

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
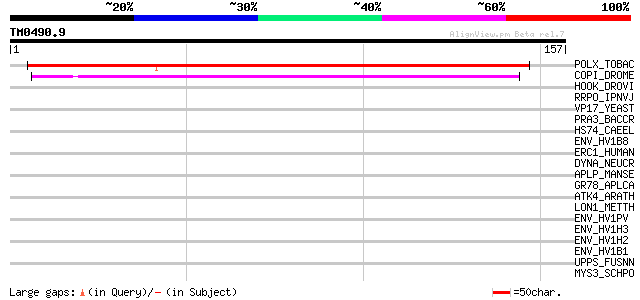
Query= TM0490.9
(157 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 134 8e-32
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 60 3e-09
HOOK_DROVI (O61493) Hook protein 34 0.15
RRPO_IPNVJ (P22173) Putative RNA-directed RNA polymerase (EC 2.7... 31 0.99
VP17_YEAST (P32913) Vacuolar protein sorting-associated protein ... 31 1.3
PRA3_BACCR (Q81DT1) Foldase protein prsA 3 precursor (EC 5.2.1.8) 31 1.3
HS74_CAEEL (P20163) Heat shock 70 kDa protein D precursor 31 1.3
ENV_HV1B8 (P04582) Envelope polyprotein GP160 precursor [Contain... 30 2.2
ERC1_HUMAN (Q8IUD2) ERC protein 1 (ELKS protein) 29 3.8
DYNA_NEUCR (Q01397) Dynactin, 150 kDa isoform (150 kDa dynein-as... 29 3.8
APLP_MANSE (Q25490) Apolipophorins precursor [Contains: Apolipop... 29 3.8
GR78_APLCA (Q16956) 78 kDa glucose-regulated protein precursor (... 29 4.9
ATK4_ARATH (O81635) Kinesin 4 (Kinesin-like protein D) 29 4.9
LON1_METTH (O26878) Putative protease La homolog type 1 (EC 3.4.... 28 6.4
ENV_HV1PV (P03376) Envelope polyprotein GP160 precursor [Contain... 28 6.4
ENV_HV1H3 (P04624) Envelope polyprotein GP160 precursor [Contain... 28 6.4
ENV_HV1H2 (P04578) Envelope polyprotein GP160 precursor [Contain... 28 6.4
ENV_HV1B1 (P03375) Envelope polyprotein GP160 precursor [Contain... 28 6.4
UPPS_FUSNN (Q8RE08) Undecaprenyl pyrophosphate synthetase (EC 2.... 28 8.4
MYS3_SCHPO (O14157) Myosin type II heavy chain 2 28 8.4
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 134 bits (337), Expect = 8e-32
Identities = 65/145 (44%), Positives = 94/145 (64%), Gaps = 3/145 (2%)
Query: 6 GAKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKAL---RDKSLDIAIVDWNEMKKKAAGL 62
G K+EV + NG F WQ R++DLL QQGLHK L K + DW ++ ++AA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHV 122
I+L +SDDV+N+ +D T +W LES YMSKTL +KL +K++ Y+L M EG + H+
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 NAFNNILADLTRLGVKVDDENKAII 147
N FN ++ L LGVK+++E+KAI+
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAIL 147
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 59.7 bits (143), Expect = 3e-09
Identities = 33/138 (23%), Positives = 70/138 (49%), Gaps = 1/138 (0%)
Query: 7 AKFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIKLC 66
AK + +G + + +W+ R++ LLA+Q + K + + W + ++ A I
Sbjct: 4 AKRNIKPFDGEK-YAIWKFRIRALLAEQDVLKVVDGLMPNEVDDSWKKAERCAKSTIIEY 62
Query: 67 VSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQDHVNAFN 126
+SD +N T + + L++ Y K+L +L +++R SLK+ L H + F+
