

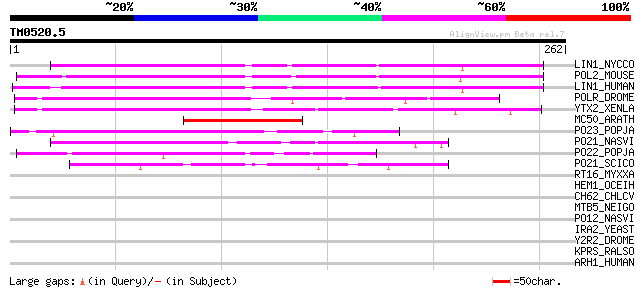
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0520.5
(262 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 90 5e-18
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 87 3e-17
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 79 1e-14
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 74 4e-13
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 68 2e-11
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 62 2e-09
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 55 2e-07
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 49 9e-06
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 49 1e-05
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 44 5e-04
RT16_MYXXA (P23072) RNA-directed DNA polymerase from retron Mx16... 34 0.41
HEM1_OCEIH (Q8EPM7) Glutamyl-tRNA reductase (EC 1.2.1.-) (GluTR) 33 0.54
CH62_CHLCV (P59698) 60 kDa chaperonin 2 (Protein Cpn60 2) (groEL... 33 0.91
MTB5_NEIGO (Q59605) Modification methylase NgoBV (EC 2.1.1.37) (... 32 2.0
PO12_NASVI (Q03270) Retrovirus-related Pol polyprotein from type... 30 4.5
IRA2_YEAST (P19158) Inhibitory regulator protein IRA2 30 4.5
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 30 5.9
KPRS_RALSO (Q8Y2E1) Ribose-phosphate pyrophosphokinase (EC 2.7.6... 30 5.9
ARH1_HUMAN (Q92888) Rho guanine nucleotide exchange factor 1 (p1... 30 7.7
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 90.1 bits (222), Expect = 5e-18
Identities = 66/235 (28%), Positives = 116/235 (49%), Gaps = 8/235 (3%)
Query: 20 KGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMWGVKCPKLSILWNCSKLP 79
K + + + ID EKA+D + F+ TL G + LI P +I+ N KL
Sbjct: 589 KNKDHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNGVKLK 648
Query: 80 SFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLFFADDV 139
SF + G RQG PLSP LF + ME LAI I+ + K + I + + +S FADD+
Sbjct: 649 SFPLRSGTRQGCPLSPLLFNIVMEVLAIAIRE---EKAIKGIHIGSEEIKLS--LFADDM 703
Query: 140 LLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAVDHRRQINLSHIAGISRAS 199
+++ + +++ + +KE+ SG K+N KS + F +++ + + +
Sbjct: 704 IVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKS-VAFIYTNNNQAEKTVKDSIPFTVVP 762
Query: 200 DLGKYLGIPLLKG--RVTKAHFASLIGKMSSRLTSWKNNLLNRAGRVCLAKSVLM 252
KYLG+ L K + K ++ +L +++ + WKN + GR+ + K ++
Sbjct: 763 KKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSWLGRINIVKMSIL 817
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 87.4 bits (215), Expect = 3e-17
Identities = 74/251 (29%), Positives = 122/251 (48%), Gaps = 9/251 (3%)
Query: 4 NIILAQEVMHTIHTQKKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMWG 63
NI + V+H I+ K ++ I +D EKA+D++ F+ L G +++I
Sbjct: 601 NIRKSINVIHYINKLKDKNHMI-ISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMIKAI 659
Query: 64 VKCPKLSILWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRI 123
P +I N KL + + G RQG PLSPYLF + +E LA I+ Q K ++I
Sbjct: 660 YSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQ---QKEIKGIQI 716
Query: 124 TKDGVGISHLFFADDVLLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAVDH 183
K+ V IS L ADD++++ K + + N + F E G K+N +KS M F +
Sbjct: 717 GKEEVKISLL--ADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKS-MAFLYTKNK 773
