

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.4
(341 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
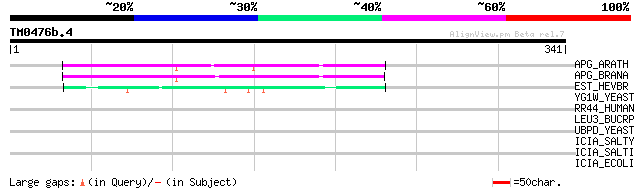
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 102 1e-21
APG_BRANA (P40603) Anter-specific proline-rich protein APG (Prot... 98 3e-20
EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early nodule... 44 8e-04
YG1W_YEAST (P53230) Hypothetical 44.2 kDa protein in RME1-TFC4 i... 33 1.3
RR44_HUMAN (Q9Y2L1) Exosome complex exonuclease RRP44 (EC 3.1.13... 31 5.1
LEU3_BUCRP (P48572) 3-isopropylmalate dehydrogenase (EC 1.1.1.85... 31 5.1
UBPD_YEAST (P38187) Ubiquitin carboxyl-terminal hydrolase 13 (EC... 30 6.7
ICIA_SALTY (P58509) Chromosome initiation inhibitor (OriC replic... 30 6.7
ICIA_SALTI (P58508) Chromosome initiation inhibitor (OriC replic... 30 6.7
ICIA_ECOLI (P24194) Chromosome initiation inhibitor (OriC replic... 30 8.7
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 102 bits (255), Expect = 1e-21
Identities = 63/210 (30%), Positives = 108/210 (51%), Gaps = 14/210 (6%)
Query: 33 VPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQLLGF 92
+P +F FGDS+ D+GN+NNL T K+N+ YG+DF T R+SNG D +A+ +G
Sbjct: 202 IPAVFFFGDSVFDTGNNNNLETKIKSNYRPYGMDFKFRVATGRFSNGMVASDYLAKYMGV 261
Query: 93 QTFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSCI-- 148
+ +P + + + +D+L GV++ASG AG + S+ + + + QL + + + +
Sbjct: 262 KEIVPAYLDPKIQPNDLLTGVSFASGGAGYNPTT-SEAANAIPMLDQLTYFQDYIEKVNR 320
Query: 149 -------AHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVL 201
+KL GL+K Q +S+ + V +ND YF Y ++
Sbjct: 321 LVRQEKSQYKLAGLEKTNQLISKGVAIVVGGSNDLIITYFGSGAQRLKN--DIDSYTTII 378
Query: 202 IHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
+ ++ LY +GAR+ ++G LGCV
Sbjct: 379 ADSAASFVLQLYGYGARRIGVIGTPPLGCV 408
>APG_BRANA (P40603) Anter-specific proline-rich protein APG (Protein
CEX) (Fragment)
Length = 449
Score = 97.8 bits (242), Expect = 3e-20
Identities = 63/206 (30%), Positives = 99/206 (47%), Gaps = 12/206 (5%)
Query: 33 VPCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQLLGF 92
+P +F FGDS+ D+GN+NNL T K N+ YG+DFP T R+SNGR D I++ LG
Sbjct: 123 IPAVFFFGDSIFDTGNNNNLDTKLKCNYRPYGMDFPMGVATGRFSNGRVASDYISKYLGV 182
Query: 93 QTFIPPFAN--------LNGSDILKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAI 144
+ +P + + L SD+L GV++ASG AG ++ V L L +
Sbjct: 183 KEIVPAYVDKKLQQNNELQQSDLLTGVSFASGGAGYLPQTSE--SWKVTTMLDQLTYFQD 240
Query: 145 VSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQ 204
KL G K + +S+ V +ND YF + + ++
Sbjct: 241 YKKRMKKLVGKKKTKKIVSKGAAIVVAGSNDLIYTYFGNGAQHLKN--DVDSFTTMMADS 298
