

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.8
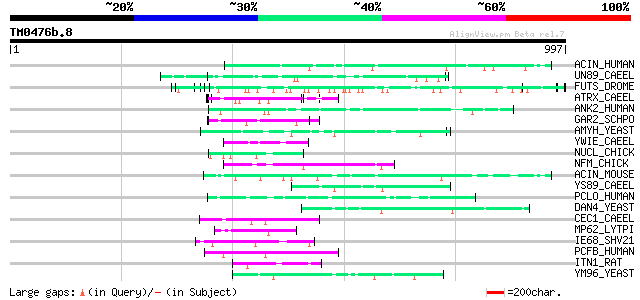
(997 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ACIN_HUMAN (Q9UKV3) Apoptotic chromatin condensation inducer in ... 62 6e-09
UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89 (Uncoo... 61 1e-08
FUTS_DROME (Q9W596) Microtubule-associated protein futsch 59 5e-08
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 57 2e-07
ANK2_HUMAN (Q01484) Ankyrin 2 (Brain ankyrin) (Ankyrin B) (Ankyr... 57 2e-07
GAR2_SCHPO (P41891) Protein gar2 57 2e-07
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 56 4e-07
YWIE_CAEEL (Q23525) Hypothetical protein ZK546.14 in chromosome II 55 1e-06
NUCL_CHICK (P15771) Nucleolin (Protein C23) 53 5e-06
NFM_CHICK (P16053) Neurofilament triplet M protein (160 kDa neur... 52 6e-06
ACIN_MOUSE (Q9JIX8) Apoptotic chromatin condensation inducer in ... 52 6e-06
YS89_CAEEL (Q09624) Hypothetical protein ZK945.9 in chromosome II 52 8e-06
PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin) 51 1e-05
DAN4_YEAST (P47179) Cell wall protein DAN4 precursor 51 1e-05
CEC1_CAEEL (P34618) Chromo domain protein cec-1 50 3e-05
MP62_LYTPI (P91753) Mitotic apparatus protein p62 49 9e-05
IE68_SHV21 (Q01042) Immediate-early protein 48 1e-04
PCFB_HUMAN (O94913) Pre-mRNA cleavage complex II protein Pcf11 (... 47 3e-04
ITN1_RAT (Q9WVE9) Intersectin 1 (EH domain and SH3 domain regula... 47 3e-04
YM96_YEAST (Q04893) Hypothetical 113.1 kDa protein in PRE5-FET4 ... 47 3e-04
>ACIN_HUMAN (Q9UKV3) Apoptotic chromatin condensation inducer in the
nucleus (Acinus)
Length = 1341
Score = 62.4 bits (150), Expect = 6e-09
Identities = 130/633 (20%), Positives = 231/633 (35%), Gaps = 68/633 (10%)
Query: 386 PAKKKAASKTVIIREASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKR 445
P K + S+ + R +R S++ +Q ++ SE + E+ E R L R
Sbjct: 203 PRKSSSISEEKGDSDDEKPRKGERRSSRVRQARAAKLSEGSQPAEEEEDQETPSRNLRVR 262
Query: 446 --RKIILEEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKE 503
R + EEEE+E+++ E +G + ++ E+ E ++P P + +
Sbjct: 263 ADRNLKTEEEEEEEEEEEEDDEEEEGDDEGQKSREAPILKEFK-EEGEEIPRVKPEEMMD 321
Query: 504 TVAEQASESASEVQRERRSKRTSDPARAVQLTR---------TSPIRSHAKVITESINSD 554
+ S+ ++R R R+ + AR L R TSP+ + I S
Sbjct: 322 ERPKTRSQEQEVLERGGRFTRSQEEARKSHLARQQQEKEMKTTSPLEEEER----EIKSS 377
Query: 555 SALVVVPEQPLPISTSMPTLTQTAPTQPETQTHTQAEHQAPISTIQTATTDIPSIPIYTS 614
L + P P + + PE QT ++ E P+ T + A++ P +++
Sbjct: 378 QGLKEKSKSPSPPRLTEDRKKASLVALPE-QTASEEETPPPLLT-KEASSPPPHPQLHSE 435
Query: 615 STSIPTE-PVPPLNSFIQGIIQSEATLKTLVQQFTSK------PASTPHTFLITQTGEIP 667
P E P PP+ + T + LV Q T + P S+P E P
Sbjct: 436 EEIEPMEGPAPPVLIQLSPPNTDADTRELLVSQHTVQLVGGLSPLSSPS----DTKAESP 491
Query: 668 AEKEKEDDDVQILEPPFNVK-PLQQIASNFEDRIPDAAETSYVSLSNYSAVNSQDLSFTS 726
AEK E+ + +++ Q+ DR L+ + + L S
Sbjct: 492 AEKVPEESVLPLVQKSTLADYSAQKDLEPESDRSAQPLPLKIEELALAKGITEECLKQPS 551
Query: 727 PEKTVTSRQRPDTISEAEHMDESDHSGVEKDHEIDSIPTEHMEESNASSSNAARTP---- 782
E+ R+ T+ + + +S S + S + S SS + + +P
Sbjct: 552 LEQK-EGRRASHTLLPSHRLKQSADSSSSRSSSSSSSSSRSRSRSPDSSGSRSHSPLRSK 610
Query: 783 -QPVLTLATSFLPQSLPQLIQKFSAEAIRRVNWLYQKAKVMNFRSSLA---GRKDSEFMM 838
+ V T P+ P++ + ++E+ R + A N R SL+ R S
Sbjct: 611 QRDVAQARTHANPRGRPKMGSRSTSESRSRSRSRSRSAS-SNSRKSLSPGVSRDSSTSYT 669
Query: 839 HLRGPEQRQRVLH------KVCEPLRRAMAERSCV----VSECTSEDVEMVSAEEEEEST 888
+ P Q V +VCEP R S V +S+ S + + + E +
Sbjct: 670 ETKDPSSGQEVATPPVPQLQVCEPKERTSTSSSSVQARRLSQPESAEKHVTQRLQPERGS 729
Query: 889 DGIVILEDADADEVQKQGPIHVDTVMTSEAEAATHP---------QASEVPPSAPAPEVA 939
E+A+ + T E+E H SE P PE
Sbjct: 730 PKKCEAEEAEPPAATQPQTSETQTSHLPESERIHHTVEEKEEVTMDTSENRPENDVPEPP 789
Query: 940 VLAARIDRIQDDQQRLFQMVEQQGQVQTEQSKQ 972
+ A D++ +D + +G V+ E+ K+
Sbjct: 790 MPIA--DQVSNDDR-------PEGSVEDEEKKE 813
>UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89
(Uncoordinated protein 89)
Length = 6632
Score = 61.2 bits (147), Expect = 1e-08
Identities = 101/473 (21%), Positives = 183/473 (38%), Gaps = 85/473 (17%)
Query: 355 ESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSERSLKRTSTQQ 414
+SPPK T+K + + KP D ++ P KK+ + I E SSE ++++T T
Sbjct: 1461 KSPPKSPTKKEK--SPEKPED-------VKSPVKKEKSPDATNIVEVSSETTIEKTETTM 1511
Query: 415 --QQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEIIQNVIQGIRD 472
+ +SE S + + + E DE +P + +K ++ E+ E I++ ++ +
Sbjct: 1512 TTEMTHESEESRTSVKKEKTPEKVDE-KPKSPTKK----DKSPEKSITEEIKSPVKKEKS 1566
Query: 473 SSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASE------VQRERRSKRTS 526
+ EE S ++ K P K K++ E S + E V E +S +
Sbjct: 1567 PEKVEEKPASP----TKKEKSPEKPASPTKKSENEVKSPTKKEKSPEKSVVEELKSPKEK 1622
Query: 527 DPARAVQLTR--TSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQPET 584
P +A + T +S K TE + S + PE+ TS PT +++PT+
Sbjct: 1623 SPEKADDKPKSPTKKEKSPEKSATEDVKSPTKKEKSPEKVEEKPTS-PTKKESSPTK--- 1678
Query: 585 QTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQSEATLKTLV 644
T E ++P ++ T + +P P + ++ K++V
Sbjct: 1679 --KTDDEVKSPTKKEKSPQT-------------VEEKPASPTK-------KEKSPEKSVV 1716
Query: 645 QQFTSKPASTPHTFLITQTGEIPAEKEKEDDDVQILEPPFNVKPLQQIASNFEDRIPDAA 704
++ S +P EK E + ++ P + + ++ + + P
Sbjct: 1717 EEVKSPKEKSPEKAEEKPKSPTKKEKSPEKSAAEEVKSPTKKEKSPEKSAEEKPKSPTKK 1776
Query: 705 ETSYVSLSNYSAVNSQDLSFTSPEK------TVTSRQRPDTISEAEHM------------ 746
E+S V +++ V S SPEK + T +++ S AE +
Sbjct: 1777 ESSPVKMAD-DEVKSPTKKEKSPEKVEEKPASPTKKEKTPEKSAAEELKSPTKKEKSPSS 1835
Query: 747 ------DESDHSGVEKDHEIDSIPTEHM------EESNASSSNAARTPQPVLT 787
DES EK E PT ++ + S A + P P LT
Sbjct: 1836 PTKKTGDESKEKSPEKPEEKPKSPTPKKSPPGSPKKKKSKSPEAEKPPAPKLT 1888
Score = 52.4 bits (124), Expect = 6e-06
Identities = 99/525 (18%), Positives = 200/525 (37%), Gaps = 32/525 (6%)
Query: 272 EDLATIVGDTLNGTKLKKMQIIEKIKVAPKEDTSDEVRQRNYLIDDFPIWSKKDNPACIL 331
E T++G G + + KI+V K D+ +VR+ +++++ P ++
Sbjct: 1311 ESATTVIGGGSGGVTEGSISV-SKIEVVSKTDSQTDVREGTP--KRRVSFAEEELPKEVI 1367
Query: 332 EYVRMLRRQGDPITLTEFVNTLPESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKA 391
+ R ++ P + E P T + S K P + PA ++
Sbjct: 1368 DSDRKKKKSPSPDKKEKSPEKTEEKPASPTKKTGEEVKSPKEKSPASPTKKEKSPAAEEV 1427
Query: 392 ASKTVIIREASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILE 451
S T + SS +++ + + + E E + +++ + ++P + + E
Sbjct: 1428 KSPTKKEKSPSSPTKKEKSPSSPTKKTGDEVKEKSPPKSPTKKEKSPEKPEDVKSPVKKE 1487
Query: 452 EEEDEQDDHEI-IQNVIQGIRDSSEAEESTDSDEVHL-VRRRKLPLKGPLQQKETVAEQA 509
+ D + E+ + I+ + E + +S+E V++ K P K + K +
Sbjct: 1488 KSPDATNIVEVSSETTIEKTETTMTTEMTHESEESRTSVKKEKTPEKVDEKPKSPTKKDK 1547
Query: 510 S-------ESASEVQRERRSKRTSDPARAVQLTRTSPIR--SHAKVITESINSDSALVVV 560
S E S V++E+ ++ + + SP + S K + S +
Sbjct: 1548 SPEKSITEEIKSPVKKEKSPEKVEEKPASPTKKEKSPEKPASPTKKSENEVKSPTKKEKS 1607
Query: 561 PEQPLPISTSMPTLTQTAPTQPETQTHTQAEHQAPISTIQTATTDIPS-IPIYTSSTSIP 619
PE+ + P + ++ T+ E S ++AT D+ S S +
Sbjct: 1608 PEKSVVEELKSPKEKSPEKADDKPKSPTKKEK----SPEKSATEDVKSPTKKEKSPEKVE 1663
Query: 620 TEPVPPLNSFIQGIIQSEATLKTLVQ-QFTSKPASTPHTFLITQTGEIPAEKEKEDDDVQ 678
+P P +S T KT + + +K +P T + + P +KEK +
Sbjct: 1664 EKPTSPTKK------ESSPTKKTDDEVKSPTKKEKSPQT--VEEKPASPTKKEKSPEK-S 1714
Query: 679 ILEPPFNVKPLQQIASNFEDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPD 738
++E + K + + + P E S S V S SPEK+ + +
Sbjct: 1715 VVEEVKSPKEKSPEKAEEKPKSPTKKEKS-PEKSAAEEVKSPTKKEKSPEKSAEEKPKSP 1773
Query: 739 TISEAEHMDESDHSGVEKDHEIDSIPTEHMEESNASSSNAARTPQ 783
T E+ + +D V+ + + P E +EE AS + +TP+
Sbjct: 1774 TKKESSPVKMADDE-VKSPTKKEKSP-EKVEEKPASPTKKEKTPE 1816
>FUTS_DROME (Q9W596) Microtubule-associated protein futsch
Length = 5412
Score = 59.3 bits (142), Expect = 5e-08
Identities = 143/699 (20%), Positives = 256/699 (36%), Gaps = 124/699 (17%)
Query: 298 VAPKEDTSDEVRQRNYLIDDFPIWSKK-DNPACILEYVRMLRRQGDPITLTEFVNTLPES 356
V + + S E +R+ + + P+ SK+ PA + E V+ + + E V
Sbjct: 3476 VKDEAEKSKEESRRDSVAEKSPLASKEASRPASVAESVQDEAEKSKEESRRESVAEKSPL 3535
Query: 357 PPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVII-REASSERSLKRTSTQQQ 415
K +R + A S K K K EE ++ A K+ + +EAS S+ + +
Sbjct: 3536 ASKEASRPASVAESIKDEAEKSK----EESRRESVAEKSPLASKEASRPTSVAESVKDEA 3591
Query: 416 QMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEIIQNVIQGIRDSSE 475
+ S ESS D+ + S +++ RP +V + ++D E
Sbjct: 3592 EKSKEESSRDSVAEKSPLASKEASRP----------------------ASVAESVQD--E 3627
Query: 476 AEESTDSDEVHLVRRRKLPLKGPLQQKE-----TVAEQASESASEVQRERRSKRTSD--P 528
AE+S + RR + K PL KE +VAE + A + + E R + ++ P
Sbjct: 3628 AEKSKEES-----RRESVAEKSPLASKEASRPASVAESVKDDAEKSKEESRRESVAEKSP 3682
Query: 529 ARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQPETQTHT 588
+ + +R + + K E +S V E+ + +P+ + PT
Sbjct: 3683 LASKEASRPASVAESVKDEAEKSKEESRRESVAEK-----SPLPSKEASRPTSVAESVKD 3737
Query: 589 QAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQSEATLKTLVQQFT 648
+AE S + +S A +L +
Sbjct: 3738 EAE-----------------------------------KSKEESRRESVAEKSSLASKKA 3762
Query: 649 SKPASTPHTFLITQTGEIPAEKEKEDDDVQIL--EPPFNVKPLQQIASNFEDRIPDAAET 706