Query: 184 RRQINLSHIAGISRASDLGKYLGIPLLK--GRVTKAHFASLIGKMSSRLTSWKNNLLNRA 241
+ + + S ++ KYLG+ L K + +F SL ++ L WK+ +
Sbjct: 774 QAEKEIRETTPFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLPCSWI 833
Query: 242 GRVCLAKSVLM 252
GR+ + K ++
Sbjct: 834 GRINIVKMAIL 844
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 78.6 bits (192), Expect = 1e-14
Identities = 67/253 (26%), Positives = 118/253 (46%), Gaps = 13/253 (5%)
Query: 2 RDNIILAQEVMHTIHTQKKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIM 61
R +I + Q + T T + I ID EKA+D++ F+ L G + +I
Sbjct: 576 RKSINIIQHINRTKDTNH-----MIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKIIR 630
Query: 62 WGVKCPKLSILWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPV 121
P +I+ N KL + + G RQG PLSP L + +E LA I+ + K +
Sbjct: 631 AIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSPLLPNIVLEVLARAIRQ---EKEIKGI 687
Query: 122 RITKDGVGISHLFFADDVLLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAV 181
++ K+ V +S FADD++++ + Q + + F + SG K+N+ KS+ F
Sbjct: 688 QLGKEEVKLS--LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQ-AFLYTN 744
Query: 182 DHRRQINLSHIAGISRASDLGKYLGIPLLKG--RVTKAHFASLIGKMSSRLTSWKNNLLN 239
+ + + + + AS KYLGI L + + K ++ L+ ++ WKN +
Sbjct: 745 NRQTESQIMSELPFTIASKRIKYLGIQLTRDVKDLFKENYKPLLNEIKEDTNKWKNIPCS 804
Query: 240 RAGRVCLAKSVLM 252
GR+ + K ++
Sbjct: 805 WVGRINIVKMAIL 817
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 73.9 bits (180), Expect = 4e-13
Identities = 69/239 (28%), Positives = 106/239 (43%), Gaps = 23/239 (9%)
Query: 3 DNIILAQEVMHTIHTQKKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMW 62
DN + V+ H+ K R +D+ KA+D +S + TL +G P VD +
Sbjct: 457 DNATIVDLVLR--HSHKHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQN 514
Query: 63 GVKCPKLSILWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVR 122
+ S+ + F P RG++QGDPLSP LF L M+RL + + +
Sbjct: 515 TYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEIG-------- 566
Query: 123 ITKDGVGISH-LFFADDVLLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAV 181
K G I++ FADD++LF + L++ TL +F G+K+N DK + K
Sbjct: 567 -AKVGNAITNAAAFADDLVLFAETRMGLQVLLDKTL-DFLSIVGLKLNADKCFTVGIKGQ 624
Query: 182 DHRR---------QINLSHIAGISRASDLGKYLGIPLLKGRVTKAHFASLIGKMSSRLT 231
++ + S I + R +D KYLGI + + A IG RLT
Sbjct: 625 PKQKCTVLEAQSFYVGSSEIPSLKR-TDEWKYLGINFTATGRVRCNPAEDIGPKLQRLT 682
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 68.2 bits (165), Expect = 2e-11
Identities = 64/252 (25%), Positives = 112/252 (44%), Gaps = 13/252 (5%)
Query: 3 DNIILAQEVMHTIHTQKKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMW 62
DN+ L ++++H ++ G L + +D EKA+DRV +L TL + F P+ V +
Sbjct: 569 DNVFLIRDLLH--FARRTGLSLAFLSLDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKT 626
Query: 63 GVKCPKLSILWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVR 122
+ + N S A RG+RQG PLS L+ L +E ++ +T +
Sbjct: 627 MYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFLCLLRKRLT-----GLV 681
Query: 123 ITKDGVGISHLFFADDVLLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAVD 182
+ + + + +ADDV+L Q L+ + + AS ++N KS L ++
Sbjct: 682 LKEPDMRVVLSAYADDVILVAQ-DLVDLERAQECQEVYAAASSARINWSKSSGLLEGSL- 739
Query: 183 HRRQINLSHIAGISRASDLGKYLGIPL-LKGRVTKAHFASLIGKMSSRLTSWKN--NLLN 239