Sbjct: 63 LSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFHIFD 122
Query: 127 NILADLTRLGVKVDDENK 144
++++L G K+++ +K
Sbjct: 123 ELISELLAAGAKIEEMDK 140
>HOOK_DROVI (O61493) Hook protein
Length = 678
Score = 33.9 bits (76), Expect = 0.15
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 9/79 (11%)
Query: 69 DDVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRR---YSLKMQEGGDLQDHVNAF 125
D++M T +LT L D D L DKL V + + Y K++E DL+ HV
Sbjct: 292 DELMKSTAELTALKDEVDVLRES------TDKLKVCEAQLETYKKKLEEYNDLKKHVKML 345
Query: 126 NNILADLTRLGVKVDDENK 144
AD + + +++ K
Sbjct: 346 EERSADYVQQNAQFEEDAK 364
>RRPO_IPNVJ (P22173) Putative RNA-directed RNA polymerase (EC
2.7.7.48) (VP1 protein) (RDRP)
Length = 845
Score = 31.2 bits (69), Expect = 0.99
Identities = 24/82 (29%), Positives = 39/82 (47%), Gaps = 5/82 (6%)
Query: 8 KFEVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIA--IVDWNEMKKKAAGLIKL 65
++E RL G N L + K + + K L K +D+ + DWN M + L +
Sbjct: 612 RYEAIRLVGGWNNPLLETAAKHMSLDK--RKRLEVKGIDVTGFLDDWNNMSEFGGDLEGI 669
Query: 66 CVSDDVMNHTL-DLTTLNDVWD 86
+S+ + N TL D+ T D +D
Sbjct: 670 TLSEPLTNQTLVDINTPLDSFD 691
>VP17_YEAST (P32913) Vacuolar protein sorting-associated protein
VPS17
Length = 551
Score = 30.8 bits (68), Expect = 1.3
Identities = 35/121 (28%), Positives = 57/121 (46%), Gaps = 4/121 (3%)
Query: 6 GAKFEVTRLNGTRNFGLWQMRV-KDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIK 64
G E R+ TRNF LW R+ +D L + A +S N+ K A+GL +
Sbjct: 183 GINSEEDRMKVTRNFQLWFNRLSQDPLIIRNEEVAFFIESDFNTYTPINKSKSLASGLKR 242
Query: 65 LCVSDDVMNHTLDLTTLNDVWDTLESQY-MSKTLMDKLL-VKQRRYSLKMQEGGDLQDHV 122
+ + ++T L + ++S Y +S++L +KLL V + R + +E QD V
Sbjct: 243 KTLKQLAPPYD-EITELAEFRPLVKSIYVVSQSLQEKLLRVSRNRKMMVQEENAFGQDFV 301
Query: 123 N 123
N
Sbjct: 302 N 302
>PRA3_BACCR (Q81DT1) Foldase protein prsA 3 precursor (EC 5.2.1.8)
Length = 283
Score = 30.8 bits (68), Expect = 1.3
Identities = 32/110 (29%), Positives = 45/110 (40%), Gaps = 25/110 (22%)
Query: 55 MKKKAA--GLIKLCV---------SDDVMNHTLDLTTLNDVWDTLESQY---------MS 94
MKKK G I CV SD+V+ + T ++ L QY +S
Sbjct: 1 MKKKKIFIGTIISCVMLALSACGSSDNVVTSKVGNVTEKELSKELRQQYGESTLYQMMLS 60
Query: 95 KTLMDKLLVKQRRYSLKMQEGGDLQDHVNAFNNILADLTRLGVKVDDENK 144
K L+DK V K++E D N + L +LG+K +DE K
Sbjct: 61 KALLDKYKVSDEEAKKKVEEAKDKMG-----ENFKSTLEQLGLKNEDELK 105
>HS74_CAEEL (P20163) Heat shock 70 kDa protein D precursor
Length = 657
Score = 30.8 bits (68), Expect = 1.3
Identities = 23/97 (23%), Positives = 44/97 (44%), Gaps = 3/97 (3%)
Query: 61 GLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTL--MDKLLVKQRRYSLKMQEGGDL 118
