

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.10
(527 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
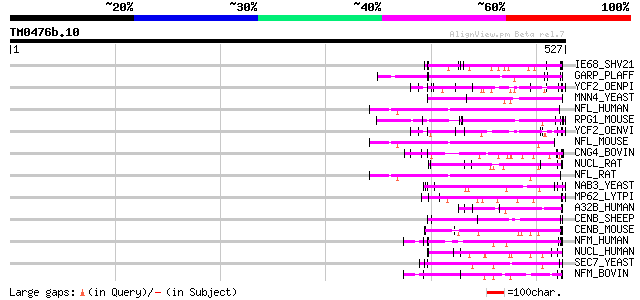
Score E
Sequences producing significant alignments: (bits) Value
IE68_SHV21 (Q01042) Immediate-early protein 87 1e-16
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 82 3e-15
YCF2_OENPI (P31568) Protein ycf2 (Fragment) 78 5e-14
MNN4_YEAST (P36044) MNN4 protein 72 3e-12
NFL_HUMAN (P07196) Neurofilament triplet L protein (68 kDa neuro... 70 1e-11
RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase regulat... 70 2e-11
YCF2_OENVI (P31569) Protein ycf2 (Fragment) 69 4e-11
NFL_MOUSE (P08551) Neurofilament triplet L protein (68 kDa neuro... 67 1e-10
CNG4_BOVIN (Q28181) 240 kDa protein of rod photoreceptor CNG-cha... 66 2e-10
NUCL_RAT (P13383) Nucleolin (Protein C23) 66 3e-10
NFL_RAT (P19527) Neurofilament triplet L protein (68 kDa neurofi... 66 3e-10
NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3 65 3e-10
MP62_LYTPI (P91753) Mitotic apparatus protein p62 65 3e-10
A32B_HUMAN (Q92688) Acidic leucine-rich nuclear phosphoprotein 3... 65 3e-10
CENB_SHEEP (P49451) Major centromere autoantigen B (Centromere p... 65 4e-10
CENB_MOUSE (P27790) Major centromere autoantigen B (Centromere p... 65 4e-10
NFM_HUMAN (P07197) Neurofilament triplet M protein (160 kDa neur... 65 6e-10
NUCL_HUMAN (P19338) Nucleolin (Protein C23) 64 7e-10
SEC7_YEAST (P11075) Protein transport protein SEC7 64 1e-09
NFM_BOVIN (O77788) Neurofilament triplet M protein (160 kDa neur... 64 1e-09
>IE68_SHV21 (Q01042) Immediate-early protein
Length = 407
Score = 87.0 bits (214), Expect = 1e-16
Identities = 48/131 (36%), Positives = 72/131 (54%), Gaps = 2/131 (1%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+EA E EE ++EE E + + E +EA E E +E EE E+E+A ++ E
Sbjct: 109 EEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEEAE 168
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATD--HEEVESEKESNKDEEAT 514
+E+EEA + EA E+ E E+ E + EEEA E EE + EE E +E+ + EEA
Sbjct: 169 EEAEEAEEAEEEAEEEAEEAEEAEEAEEAEEEAEEAEEEAEEAEEEAEEAEEAEEAEEAE 228
Query: 515 EDEEEESSEDD 525
E+ EE E++
Sbjct: 229 EEAEEAEEEEE 239
Score = 84.0 bits (206), Expect = 9e-16
Identities = 53/135 (39%), Positives = 73/135 (53%), Gaps = 5/135 (3%)
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEV---EAAEDEEEETTEDEDA 451
G +EA E E K+ EEE + + + E EA EE E AE+EE E E+E+A
Sbjct: 92 GREEAEEEEAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEA 151
Query: 452 SDDKEDESEEASTEDGE--ATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNK 509
+ +E+ EE + E+ E A E EEA E+ E +E EEA E EEA + E E+
Sbjct: 152 EEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEEEAEEAEEEAEEA 211
Query: 510 DEEATEDEEEESSED 524
+EEA E EE E +E+
Sbjct: 212 EEEAEEAEEAEEAEE 226
Score = 81.3 bits (199), Expect = 6e-15
Identities = 51/132 (38%), Positives = 71/132 (53%), Gaps = 5/132 (3%)
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDD 454
G +E EEA EEEEA ++ E + EE E AE EEEE E+E ++
Sbjct: 86 GGEEGEGREEA---EEEEAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEE 142
Query: 455 KEDESEEASTEDGEATEDEEATEDVEATVDEEEEATE--VEEATDHEEVESEKESNKDEE 512
E+ EE + E E E+EEA E+ E +E EEA E EEA + EE E +E+ ++ E
Sbjct: 143 AEEAEEEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEEEAE 202
Query: 513 ATEDEEEESSED 524
E+E EE+ E+
Sbjct: 203 EAEEEAEEAEEE 214
Score = 77.0 bits (188), Expect = 1e-13
Identities = 52/134 (38%), Positives = 68/134 (49%), Gaps = 6/134 (4%)
Query: 397 KEANEVEEATKDEEEEASG----LSLHSVKSLTCEFKEATEE-VEAAEDEEEETTEDEDA 451
+E E E +EE E G + E KEA EE E AE+E EE +E
Sbjct: 68 EEEGEERERRGEEEREGEGGEEGEGREEAEEEEAEEKEAEEEEAEEAEEEAEEEEAEEAE 127