S+PAS + ++ + AEK KE+ + + + P K + AS E + D AE
Sbjct: 3763 SRPAS------VAESVKDEAEKSKEESRRESVAEKSPLASKEASRPASVAES-VKDEAEK 3815
Query: 707 SYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPDTISEAEHMDESDHSGVEKDHEIDSIPTE 766
S S L P K + RP +++E+ DE+D S E E +
Sbjct: 3816 SKEESRRESVAEKSPL----PSKEAS---RPTSVAESV-KDEADKSKEESRRESGA---- 3863
Query: 767 HMEESNASSSNAARTPQPVLTLATSFLPQSLPQLIQKFSAEAIRRVNWLYQKAKVMNFRS 826
E+S +S A+R T + + ++ E++ + L K
Sbjct: 3864 --EKSPLASMEASRP-----TSVAESVKDETEKSKEESRRESVTEKSPLPSKEASRPTSV 3916
Query: 827 SLAGRKDSEFMMHLRGPEQRQRVLHKVCEPLRRAMAERSCVVSECTSEDVEMVSAEEEEE 886
+ + + ++E + E+ +R PL + R V+E ++ E E E
Sbjct: 3917 AESVKDEAE-----KSKEESRRESVAEKSPLASKESSRPASVAESIKDEAEGTKQESRRE 3971
Query: 887 S--TDGIVILEDADADEVQKQGPIHVDTVMTSEAEAATHPQASEVPPS--APAPEVAVLA 942
S G D + + D+V+ S + P+ S + S A PE ++
Sbjct: 3972 SMPESGKAESIKGDQSSLASKETSRPDSVVESVKDETEKPEGSAIDKSQVASRPESVAVS 4031
Query: 943 ARIDRIQDDQQRLFQMVEQQGQVQTEQSKQIAESSSKME 981
A+ D++ L E + SK+ + S S E
Sbjct: 4032 AK-----DEKSPLHSRPESVADKSPDASKEASRSLSVAE 4065
Score = 57.4 bits (137), Expect = 2e-07
Identities = 136/727 (18%), Positives = 261/727 (35%), Gaps = 109/727 (14%)
Query: 298 VAPKEDTSDEVRQRNYLIDDFPIWSKK-DNPACILEYVRMLRRQGDPITLTEFVNTLPES 356
V + + S E +R + + P+ SK+ PA + E V+ + + E V
Sbjct: 3772 VKDEAEKSKEESRRESVAEKSPLASKEASRPASVAESVKDEAEKSKEESRRESVAEKSPL 3831
Query: 357 PPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIR-EASSERSLKRTSTQQQ 415
P K +R + A S K K K EE ++ A K+ + EAS S+ + +
Sbjct: 3832 PSKEASRPTSVAESVKDEADKSK----EESRRESGAEKSPLASMEASRPTSVAESVKDET 3887
Query: 416 QMSDSESSEDTWEDFSSEETEDEDRPLA---------------KRRKIILEEEEDEQDDH 460
+ S ES ++ + S +++ RP + RR+ + E+ +
Sbjct: 3888 EKSKEESRRESVTEKSPLPSKEASRPTSVAESVKDEAEKSKEESRRESVAEKSPLASKES 3947
Query: 461 EIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRER 520
+V + I+D +E + RR +P G +ES Q
Sbjct: 3948 SRPASVAESIKDEAEGTKQES-------RRESMPESG-----------KAESIKGDQSSL 3989
Query: 521 RSKRTSDPARAVQLTRTSPIRSHAKVITES---INSDSALVVVPEQPLPISTSMPTLTQT 577
SK TS P V+ + + I +S +S V ++ P+ + ++
Sbjct: 3990 ASKETSRPDSVVESVKDETEKPEGSAIDKSQVASRPESVAVSAKDEKSPLHSRPESVADK 4049
Query: 578 APTQPETQTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQSE 637
+P + + + + + S I+ I + + + T +P L+S I +E
Sbjct: 4050 SPDASKEASRSLSVAETASSPIEEGPRSIADLSLPLNLTGEAKGKLPTLSSPID---VAE 4106
Query: 638 ATLKTLVQQFTSKPA--STPHTFLITQTGEIPAEKEKEDDDVQILEPPFNVKPLQQIASN 695
+ + + +PA S P F TG D+ +LE ++ ++Q ++
Sbjct: 4107 GDFLEVKAESSPRPAVLSKPAEFSQPDTGH--TASTPVDEASPVLE---EIEVVEQHTTS 4161
Query: 696 FEDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPDTISEAEHMDESDHSGVE 755
AET + L+ + S T+ +T+TS+ VE
Sbjct: 4162 GVGATGATAETDLLDLTETKSETVTKQSETTLFETLTSK-------------------VE 4202
Query: 756 KDHEIDSIPTEHMEESNASSSNAARTPQPVLTLATSFLPQSLPQLIQKFSAE-----AIR 810
E+ + +EE +S A T + SL QL +K S + ++
Sbjct: 4203 SKVEVLESSVKQVEEKVQTSVKQAETT----------VTDSLEQLTKKSSEQLTEIKSVL 4252
Query: 811 RVNWLYQKAKVMNFRSSLAGRKDSEFMMHLRGPEQRQRVLHKVCEPLRRAMAERSCVVSE 870
N+ V + L KD ++ Q + L + + S
Sbjct: 4253 DTNFEEVAKIVADVAKVLKSDKDITDIIPDFDERQLEEKLKSTADTEEESDKSTRDEKSL 4312
Query: 871 CTSEDVEMVSAEEEEESTDGIVILEDADADEVQKQGPIHVDTVMTSEAEAATHPQASEVP 930
S VE+ S + + G + +E+ D E ++ + + +S E+ + P
Sbjct: 4313 EISVKVEIESEKSSPDQKSGPISIEEKDKIEQSEKAQLRQGILTSSRPES-----VASQP 4367
Query: 931 PSAPAPEVAVLAARIDRIQDDQQRLFQMVEQQGQVQTEQSKQIAESSSKMEAIMKYLMEN 990
S P+P + + + +V+ +S + AE SS+ E++ + E
Sbjct: 4368 ESVPSPSQSAAS-----------------HEHKEVELSESHK-AEKSSRPESVASQVSEK 4409
Query: 991 LPSSSKP 997
+S+P
Sbjct: 4410 DMKTSRP 4416
Score = 54.3 bits (129), Expect = 2e-06
Identities = 129/663 (19%), Positives = 239/663 (35%), Gaps = 97/663 (14%)
Query: 343 PITLTEFVNTLPESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAA-SKTVIIREA 401
P ++ E V E + + R+S S PS + + E K +A SK RE+
Sbjct: 3876 PTSVAESVKDETEKSKEESRRESVTEKSPLPSKEASRPTSVAESVKDEAEKSKEESRRES 3935
Query: 402 SSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHE 461
+E+S + + S +ES +D E E + K I ++ +
Sbjct: 3936 VAEKSPLASKESSRPASVAESIKDEAEGTKQESRRESMPESGKAESIKGDQSSLASKETS 3995
Query: 462 IIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPL------KGPLQQK-ETVAEQASESAS 514
+V++ ++D +E E + D+ + R + K PL + E+VA+++ +++
Sbjct: 3996 RPDSVVESVKDETEKPEGSAIDKSQVASRPESVAVSAKDEKSPLHSRPESVADKSPDASK 4055
Query: 515 EVQRERRSKRTSD------PARAVQLTRTSPIRSHAKVITESINS-------DSALVVVP 561
E R T+ P L+ + AK +++S D V
Sbjct: 4056 EASRSLSVAETASSPIEEGPRSIADLSLPLNLTGEAKGKLPTLSSPIDVAEGDFLEVKAE 4115
Query: 562 EQPLPISTSMPT--------------LTQTAPTQPETQTHTQAEHQAPISTIQTATTDIP 607
P P S P + + +P E + Q +T TA TD+
Sbjct: 4116 SSPRPAVLSKPAEFSQPDTGHTASTPVDEASPVLEEIEVVEQHTTSGVGATGATAETDLL 4175
Query: 608 SI--------------PIYTSSTSIPTEPVPPLNSFIQ--------GIIQSEATLKTLVQ 645
+ ++ + TS V L S ++ + Q+E T+ ++
Sbjct: 4176 DLTETKSETVTKQSETTLFETLTSKVESKVEVLESSVKQVEEKVQTSVKQAETTVTDSLE 4235
Query: 646 QFTSKPA---STPHTFLITQTGEIP------AEKEKEDDDVQILEPPFNVKPLQQIASNF 696
Q T K + + + L T E+ A+ K D D+ + P F+ + L
Sbjct: 4236 QLTKKSSEQLTEIKSVLDTNFEEVAKIVADVAKVLKSDKDITDIIPDFDERQL------- 4288
Query: 697 EDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPDTISEAEHMDESDH-SGVE 755
E+++ A+T S+ S + + L + + + + PD S ++E D E
Sbjct: 4289 EEKLKSTADTE--EESDKSTRDEKSLEISVKVEIESEKSSPDQKSGPISIEEKDKIEQSE 4346
Query: 756 KDHEIDSIPTEHMEESNASSSNAARTPQPVLTLATSFLPQSLPQLIQKFSAEAIRRVNWL 815
K I T ES AS + P P + A+ +L + AE R +
Sbjct: 4347 KAQLRQGILTSSRPESVASQPES--VPSPSQSAASH--EHKEVELSESHKAEKSSRPESV 4402
Query: 816 YQKAKVMNFRSSLAGRKDSEFMMHLRGPEQRQRVLHKVCEP---LRRAMAERSCVVSECT 872
+ + ++S S+F E + +LH + + M E+S S T
Sbjct: 4403 ASQVSEKDMKTSRPASSTSQFSTKEGDEETTESLLHSLTTTETVETKQMEEKSSFESVST 4462
Query: 873 ---------SEDVEMVSAEEEEESTDGIVILEDADADE-----VQKQGPIHVDTVMTSEA 918
S+ + E ES + +ED+ E + ++G I +T + +
Sbjct: 4463 SVTKSTVLSSQSTVQLREESTSESLSSSLKVEDSSRRESLSSLLAEKGGIATNTSLKEDT 4522
Query: 919 EAA 921
A+
Sbjct: 4523 SAS 4525
Score = 52.4 bits (124), Expect = 6e-06
Identities = 139/694 (20%), Positives = 263/694 (37%), Gaps = 101/694 (14%)
Query: 359 KLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSERSLKRTSTQQQQMS 418
K T+K + S + S P L+++ +K+ + + I + ++ S +
Sbjct: 3022 KADTKKDGK--SQEASRPSSVDELLKDDDEKQESRRQSITGSHKAMSTMGDESPMDKADK 3079
Query: 419 DSESS--EDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEIIQNVIQGIRDSSEA 476
E S E E E T+DE+ PL RR + E I ++ +G + +
Sbjct: 3080 SKEPSRPESVAESIKHENTKDEESPLGSRRDSVAE---------SIKSDITKGEKSPLPS 3130
Query: 477 EE-STDSDEVHLVRRRKLPLKGPLQQKETVAEQAS-ESASEVQRERRSKRTSDPARAVQL 534
+E S V ++ K ++E+VAE ES+ + SK S P +
Sbjct: 3131 KEVSRPESVVGSIKDEKAE-----SRRESVAESVKPESSKDATSAPPSKEHSRPESVLGS 3185
Query: 535 TRTSPIRSHAK--VITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQ----PETQTHT 588
+ ++ ++ + +SI + +L+V E P S + AP+Q PE+ T +
Sbjct: 3186 LKDEGDKTTSRRVSVADSIKDEKSLLVSQEASRPESEAESLKDAAAPSQETSRPESVTES 3245
Query: 589 QAEHQAPISTIQT----------------ATTDIPSIPIYTSSTSIPTEPVPPLNSFIQG 632
+ ++P+++ + + P + + SI E P +
Sbjct: 3246 VKDGKSPVASKEASRPASVAENAKDSADESKEQRPESLPQSKAGSIKDEKSPLASKDEAE 3305
Query: 633 IIQSEATLKTLVQQF--TSKPASTPHTFLITQTGEIPAEKEKEDDDVQILEPPFNVKPLQ 690
+ E+ +++ +QF SK S P + + ++ + AEK KE E P K
Sbjct: 3306 KSKEESRRESVAEQFPLVSKEVSRPAS--VAESVKDEAEKSKE-------ESPLMSKEAS 3356
Query: 691 QIASNFEDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPDTISEAEHMDESD 750
+ AS + D AE S S L P K + RP +++E+ DE+D
Sbjct: 3357 RPAS-VAGSVKDEAEKSKEESRRESVAEKSPL----PSKEAS---RPASVAESV-KDEAD 3407
Query: 751 HSGVEKDHE-------------------IDSI--PTEHMEESNASSSNAARTPQPVLTLA 789
S E E +SI E +E + S A ++P P +
Sbjct: 3408 KSKEESRRESGAEKSPLASKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEAS 3467
Query: 790 TSFLPQSLPQLIQ---KFSAEAIRRVNWLYQKAKVMNFRSSLAGRKDSEFMMHLRGPEQR 846
P S+ + ++ + S E RR + + S A +S + E+
Sbjct: 3468 R---PTSVAESVKDEAEKSKEESRRDSVAEKSPLASKEASRPASVAESVQDEAEKSKEES 3524
Query: 847 QRVLHKVCEPLRRAMAERSCVVSECTSEDVEMVSAEEEEESTDGIVILEDADADEVQKQG 906
+R PL A R V+E ++ E E ES + E + +
Sbjct: 3525 RRESVAEKSPLASKEASRPASVAESIKDEAEKSKEESRRES-----VAEKSPLASKEASR 3579
Query: 907 PIHVDTVMTSEA----EAATHPQASEVPPSAPAPEVAVLAARIDRIQDDQQRLFQMVEQQ 962
P V + EA E ++ +E P A + E + A+ + +QD+ ++ E +
Sbjct: 3580 PTSVAESVKDEAEKSKEESSRDSVAEKSPLA-SKEASRPASVAESVQDEAEK--SKEESR 3636
Query: 963 GQVQTEQSKQIAESSSKMEAIMKYLMENLPSSSK 996
+ E+S ++ +S+ ++ + + ++ S +
Sbjct: 3637 RESVAEKSPLASKEASRPASVAESVKDDAEKSKE 3670
Score = 50.8 bits (120), Expect = 2e-05
Identities = 133/657 (20%), Positives = 247/657 (37%), Gaps = 68/657 (10%)
Query: 350 VNTLPESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSERSLKR 409
+ +L + K T+R+ A S K K +L+ + A R S SLK
Sbjct: 3183 LGSLKDEGDKTTSRRVSVADSIKDE----KSLLVSQEAS----------RPESEAESLKD 3228
Query: 410 TSTQQQQMSDSESSEDTWEDFSSEETEDE-DRP--LAKRRKIILEEEEDEQDDHEIIQNV 466
+ Q+ S ES ++ +D S E RP +A+ K +E + EQ + Q+
Sbjct: 3229 AAAPSQETSRPESVTESVKDGKSPVASKEASRPASVAENAKDSADESK-EQRPESLPQSK 3287
Query: 467 IQGIRDSSEAEESTDSDEVHLV--RRRKLPLKGPLQQKETVAEQASESASEVQRERRSKR 524