G++ + D + LT ND + L + + + + DK + K++ +L
Sbjct: 509 GILHVSAEDKGTGNKNKLTITND-HNRLSPEDIERMINDADKFAADDQAQKEKVESRNEL 567
Query: 119 QDHVNAFNNILADLTRLGVKVDDENKAIIFCALYRAL 155
+ + +AD +LG K+ DE+K I A+ RA+
Sbjct: 568 EAYAYQMKTQIADKEKLGGKLTDEDKVSIESAVERAI 604
>ENV_HV1B8 (P04582) Envelope polyprotein GP160 precursor [Contains:
Exterior membrane glycoprotein (GP120); Transmembrane
glycoprotein (GP41)]
Length = 851
Score = 30.0 bits (66), Expect = 2.2
Identities = 28/110 (25%), Positives = 49/110 (44%), Gaps = 22/110 (20%)
Query: 10 EVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIKLCVSD 69
EV +N T NF +W+ D++ Q +H+ + I W++ K L LCVS
Sbjct: 83 EVVLVNVTENFNMWK---NDMVEQ--MHEDI--------ISLWDQSLKPCVKLTPLCVS- 128
Query: 70 DVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQ 119
L T L + DT + + +M+K +K +++ + G +Q
Sbjct: 129 ------LKCTDLKN--DTNTNSSSGRMIMEKGEIKNCSFNISTSKRGKVQ 170
>ERC1_HUMAN (Q8IUD2) ERC protein 1 (ELKS protein)
Length = 1116
Score = 29.3 bits (64), Expect = 3.8
Identities = 20/70 (28%), Positives = 33/70 (46%), Gaps = 7/70 (10%)
Query: 52 WNEMKKKAAGLIKLC--VSDDVMNHTLDLTTLNDVWDTLESQYMS-----KTLMDKLLVK 104
W E+KKKAAGL V ++ +L L +TL +Q+ + L + L K
Sbjct: 454 WEELKKKAAGLQAEIGQVKQELSRKDTELLALQTKLETLTNQFSDSKQHIEVLKESLTAK 513
Query: 105 QRRYSLKMQE 114
++R ++ E
Sbjct: 514 EQRAAILQTE 523
>DYNA_NEUCR (Q01397) Dynactin, 150 kDa isoform (150 kDa
dynein-associated polypeptide) (DP-150) (DAP-150)
(p150-glued)
Length = 1300
Score = 29.3 bits (64), Expect = 3.8
Identities = 28/102 (27%), Positives = 46/102 (44%), Gaps = 7/102 (6%)
Query: 37 HKALRDK--SLDIAIVDWNEMKKKAAGLIKLCVS--DDVMNHTLDLTTLNDVWDTLESQY 92
H ++K + + D E A G ++ S + MN + ++ L V D LES
Sbjct: 452 HTTAKEKLAQTEAIVEDLREQLNNALGAEEIIESLTEQTMNQSEEIKELRAVIDDLESL- 510
Query: 93 MSKTLMDKLLVKQRRYSLKMQEGGDLQDHVNAFNNILADLTR 134
K + D+L + + +MQE DL+D + A A+L R
Sbjct: 511 --KEINDELEINHVQNEKEMQEEIDLKDSIIAEQFRQANLQR 550
>APLP_MANSE (Q25490) Apolipophorins precursor [Contains:
Apolipophorin-I (APOLP-1); Apolipophorin-II (APOLP-2)]
Length = 3305
Score = 29.3 bits (64), Expect = 3.8
Identities = 15/60 (25%), Positives = 31/60 (51%), Gaps = 2/60 (3%)
Query: 21 GLWQMRVKDLLAQQGLHKALR--DKSLDIAIVDWNEMKKKAAGLIKLCVSDDVMNHTLDL 78
G +++ +KD +A G +K K + I+D+ ++ +K G ++ + V N +DL
Sbjct: 1422 GNFKLVIKDSIAANGQYKVTDADGKGNGLIIIDFKKINRKIKGDVRFTAKEPVFNADIDL 1481
>GR78_APLCA (Q16956) 78 kDa glucose-regulated protein precursor (GRP
78) (BiP) (Protein 1603)
Length = 667
Score = 28.9 bits (63), Expect = 4.9
Identities = 21/97 (21%), Positives = 44/97 (44%), Gaps = 3/97 (3%)