Query: 205 LSHYLQTLYHFGARKSVLVGLDRLGC 230
+ ++ LY +GAR+ ++G +GC
Sbjct: 299 AASFVLQLYGYGARRIGVIGTPPIGC 324
>EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early
nodule-specific protein homolog) (Latex allergen Hev b
13)
Length = 391
Score = 43.5 bits (101), Expect = 8e-04
Identities = 52/211 (24%), Positives = 83/211 (38%), Gaps = 27/211 (12%)
Query: 34 PCIFVFGDSLSDSGNDNNLPTSAKANFLSYGIDFPATF---PTERYSNGRNPIDKIAQLL 90
P IF FGDS SD+G A F + TF T RYS+GR ID IA+
Sbjct: 33 PAIFNFGDSNSDTG-------GKAAAFYPLNPPYGETFFHRSTGRYSDGRLIIDFIAESF 85
Query: 91 GFQTFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHN----VNLGLQLLHHRAIV- 145
++ P+ + GS+ G ++A+ + I+ + H L +Q R +
Sbjct: 86 NL-PYLSPYLSSLGSNFKHGADFATAGSTIKLPTTIIPAHGGFSPFYLDVQYSQFRQFIP 144
Query: 146 -SCIAHKLGG----LDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKV 200
S + GG L + LY +I ND + + V +
Sbjct: 145 RSQFIRETGGIFAELVPEEYYFEKALYTFDIGQNDLTEGFLNLTVEEVNATVPD------ 198
Query: 201 LIHQLSHYLQTLYHFGARKSVLVGLDRLGCV 231
L++ S ++ +Y GAR + +GC+
Sbjct: 199 LVNSFSANVKKIYDLGARTFWIHNTGPIGCL 229
>YG1W_YEAST (P53230) Hypothetical 44.2 kDa protein in RME1-TFC4
intergenic region
Length = 385
Score = 32.7 bits (73), Expect = 1.3
Identities = 26/97 (26%), Positives = 42/97 (42%), Gaps = 13/97 (13%)
Query: 27 VLGESQVPCIFVFGDSLSDSGNDNNLPTSAKANF-LSYGIDFPATFPT-------ERYSN 78
V+ + PC FVFG + +K + G+ +P+ F + + YS+
Sbjct: 116 VMSSFKAPCRFVFGYGSGVFEQAGYSKSHSKPQIDIILGVTYPSHFHSINMRQNPQHYSS 175
Query: 79 ----GRNPIDKIAQLLGFQTFIPPFANLNGSDILKGV 111
G + K Q+ G + PFAN+NG D+ GV
Sbjct: 176 LKYFGSEFVSKFQQI-GAGVYFNPFANINGHDVKYGV 211
>RR44_HUMAN (Q9Y2L1) Exosome complex exonuclease RRP44 (EC 3.1.13.-)
(Ribosomal RNA processing protein 44) (DIS3 protein
homolog)
Length = 928
Score = 30.8 bits (68), Expect = 5.1
Identities = 16/48 (33%), Positives = 28/48 (58%), Gaps = 1/48 (2%)
Query: 158 ATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYTPKQYAKVLIHQL 205
AT+C+ Q +Y+ + NDF +Y L + T ++YA V++H+L
Sbjct: 714 ATRCMMQAVYFCSGMDNDF-HHYGLASPIYTHFTSPIRRYADVIVHRL 760
>LEU3_BUCRP (P48572) 3-isopropylmalate dehydrogenase (EC 1.1.1.85)
(Beta-IPM dehydrogenase) (IMDH) (3-IPM-DH)
Length = 363
Score = 30.8 bits (68), Expect = 5.1
Identities = 30/93 (32%), Positives = 42/93 (44%), Gaps = 8/93 (8%)
Query: 35 CIFVFGDSLSD-----SGNDNNLPTSAKANFLSYGIDFPATFPTERYSNGRNPIDKIAQL 89
C +FGD +SD +G+ LP SA N +G+ PA G+N + IAQ+
Sbjct: 245 CSNIFGDIISDECAMITGSIGMLP-SASLNSKKFGLYEPAGGSAPDIE-GKNIANPIAQI 302
Query: 90 LGFQTFIPPFANLNG-SDILKGVNYASGSAGIR 121
L I NLN +D + Y + AG R
Sbjct: 303 LSLSMLIRHSMNLNEIADKIDSSVYRALKAGYR 335
>UBPD_YEAST (P38187) Ubiquitin carboxyl-terminal hydrolase 13 (EC