I+D S D E RR + + PL KE V+ AS + S +SK
Sbjct: 3288 AGSIKDEKSPLASKDEAEKSKEESRRESVAEQFPLVSKE-VSRPASVAESVKDEAEKSKE 3346
Query: 525 TSDPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQPET 584
S P + + +R + + K E +S V E+ + +P+ + P
Sbjct: 3347 ES-PLMSKEASRPASVAGSVKDEAEKSKEESRRESVAEK-----SPLPSKEASRPASVAE 3400
Query: 585 QTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQSEATLKTLV 644
+A+ S ++ P+ + S P + + + E+ +++
Sbjct: 3401 SVKDEADKSKEESRRESGAE---KSPLASKEASRPASVAESIKDEAEKS-KEESRRESVA 3456
Query: 645 QQ--FTSKPASTPHTFLITQTGEIPAEKEKEDDDVQILEPPFNVKPLQQIASNFEDRIPD 702
++ SK AS P + + E KE+ D + P K + AS E + D
Sbjct: 3457 EKSPLPSKEASRPTSVAESVKDEAEKSKEESRRDSVAEKSPLASKEASRPASVAES-VQD 3515
Query: 703 AAETSYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPDTISEAEHMDESDHSGVEKDHEIDS 762
AE S S L+ S E + RP +++E+ DE++ S E E +
Sbjct: 3516 EAEKSKEESRRESVAEKSPLA--SKEAS-----RPASVAESIK-DEAEKSKEESRRESVA 3567
Query: 763 IPTEHMEESNASSSNAARTPQPVLTLATSFLPQSLPQLIQKFSAEAIRRVNWLYQKAKVM 822
E+S +S A+R T + + ++ S +++ + L K
Sbjct: 3568 ------EKSPLASKEASRP-----TSVAESVKDEAEKSKEESSRDSVAEKSPLASKEASR 3616
Query: 823 NFRSSLAGRKDSEFMMHLRGPEQRQRVLHKVCEPLRRAMAERSCVVSECTSEDVEMVSAE 882
+ + + ++E + E+ +R PL A R V+E +D E E
Sbjct: 3617 PASVAESVQDEAE-----KSKEESRRESVAEKSPLASKEASRPASVAESVKDDAEKSKEE 3671
Query: 883 EEEESTDGIVILEDADADEVQKQGPIHVDTVMTSEAEAATHPQASE-VPPSAPAP--EVA 939
ES + E + + P V + EAE + E V +P P E +
Sbjct: 3672 SRRES-----VAEKSPLASKEASRPASVAESVKDEAEKSKEESRRESVAEKSPLPSKEAS 3726
Query: 940 VLAARIDRIQDDQQRLFQMVEQQGQVQTEQSKQIAESSSKMEAIMKYLMENLPSSSK 996
+ + ++D+ ++ E + + E+S ++ +S+ ++ + + + S +
Sbjct: 3727 RPTSVAESVKDEAEK--SKEESRRESVAEKSSLASKKASRPASVAESVKDEAEKSKE 3781
Score = 49.7 bits (117), Expect = 4e-05
Identities = 138/729 (18%), Positives = 262/729 (35%), Gaps = 102/729 (13%)
Query: 332 EYVRMLRRQGDPITLTEFVNTLPESPPKLTTRKSRRATSSKPSDPKGKGI-LIEEPAKKK 390
+YV+ ++ + I + V TLPE+ P AT P D + + + + ++
Sbjct: 1135 KYVKEETKEAEAIVVAT-VQTLPEAAPLAIDTILASATKDAPKDANAEALGELPDSGERV 1193
Query: 391 AASKTVIIREASSERSLKRTSTQQQQMSDSE---------SSEDTW-------EDFSSEE 434
K + + R + +T + + E S+D E+ + EE
Sbjct: 1194 LPMKMTFEAQQNLLRDVIKTPDEVADLPVHEEADLGLYEKDSQDAGAKSISHKEESAKEE 1253
Query: 435 TEDEDRPLAKRRKIILEEEEDEQD-DHEIIQNVIQGIRD---------SSEAEESTDSDE 484
E +D K +I L +E ++ D H +++ +Q + + + EE ++
Sbjct: 1254 KETDDEKENKVGEIELGDEPNKVDISHVLLKESVQEVAEKVVVIETTVEKKQEEIVEATT 1313
Query: 485 VHLVRRRKLPLKGPLQQKE----------TVAEQASESASEVQRERRSKRTSD---PARA 531
V + + + L ++ KE ++A +SAS + + S TSD PA+
Sbjct: 1314 V-ITQENQEDLMEQVKDKEEHEQKIESGIITEKEAKKSASTPEEKETSDITSDDELPAQL 1372
Query: 532 VQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQPETQTHTQAE 591
T P + + T SI S + E+ + + Q PE T+
Sbjct: 1373 ADPTTVPPKSAKDREDTGSIESPPTI----EEAIEVEVQAKQEAQKPVPAPEEAIKTE-- 1426
Query: 592 HQAPISTIQTATTD--IPSIPIYTSSTSIPTEPVP---------PLNSFIQGIIQSEATL 640
++P+++ +T+ + S+ T T PVP + F G ++
Sbjct: 1427 -KSPLASKETSRPESATGSVKEDTEQTKSKKSPVPSRPESEAKDKKSPFASGEASRPESV 1485
Query: 641 KTLVQQFTSKPASTPHTFLITQTGEIPAEKEKEDDD-----VQILEPPFNVKPLQQIASN 695
V+ K S + T E +K KE + + ++P + +AS
Sbjct: 1486 AESVKDEAGKAESRRESIAKTHKDESSLDKAKEQESRRESLAESIKPESGIDEKSALASK 1545
Query: 696 FEDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKTV---TSRQRPDTISEA---EHMDES 749
R + S S S T EK+ RP ++ E+ E
Sbjct: 1546 EASRPESVTDKSKEPSRRESIAESLKAESTKDEKSAPPSKEASRPGSVVESVKDETEKSK 1605
Query: 750 DHSGVEKDHEIDSIPTEHMEES----------NASSSNAARTPQPVLTLATSFLPQSLPQ 799
+ S E E P E E S + S+ +R P+ + S P+S
Sbjct: 1606 EPSRRESIAESAKPPIEFREVSRPESVIDGIKDESAKPESRRDSPLASKEAS-RPES--- 1661
Query: 800 LIQKFSAEAIRRVNWLYQKAKVMNFRSSLAGRKDSEFMMHLRGPEQRQRVLHKVCEPLRR 859
+++ E I+ +++ +F++ KD + + + + + + V + +
Sbjct: 1662 VLESVKDEPIKSTEKSRRESVAESFKAD--STKDEKSPLTSKDISRPESAVENVMDAV-- 1717
Query: 860 AMAERSCVVSECTSEDVEMVSAEEEEESTDGIVILEDADADEVQKQGPIHVDTVMTSEAE 919
AERS S S DV + E E +D D E + I V+ E
Sbjct: 1718 GSAERSQPESVTASRDVSRPESVAESEK-------DDTDKPESVVESVIPASDVVEIEKG 1770
Query: 920 AATHPQASEV-----PPSAPAPEVAVLAARIDRIQDDQQRLFQMVEQQGQVQTEQSKQIA 974
AA + V P +P+ ++ ++ ++ + +R E E SK+ +
Sbjct: 1771 AADKEKGVFVSLEIGKPDSPSEVISRPGPVVESVKPESRR-ESSTEIVLPCHAEDSKEPS 1829
Query: 975 ESSSKMEAI 983
SK+E +
Sbjct: 1830 RPESKVECL 1838
Score = 48.1 bits (113), Expect = 1e-04
Identities = 146/743 (19%), Positives = 271/743 (35%), Gaps = 89/743 (11%)
Query: 292 IIEKIKVAPKE--------DTSDEVRQRNYLIDDFPIWSKKDNPACILEYVRMLRRQGDP 343
I EK +A KE D S E +R + + S KD + + P
Sbjct: 1536 IDEKSALASKEASRPESVTDKSKEPSRRESIAESLKAESTKDEKSAPPS-----KEASRP 1590
Query: 344 ITLTEFVNTLPESPPKLTTRKSRRATSSKP-------SDPKG--KGILIEEPAKKKAASK 394
++ E V E K +R+ A S+KP S P+ GI E +
Sbjct: 1591 GSVVESVKDETEKS-KEPSRRESIAESAKPPIEFREVSRPESVIDGIKDESAKPESRRDS 1649
Query: 395 TVIIREASSERSLKRTSTQQQQMSDSESS--EDTWEDFSSEETEDEDRPLAKRRKIILEE 452
+ +EAS S+ S + + + +E S E E F ++ T+DE PL +
Sbjct: 1650 PLASKEASRPESVLE-SVKDEPIKSTEKSRRESVAESFKADSTKDEKSPLTSK------- 1701
Query: 453 EEDEQDDHEIIQNVIQ--GIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQAS 510
D ++NV+ G + S+ E T S +V K + E+V E
Sbjct: 1702 --DISRPESAVENVMDAVGSAERSQPESVTASRDVSRPESVAESEKDDTDKPESVVESVI 1759
Query: 511 ESASEVQRERRSKRTSDPARAVQLT-------RTSPIRSHAKVITESINSDSALVVVPEQ 563
++ V+ E K +D + V ++ S + S + ES+ +S E
Sbjct: 1760 PASDVVEIE---KGAADKEKGVFVSLEIGKPDSPSEVISRPGPVVESVKPESRRESSTEI 1816
Query: 564 PLPISTSMPTLTQTAPTQPETQTHTQAEHQAPIS----TIQTATTDIPSIPIYTSSTSIP 619
LP P++PE++ + + A +D S P +S
Sbjct: 1817 VLPCHAE----DSKEPSRPESKVECLKDESEVLKGSTRRESVAESDKSSQPFKETS---- 1868
Query: 620 TEPVPPLNSFIQGIIQSEATLKTLVQQFTSKPASTPHTFLITQTGEIPAEKEKEDDDVQI 679
P + S + E + + V+ ++ T + ++ + A DD++
Sbjct: 1869 -RPESAVGSMKDESMSKEPSRRESVKDGAAQSRETSRPASVAESAKDGA------DDLKE 1921
Query: 680 LEPPFNVKPLQQIASNFEDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPDT 739
L P + ++ S +++ P A+E + S +V + EK+ R ++
Sbjct: 1922 LSRPESTTQSKEAGSIKDEKSPLASEEASRPASVAESVKDE------AEKS-KEESRRES 1974
Query: 740 ISEAEHMDESDHSGVEKDHEIDSIPTEHMEESNASSSNAARTPQPVLTLATSFLPQSLPQ 799
++E + + S E E +E + S A ++P P + P S+ +
Sbjct: 1975 VAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASR---PASVAE 2031
Query: 800 LIQ---KFSAEAIRRVNWLYQKAKVMNFRSSLAGRKDSEFMMHLRGPEQRQRVLHKVCEP 856
I+ + S E RR + + S A +S + E+ +R P
Sbjct: 2032 SIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSP 2091
Query: 857 LRRAMAERSCVVSECTSEDVEMVSAEEEEESTDGIVILEDADADEVQKQGPIHVDTVMTS 916
L A R V+E ++ E E ES + E + + P V +
Sbjct: 2092 LPSKEASRPASVAESIKDEAEKSKEESRRES-----VAEKSPLPSKEASRPASVAESIKD 2146
Query: 917 EAEAATHPQASE-VPPSAPAP--EVAVLAARIDRIQDDQQRLFQMVEQQGQVQTEQSKQI 973
EAE + E V +P P E + A+ + I+D+ ++ E + + E+S
Sbjct: 2147 EAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEK--SKEESRRESVAEKSPLP 2204
Query: 974 AESSSKMEAIMKYLMENLPSSSK 996
++ +S+ ++ + + + S +
Sbjct: 2205 SKEASRPASVAESIKDEAEKSKE 2227
Score = 38.9 bits (89), Expect = 0.069
Identities = 115/623 (18%), Positives = 232/623 (36%), Gaps = 68/623 (10%)
Query: 397 IIREASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETED-EDRPLAKRRKIILEEEED 455
++RE + S QQ Q +E E +E T D E+ P + +I+E+EE
Sbjct: 895 VVREIEAVFSRDEMKRQQHQQIKAELRE-----MPAEGTGDGENEPDEEEEYLIIEKEEV 949
Query: 456 EQ-DDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQ---QKETVAEQASE 511
EQ + I++ ++ + DS E ++RK + ++ K AE+ +
Sbjct: 950 EQYTEDSIVEQESSMTKEEEIQKHQRDSQESE--KKRKKSAEEEIEAAIAKVEAAERKAR 1007
Query: 512 SASEVQRERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSM 571
R+ S+ +P ++ I + AK I +S ++ L E+ L T
Sbjct: 1008 LEGASARQDESELDVEPEQSKIKAEVQDIIATAKDIAKS-RTEEQLAKPAEEELSSPTPE 1066
Query: 572 PTLTQTAPTQPETQTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVP---PLNS 628
L++ + Q + P++ ++ + S I + +T+ PT P PL+
Sbjct: 1067 EKLSKKTSDTKDDQIGAPVD-VLPVNLQESLPEEKFSATIESGATTAPTLPEDERIPLDQ 1125
Query: 629 FIQGII-----------QSEATLKTLVQQFTSKPASTPHTFLITQTGEIPAEKEKE---- 673
+ ++ ++EA + VQ T L + T + P + E
Sbjct: 1126 IKEDLVIEEKYVKEETKEAEAIVVATVQTLPEAAPLAIDTILASATKDAPKDANAEALGE 1185
Query: 674 --DDDVQILEPPFNVKPLQQIASNF---EDRIPDAAETSYVSLSNYSAVNSQDLSFTS-- 726
D ++L + Q + + D + D L Y +SQD S
Sbjct: 1186 LPDSGERVLPMKMTFEAQQNLLRDVIKTPDEVADLPVHEEADLGLYEK-DSQDAGAKSIS 1244
Query: 727 ------PEKTVTSRQRPDTISEAEHMDESDHSGVEKDHEIDSIPTEHMEESNASSSNAAR 780
E+ T ++ + + E E DE + V+ H + + + E
Sbjct: 1245 HKEESAKEEKETDDEKENKVGEIELGDEPNK--VDISHVLLKESVQEVAEKVVVIETTVE 1302
Query: 781 TPQPVLTLATSFLPQSLPQLIQKFSAEAIRRVNWLYQKAKVMNFRSSLAGRKDSEFMMHL 840
Q + AT+ + Q Q+ E ++ QK S + K+++
Sbjct: 1303 KKQEEIVEATTVITQE----NQEDLMEQVKDKEEHEQK-----IESGIITEKEAK--KSA 1351
Query: 841 RGPEQRQRVLHKVCEPLRRAMAERSCVVSECTSEDVEMVSAEEEEESTDGIVILEDADAD 900
PE+++ + L +A+ + V + +++D E + E + + + +E
Sbjct: 1352 STPEEKETSDITSDDELPAQLADPTTVPPK-SAKDREDTGSIESPPTIEEAIEVEVQAKQ 1410
Query: 901 EVQKQGPIHVDTVMTSEAEAATHPQASEVPPSAPAPEVAVLAARIDRIQDDQQRLFQMVE 960
E QK P + + T ++ P AS+ PE A + + D Q ++
Sbjct: 1411 EAQKPVPAPEEAIKTEKS-----PLASK---ETSRPESATGSVKEDTEQTKSKKSPVPSR 1462
Query: 961 QQGQVQTEQSKQIAESSSKMEAI 983
+ + + ++S + +S+ E++
