEMNTISTSFSGVGCVTTELHRNILSHYGKLLGLDSIRVPAN 255
RSDY++D+K T+++ Q+E NT+S SF+G+ LH + L+ K I N
Sbjct: 127 FRSDYLIDKKKGTEQIKQVEFNTVSVSFAGLSEKVDRLH-SYLNRANKYDPKGPIYNDQN 185
Query: 256 SATNK----FAETLAKAWLEYNNPRA-------VIMIMVQPEEHNMYDQHMISAVLRDTY 304
+ ++ LAKA Y + ++ ++ +VQ E N++DQ ++ L + +
Sbjct: 186 MVISDSGYLLSKALAKAVESYKSQQSSSTTSDPIVAFIVQRNERNVFDQKVLELNLLEKF 245
Query: 305 HIPTIRKTLAEVDQEGAVLPDGT----LSVDGQAIAIVYFRAGYTPKDYPSESEWRARLL 360
++R T +V+ + + D T + Q IA+VY+R GYT DY SE +W ARL
Sbjct: 246 GTKSVRLTFDDVNDK-LFIDDKTGKLFIRDTEQEIAVVYYRTGYTTTDYTSEKDWEARLF 304
Query: 361 MEQSSAIKCPPISYHLVGTKKIQQELAKPNVLERFFENKDDVAKLRECFAGLWSLDDSDI 420
+E+S AIK P + L G+KKIQQ L VL ++ + + + L + F ++ LDD+ +
Sbjct: 305 LEKSFAIKAPDLLTQLSGSKKIQQLLTDEGVLGKYISDAEKKSSLLKTFVKIYPLDDTKL 364
Query: 421 VKQ----AIEKPELFVMKPQREGGGNNIYGDDLRETLIKLQKSGSEEDAAYILMQRIFP- 475
++ A+ +P +V+KPQREGGGNN+Y +++ L +++ + AYILM+ I P
Sbjct: 365 GREGKRLALSEPSKYVLKPQREGGGNNVYKENIPNFLKGIEERHWD---AYILMELIEPE 421
Query: 476 TSTAAILMRNGCWHKDHVLTEFGIFGTYLRNKDKVIINSESGYMVRTKMSSSDEGGVLPG 535
+ I++R+ + + +++E GI+G L N ++V+ N SG ++R+K ++S+EGGV G
Sbjct: 422 LNENNIILRDNKSYNEPIISELGIYGCVLFNDEQVLSNEFSGSLLRSKFNTSNEGGVAAG 481
Query: 536 FGVIDSVYL 544
FG +DS+ L
Sbjct: 482 FGCLDSIIL 490
>AMIC_PSEAE (P27017) Aliphatic amidase expression-regulating protein
Length = 384
Score = 32.0 bits (71), Expect = 4.2
Identities = 24/73 (32%), Positives = 34/73 (45%), Gaps = 7/73 (9%)
Query: 191 EEIRMGLHRSDYMLDEKTKRLLQIEMNTISTSFSGVGCVTTELHRNILSHYGKLLGLDSI 250
EEI + L+ SD + +R+ Q + + ++ G G T EL+R I YG D
Sbjct: 172 EEIYIPLYPSDDDVQRAVERIYQARADVVFSTVVGTG--TAELYRAIARRYG-----DGR 224
Query: 251 RVPANSATNKFAE 263
R P S T AE
Sbjct: 225 RPPIASLTTSEAE 237
>RRPL_MUMPM (P30929) RNA polymerase beta subunit (EC 2.7.7.48) (Large
structural protein) (L protein)
Length = 2261
Score = 31.6 bits (70), Expect = 5.5
Identities = 11/30 (36%), Positives = 18/30 (59%)
Query: 124 FSLLPPPLPESYWKQACELAPLFNELVDRV 153
FSL+PP L YW++ E + + +D+V
Sbjct: 2021 FSLIPPELVSEYWRRRVEQGQIIQDCIDKV 2050
>ARH7_RAT (O55043) Rho guanine nucleotide exchange factor 7
(PAK-interacting exchange factor beta) (Beta-Pix)
Length = 646
Score = 31.6 bits (70), Expect = 5.5
Identities = 19/84 (22%), Positives = 35/84 (41%), Gaps = 2/84 (2%)
Query: 341 RAGYTPKDYPSESEWRARLLMEQSSAIKCPPISYHLVGTKKIQQELAKPNVLERFFENKD 400
R G+ P +Y E + + + +S +K PP + K + N+LE E
Sbjct: 51 RTGWFPSNYVREIKPSEKPVSPKSGTLKSPPKGFDTTAINKSYYNVVLQNILETEHEYSK 110