+ IS S + KYLG+ L + +F L + +RL WK +L+
Sbjct: 740 -KVDFLPPAFRDISWESKIIKYLGVYLSAEEYPVSQNFIELEECVLTRLGKWKGFAKVLS 798
Query: 240 RAGRVCLAKSVL 251
GR + ++
Sbjct: 799 MRGRALVINQLV 810
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 61.6 bits (148), Expect = 2e-09
Identities = 29/56 (51%), Positives = 37/56 (65%)
Query: 83 PQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLFFADD 138
P RGLRQGDPLSPYLF+LC E L+ + QG +R++ + I+HL FADD
Sbjct: 24 PSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRINHLLFADD 79
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 55.1 bits (131), Expect = 2e-07
Identities = 50/188 (26%), Positives = 83/188 (43%), Gaps = 17/188 (9%)
Query: 1 TRDNIILAQEVMHTIHTQK-KGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDL 59
T NII+ + H I ++ KG+ + +D+ KA+D VS + + FG + D
Sbjct: 2 TLANIIMLE---HYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDF 58
Query: 60 IMWGVKCPKLSILWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWK 119
IM + +I+ + G++QGDPLSP LF + ++ L ++ + +
Sbjct: 59 IMSTITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASMT 118
Query: 120 PVRITKDGVGISHLFFADDVLLFCQASKEQLQLVNNTLKEFC---EASGMKVNLDKSRML 176
P I+ L FADD+LL + V N+L C GM +N +K +
Sbjct: 119 P------ACKIASLAFADDLLLLEDRDID----VPNSLATTCAYFRTRGMTLNPEKCASI 168
Query: 177 FSKAVDHR 184
+ V R
Sbjct: 169 SAATVSGR 176
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 49.3 bits (116), Expect = 9e-06
Identities = 52/197 (26%), Positives = 86/197 (43%), Gaps = 18/197 (9%)
Query: 20 KGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMWGVKCPKLSILWNCSKLP 79
K +GL +D++KA+D V + L P + + IMW + K + +K
Sbjct: 446 KIKGLYIAILDVKKAFDSVEHRSILDALRRKKLPLEMRNYIMWVYRNSKTRLEVVKTKGR 505
Query: 80 SFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLFFADDV 139
P RG+RQGDPLSP LF M+ + + + T + K G L FADD+
Sbjct: 506 WIRPARGVRQGDPLSPLLFNCVMD----AVLRRLPENTGFLMGAEKIGA----LVFADDL 557
Query: 140 LLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLFSKAVDHRRQINLS----HIAGI 195
+L + ++E LQ + ++ + G+++ K L ++I + G
Sbjct: 558 VLLAE-TREGLQASLSRIEAGLQEQGLEMMPRKCHTLALVPSGKEKKIKVETHKPFTVGN 616
Query: 196 SRASDLG-----KYLGI 207
+ LG KYLG+
Sbjct: 617 QEITQLGHADQWKYLGV 633
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 48.9 bits (115), Expect = 1e-05
Identities = 48/171 (28%), Positives = 77/171 (44%), Gaps = 10/171 (5%)
Query: 4 NIILAQEVMHTIHTQKKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMWG 63
N +L + + Q+K +V++ D+ KA+D VS + L G + I
Sbjct: 119 NSLLLDTYISSRREQRKTYNVVSL--DVRKAFDTVSHSSICRALQRLGIDEGTSNYITGS 176
Query: 64 VKCPKLSI-LWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVR 122
+ +I + S+ +RG++QGDPLSP+LF ++ L +Q+ T G +
Sbjct: 177 LSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQS--TPGIGGTIG 234
Query: 123 ITKDGVGISHLFFADDVLLFCQASKEQLQLVNNTLKEFCEASGMKVNLDKS 173
K I L FADD+LL + + L T+ F GM +N KS
Sbjct: 235 EEK----IPVLAFADDLLLL-EDNDVLLPTTLATVANFFRLRGMSLNAKKS 280
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 43.5 bits (101), Expect = 5e-04
Identities = 52/193 (26%), Positives = 80/193 (40%), Gaps = 29/193 (15%)
Query: 29 IDLEKAYDRVSWDFLRATLYDFGFPPRIVDLI--MWGVKCPKLSILWNCSKLPSFAPQRG 86
+DL KA++ V L + + G PP +VD I M+ ++ C A G