Sbjct: 1463 PESEAKDKKSPFASGEASRPESV 1485
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog
(X-linked nuclear protein-1)
Length = 1359
Score = 57.4 bits (137), Expect = 2e-07
Identities = 45/190 (23%), Positives = 93/190 (48%), Gaps = 23/190 (12%)
Query: 355 ESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSERSLKRT---- 410
ESP K + + +RA S SD EE +KK+ SK + ++ + KRT
Sbjct: 73 ESPRKSSKKSRKRAKSESESDESD-----EEEDRKKSKSKKKVDQKKKEKSKKKRTTSSS 127
Query: 411 ----STQQQQMSDSESSEDTWEDFSSEETE--DEDRPLAKRR-------KIILEEEEDEQ 457
S ++++ + S+ T + SSE +E +E+R + K + K E E+
Sbjct: 128 EDEDSDEEREQKSKKKSKKTKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAETSEESD 187
Query: 458 DDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQ 517
+D + + +G++ +++E ++S++ V++ K K + +KE+ +E + + +
Sbjct: 188 EDEKPSKKSKKGLKKKAKSESESESEDEKEVKKSKKKSK-KVVKKESESEDEAPEKKKTE 246
Query: 518 RERRSKRTSD 527
+ +RSK +S+
Sbjct: 247 KRKRSKTSSE 256
Score = 56.2 bits (134), Expect = 4e-07
Identities = 50/217 (23%), Positives = 94/217 (43%), Gaps = 28/217 (12%)
Query: 354 PESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSE--------- 404
P K +K ++ S S+ + +E K K SK V+ +E+ SE
Sbjct: 192 PSKKSKKGLKKKAKSESESESEDE------KEVKKSKKKSKKVVKKESESEDEAPEKKKT 245
Query: 405 RSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEII- 463
KR+ T ++ S+SE S++ E+ S + +PLA ++ + +EE E+ D E++
Sbjct: 246 EKRKRSKTSSEESSESEKSDEEEEEKESSPKPKKKKPLAVKK--LSSDEESEESDVEVLP 303
Query: 464 ----QNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRE 519
+ + I DS + ++ E V + K Q+ +SE + V
Sbjct: 304 QKKKRGAVTLISDSEDEKDQKSESEASDVEEKVSKKKAKKQESSESGSDSSEGSITV--N 361
Query: 520 RRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSA 556
R+SK+ P + + I +K+ E+I+++ A
Sbjct: 362 RKSKKKEKPEK----KKKGIIMDSSKLQKETIDAERA 394
Score = 50.4 bits (119), Expect = 2e-05
Identities = 52/230 (22%), Positives = 103/230 (44%), Gaps = 44/230 (19%)
Query: 363 RKSRRATSSKPS-DPKGKGILIEEPAKKKAASKTVIIREASSERSLKRTSTQQQQMSDSE 421
RK +RA K + +GK + PAKK+ AS + + E S +++S + ++ + SE
Sbjct: 31 RKEKRAQKLKEKREREGKPPPKKRPAKKRKASSSE-EDDDDEEESPRKSSKKSRKRAKSE 89
Query: 422 SSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEIIQNVIQGIRDSSEAEESTD 481
S D E E+EDR +K +K + ++++++ + ++ + E D
Sbjct: 90 SESD-------ESDEEEDRKKSKSKKKVDQKKKEKSKK-----------KRTTSSSEDED 131
Query: 482 SDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRERRSKRTSDPARAVQLTRTSPIR 541
SDE + +K + K+T + +SES+ E + ER+ K++ + + ++
Sbjct: 132 SDEEREQKSKK-------KSKKTKKQTSSESSEESEEERKVKKS-------KKNKEKSVK 177
Query: 542 SHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQPETQTHTQAE 591
A+ ES E P S L + A ++ E+++ + E
Sbjct: 178 KRAETSEES----------DEDEKPSKKSKKGLKKKAKSESESESEDEKE 217
Score = 49.7 bits (117), Expect = 4e-05
Identities = 44/185 (23%), Positives = 84/185 (44%), Gaps = 25/185 (13%)
Query: 357 PPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSE-----------R 405
PPK K R+A+SS+ D + E P K S+ E+ S+ +
Sbjct: 50 PPKKRPAKKRKASSSEEDDDDEE----ESPRKSSKKSRKRAKSESESDESDEEEDRKKSK 105
Query: 406 SLKRTSTQQQQMSDSESSEDTWED-FSSEETEDEDRPLAKRRK-----IILEEEEDEQDD 459
S K+ ++++ S + + + ED S EE E + + +K+ K EE E+E+
Sbjct: 106 SKKKVDQKKKEKSKKKRTTSSSEDEDSDEEREQKSKKKSKKTKKQTSSESSEESEEERKV 165
Query: 460 HEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRE 519
+ +N + ++ +E E +D DE + K KG ++ ++ +E SE EV++
Sbjct: 166 KKSKKNKEKSVKKRAETSEESDEDE----KPSKKSKKGLKKKAKSESESESEDEKEVKKS 221
Query: 520 RRSKR 524
++ +
Sbjct: 222 KKKSK 226
Score = 43.5 bits (101), Expect = 0.003
Identities = 39/174 (22%), Positives = 79/174 (44%), Gaps = 6/174 (3%)
Query: 355 ESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSERSLKRTSTQQ 414
E K ++K+++ TSS+ S+ + +++ K K K+V R +SE S + +
Sbjct: 137 EQKSKKKSKKTKKQTSSESSEESEEERKVKKSKKNK--EKSVKKRAETSEESDEDEKPSK 194
Query: 415 QQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEE--DEQDDHEIIQNVIQGIRD 472
+ + + + SE+ ++ + K +K++ +E E DE + + + +
Sbjct: 195 KSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPEKKKTEKRKRSKTS 254
Query: 473 SSEAEESTDSDEVHLVRRR--KLPLKGPLQQKETVAEQASESASEVQRERRSKR 524
S E+ ES SDE + K K PL K+ +++ SE + ++ KR
Sbjct: 255 SEESSESEKSDEEEEEKESSPKPKKKKPLAVKKLSSDEESEESDVEVLPQKKKR 308
Score = 32.7 bits (73), Expect = 5.0
Identities = 28/90 (31%), Positives = 45/90 (49%), Gaps = 11/90 (12%)
Query: 400 EASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKR------RKIILEEE 453
E+ SE S K S S SS+D + SS++ ++ ++P K+ RK + ++
Sbjct: 829 ESESEGSFKSGSESDSGKSVVLSSDD---EGSSKKKKNGNKPEIKKTAPQKKRKFLNSDD 885
Query: 454 EDEQDDHEIIQNVIQ-GIRDSSE-AEESTD 481
EDE+D + ++Q GIR S A E D
Sbjct: 886 EDEEDGEDTAMAILQDGIRQSKRLAGEEAD 915
>ANK2_HUMAN (Q01484) Ankyrin 2 (Brain ankyrin) (Ankyrin B) (Ankyrin,
nonerythroid)
Length = 3924
Score = 57.4 bits (137), Expect = 2e-07
Identities = 118/587 (20%), Positives = 220/587 (37%), Gaps = 84/587 (14%)
Query: 358 PKLTTRKSRRATSSKPSDPK--------GKGILIEEPAKKKAASKTVIIREASSERSLKR 409
P L T +SR+ +SS S+P+ G+L E + + S +++S +
Sbjct: 2617 PVLVTSESRKVSSSSESEPELAQLKKGADSGLLPEPVIRVQPPSPLPSSMDSNSSPEEVQ 2676
Query: 410 TSTQQQQMSDSESSEDTWED-FSSEETEDEDRPLAKRRKIILEEEED--------EQDDH 460
+ + +EDT E+ SEE +D + LA+ R + E ED + D
Sbjct: 2677 FQPVVSKQYTFKMNEDTQEEPGKSEEEKDSESHLAEDRHAVSTEAEDRSYDKLNRDTDQP 2736
Query: 461 EII----------QNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQAS 510
+I + + + ++ D DE ++ + L L+G +K+T E+
Sbjct: 2737 KICDGHGCEAMSPSSSARPVSSGLQSPTGDDVDEQPVIYKESLALQG-THEKDTEGEELD 2795
Query: 511 ESASEVQRERRSKRTSDPARAVQLTRTSP--IRSHAKVITESINSDSALVVVPEQPLPIS 568
S +E S + P+ + + + P + S K + E I++ S+ + +
Sbjct: 2796 VSRAE------SPQADCPSESFSSSSSLPHCLVSEGKELDEDISATSS--IQKTEVTKTD 2847
Query: 569 TSMPTLTQTAPTQPETQTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNS 628
+ L + P+Q + T + A D IY + P E VP +
Sbjct: 2848 ETFENLPKDCPSQDSSITTQTDRFSMDVPVSDLAEND----EIYDPQITSPYENVPSQSF 2903
Query: 629 FIQGIIQSEATLKTLVQQFTSKPASTPHTFLITQTGEIPAEKEKEDDDVQILEPPF---N 685
F E+ +T TS +S ++ IT E V E F +
Sbjct: 2904 F----SSEESKTQTDANHTTSFHSSEVYSVTITSPVEDVVVASSSSGTVLSKESNFEGQD 2959
Query: 686 VKPLQQIASNFEDRIPDAAETSY-VSLSNYSAVNSQDLSFTSPEKTVTSRQRPDTISEAE 744
+K Q+ S + D+ +S+ ++S + V + +S KT D+ SE
Sbjct: 2960 IKMESQLESTLWEMQSDSVSSSFEPTMSATTTVVGEQISKVIITKTDVD---SDSWSEIR 3016
Query: 745 HMDESDHSGV-EKDHEIDSIPTEHMEESNASSSNAARTPQPVLTLATSFLPQSLPQLIQK 803
DE+ + V E++ +I + + + + ARTP T + P
Sbjct: 3017 EDDEAFEARVKEEEQKIFGLMVDRQSQGTTPDTTPARTPTEEGTPTSEQNP--------- 3067
Query: 804 FSAEAIRRVNWLYQKAKVMNFRSSLA------GRKDSEFMMHLRGPEQRQRVLHKVCEPL 857
+L+Q+ K+ S A D F G E R+ L E +
Sbjct: 3068 ----------FLFQEGKLFEMTRSGAIDMTKRSYADESFHFFQIGQESREETL---SEDV 3114
Query: 858 RRAMAERSCVVSECTSEDVEMVSAEEEEESTDGIVILEDADADEVQK 904
+ + E ++E + + +E +E D +L D+ ++EV++
Sbjct: 3115 KEGATGADPLPLETSAESLAL--SESKETVDDEADLLPDSVSEEVEE 3159
>GAR2_SCHPO (P41891) Protein gar2
Length = 500
Score = 57.0 bits (136), Expect = 2e-07
Identities = 52/214 (24%), Positives = 93/214 (43%), Gaps = 41/214 (19%)
Query: 358 PKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSERSLKRTSTQQQQM 417
PK + ++++RA+S +PS K + ++ +KKK S + E+SS S +S +
Sbjct: 51 PKKSKKEAKRASSPEPSK---KSVKKQKKSKKKEESSSESESESSSSESESSSSESESSS 107
Query: 418 SDSESS---------------------EDTWEDFSSEETEDEDRPLAKRRKIILEEEEDE 456
S+SESS E + E SS E+E+E+ + K +EE+++
Sbjct: 108 SESESSSSESSSSESEEEVIVKTEEKKESSSESSSSSESEEEEEAVVK-----IEEKKES 162
Query: 457 QDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESA--- 513
D + + +SS +E + + V +K E+ ++ +SES
Sbjct: 163 SSDSSSESSSSESESESSSSESEEEEEVVEKTEEKKEGSSESSSDSESSSDSSSESGDSD 222
Query: 514 ------SEVQRERRSKRTSDPA---RAVQLTRTS 538
SE E KR ++PA R ++T+ S
Sbjct: 223 SSSDSESESSSEDEKKRKAEPASEERPAKITKPS 256
Score = 50.8 bits (120), Expect = 2e-05
Identities = 47/205 (22%), Positives = 88/205 (42%), Gaps = 18/205 (8%)
Query: 355 ESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKK--KAASKTVIIREASSERSLKRTST 412
E+ ++ + S+ S K S + K EP+KK K K+ E+SSE + +S+
Sbjct: 35 EAAKEIAKQSSKTDVSPKKSKKEAKRASSPEPSKKSVKKQKKSKKKEESSSESESESSSS 94
Query: 413 Q-QQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEIIQNVIQGIR 471
+ + S+SESS E SSE + E ++I++ EE ++ E
Sbjct: 95 ESESSSSESESSSSESESSSSESSSSE-----SEEEVIVKTEEKKESSSE---------- 139
Query: 472 DSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRERRSKRTSDPARA 531
SS +E + + V + +K + + ++ S+SE + E ++ +
Sbjct: 140 SSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESESESSSSESEEEEEVVEKTEEKKE 199
Query: 532 VQLTRTSPIRSHAKVITESINSDSA 556
+S S + +ES +SDS+
Sbjct: 200 GSSESSSDSESSSDSSSESGDSDSS 224
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 56.2 bits (134), Expect = 4e-07
Identities = 100/462 (21%), Positives = 160/462 (33%), Gaps = 52/462 (11%)
Query: 343 PITLTEFVNTLPESPPKLTTRKSRRATSSKP---SDPKGKGILIEEPAKKKAASKTVIIR 399
P+ T S P T S +SS P S + + P+ S + +
Sbjct: 511 PVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSTPVT 570
Query: 400 EASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDD 459