Query: 401 DVAKLRECFAGLWSLDDSDIVKQA 424
++ + + LW L S+ + A
Sbjct: 111 ELQSVLSTY--LWPLQTSEKLSSA 132
>YJ9C_YEAST (P47166) Hypothetical 81.2 kDa protein in NMD5-HOM6
intergenic region
Length = 707
Score = 31.2 bits (69), Expect = 7.2
Identities = 25/86 (29%), Positives = 46/86 (53%), Gaps = 19/86 (22%)
Query: 149 LVDRVSLDGKFLQESLSRTK-----------KADEFTSRLLDIHSKMLEINKK------- 190
LV+++S + K L+ LS +K KA++ RLL+ + K E+NK+
Sbjct: 597 LVNKLSTELKRLEGELSASKELYDNLLKEKTKANDEILRLLEENDKFNEVNKQKDDLLKR 656
Query: 191 -EEIRMGLHRSDYMLDEKTKRLLQIE 215
E+++ L S +L EKT+++ ++E
Sbjct: 657 VEQMQSKLETSLQLLGEKTEQVEELE 682
>ENGA_MYCGE (P47571) GTP-binding protein engA
Length = 448
Score = 31.2 bits (69), Expect = 7.2
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Query: 375 HLVGTKKIQQELAKPNVLERFFENKDDVAKLRECFAGLWSLDDSDIVKQAIEKPELFV 432
H +G + L K N L EN DD+AK+R C G ++ S ++ Q +++ + V
Sbjct: 150 HGIGIGDLMDLLVKQNQLLPN-ENNDDLAKIRFCVIGKPNVGKSSLINQLVKQNRVLV 206
>SYGB_XYLFT (Q87D45) Glycyl-tRNA synthetase beta chain (EC 6.1.1.14)
(Glycine--tRNA ligase beta chain) (GlyRS)
Length = 722
Score = 30.8 bits (68), Expect = 9.4
Identities = 26/95 (27%), Positives = 49/95 (51%), Gaps = 7/95 (7%)
Query: 360 LMEQSSAIKCPPISYHLVGTKKIQQELAKP-NVLERFFENKDDVAKLRECFAGLWSLDDS 418
L+E SA+ C S+ V + Q+ L + V ++FF D + KL E F G+ +++ +
Sbjct: 256 LVEWPSAVLC---SFERVFLEVPQEALIQTMEVNQKFFPVLDSLGKLTEKFIGIANIESN 312
Query: 419 DIVKQAIEKPELFVMKPQREGGGNNIYGDDLRETL 453
D+ + + K V++P R + +DL++ L
Sbjct: 313 DVAE--VAKGYERVIRP-RFSDAKFFFNEDLKQGL 344
>SYGB_XYLFA (Q9PC26) Glycyl-tRNA synthetase beta chain (EC 6.1.1.14)
(Glycine--tRNA ligase beta chain) (GlyRS)
Length = 722
Score = 30.8 bits (68), Expect = 9.4
Identities = 26/95 (27%), Positives = 49/95 (51%), Gaps = 7/95 (7%)
Query: 360 LMEQSSAIKCPPISYHLVGTKKIQQELAKP-NVLERFFENKDDVAKLRECFAGLWSLDDS 418
L+E SA+ C S+ V + Q+ L + V ++FF D + KL E F G+ +++ +
Sbjct: 256 LVEWPSAVLC---SFERVFLEVPQEALIQTMEVNQKFFPVLDSLGKLTEKFIGIANIESN 312
Query: 419 DIVKQAIEKPELFVMKPQREGGGNNIYGDDLRETL 453
D+ + + K V++P R + +DL++ L
Sbjct: 313 DVAE--VAKGYERVIRP-RFSDAKFFFNEDLKQGL 344
>PUR4_YERPE (Q8ZCQ2) Phosphoribosylformylglycinamidine synthase (EC
6.3.5.3) (FGAM synthase) (FGAMS) (Formylglycinamide
ribotide amidotransferase) (FGARAT) (Formylglycinamide
ribotide synthetase)
Length = 1296
Score = 30.8 bits (68), Expect = 9.4
Identities = 29/112 (25%), Positives = 53/112 (46%), Gaps = 17/112 (15%)