Sbjct: 296 LDLVKAFNSVYHSALIDAITEAGCPPGVVDYIADMYNNVITEMQFEGKCELASILA---G 352
Query: 87 LRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLFFADDVLLFCQ-- 144
+ QGDPLS LF L E+ + N +G R V ++ ++DD LL
Sbjct: 353 VYQGDPLSGPLFTLAYEKALRALNN---EG-----RFDIADVRVNASAYSDDGLLLAMTV 404
Query: 145 -ASKEQLQLVNNTLKEFCEASGMKVNLDKSRMLF---------SKAVDHRRQINLSHIAG 194
+ L TL + G+++N KS+ + K V +RR + +
Sbjct: 405 IGLQHNLDKFGETLAKI----GLRINSRKSKTVSLVPSGREKKMKIVSNRRLLLKASELK 460
Query: 195 ISRASDLGKYLGI 207
SDL KYLG+
Sbjct: 461 PLTISDLWKYLGV 473
>RT16_MYXXA (P23072) RNA-directed DNA polymerase from retron
Mx162-RT (EC 2.7.7.49) (Reverse transcriptase)
(Mx162-RT)
Length = 485
Score = 33.9 bits (76), Expect = 0.41
Identities = 34/161 (21%), Positives = 67/161 (41%), Gaps = 22/161 (13%)
Query: 25 VAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLI-MWGVKCPKLSILWNCSKLPSFAP 83
V +K+DL+ + V+W ++ L G L+ + + P+ ++ + L
Sbjct: 245 VVVKVDLKDFFPSVTWRRVKGLLRKGGLREGTSTLLSLLSTEAPREAVQFRGKLLHVAKG 304
Query: 84 QRGLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLFFADDVLLFC 143
R L QG P SP I N + K + +G ++ +ADD+
Sbjct: 305 PRALPQGAPTSP------------GITNALCLKLDKRLSALAKRLGFTYTRYADDLTFSW 352
Query: 144 QASKE---------QLQLVNNTLKEFCEASGMKVNLDKSRM 175
+K+ + ++ + ++E EA G +V+ DK+R+
Sbjct: 353 TKAKQPKPRRTQRPPVAVLLSRVQEVVEAEGFRVHPDKTRV 393
>HEM1_OCEIH (Q8EPM7) Glutamyl-tRNA reductase (EC 1.2.1.-) (GluTR)
Length = 457
Score = 33.5 bits (75), Expect = 0.54
Identities = 17/56 (30%), Positives = 28/56 (49%)
Query: 3 DNIILAQEVMHTIHTQKKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVD 58
D+I++ +E+M + Q+KGR L + I + + D + LYD IVD
Sbjct: 253 DSIVVTKEMMEKVQKQRKGRALFLVDIAVPRDMDPAISELENVFLYDIDNLQHIVD 308
>CH62_CHLCV (P59698) 60 kDa chaperonin 2 (Protein Cpn60 2) (groEL
protein 2)
Length = 536
Score = 32.7 bits (73), Expect = 0.91
Identities = 21/73 (28%), Positives = 31/73 (41%), Gaps = 13/73 (17%)
Query: 60 IMWGVKC-------PKLSILWNCSKLPSFAPQRGLRQGDPLSPYLFVLCMERLAIQIQNL 112
+M+G KC P + NC K P + L+ DP Y C+ +NL
Sbjct: 437 VMFGCKCMLQSAEEPLRVLATNCGKDPEYVVDTVLKHADP---YFGYNCIND---SFENL 490
Query: 113 VTQGTWKPVRITK 125
+T G + P +TK
Sbjct: 491 ITSGVFDPFSVTK 503
>MTB5_NEIGO (Q59605) Modification methylase NgoBV (EC 2.1.1.37)
(Cytosine-specific methyltransferase NgoBV) (M.NgoBV)
(M.NgoV)
Length = 423
Score = 31.6 bits (70), Expect = 2.0
Identities = 42/158 (26%), Positives = 66/158 (41%), Gaps = 17/158 (10%)
Query: 17 TQKKGRGLVAIKIDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMWGVKCPKLSILWNCS 76
TQK GR L I LE VSW L A +FG P + + G K + + S
Sbjct: 132 TQKIGRTLTVILETLEALGYYVSWKVLNAK--EFGIPQNRKRIYLTGSLKSKPDLSFETS 189
Query: 77 KLPSFAPQRGLRQGDPL--SPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLF 134
P + L G P SP++ L + ++ + KD G +
Sbjct: 190 PSPKL--KNILESGLPTESSPFIKKLLKKFPPSELYG----------KSVKDKRGGKNNI 237
Query: 135 FADDVLLFCQASKEQLQLVNNTLKEFCEASGM-KVNLD 171
+ D+ L ++E+ QL+N LKE + +G+ K+ +D
Sbjct: 238 HSWDIELKGAVTEEEKQLLNILLKERRKKNGLQKIGID 275
>PO12_NASVI (Q03270) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 410
Score = 30.4 bits (67), Expect = 4.5
Identities = 36/162 (22%), Positives = 68/162 (41%), Gaps = 26/162 (16%)
Query: 86 GLRQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLFFADDVLLFCQA 145
G QG P L+ L M + I++ G G+S FADDV + +