+++E S T S +ESS SS TE P E
Sbjct: 571 SSTTESSSAPVPTPSS--STTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAP--- 625
Query: 460 HEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRE 519
V +SS A T S +P T + +ES+S
Sbjct: 626 ------VTSSTTESSSAPVPTPSSSTTESSSAPVP---------TPSSSTTESSSAPVPT 670
Query: 520 RRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAP 579
S T + V + T S A V + + S SA P+P +S T + +AP
Sbjct: 671 PSSSTTESSSAPVTSSTTES--SSAPVTSSTTESSSA-------PVPTPSSSTTESSSAP 721
Query: 580 TQPETQTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVP-PLNSFIQGIIQSEA 638
P + T AP+ T ++TT+ S P+ +S+T + PVP P +S +
Sbjct: 722 V-PTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVP 780
Query: 639 TLKTLVQQFTSKPASTPHTFLITQTGEIPAEKEKEDDDVQILEP---PFNVKPLQQIASN 695
T + + +S P TP + T++ P ++ P PF+ +S
Sbjct: 781 TPSSSTTESSSAPVPTPSS-STTESSVAPVPTPSSSSNITSSAPSSTPFS-------SST 832
Query: 696 FEDRIPDAAETSYVSLSNYSAVNSQDL-SFTSPEKTVTSRQR-----PDTISEAEHMDES 749
+P +S + S+ + V+S S +P T +S P +I + ES
Sbjct: 833 ESSSVPVPTPSSSTTESSSAPVSSSTTESSVAPVPTPSSSSNITSSAPSSIPFSS-TTES 891
Query: 750 DHSGVEKDHEIDSIPTEHMEESNASSSNAARTPQPVLTLATS 791
+G P E S +S++ P T T+
Sbjct: 892 FSTGTTVTPSSSKYPGSQTETSVSSTTETTIVPTKTTTSVTT 933
Score = 53.9 bits (128), Expect = 2e-06
Identities = 86/455 (18%), Positives = 154/455 (32%), Gaps = 31/455 (6%)
Query: 343 PITLTEFVNTLPESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREAS 402
P T T P P TT +++ T++ + K + P+ S + + S
Sbjct: 274 PPTTTSCTKEKPTPPHHDTTPCTKKKTTTSKTCTKKTTTPVPTPSSSTTESSSAPVPTPS 333
Query: 403 SERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEI 462
S S +S+ S +ESS SS TE P+ E +
Sbjct: 334 S--STTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTS--------STTESSSAPV 383
Query: 463 IQNVIQGIRDS--SEAEESTDSDEVHLVRRRKLPLKGPLQQKET------VAEQASESAS 514
+ + + + +T+S + P+ T V +ES+S
Sbjct: 384 TSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSS 443
Query: 515 EVQRERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTL 574
++ +S P + T S A V + + S SA V P S+S P
Sbjct: 444 APVTSSTTESSSAPVPTPSSSTTES--SSAPVTSSTTESSSAPVPTPSSSTTESSSAPVT 501
Query: 575 TQTAPTQ----PETQTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFI 630
+ T + P + T AP T ++TT+ S P+ +S+T + PVP +S
Sbjct: 502 SSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSST 561
Query: 631 QGIIQSEATLKTLVQQFTSKPASTPHTFLI-TQTGEIPAEKEKEDDDVQILEPPFNVKPL 689
+ T T + +S P TP + + + +P + P +
Sbjct: 562 TESSSTPVTSST--TESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTT 619
Query: 690 QQIASNFEDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPDTISEAEHMDES 749
+ ++ +++ + S+ + +S T T S P + + S
Sbjct: 620 ESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESS 679
Query: 750 DHSGVEKDHEIDSIPTEHMEESNASSSNAARTPQP 784
E S P S+ + S++A P P
Sbjct: 680 SAPVTSSTTESSSAPV----TSSTTESSSAPVPTP 710
Score = 42.0 bits (97), Expect = 0.008
Identities = 57/309 (18%), Positives = 101/309 (32%), Gaps = 54/309 (17%)
Query: 501 QKETVAEQASESASEVQRERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSALVVV 560
+ T SES++ S T+ T +S S T + + +
Sbjct: 209 KSSTTTSSTSESSTTTSSTSESSTTTSSTSESSTTTSSTSESSTSSSTTAPATPTTTSCT 268
Query: 561 PEQPLPISTSMPTLTQTAPTQPET-------------------------QTHTQAEHQAP 595
E+P P +T+ T + P +T + T AP
Sbjct: 269 KEKPTPPTTTSCTKEKPTPPHHDTTPCTKKKTTTSKTCTKKTTTPVPTPSSSTTESSSAP 328
Query: 596 ISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQSEATLKTLVQQFTSKPASTP 655
+ T ++TT+ S P+ +S+T + PVP +S + + V T++ +S P
Sbjct: 329 VPTPSSSTTESSSAPVTSSTTESSSAPVPTPSS------STTESSSAPVTSSTTESSSAP 382
Query: 656 HTFLITQTGEIPAEKEKEDDDVQILEPPFNVKPLQQIASNFEDRIPDAAETSYVSLSNYS 715
T T++ P P ++ P + T+ S + +
Sbjct: 383 VTSSTTESSSAPV-------------------PTPSSSTTESSSAPVTSSTTESSSAPVT 423
Query: 716 AVNSQDLSFTSPEKTVTSRQRPDTISEAEHMDESDHSGVEKDHEIDSIPTEHMEESNASS 775
+ ++ S T S P T S E + E S P S+ +
Sbjct: 424 SSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPV----TSSTTE 479
Query: 776 SNAARTPQP 784
S++A P P
Sbjct: 480 SSSAPVPTP 488
Score = 40.0 bits (92), Expect = 0.031
Identities = 63/316 (19%), Positives = 106/316 (32%), Gaps = 35/316 (11%)
Query: 351 NTLPESPPKLTTRKSRRATSSKPSDP--KGKGILIEEPAKKKAASKTVIIREASSERSLK 408
++ P P +T +S A PS + + P+ S + + +++E S
Sbjct: 703 SSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSA 762
Query: 409 RTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEIIQNVIQ 468
T S +ESS SS TE P+ E N+
Sbjct: 763 PVPTPSS--STTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSVAPVPTPSSSSNITS 820
Query: 469 GIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRERRSKRTSDP 528
S+ ST+S V P+ P + E +S S E P
Sbjct: 821 SAPSSTPFSSSTESSSV--------PVPTP---SSSTTESSSAPVSSSTTESSVAPVPTP 869
Query: 529 ARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQPETQTHT 588
+ + +T ++P TES ++ + T+T ++ P +QT T
Sbjct: 870 SSSSNITSSAPSSIPFSSTTESFSTGT-----------------TVTPSSSKYPGSQTET 912
Query: 589 QAEHQAPISTIQTATTDIPSIPIYTS-STSIPTEPVPPLNSFIQGIIQSEATLKTLVQQF 647
+ + T TT + P T+ +T++ + G S T+ T V
Sbjct: 913 SVSSTTETTIVPTKTTTSVTTPSTTTITTTVCSTGTNSAGETTSGC--SPKTVTTTVPTT 970
Query: 648 TSKPASTPHTFLITQT 663
T+ +T T IT T
Sbjct: 971 TTTSVTTSSTTTITTT 986
>YWIE_CAEEL (Q23525) Hypothetical protein ZK546.14 in chromosome II
Length = 472
Score = 54.7 bits (130), Expect = 1e-06
Identities = 40/152 (26%), Positives = 74/152 (48%), Gaps = 14/152 (9%)
Query: 385 EPAKKKAASKTVIIREASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAK 444
+P K A K + + E+ +K+ +Q + +DS+S ED D EE E+ D P+AK
Sbjct: 97 QPQKVVAPVKRPADQNKNKEKVVKKDQKKQDKKADSDSEEDDSSD--DEEKEETDEPVAK 154
Query: 445 RRKIILEEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKET 504
++K EE D+ +D E + +G + EAE+S +DE + P + +T
Sbjct: 155 KQK--KEESSDDDEDSEDGEEP-EGNNGAVEAEDSDSTDEEE---------ETPSKPNKT 202
Query: 505 VAEQASESASEVQRERRSKRTSDPARAVQLTR 536
VA+ +S ++ +E + + + ++ R
Sbjct: 203 VAQSTLKSNGKIDKEIQKLEDDEDNESPEIRR 234
>NUCL_CHICK (P15771) Nucleolin (Protein C23)
Length = 694
Score = 52.8 bits (125), Expect = 5e-06
Identities = 45/198 (22%), Positives = 79/198 (39%), Gaps = 37/198 (18%)
Query: 357 PPK------LTTRKSRRATSSKPSDPKGKGIL-----IEEPAKKKAASK----TVIIREA 401
PPK +T K + K + P K + + PAKK A V+ + A
Sbjct: 46 PPKKQQKAAVTPAKKAATPAKKAATPAKKAVTPAKKAVATPAKKAVAPSPKKAAVVGKGA 105
Query: 402 SSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPL------------AKRRKII 449
+ ++ K+ ++++ D + ED E+ S+E E+ P+ AK+ ++
Sbjct: 106 KNGKNAKKEESEEEDEDDEDDEEDEDEEEESDEEEEPAVPVKPAAKKSAAAVPAKKPAVV 165
Query: 450 LEEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQA 509
++E E+++ E D E +E D E + P+K P K T A+
Sbjct: 166 PAKQESEEEEEE----------DDEEEDEEDDESEDEAMDTTPAPVKKPTPAKATPAKAK 215
Query: 510 SESASEVQRERRSKRTSD 527
+ES E E + D
Sbjct: 216 AESEDEEDEEDEDEDEED 233
Score = 42.4 bits (98), Expect = 0.006
Identities = 43/175 (24%), Positives = 71/175 (40%), Gaps = 41/175 (23%)
Query: 374 SDPKGKGILIEEPAKKKAASKTVIIREASSERSLKRTSTQQQQMSDSESSEDTWEDFSSE 433
SD + + + +PA KK+A+ + A +Q+ + E +D ED +
Sbjct: 136 SDEEEEPAVPVKPAAKKSAAAVPAKKPA--------VVPAKQESEEEEEEDDEEEDEEDD 187
Query: 434 ETEDE-----DRPLAK---------RRKIILEEEEDEQDDHEIIQNVIQGIRDSSEAEES 479
E+EDE P+ K + K E+EEDE+D+ E ++ D E +E
Sbjct: 188 ESEDEAMDTTPAPVKKPTPAKATPAKAKAESEDEEDEEDEDEDEED-----EDDEEEDEE 242
Query: 480 TDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRERRSKRTSDPARAVQL 534
DE + KE ++ E A++ E + K+T PA A L
Sbjct: 243 ESEDEKPV--------------KEAPGKRKKEMANKSAPEAKKKKTETPASAFSL 283
>NFM_CHICK (P16053) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M)
Length = 857
Score = 52.4 bits (124), Expect = 6e-06
Identities = 63/330 (19%), Positives = 136/330 (41%), Gaps = 43/330 (13%)
Query: 384 EEPAKKKAASKTVIIREASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLA 443
EE +++ A + + EA SE++ ++ + ++++ + E+ + ++EE
Sbjct: 482 EEEQEEEKAEEEAVEEEAVSEKAAEQAAEEEEKEEEEAEEEEAAKSDAAEEG-------G 534
Query: 444 KRRKIILEEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQ-- 501
+++ I E+EE E+ + E +EA+ + + + + P K P +
Sbjct: 535 SKKEEIEEKEEGEEAEEE-----------EAEAKGKAEEAGAKVEKVKSPPAKSPPKSPP 583
Query: 502 KETVAEQA---SESASEVQRERRSKRTS------DPARAVQLTRTSPIRSHAKVITESIN 552
K V EQA ++A+EV +++++++ + + A + + T + S K T
Sbjct: 584 KSPVTEQAKAVQKAAAEVGKDQKAEKAAEKAAKEEKAASPEKPATPKVTSPEKPATPEKP 643
Query: 553 SDSALVVVPEQ---PLPISTSMPTLTQTAPTQPETQTHTQ--AEHQAPISTIQTATTDIP 607
+ PE+ P +T ++ P PE + A + P + + T + P
Sbjct: 644 PTPEKAITPEKVRSPEKPTTPEKVVSPEKPASPEKPRTPEKPASPEKPATPEKPRTPEKP 703
Query: 608 SIPIYTSSTSIPTEPVPPLNSFIQGIIQSEATLKTLVQQFTSKPASTPHTFLITQTGEI- 666
+ P S P+ P+ + ++ I K + SK A I GE+
Sbjct: 704 ATPEKPRSPEKPSSPLKDEKAVVEESITVTKVTKVTAEVEVSKEARKED---IAVNGEVE 760
Query: 667 -----PAEKEKEDDDVQILEPPFNVKPLQQ 691
EKE E+++ ++ +V P+ +
Sbjct: 761 EKKDEAKEKEAEEEEKGVVTNGLDVSPVDE 790
>ACIN_MOUSE (Q9JIX8) Apoptotic chromatin condensation inducer in the
nucleus (Acinus)
Length = 1338
Score = 52.4 bits (124), Expect = 6e-06
Identities = 128/681 (18%), Positives = 255/681 (36%), Gaps = 109/681 (16%)