Query: 61 GLIKLCVSDDVMNHTLDLTTLNDVWDTLESQYMSKTLMD--KLLVKQRRYSLKMQEGGDL 118
G++K+ D + ND + L + + + + D K + ++ K+ +L
Sbjct: 516 GILKVTAEDKGTGSKNQIVIQNDQ-NRLSPEDIERMINDAEKYADEDKKVKEKVDAKNEL 574
Query: 119 QDHVNAFNNILADLTRLGVKVDDENKAIIFCALYRAL 155
+ + + N + D +LG K+ DE+K I A+ A+
Sbjct: 575 ESYAYSLKNQIGDKEKLGAKLSDEDKEKITEAVDEAI 611
>ATK4_ARATH (O81635) Kinesin 4 (Kinesin-like protein D)
Length = 987
Score = 28.9 bits (63), Expect = 4.9
Identities = 21/70 (30%), Positives = 33/70 (47%), Gaps = 13/70 (18%)
Query: 22 LWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIKLCVSDDV--------MN 73
++ +V+DLLAQ G + K L+I N + A L+ + +DDV MN
Sbjct: 531 IYNEQVRDLLAQDG-----QTKRLEIRNNSHNGINVPEASLVPVSSTDDVIQLMDLGHMN 585
Query: 74 HTLDLTTLND 83
+ T +ND
Sbjct: 586 RAVSSTAMND 595
>LON1_METTH (O26878) Putative protease La homolog type 1 (EC
3.4.21.-)
Length = 644
Score = 28.5 bits (62), Expect = 6.4
Identities = 19/91 (20%), Positives = 45/91 (48%), Gaps = 7/91 (7%)
Query: 32 AQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIKLCVSDDVMNHTLDLTTLNDVWDTLESQ 91
+++ + + +R+ D +K + G + D + +L DV +E++
Sbjct: 354 SREAVEEIIREAQRRAGKKDSLTLKLRELGGLVRAAGDIAKSRGAELVETEDV---IEAK 410
Query: 92 YMSKTL----MDKLLVKQRRYSLKMQEGGDL 118
+S+TL D+ +V++++YS+ EGG++
Sbjct: 411 KLSRTLEQQIADRYIVQKKKYSVFKSEGGEV 441
>ENV_HV1PV (P03376) Envelope polyprotein GP160 precursor [Contains:
Exterior membrane glycoprotein (GP120); Transmembrane
glycoprotein (GP41)]
Length = 856
Score = 28.5 bits (62), Expect = 6.4
Identities = 28/110 (25%), Positives = 48/110 (43%), Gaps = 22/110 (20%)
Query: 10 EVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIKLCVSD 69
EV +N T NF +W+ D++ Q +H+ + I W++ K L LCVS
Sbjct: 83 EVVLVNVTENFNMWK---NDMVEQ--MHEDI--------ISLWDQSLKPCVKLTPLCVS- 128
Query: 70 DVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQ 119
L T L + DT + + +M+K +K +++ G +Q
Sbjct: 129 ------LKCTDLKN--DTNTNSSSGRMIMEKGEIKNCSFNISTSIRGKVQ 170
>ENV_HV1H3 (P04624) Envelope polyprotein GP160 precursor [Contains:
Exterior membrane glycoprotein (GP120); Transmembrane
glycoprotein (GP41)]
Length = 856
Score = 28.5 bits (62), Expect = 6.4
Identities = 28/110 (25%), Positives = 48/110 (43%), Gaps = 22/110 (20%)
Query: 10 EVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIKLCVSD 69
EV +N T NF +W+ D++ Q +H+ + I W++ K L LCVS
Sbjct: 83 EVVLVNVTENFNMWK---NDMVEQ--MHEDI--------ISLWDQSLKPCVKLTPLCVS- 128
Query: 70 DVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQ 119
L T L + DT + + +M+K +K +++ G +Q
Sbjct: 129 ------LKCTDLKN--DTNTNSSSGRMIMEKGEIKNCSFNISTSIRGKVQ 170