3.1.2.15) (Ubiquitin thiolesterase 13)
(Ubiquitin-specific processing protease 13)
(Deubiquitinating enzyme 13)
Length = 688
Score = 30.4 bits (67), Expect = 6.7
Identities = 36/142 (25%), Positives = 61/142 (42%), Gaps = 20/142 (14%)
Query: 86 IAQLLGFQTFIPPFANLNGSDILKGVNYASGSAGI------RKESGSQLGHNVNLGLQLL 139
+ ++L ++ +N N D LKG + +S S + R S SQ+ L+
Sbjct: 312 VGRVLNYENPSRGSSNSNNLD-LKGESNSSLSTPLDKKDTRRSSSSSQISPEHRKKSALI 370
Query: 140 HHRAIVSCIAHKLGGLDKAT---------QCLSQCLYYVNI-DTNDFEQNYFLPNV-FNT 188
R V I H L G DKAT +C+++ Y + + F NV FNT
Sbjct: 371 --RGPVLNIDHSLNGSDKATLYSSLRDIFECITENTYLTGVVSPSSFVDVLKRENVLFNT 428
Query: 189 SRMYTPKQYAKVLIHQLSHYLQ 210
+ ++ L+++LS Y++
Sbjct: 429 TMHQDAHEFFNFLLNELSEYIE 450
>ICIA_SALTY (P58509) Chromosome initiation inhibitor (OriC
replication inhibitor)
Length = 297
Score = 30.4 bits (67), Expect = 6.7
Identities = 17/61 (27%), Positives = 28/61 (45%), Gaps = 11/61 (18%)
Query: 134 LGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYT 193
+G + H+A+ SC+ KLG LD Y+ + + F + YF V +S +
Sbjct: 141 VGAVSIQHQALPSCLVDKLGALD-----------YLFVASKPFAERYFPNGVTRSSLLKA 189
Query: 194 P 194
P
Sbjct: 190 P 190
>ICIA_SALTI (P58508) Chromosome initiation inhibitor (OriC
replication inhibitor)
Length = 297
Score = 30.4 bits (67), Expect = 6.7
Identities = 17/61 (27%), Positives = 28/61 (45%), Gaps = 11/61 (18%)
Query: 134 LGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYT 193
+G + H+A+ SC+ KLG LD Y+ + + F + YF V +S +
Sbjct: 141 VGAVSIQHQALPSCLVDKLGALD-----------YLFVASKPFAERYFPNGVTRSSLLKA 189
Query: 194 P 194
P
Sbjct: 190 P 190
>ICIA_ECOLI (P24194) Chromosome initiation inhibitor (OriC
replication inhibitor)
Length = 297
Score = 30.0 bits (66), Expect = 8.7
Identities = 16/61 (26%), Positives = 28/61 (45%), Gaps = 11/61 (18%)
Query: 134 LGLQLLHHRAIVSCIAHKLGGLDKATQCLSQCLYYVNIDTNDFEQNYFLPNVFNTSRMYT 193
+G + H+A+ SC+ KLG LD Y+ + + F + YF V ++ +
Sbjct: 141 VGAVSIQHQALPSCLVDKLGALD-----------YLFVSSKPFAEKYFPNGVTRSALLKA 189
Query: 194 P 194
P
Sbjct: 190 P 190
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.336 0.145 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,114,973
Number of Sequences: 164201
Number of extensions: 1435889
Number of successful extensions: 4103
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 4092
Number of HSP's gapped (non-prelim): 10
length of query: 341
length of database: 59,974,054
effective HSP length: 111
effective length of query: 230
effective length of database: 41,747,743
effective search space: 9601980890
effective search space used: 9601980890
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0476b.4