Query: 348 EFVNTLPESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSE--- 404
+F ++L +L+T R+++S S+ KG+ ++ +K ++ +R+A S+
Sbjct: 186 DFQSSLNRPELELSTHSPRKSSSF--SEEKGES---DDEKPRKGERRSSRVRQAKSKLPE 240
Query: 405 ---------------RSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKII 449
R+L+ + + ++ + E E+ ED EE E+ D R
Sbjct: 241 YSQTAEEEEDQETPSRNLRVRADRNLKIEEEEEEEEEEEDDDDEEEEEVDEAQKSREAEA 300
Query: 450 -----LEEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRR-----RKLPLKGPL 499
E+EE E+ + V+ + ++E + ++ V R R+ L
Sbjct: 301 PTLKQFEDEEGEERTRAKPEKVVDEKPLNIRSQEKGELEKGGRVTRSQEEARRSHLARQQ 360
Query: 500 QQKET----VAEQASESASEVQRERRSKRTSDPARAVQLTRTSPIRSHAKVITES----- 550
Q+KET + ++ +E S E +S+ S P L + + ++++E
Sbjct: 361 QEKETQIVSLPQEENEVKSSQSLEEKSQSPSPPPLPEDLEKAPVVLQPEQIVSEEETPPP 420
Query: 551 -----INSDSALVVVPEQPLPIS-TSMPTLTQTAPTQPETQTHTQAEHQAPISTIQTATT 604
+S + + E+ P+ + P L Q +P +T A + P+++ A
Sbjct: 421 LLTKEASSPPTHIQLQEEMEPVEGPAPPVLIQLSPP------NTDAGAREPLASPHPAQL 474
Query: 605 DIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQSEATLKTLVQQFTSKPASTPHTFLITQTG 664
P+ S + T+ P + A +L + T K + ++
Sbjct: 475 LRSLSPL---SGTTDTKAESPAGRVSDESVLPLAQKSSLPECSTQKGVESERE----KSA 527
Query: 665 EIPAEKEKEDDDVQILEPPFNVKPLQQIASNFEDRIPDAAETSYVSLSNYSAVNSQDLSF 724
+P E+ I E P + L+Q + S + S+ +S S
Sbjct: 528 PLPLTVEEFAPAKGITEEPMKKQSLEQKEGRRASHALFPEHSGKQSADSSSSRSSSPSSS 587
Query: 725 TSPEKTVTS---RQRPDTISEAEHMDESDHSGVEKDHEIDSIPTEHMEESNA-------- 773
+SP ++ + RP + ++ D + HE + + ES +
Sbjct: 588 SSPSRSPSPDSVASRPQSSPGSKQRDGAQARVHANPHERPKMGSRSTSESRSRSRSRSRS 647
Query: 774 --SSSNAARTPQPVLTLATSFLPQSLPQLIQKFSAEAIRRVNWLYQKAKVMNFRSSLAGR 831
SSS + +P TS+ P Q+ +A + ++ L K K F +S+ GR
Sbjct: 648 ASSSSRKSLSPGVSRDSNTSYTETKDPSCGQEAAAPSGPQLQVLEPKEKAPTFSASVRGR 707
Query: 832 KDSEFMMHLRGPEQRQRVLHKVCEPLRRAMAERSCVVSECTSEDVEMVSAEEEEESTDGI 891
HL PE Q+ + + +P + + +C +E+ E +A + + S I
Sbjct: 708 -------HLSHPEPEQQHVIQRLQPEQGS-------PKKCEAEEAEPPAATQPQTSETQI 753
Query: 892 VILEDADADEVQKQGPIHVDTVMTSEAEAATHPQASEVPPSAPAPEVAVLAARIDRIQDD 951
L +++ T T E + SE P PE + A D++ +D
Sbjct: 754 SHLLESER------------THHTVEEKEEVTMDTSENRPENEVPEPPLPVA--DQVSND 799
Query: 952 QQRLFQMVEQQGQVQTEQSKQ 972
++ +G + E+ K+
Sbjct: 800 ER-------PEGGAEEEEKKE 813
>YS89_CAEEL (Q09624) Hypothetical protein ZK945.9 in chromosome II
Length = 3178
Score = 52.0 bits (123), Expect = 8e-06
Identities = 74/306 (24%), Positives = 118/306 (38%), Gaps = 27/306 (8%)
Query: 506 AEQASESASEVQRERRSKRTSDPARA---VQLTRTSPIRSHAKVITESINSDSALVVVPE 562
A+Q S S RR KR + ++ T TS +T ++ S + VP
Sbjct: 242 AKQFSMRTSGSPTLRRMKRDAGDNTCDYTIESTSTSTTTPTTTTVTSTVTSTTT---VPT 298
Query: 563 QPLPISTSMPTLTQTAPTQ------PETQTHTQAEHQAPISTIQTATTDIPSIPIYTS-S 615
++T+M T T T T T T T + + ST Q +++ I S P T+ S
Sbjct: 299 STSTVTTAMSTSTSTPSTSTTIESTSTTFTSTASTSTSSTSTTQQSSSTITSSPSSTTLS 358
Query: 616 TSIPTEPVPPLNSFIQGIIQSEATLKTLVQQFTSKPASTPHTFLITQTGEIPA------E 669
TSIPT P + S + + A L + TS +T T L + T E P+ E
Sbjct: 359 TSIPTTTTPEITSTLSS-LPDNAICSYLDETTTSTTFTT--TMLTSTTTEEPSTSTTTTE 415
Query: 670 KEKEDDDVQILEPPFNVKPLQQIASNFE---DRIPDAAETSYVSLSNYSAVNSQDLSFTS 726
V EP + S E + + TS V+ S ++ +S + T+
Sbjct: 416 VTSTSSTVTTTEPTTTLTTSTASTSTTEPSTSTVTTSPSTSPVT-STVTSSSSSSTTVTT 474
Query: 727 PEKTVTSRQRP-DTISEAEHMDESDHSGVEKDHEIDSIPTEHMEESNASSSNAARTPQPV 785
P T ++ P T++ + + +G S S ASS+ ++ + Q
Sbjct: 475 PTSTESTSTSPSSTVTTSTTAPSTSTTGPSSSSSTPSSTASSSVSSTASSTQSSTSTQQS 534
Query: 786 LTLATS 791
T S
Sbjct: 535 STTTKS 540
Score = 37.4 bits (85), Expect = 0.20
Identities = 53/248 (21%), Positives = 89/248 (35%), Gaps = 29/248 (11%)
Query: 504 TVAEQASESASEVQRERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQ 563
T S S++ ++ S TS P+ T + T S D+A+ ++
Sbjct: 328 TSTASTSTSSTSTTQQSSSTITSSPSSTTLSTSIPTTTTPEITSTLSSLPDNAICSYLDE 387
Query: 564 PLPISTSMPT-LTQTAPTQPETQTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEP 622
+T T LT T +P T T TT++ S TSST TEP
Sbjct: 388 TTTSTTFTTTMLTSTTTEEPST---------------STTTTEVTS----TSSTVTTTEP 428
Query: 623 VPPLNSFIQGIIQSEATLKTLVQQFTSKPASTPHTFLITQTGEIPAEKEKEDDDVQILEP 682
L + +E + T+ T+ P+++P T +T +
Sbjct: 429 TTTLTTSTASTSTTEPSTSTV----TTSPSTSPVTSTVTSSSSSSTTVTTPTSTESTSTS 484
Query: 683 PFNVKPLQQIASNFEDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPDTISE 742
P + A + P ++ S + S +S S S ++ TS Q+ T ++
Sbjct: 485 PSSTVTTSTTAPSTSTTGPSSS-----SSTPSSTASSSVSSTASSTQSSTSTQQSSTTTK 539
Query: 743 AEHMDESD 750
+E SD
Sbjct: 540 SETTTSSD 547
>PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin)
Length = 5183
Score = 51.2 bits (121), Expect = 1e-05
Identities = 99/490 (20%), Positives = 182/490 (36%), Gaps = 62/490 (12%)
Query: 356 SPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVI--IREASSERSLKRTSTQ 413
SP + T S + T+ P + K + +E K A K ++ ++E S + T
Sbjct: 1074 SPMPVPTESSSQKTAVPP---QVKLVKKQEQEVKTEAEKVILEKVKETLSMEKIPPMVTT 1130
Query: 414 QQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEIIQNVIQGIRDS 473
Q+ +S+ +D E +PL + +K+I EEE+ ++ + +
Sbjct: 1131 DQKQEESKLEKD------KASALQEKKPLPEEKKLIPEEEKIRSEEKKPL---------- 1174
Query: 474 SEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRERRSKRTSDPARAVQ 533
E ++ T D+ L + P +QK + + + A E R + +T + Q
Sbjct: 1175 LEEKKPTPEDKKLLPEAK---TSAPEEQKHDLLKSQVQIAEEKLEGRVAPKTVQEGKQPQ 1231
Query: 534 LTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQPETQTHTQAEHQ 593
T+ + S D + EQP P T+ P + E + Q
Sbjct: 1232 -TKMEGLPSGTPQSLPK-EDDKTTKTIKEQPQPPCTAKP------DQEKEDDKSDTSSSQ 1283
Query: 594 APISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQSEATLKTLVQQFT--SKP 651
P S + T S I +S IP+ ++G+ + + ++ + +K
Sbjct: 1284 QPKSPQGLSDTGYSSDGISSSLGEIPSLIPTDEKDILKGLKKDSFSQESSPSSPSDLAKL 1343
Query: 652 ASTPHTFLITQTGEIPAEKEKEDDDVQILEPPFNVKPLQQIASNFEDRIPDAAETSYVSL 711
ST + L Q + EK ++ P V P Q +++ +ET +++
Sbjct: 1344 ESTVLSILEAQASTLADEKSEKKTQ------PHEVSPEQ---PKDQEKTQSLSETLEITI 1394
Query: 712 SNYSAVNSQDLSFTSPEKTVTSRQRPDTISEAEHMD---ESDHSGVEKDHEIDSIPTEHM 768
S SQ+ +R DT + D DH EK +D I T
Sbjct: 1395 SEEEIKESQE-------------ERKDTFKKDSQQDIPSSKDHK--EKSEFVDDITTRRE 1439
Query: 769 E-ESNASSSNAARTPQPVLTLATSFLPQSLPQLIQKFSAEAIRRVNWLYQKAKVMNFRSS 827
+S SS + +P P TS S + Q+ S + +++ ++ M+
Sbjct: 1440 PYDSVEESSESENSPVPQRKRRTSVGSSSSDEYKQEDSQGSGEEEDFIRKQIIEMSADED 1499
Query: 828 LAGRKDSEFM 837
+G +D EF+
Sbjct: 1500 ASGSEDDEFI 1509
Score = 44.3 bits (103), Expect = 0.002
Identities = 76/353 (21%), Positives = 133/353 (37%), Gaps = 48/353 (13%)
Query: 336 MLRRQGDPITLTEFVNTLPESPPKLT---TRKSRRATSSKPSDPKGKGILIEEPAKK--K 390
M R DP L+ + +P + T T K + +S + K K L P ++
Sbjct: 3606 MQRSMSDPKPLSPTADESSRAPFQYTEGYTTKGSQTMTSSGAQKKVKRTLPNPPPEEIST 3665
Query: 391 AASKTVIIREASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIIL 450
T S R + RT+T M+ ++ +D E + +R AK RK
Sbjct: 3666 GTQSTFSTMGTVSRRRICRTNT----MARAKILQDI-----DRELDLVERESAKLRKKQA 3716
Query: 451 EEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQAS 510
E +E+E++ ++ + GI EA +R + L+G + ++ +++
Sbjct: 3717 ELDEEEKEIDAKLRYLEMGINRRKEALLKERE------KRERAYLQGVAEDRDYMSD--- 3767
Query: 511 ESASEVQRERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTS 570
SEV R ++ S ++ RT+P ++ I ++S LV S
Sbjct: 3768 ---SEVSSTRPTRIESQ--HGIERPRTAPQTEFSQFIPPQTQTESQLVPPTSPYTQYQYS 3822
Query: 571 MPTLTQTAPTQPETQTH--TQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNS 628
P L APT Q+H Q + +S QT T ++ + + + P P +
Sbjct: 3823 SPALPTQAPTSYTQQSHFEQQTLYHQQVSPYQTQPT-FQAVATMSFTPQVQPTPTPQPSY 3881
Query: 629 FIQG---IIQSEATLKTLV--------------QQFTSKPASTPHTFLITQTG 664
+ +IQ + TL Q P ST +TF ++ G
Sbjct: 3882 QLPSQMMVIQQKPRQTTLYLEPKITSNYEVIRNQPLMIAPVSTDNTFAVSHLG 3934
Score = 37.4 bits (85), Expect = 0.20
Identities = 40/174 (22%), Positives = 75/174 (42%), Gaps = 13/174 (7%)
Query: 344 ITLTEFVNTLPESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASS 403
IT++E + K T +K + D K K +++ ++ +V E SS
Sbjct: 1392 ITISEEEIKESQEERKDTFKKDSQQDIPSSKDHKEKSEFVDDITTRREPYDSV---EESS 1448
Query: 404 ERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQ----DD 459
E ++++ S SS D ++ S+ + +E+ + RK I+E DE +D
Sbjct: 1449 ESENSPVPQRKRRTSVGSSSSDEYKQEDSQGSGEEEDFI---RKQIIEMSADEDASGSED 1505
Query: 460 HEIIQNVIQGIRDSSEA---EESTDSDEVHLVRRRKLPLKGPLQQKETVAEQAS 510
E I+N ++ I S+E+ EE+ ++ + R+L K E + S
Sbjct: 1506 DEFIRNQLKEISSSTESQKKEETKGKGKITAGKHRRLTRKSSTSIDEDAGRRHS 1559
Score = 37.4 bits (85), Expect = 0.20
Identities = 101/520 (19%), Positives = 199/520 (37%), Gaps = 81/520 (15%)
Query: 355 ESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSERSLKRTST-- 412
+ P+L R+++ S + G G+ K + I + S+E S K+TS
Sbjct: 1569 DESPELKYRETKSQESEELVVTGGGGLR----RFKTIELNSTIADKYSAESSQKKTSLYF 1624
Query: 413 ------QQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEIIQN- 465
+ + ++DS E SS + L+E+ D H+ ++
Sbjct: 1625 DEEPELEMESLTDSPEDRSRGEGSSSLHASSFTPGTSPTSVSSLDEDSDSSPSHKKGESK 1684
Query: 466 ------------VIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESA 513
++ I DSSE EE + +E L++ ++ + QQ+++ ++++ +