>ENV_HV1H2 (P04578) Envelope polyprotein GP160 precursor [Contains:
Exterior membrane glycoprotein (GP120); Transmembrane
glycoprotein (GP41)]
Length = 856
Score = 28.5 bits (62), Expect = 6.4
Identities = 28/110 (25%), Positives = 48/110 (43%), Gaps = 22/110 (20%)
Query: 10 EVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIKLCVSD 69
EV +N T NF +W+ D++ Q +H+ + I W++ K L LCVS
Sbjct: 83 EVVLVNVTENFNMWK---NDMVEQ--MHEDI--------ISLWDQSLKPCVKLTPLCVS- 128
Query: 70 DVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQ 119
L T L + DT + + +M+K +K +++ G +Q
Sbjct: 129 ------LKCTDLKN--DTNTNSSSGRMIMEKGEIKNCSFNISTSIRGKVQ 170
>ENV_HV1B1 (P03375) Envelope polyprotein GP160 precursor [Contains:
Exterior membrane glycoprotein (GP120); Transmembrane
glycoprotein (GP41)]
Length = 856
Score = 28.5 bits (62), Expect = 6.4
Identities = 28/110 (25%), Positives = 48/110 (43%), Gaps = 22/110 (20%)
Query: 10 EVTRLNGTRNFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGLIKLCVSD 69
EV +N T NF +W+ D++ Q +H+ + I W++ K L LCVS
Sbjct: 83 EVVLVNVTENFNMWK---NDMVEQ--MHEDI--------ISLWDQSLKPCVKLTPLCVS- 128
Query: 70 DVMNHTLDLTTLNDVWDTLESQYMSKTLMDKLLVKQRRYSLKMQEGGDLQ 119
L T L + DT + + +M+K +K +++ G +Q
Sbjct: 129 ------LKCTDLKN--DTNTNSSSGRMIMEKGEIKNCSFNISTSIRGKVQ 170
>UPPS_FUSNN (Q8RE08) Undecaprenyl pyrophosphate synthetase (EC
2.5.1.31) (UPP synthetase)
(Di-trans-poly-cis-decaprenylcistransferase)
(Undecaprenyl diphosphate synthase) (UDS)
Length = 230
Score = 28.1 bits (61), Expect = 8.4
Identities = 13/44 (29%), Positives = 23/44 (51%)
Query: 19 NFGLWQMRVKDLLAQQGLHKALRDKSLDIAIVDWNEMKKKAAGL 62
NF LWQ+ +L L +K +D AI +N+ +++ G+
Sbjct: 184 NFLLWQIAYSELYITDVLWPDFDEKEIDKAIESYNQRERRFGGV 227
>MYS3_SCHPO (O14157) Myosin type II heavy chain 2
Length = 2104
Score = 28.1 bits (61), Expect = 8.4
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 4/40 (10%)
Query: 97 LMDKLLVKQRRYSLKMQ----EGGDLQDHVNAFNNILADL 132
L D+LL+KQR Y K+Q E L+D + + LA L
Sbjct: 1012 LDDELLLKQRSYDTKVQELREENASLKDQCRTYESQLASL 1051
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,712,070
Number of Sequences: 164201
Number of extensions: 643300
Number of successful extensions: 1822
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 1814
Number of HSP's gapped (non-prelim): 24
length of query: 157
length of database: 59,974,054
effective HSP length: 101
effective length of query: 56
effective length of database: 43,389,753
effective search space: 2429826168
effective search space used: 2429826168
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0490.9