Query: 122 LPFSLLPPPLPESYWKQACELAPLFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIH 181
L FS+ P L ES WKQ L L + +++ V D + ++ S + A + +DI
Sbjct: 109 LAFSIQGPDLNESQWKQLAAL--LHDRMMEAVFTDLQQAEQLFSHHQPA---PVQRVDIL 163
Query: 182 SKMLEINKKEEIRMGLHRSDYMLDEKTKRLLQIEMNTISTSFSGVGCVTTEL 233
+ ++ I++GL L Q E++ + T+F+G+G T++
Sbjct: 164 GQGRSALEQANIKLGL------------ALAQDEIDYLLTAFTGLGRNPTDI 203
>GTA2_CHICK (Q08393) Glutathione S-transferase (EC 2.5.1.18)
(Class-alpha)
Length = 222
Score = 30.8 bits (68), Expect = 9.4
Identities = 35/144 (24%), Positives = 59/144 (40%), Gaps = 26/144 (18%)
Query: 393 ERFFENKDDVAKLRECFAGLWSLDDSDIVKQAIEKPELFVMKPQRE-------GGGNNIY 445
E F E K+D+ KLR +D ++ Q + E+ MK + G N+Y
Sbjct: 32 EEFIEKKEDLEKLR---------NDGSLLFQQVPMVEIDGMKMVQSRAILCYIAGKYNLY 82
Query: 446 GDDLRE-TLIKLQKSGSEEDAAYILMQRIFPTSTAAILMRNGCWHKDHVLTEFGIFGTYL 504
G DL+E I + G+ + I+ P A + +N + T + F Y
Sbjct: 83 GKDLKERAWIDMYVEGTTDLMGMIM---ALPFQAADVKEKNIALITERATTRY--FPVY- 136
Query: 505 RNKDKVIINSESGYMVRTKMSSSD 528
+K + + Y+V K+S +D
Sbjct: 137 ---EKALKDHGQDYLVGNKLSWAD 157
>CPCM_RAT (P19225) Cytochrome P450 2C22 (EC 1.14.14.1) (CYPIIC22)
(P450 MD) (P450 P49)
Length = 489
Score = 30.8 bits (68), Expect = 9.4
Identities = 46/200 (23%), Positives = 74/200 (37%), Gaps = 31/200 (15%)
Query: 134 SYWKQACELAPLFNELVDRVSLDGKFLQESLSRTKKADEFTSRLLDIHSKMLEINKKEEI 193
S W Q C P+ L + KFL++ + K F ++ H L ++ ++
Sbjct: 210 SPWAQLCSAYPVLYYLP---GIHNKFLKDVTEQKK----FILMEINRHRASLNLSNPQDF 262
Query: 194 RMGLHRSDYML----DEKTKRLLQIEMNTISTSFSGVGCVTTELHRNILSHYGKLLGLDS 249
DY L EK + M+ + + + TE + + YG LL L
Sbjct: 263 ------IDYFLIKMEKEKHNEKSEFTMDNLIVTIGDLFGAGTETTSSTIK-YGLLLLLKY 315
Query: 250 IRVPANSATNKFAETLAKAWLEYNNPRAVIMIMVQPEEHNMYDQHMISAVLR--DTYHIP 307
V A K E + + + P +Q H Y ++ + R D IP
Sbjct: 316 PEVTA-----KIQEEITRVIGRHRRP------CMQDRNHMPYTDAVLHEIQRYIDFVPIP 364
Query: 308 TIRKTLAEVDQEGAVLPDGT 327
RKT +V+ G +P GT
Sbjct: 365 LPRKTTQDVEFRGYHIPKGT 384
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,564,247
Number of Sequences: 164201
Number of extensions: 2749148
Number of successful extensions: 7200
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 7134
Number of HSP's gapped (non-prelim): 22
length of query: 545
length of database: 59,974,054
effective HSP length: 115
effective length of query: 430
effective length of database: 41,090,939
effective search space: 17669103770
effective search space used: 17669103770
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0544.13