Sbjct: 1685 QQRKARHRPHGPLLPTIEDSSEEEELREEEE--LLKEQEKQREIEQQQRKSSSKKSKKDK 1742
Query: 514 SEVQRERRSKR-TSDPARAVQLTRTSPI-------------RSHAKVITESINSD-SALV 558
E++ +RR +R + P+ + SP RS + SI SD
Sbjct: 1743 DELRAQRRRERPKTPPSNLSPIEDASPTEELRQAAEMEELHRSSCSEYSPSIESDPEGFE 1802
Query: 559 VVPEQPLPIST--SMPT-LTQTAPTQPETQTHTQAEHQAPISTIQTATTDIPSIPIYTS- 614
+ PE+ + + +PT ++ +PT ++ + +A S + + Y +
Sbjct: 1803 ISPEKIIEVQKVYKLPTAVSLYSPTDEQSIMQKEGSQKALKSAEEMYEEMMHKTHKYKAF 1862
Query: 615 STSIPTEPVPPLNSFIQGIIQSEATLKTLVQQFTSKPASTPHTFLITQTGEIPAEKEKED 674
+ + V G++ + ++LV+ T+ + G + +E+E+
Sbjct: 1863 PAANERDEVFEKEPLYGGMLIEDYIYESLVED----------TYNGSVDGSLLTRQEEEN 1912
Query: 675 DDVQILEPPFNVKPLQQIASNFEDRIPDAAETSYVSLSNYSAVNSQDL---SFTSPEK-- 729
+Q ++ +QI + +I D + Y S +S V +D+ SF PE
Sbjct: 1913 GFMQQKGREQKIRLSEQIYEDPMQKITDLQKEFYELESLHSVVPQEDIVSSSFIIPESHE 1972
Query: 730 ------TVTSRQRPDTISEAEHMDESDHSGVEKDHEIDSIPTEHM--EESNASSSNAART 781
VTS + + +A+ E + S PT+ E+ S+ + R
Sbjct: 1973 IVDLGTMVTSTEEERKLLDADAAYEELMKRQQMQLTPGSSPTQAPIGEDMTESTMDFDRM 2032
Query: 782 PQPVLTL----------ATSFLPQSLP--QLIQKFSAEAI 809
P LT +TS S+P ++ Q FS E I
Sbjct: 2033 PDASLTSSVLSGASLTDSTSSATLSIPDVKITQHFSTEEI 2072
>DAN4_YEAST (P47179) Cell wall protein DAN4 precursor
Length = 1161
Score = 51.2 bits (121), Expect = 1e-05
Identities = 91/433 (21%), Positives = 156/433 (36%), Gaps = 35/433 (8%)
Query: 525 TSDPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQPET 584
T+ T T+P S + + + P +++ PT + T+ T +
Sbjct: 184 TTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTPTTSTTSTT---S 240
Query: 585 QTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQSEATLKTLV 644
QT T++ ST T TT + P +++++ PT S I + T T
Sbjct: 241 QTSTKSTTPTTSSTSTTPTTS--TTPTTSTTSTAPTTSTTSTTS-TTSTISTAPTTSTTS 297
Query: 645 QQFTSKPASTPHTFLITQTGE-------IPAEKEKEDDDVQILEPPFNVKPLQQIASNFE 697
F++ AS T T PA D N +
Sbjct: 298 STFSTSSASASSVISTTATTSTTFASLTTPATSTASTDHTTSSVSTTNAFTTSATTTTTS 357
Query: 698 DRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKT--VTSRQRPDTISE-AEHMDESDHSGV 754
D ++ S V+ S S+ S P ++ VTS P T+SE ++ + S V
Sbjct: 358 DTYISSSSPSQVTSSAEPTTVSEVTSSVEPTRSSQVTSSAEPTTVSEFTSSVEPTRSSQV 417
Query: 755 EKDHEIDSIP--TEHMEESNASSSNAARTPQPVLTLATSFLP--------QSLPQLIQKF 804
E ++ T +E + +S ++ P V +S P + P + +F
Sbjct: 418 TSSAEPTTVSEFTSSVEPTRSSQVTSSAEPTTVSEFTSSVEPTRSSQVTSSAEPTTVSEF 477
Query: 805 --SAEAIR--RVNWLYQKAKVMNFRSSLAGRKDSEFMMHLRGPEQRQRVLHKVCEPLRRA 860
S E R +V + V F SS+ + S+ P V V EP+R +
Sbjct: 478 TSSVEPTRSSQVTSSAEPTTVSEFTSSVEPIRSSQVTSSAE-PTTVSEVTSSV-EPIRSS 535
Query: 861 MAERSCVVSECTSEDVEMVSAEEEEESTDGIVILEDADADEVQKQGPIHVDTVMTSEAEA 920
+ VS S E+ S+ E + E ++++ + V +V+TS +E
Sbjct: 536 QVTTTEPVSSFGSTFSEITSSAEPLSFSKATTSAESISSNQITISSELIVSSVITSSSEI 595
Query: 921 ATHPQASEVPPSA 933
P + EV S+
Sbjct: 596 ---PSSIEVLTSS 605
Score = 45.1 bits (105), Expect = 0.001
Identities = 86/446 (19%), Positives = 151/446 (33%), Gaps = 54/446 (12%)
Query: 567 ISTSMPTLTQTAPTQPETQTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPL 626
I T++PT T T T+ T T P +TI + T+ + P +++++ PT
Sbjct: 117 IYTAIPTSTSTTTTKSSTST-------TPTTTITSTTSTTSTTPTTSTTSTTPTTSTTST 169
Query: 627 NSFIQGIIQSEATLKTLVQQFTSKPASTPHTFLITQTGEIPAEKEKEDDDVQILEPPFNV 686
+ T T TS ++TP T + T P +
Sbjct: 170 TPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTST 229
Query: 687 KPLQQIAS-----NFEDRIPDAAETSY--------VSLSNYSAVNSQDLSFTSPEKTVTS 733
P S + + P + TS + + +A + S TS T+++
Sbjct: 230 TPTTSTTSTTSQTSTKSTTPTTSSTSTTPTTSTTPTTSTTSTAPTTSTTSTTSTTSTIST 289
Query: 734 RQRPDTISEAEHMDESDHSGVEKDHEIDS-------------IPTEHMEESNASSSNAAR 780
T S + S V S T+H S+ S++NA
Sbjct: 290 APTTSTTSSTFSTSSASASSVISTTATTSTTFASLTTPATSTASTDH-TTSSVSTTNAFT 348
Query: 781 TPQPVLTLATSFLPQSLP-QLIQKFSAEAIRRVNWLYQKAKVMNFRSSLAGRKDSEFMMH 839
T T + +++ S P Q+ + V + + SS SEF
Sbjct: 349 TSATTTTTSDTYISSSSPSQVTSSAEPTTVSEVTSSVEPTRSSQVTSSAEPTTVSEFTSS 408
Query: 840 LRGPEQRQRVLHKV-----------CEPLRRAMAERSC---VVSECTSEDVEMVSAEEEE 885
+ P + +V EP R + S VSE TS VE + +
Sbjct: 409 VE-PTRSSQVTSSAEPTTVSEFTSSVEPTRSSQVTSSAEPTTVSEFTS-SVEPTRSSQVT 466
Query: 886 ESTDGIVILEDADADEVQKQGPI--HVDTVMTSEAEAATHP-QASEVPPSAPAPEVAVLA 942
S + + E + E + + + SE ++ P ++S+V SA V+ +
Sbjct: 467 SSAEPTTVSEFTSSVEPTRSSQVTSSAEPTTVSEFTSSVEPIRSSQVTSSAEPTTVSEVT 526
Query: 943 ARIDRIQDDQQRLFQMVEQQGQVQTE 968
+ ++ I+ Q + V G +E
Sbjct: 527 SSVEPIRSSQVTTTEPVSSFGSTFSE 552
Score = 36.2 bits (82), Expect = 0.45
Identities = 73/372 (19%), Positives = 136/372 (35%), Gaps = 65/372 (17%)
Query: 473 SSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAE--------QASESASEVQRERRSKR 524
+S AE +T S+ V + + TV+E ++S+ S + S+
Sbjct: 466 TSSAEPTTVSEFTSSVEPTRSSQVTSSAEPTTVSEFTSSVEPIRSSQVTSSAEPTTVSEV 525
Query: 525 TS--DPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAPTQP 582
TS +P R+ Q+T T P+ S +E +S +PL S + T +
Sbjct: 526 TSSVEPIRSSQVTTTEPVSSFGSTFSEITSS--------AEPLSFSKA------TTSAES 571
Query: 583 ETQTHTQAEHQAPISTIQTATTDIP-SIPIYTSS--------TSI----------PTEPV 623
+ + +S++ T++++IP SI + TSS TS+ TE +
Sbjct: 572 ISSNQITISSELIVSSVITSSSEIPSSIEVLTSSGISSSVEPTSLVGPSSDESISSTESL 631
Query: 624 PPLNSFIQGIIQSEATLKTLVQQFTSKPA--------------STPHTFLITQTGEIPAE 669
++F ++ S + S + +TP T L + + +
Sbjct: 632 SATSTFTSAVVSSSKAADFFTRSTVSAKSDVSGNSSTQSTTFFATPSTPLAVSSTVVTSS 691
Query: 670 KEKEDDDVQILEPPFNVKPLQQIASNFEDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEK 729
+ ++ E + + I S I + + Y+S SN S+ +FT +
Sbjct: 692 TDSVSPNIPFSEISSSPESSTAITSTSTSFIAERTSSLYLSSSNMSSFTLS--TFTVSQS 749
Query: 730 TVTSRQRPDTISEAEHMDESDHSGVEKDHEIDSIPTEHMEESNASSSNAARTPQPVLTLA 789
V+S T S A S + + D+ SN SS T + V
Sbjct: 750 IVSSFSMEPTSSVASFASSSPLLVSSRSNCSDA------RSSNTISSGLFSTIENVRNAT 803
Query: 790 TSFLPQSLPQLI 801
++F S +++
Sbjct: 804 STFTNLSTDEIV 815
>CEC1_CAEEL (P34618) Chromo domain protein cec-1
Length = 304
Score = 50.1 bits (118), Expect = 3e-05
Identities = 54/235 (22%), Positives = 98/235 (40%), Gaps = 25/235 (10%)
Query: 342 DPITLTEFVNTLPESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREA 401
DP + F ++ +K + A K + K + K +AS +A
Sbjct: 47 DPTLIEAFFTREAARKAEIKAKKDKMAAGKKGASSKASASV-----SKASASTPARGAKA 101
Query: 402 SSERSLKRTSTQQQQMS--DSESSEDTWEDFSS----------EETEDEDRPLAKRRKII 449
+ + K++ ++Q+++ D DT E+ SS EE ED++ P+ K++K +
Sbjct: 102 APKPPPKKSPPKRQRLAGGDIRPDSDTDEEHSSADKKSKAEDEEEVEDDEEPVPKKKKEV 161
Query: 450 LEEEEDEQ----DDHEIIQNVIQGIRDSS-EAEESTDSDEVHLVRRRKLPLKGPLQQKET 504
EE E+E+ +D E Q V D E +E + ++V L + + + +E
Sbjct: 162 QEEPEEEESVEGEDEEESQEVEDLKEDEKMEEDEKEEEEDVQLESEKNEKEEEEEKVEEK 221
Query: 505 VAEQASESASEVQR---ERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSA 556
E+ E E+Q E+ T+ AV S S K + E+ +S +A
Sbjct: 222 KEEEEEEEEEEIQLVIVEKTVIETTIVEPAVATPEPSEPSSSEKAVVENGSSSAA 276
>MP62_LYTPI (P91753) Mitotic apparatus protein p62
Length = 411
Score = 48.5 bits (114), Expect = 9e-05
Identities = 41/161 (25%), Positives = 68/161 (41%), Gaps = 23/161 (14%)
Query: 368 ATSSKPSDPKGKGILIEEPAKKKAASKTVIIREASSERSLKRTSTQQQQMSDSESSEDTW 427
A K P KG PAKK A + ++ +S+R + S + D + ++
Sbjct: 204 AKGKKRPAPSAKG-----PAKKLAK----VDKDGTSKRKVPNGSVENGHAIDDDEDDEED 254
Query: 428 EDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHEII-------------QNVIQGIRDSS 474
ED+ + E+E+ + + EEEE+E+DD E+ +G+ D
Sbjct: 255 EDYKVGDEEEEEEATSGEEEEEDEEEEEEEDDEEMALGDDDDEDDDEEDDEDEEGMDDED 314
Query: 475 EAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASE 515
E EE DS V ++ K + G + K T QA++ E
Sbjct: 315 E-EEEEDSSPVKPAKKAKGKVNGTAKPKGTPKSQANKGMKE 354
>IE68_SHV21 (Q01042) Immediate-early protein
Length = 407
Score = 47.8 bits (112), Expect = 1e-04
Identities = 48/235 (20%), Positives = 95/235 (40%), Gaps = 40/235 (17%)
Query: 335 RMLRRQGDPITLTEFVNTLPESPPKLTTR------------KSRRATSSKPSDPKGKGIL 382
R L+ QGD + +NT + LT + RR + + +G
Sbjct: 38 RKLKPQGD-----DDINTTHQQQAALTEEQRREEVEEEGEERERRGEEEREGEGGEEGEG 92
Query: 383 IEEPAKKKAASKTVIIREAS-SERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDED-- 439
EE +++A K EA +E + ++ + + E+ E+ E+ +EE E+E+
Sbjct: 93 REEAEEEEAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAE 152
Query: 440 --RPLAKRRKIILEEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKG 497
A+ + E EE+ ++ E + + ++ EAEE+ +++E
Sbjct: 153 EAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEE------------- 199
Query: 498 PLQQKETVAEQASESASEVQRERRSKRTSDPARAVQLTR-----TSPIRSHAKVI 547
++ E AE+A E A E + ++ + A + ++P H KV+
Sbjct: 200 EAEEAEEEAEEAEEEAEEAEEAEEAEEAEEEAEEAEEEEEEAGPSTPRLPHYKVV 254
Score = 37.7 bits (86), Expect = 0.15
Identities = 31/148 (20%), Positives = 62/148 (40%), Gaps = 2/148 (1%)
Query: 386 PAKKKAASKTVIIREASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKR 445
P K++ K + ++ + T++Q+ + E + E EE E E +
Sbjct: 33 PRKRRRKLKPQGDDDINTTHQQQAALTEEQRREEVEEEGEERERRGEEEREGEGGEEGEG 92
Query: 446 RKIILEEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETV 505
R+ EEE +E++ E + + EAEE+ +E + + + +E
Sbjct: 93 REEAEEEEAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEE--AEEEEAEEEEAEEAEEEE 150
Query: 506 AEQASESASEVQRERRSKRTSDPARAVQ 533