Sbjct: 1 GCPQGSIGGPILWNLSMNGMLIELA----------------GSGVSVCAFADDVAILAEG 44
Query: 146 S--KEQLQLVNNTLK---EFCEASGMKVNLDKS--RMLFSKAVDHRRQINLSHIAGISRA 198
+ KE +++N ++ ++ E G+ V+ +K+ +L K V R + +S + +
Sbjct: 45 NTRKEVERVINEKMEIVYKWGEKMGVSVSEEKTVCMLLKGKYVTSNRAVRVSKTINVYKR 104
Query: 199 ---SDLGKYLGIPLLKGRVTKAHFASLIGKMSSRLTSWKNNL 237
+YLG+ + +G H + K++ + K L
Sbjct: 105 IKYVSCFRYLGVNVTEGMGFGEHVRGMKVKVAKGVQKLKRVL 146
>IRA2_YEAST (P19158) Inhibitory regulator protein IRA2
Length = 3079
Score = 30.4 bits (67), Expect = 4.5
Identities = 22/63 (34%), Positives = 33/63 (51%), Gaps = 8/63 (12%)
Query: 166 MKVNLDKSRMLFSKAVDHRRQINLSHIAGISRASDLGKY------LGIPLLKGRVTKAHF 219
+K+N +K ++L V + SH+AGI+ AS +Y L PL+ G V +A F
Sbjct: 1151 LKINNEKRQILLDITVKFMKV--RSHLAGIAEASHHMEYISDSEKLTFPLIMGTVGRALF 1208
Query: 220 ASL 222
SL
Sbjct: 1209 VSL 1211
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 30.0 bits (66), Expect = 5.9
Identities = 44/204 (21%), Positives = 78/204 (37%), Gaps = 26/204 (12%)
Query: 29 IDLEKAYDRVSWDFLRATLYDFGFPPRIVDLIMW-GVKCPKLSILWNCSKLPSFAPQRGL 87
+D + A+D V W + L D G ++ +W + +++ + S RG
Sbjct: 577 VDFKGAFDNVEWSAALSRLADLG----CREMGLWQSFFSGRRAVIRSSSGTVEVPVTRGC 632
Query: 88 RQGDPLSPYLFVLCMERLAIQIQNLVTQGTWKPVRITKDGVGISHLFFADDVLLFCQA-S 146
QG P+++ + M+ L ++Q +ADD+LL + S
Sbjct: 633 PQGSISGPFIWDILMDVLLQRLQPYCQLSA-----------------YADDLLLLVEGNS 675
Query: 147 KEQLQLVNNTLKEFCEASGMKVN--LDKSRMLFSKAVDHRRQINLSHIAGISRASDLG-K 203
+ L+ L E G +V L S+ + R+ AG + +
Sbjct: 676 RAVLEEKGAQLMSIVETWGAEVGDCLSTSKTVIMLLKGALRRAPTVRFAGRNLPYVRSCR 735
Query: 204 YLGIPLLKGRVTKAHFASLIGKMS 227
YLGI + +G H ASL +M+
Sbjct: 736 YLGITVSEGMKFLTHIASLRQRMT 759
>KPRS_RALSO (Q8Y2E1) Ribose-phosphate pyrophosphokinase (EC 2.7.6.1)
(RPPK) (Phosphoribosyl pyrophosphate synthetase)
(P-Rib-PP synthetase) (PRPP synthetase)
Length = 316
Score = 30.0 bits (66), Expect = 5.9
Identities = 20/65 (30%), Positives = 35/65 (53%), Gaps = 5/65 (7%)
Query: 192 IAGISRASDLGKYLGIPL--LKGRVTKAHFA---SLIGKMSSRLTSWKNNLLNRAGRVCL 246
+ G+ RA L K LG+ L + R KA+ A ++IG++ R +++++ G +C
Sbjct: 173 VGGVVRARALAKQLGVDLAIIDKRRPKANVAEVMNIIGEVEGRNCVIMDDMIDTGGTLCK 232
Query: 247 AKSVL 251
A VL
Sbjct: 233 AAQVL 237
>ARH1_HUMAN (Q92888) Rho guanine nucleotide exchange factor 1
(p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide
exchange factor) (Sub1.5)
Length = 912
Score = 29.6 bits (65), Expect = 7.7
Identities = 20/56 (35%), Positives = 30/56 (52%), Gaps = 5/56 (8%)
Query: 103 ERLAIQIQNLVTQG--TWKPVRITKDGVGISHLFFADDVLLFCQASKEQLQLVNNT 156
+ L I + LV +G TW R+TKD H+ DD+LL Q E+L L +++
Sbjct: 639 KNLDITKKKLVHEGPLTW---RVTKDKAVEVHVLLLDDLLLLLQRQDERLLLKSHS 691
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.141 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,318,257
Number of Sequences: 164201
Number of extensions: 1171893
Number of successful extensions: 3215
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 3191
Number of HSP's gapped (non-prelim): 20
length of query: 262
length of database: 59,974,054
effective HSP length: 108
effective length of query: 154
effective length of database: 42,240,346
effective search space: 6505013284
effective search space used: 6505013284
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0520.5