AE+A E A E + E ++ ++ A +
Sbjct: 151 AEEAEEEAEEEEAEEEAEEEAEEAEEAE 178
>PCFB_HUMAN (O94913) Pre-mRNA cleavage complex II protein Pcf11
(Fragment)
Length = 1654
Score = 47.0 bits (110), Expect = 3e-04
Identities = 47/259 (18%), Positives = 104/259 (40%), Gaps = 18/259 (6%)
Query: 350 VNTLPESPPKLT-TRKSRRATSSKPSDPK-GKGIL------IEEPAKKKAASKTVIIREA 401
+NTL +S K + T S + SSK K G+ I ++ +K K+ S + + +
Sbjct: 418 MNTLNQSDTKTSKTIPSEKLNSSKQEKSKSGEKITKKELDQLDSKSKSKSKSPSPLKNKL 477
Query: 402 SSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQ---- 457
S + LK ++ ++SD + + ++T+ +D + ++RK ++++DE
Sbjct: 478 SHTKDLKNQESESMRLSDMNKRDPRLKKHLQDKTDGKDDDVKEKRKTAEKKDKDEHMKSS 537
Query: 458 ------DDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASE 511
++II ++Q +E E + R++ + P + +
Sbjct: 538 EHRLAGSRNKIINGIVQKQDTITEESEKQGTKPGRSSTRKRSRSRSPKSRSPIIHSPKRR 597
Query: 512 SASEVQRERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSM 571
+R +RS + +A ++ ++ +SH + T D +Q + M
Sbjct: 598 DRRSPKRRQRSMSPTSTPKAGKIRQSGAKQSHMEEFTPPSREDRNAKRSTKQDIRDPRRM 657
Query: 572 PTLTQTAPTQPETQTHTQA 590
+ P + Q T++
Sbjct: 658 KKTEEERPQETTNQHSTKS 676
>ITN1_RAT (Q9WVE9) Intersectin 1 (EH domain and SH3 domain regulator
of endocytosis 1)
Length = 1217
Score = 47.0 bits (110), Expect = 3e-04
Identities = 40/171 (23%), Positives = 78/171 (45%), Gaps = 24/171 (14%)
Query: 401 ASSERSLKRTSTQQQQMSDSESSED----------TWEDFSSEETEDEDRPLAKRRKIIL 450
+ S S+ +S+ Q++ + SSED T+ED E E + L KRR+ +L
Sbjct: 313 SGSGMSVISSSSADQRLPEEPSSEDEQQVEKKLPVTFEDKKRENFERGNLELEKRRQALL 372
Query: 451 EEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQ-QKETVAEQA 509
E++ EQ+ ++ Q ++ E+ R+R+L L+ L+ Q+E ++
Sbjct: 373 EQQRKEQERLAQLERAEQERKERERQEQE---------RKRQLELEKQLEKQRELERQRE 423
Query: 510 SESASEVQRERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSALVVV 560
E E++R +KR + R ++ R +++T+ +VV+
Sbjct: 424 EERRKEIERREAAKRELERQRQLEWER----NRRQELLTQRNKDQEGIVVL 470
Score = 32.7 bits (73), Expect = 5.0
Identities = 23/91 (25%), Positives = 42/91 (45%), Gaps = 2/91 (2%)
Query: 433 EETEDEDRPLAKRRKIILEEEEDEQDDHEIIQNVIQGIRDSSEAEESTDSDEVHLVRRRK 492
+E E ++ R I ++ Q+ +++ +I + S+ + + +H R
Sbjct: 507 QEIESTNKSRELRIAEITHLQQQLQESQQMLGRLIPEKQILSDQLKQVQQNSLH--RDSL 564
Query: 493 LPLKGPLQQKETVAEQASESASEVQRERRSK 523
L LK L+ KE +Q E EV++E RSK
Sbjct: 565 LTLKRALEAKELARQQLREQLDEVEKETRSK 595
>YM96_YEAST (Q04893) Hypothetical 113.1 kDa protein in PRE5-FET4
intergenic region
Length = 1140
Score = 46.6 bits (109), Expect = 3e-04
Identities = 84/420 (20%), Positives = 157/420 (37%), Gaps = 59/420 (14%)
Query: 401 ASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDH 460
+S+ L ++T+ +S S + + SS PL K +LE D
Sbjct: 123 SSTNAQLSSSTTETDSISSSAIQTSSPQTSSSNGGGSSSEPLGKSS--VLETTASSSDTT 180
Query: 461 EIIQNVIQGIRD-----------------------SSEAEESTDSDEVHLVRRRKLPLKG 497
+ + + D S E S+ SD L+ P
Sbjct: 181 AVTSSTFTTLTDVSSSPKISSSGSAVTSVGTTSDASKEVFSSSTSDVSSLLSSTSSPASS 240
Query: 498 PLQQKETVAEQA-SESASEVQRERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSD-- 554
+ + + S ++S V E S +S + + +S + S A + T S+ S
Sbjct: 241 TISETLPFSSTILSITSSPVSSEAPSATSSSVSSEASSSTSSSVSSEAPLATSSVVSSEA 300
Query: 555 -SALVVVPEQPLPISTSMPTLTQTAPTQPETQTHTQAEHQAPISTIQTATTDIPSIPIYT 613
S+ V P STS ++ + T T + +AP++T +++ PS +
Sbjct: 301 PSSTSSVVSSEAPSSTSSSVSSEISST-----TSSSVSSEAPLATSSVVSSEAPS----S 351
Query: 614 SSTSIPTEPVPPLNSFIQGIIQSEATLKTLVQQFTSKPAST--------PHTFLITQTGE 665
+S+S+ +E ++S + SEA L T + P+ST P + + + E
Sbjct: 352 TSSSVSSE----ISSTTSSSVSSEAPLATSSVVSSEAPSSTSSSVSSEAPSSTSSSVSSE 407
Query: 666 IPAEKEKEDDDVQILEPPFNVKPLQQIASNFEDRIPDAAETSYVSLSNYSAVNSQDLSFT 725
P+ +I +V +++S + A ++ SL++ +S++ S T
Sbjct: 408 APSSTSSSVSS-EISSTKSSVMS-SEVSSATSSLVSSEAPSAISSLASSRLFSSKNTSVT 465
Query: 726 S-----PEKTVTSRQRP--DTISEAEHMDESDHSGVEKDHEIDSIPTEHMEESNASSSNA 778
S +VTS RP +T++ ++ S +G + S SSSN+
Sbjct: 466 STLVATEASSVTSSLRPSSETLASNSIIESSLSTGYNSTVSTTTSAASSTLGSKVSSSNS 525
Score = 40.0 bits (92), Expect = 0.031
Identities = 79/361 (21%), Positives = 134/361 (36%), Gaps = 50/361 (13%)
Query: 583 ETQTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQSEATL-K 641
++ T T H + +T T +T P+ TS+TS P + I+ S +TL
Sbjct: 7 KSTTATTTSHSSTTTTSSTTSTTTPTTTSTTSTTSTKVTTSPEI------IVSSSSTLVS 60
Query: 642 TLVQQFTSKPASTPHTFLITQTGEIPAEKEKEDDDVQILEPPFNVKPLQQIASNFEDRIP 701
++V +FTS + + T + E V I L +S+
Sbjct: 61 SVVPEFTSSSSLSSDTIASILSSE---------SLVSIFSS------LSYTSSDISSTSV 105
Query: 702 DAAETSYVSLSN-YSAVNSQDLSFTSPEKTVTSRQRPDTISEAEHMDESDHSGVEKDHEI 760
+ E+S SN YSA++S + +S S D+IS + S +
Sbjct: 106 NDVESSTSGPSNSYSALSSTNAQLSS------STTETDSISSSAIQTSSPQTSSSNGGGS 159
Query: 761 DSIPT--EHMEESNASSSNAARTPQPVLTLATSFLPQSLPQLIQKFSAEAIRRVNWLYQK 818
S P + E+ ASSS+ T T S P++ S A+ V
Sbjct: 160 SSEPLGKSSVLETTASSSDTTAVTSSTFTTLTDV--SSSPKISS--SGSAVTSVGTTSDA 215
Query: 819 AKVMNFRSSLAGRKDSEFMMHLRGPEQRQRVLHKVCEPLRRAMAERSCVVSECTSEDVEM 878
+K + F SS + S + P + E L + S S +SE
Sbjct: 216 SKEV-FSSSTSDV--SSLLSSTSSPASST-----ISETLPFSSTILSITSSPVSSEAPSA 267
Query: 879 VSAEEEEESTDGIVILEDADADEVQKQGPIHVDTVMTSEAEAATHPQASEVPPSAPAPEV 938
S+ E++ + + V + P+ +V++SEA ++T S PS+ + V
Sbjct: 268 TSSSVSSEASS-------STSSSVSSEAPLATSSVVSSEAPSSTSSVVSSEAPSSTSSSV 320
Query: 939 A 939
+
Sbjct: 321 S 321
Score = 37.7 bits (86), Expect = 0.15
Identities = 90/454 (19%), Positives = 158/454 (33%), Gaps = 73/454 (16%)
Query: 343 PITLTEFVNT-LPESPPKLTTRKSRRATSSKPSDPKGKGILIEEPAKKKAASKTVIIREA 401
P+ + V++ P S + + ++ +TSS S ++ A+ +V+ EA
Sbjct: 289 PLATSSVVSSEAPSSTSSVVSSEAPSSTSSSVSSEISSTTSSSVSSEAPLATSSVVSSEA 348
Query: 402 SSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDHE 461
S S +S S S SSE ++ + P + + E
Sbjct: 349 PSSTSSSVSSEISSTTSSSVSSE---APLATSSVVSSEAPSSTSSSVSSEAPSSTSSS-- 403
Query: 462 IIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEV----- 516
SSEA ST S + K + T + +SE+ S +
Sbjct: 404 ----------VSSEAPSSTSSSVSSEISSTKSSVMSSEVSSATSSLVSSEAPSAISSLAS 453
Query: 517 QRERRSKRTSDPARAVQLTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQ 576
R SK TS + V T S + S + +E++ S+S ++ ++++ T T
Sbjct: 454 SRLFSSKNTSVTSTLV-ATEASSVTSSLRPSSETLASNS--IIESSLSTGYNSTVSTTTS 510
Query: 577 TAPTQPETQTHTQAEHQAPISTIQTATTDIPSIPIYTSSTSIPTEP-------VPPLNSF 629
A + ++ + A T T++ S I+ +S+++ T P PL S
Sbjct: 511 AASSTLGSKVSSSNSRMATSKTSSTSSDLSKSSVIFGNSSTVTTSPSASISLTASPLPSV 570
Query: 630 IQGIIQSEATLKTLVQQFTSKPASTPHT-----FLITQT--------------GEIPAEK 670
I SEA+ + +S P+ T ++T+T E E
Sbjct: 571 WSDITSSEASSISSNLASSSAPSDNNSTIASASLIVTKTKNSVVSSIVSSITSSETTNES 630
Query: 671 EKEDDDVQILEPPFNVKPLQQIASNFEDRIPDAA---------------ETSYVSLSNYS 715
+L + L + +P TS SL+ S
Sbjct: 631 NLATSSTSLLSNKATARSLSTSNATSASNVPTGTFSSMSSHTSVITPGFSTSSASLAINS 690
Query: 716 AVNSQDL---SFTSPE-----KTVTSRQRPDTIS 741
V S L SF++PE T+ + + P T+S
Sbjct: 691 TVVSSSLAGYSFSTPESSPTTSTLVTSEAPSTVS 724
Score = 35.0 bits (79), Expect = 1.0
Identities = 79/400 (19%), Positives = 146/400 (35%), Gaps = 55/400 (13%)
Query: 401 ASSERSLKRTSTQQQQMSDSESSEDTWEDFSSEETEDEDRPLAKRRKIILEEEEDEQDDH 460
+S +S T+T + + S+ T ++ T + +II+
Sbjct: 3 SSGSKSTTATTTSHSSTTTTSSTTSTTTPTTTSTTSTTSTKVTTSPEIIVSSSST----- 57
Query: 461 EIIQNVIQGIRDSSEAEESTDSDEVHLVRRRKLPLKGPLQQKETVAEQASESASEVQRER 520
++ +V+ SS S SD + + + + T ++ +S S ++V+
Sbjct: 58 -LVSSVVPEFTSSS----SLSSDTIASILSSESLVSIFSSLSYTSSDISSTSVNDVESS- 111
Query: 521 RSKRTSDPARAVQ-LTRTSPIRSHAKVITESINSDSALVVVPEQPLPISTSMPTLTQTAP 579
TS P+ + L+ T+ S + T+SI+S + P+ S+S + + P
Sbjct: 112 ----TSGPSNSYSALSSTNAQLSSSTTETDSISSSAIQTSSPQT----SSSNGGGSSSEP 163
Query: 580 TQPETQTHTQAEHQ---APISTIQTATTDIPSIPIYTSSTSIPTEPVPPLNSFIQGIIQS 636
+ T A A S+ T TD+ S P +SS S T ++ + S
Sbjct: 164 LGKSSVLETTASSSDTTAVTSSTFTTLTDVSSSPKISSSGSAVTSVGTTSDASKEVFSSS 223
Query: 637 EATLKTLVQQFTSKPASTPHTFLITQTGEIPAEKEKEDDDVQILEPPFNVKPLQQIASNF 696
+ + +L+ TS PAS+ I++T PF+ L +S
Sbjct: 224 TSDVSSLLSS-TSSPASST----ISET------------------LPFSSTILSITSSPV 260
Query: 697 EDRIPDAAETSYVSLSNYSAVNSQDLSFTSPEKTVTSRQRPDTISEAEHMDESDHSGVEK 756
P A +S S ++ S +S +V S + P + S + +
Sbjct: 261 SSEAPSATSSSVSSEASSSTSSSVSSEAPLATSSVVSSEAPSSTSSVVSSEAPSSTSSSV 320
Query: 757 DHEIDSIPTEHMEESNASSSNAARTPQPVLTLATSFLPQS 796
EI S SSS ++ P ++ +S P S
Sbjct: 321 SSEI---------SSTTSSSVSSEAPLATSSVVSSEAPSS 351
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.127 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 110,042,717
Number of Sequences: 164201
Number of extensions: 4693513
Number of successful extensions: 22485
Number of sequences better than 10.0: 679
Number of HSP's better than 10.0 without gapping: 75
Number of HSP's successfully gapped in prelim test: 631
Number of HSP's that attempted gapping in prelim test: 18868
Number of HSP's gapped (non-prelim): 2498
length of query: 997
length of database: 59,974,054
effective HSP length: 120
effective length of query: 877
effective length of database: 40,269,934
effective search space: 35316732118
effective search space used: 35316732118
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0476b.8