Query: 452 SDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDE 511
++++E E EEA E+ E E+EEA E E +EE E EEA + EE E E E E
Sbjct: 128 AEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEA-E 186
Query: 512 EATEDEEEESSEDD 525
EA E EE E +E++
Sbjct: 187 EAEEAEEAEEAEEE 200
Score = 66.2 bits (160), Expect = 2e-10
Identities = 39/99 (39%), Positives = 53/99 (53%), Gaps = 2/99 (2%)
Query: 427 EFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEE 486
E +E EE E +EE E E+ +E E EEA ++ E E EEA E EA +E
Sbjct: 66 EVEEEGEERERRGEEEREGEGGEEGEGREEAEEEEAEEKEAEEEEAEEAEE--EAEEEEA 123
Query: 487 EEATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
EEA EE + EE E E+ +EE E+ EEE+ E++
Sbjct: 124 EEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEE 162
Score = 65.5 bits (158), Expect = 3e-10
Identities = 38/100 (38%), Positives = 59/100 (59%), Gaps = 6/100 (6%)
Query: 432 TEEVEAAEDEEEETTEDEDASDDKE----DESEEASTEDGEATED--EEATEDVEATVDE 485
T + +AA EE+ E E+ +++E +E E E+GE E+ EE E+ EA +E
Sbjct: 51 THQQQAALTEEQRREEVEEEGEERERRGEEEREGEGGEEGEGREEAEEEEAEEKEAEEEE 110
Query: 486 EEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
EEA E E + EE E+E+E ++EEA E+E EE+ E++
Sbjct: 111 AEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEE 150
Score = 62.4 bits (150), Expect = 3e-09
Identities = 45/116 (38%), Positives = 62/116 (52%), Gaps = 5/116 (4%)
Query: 398 EANEVEEATKD-EEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
E E EEA ++ EEEEA + + +EA EE E AE E EE E E+ +++ E
Sbjct: 147 EEEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAE-EAEEAEEAEEEAEEAE 205
Query: 457 DESEEA---STEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNK 509
+E+EEA + E EA E EEA E+ E +EEEEA H +V +K S +
Sbjct: 206 EEAEEAEEEAEEAEEAEEAEEAEEEAEEAEEEEEEAGPSTPRLPHYKVVGQKPSTQ 261
Score = 60.5 bits (145), Expect = 1e-08
Identities = 37/95 (38%), Positives = 54/95 (55%), Gaps = 2/95 (2%)
Query: 429 KEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEE 488
++ EEVE E EE E +E+ + +E E + E E++EA E+ +EE E
Sbjct: 61 EQRREEVEE-EGEERERRGEEEREGEGGEEGEGREEAEEEEAEEKEAEEEEAEEAEEEAE 119
Query: 489 ATEVEEATDHEEVESEKESNKDEEATEDEEEESSE 523
E EEA + EE E+E+E ++EEA E EEEE+ E
Sbjct: 120 EEEAEEA-EAEEEEAEEEEAEEEEAEEAEEEEAEE 153
Score = 34.3 bits (77), Expect = 0.82
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 4/61 (6%)
Query: 468 EATEDEEATEDVEATVDEEEEATEVEEATDHE----EVESEKESNKDEEATEDEEEESSE 523
+ +D T +A + EE+ EVEE + E E E E ++ E E+ EEE +E
Sbjct: 43 QGDDDINTTHQQQAALTEEQRREEVEEEGEERERRGEEEREGEGGEEGEGREEAEEEEAE 102
Query: 524 D 524
+
Sbjct: 103 E 103
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 82.4 bits (202), Expect = 3e-15
Identities = 38/128 (29%), Positives = 74/128 (57%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKED 457
E + + K+E+++ + E +E EE E E+EEEE E+E+ +++E+
Sbjct: 538 ELQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEE 597
Query: 458 ESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDE 517
E EE ED + ++++A ED + ++E++A E ++ D +E + +++ ++DEE E+E
Sbjct: 598 EEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEEEE 657
Query: 518 EEESSEDD 525
EEE E +
Sbjct: 658 EEEEEESE 665
Score = 81.3 bits (199), Expect = 6e-15
Identities = 48/129 (37%), Positives = 74/129 (57%), Gaps = 2/129 (1%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKED 457
E E+++ ++EE V+ + E +E EEVE E+EEEE E+E+ +++E+
Sbjct: 535 EEAELQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEE 594
Query: 458 ESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDE-EATED 516
E EE E+ E EDEE +D E D+ EE + E D EE + E++ ++DE E ED
Sbjct: 595 EEEEEEEEE-EEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEED 653
Query: 517 EEEESSEDD 525
EEEE E++
Sbjct: 654 EEEEEEEEE 662
Score = 77.4 bits (189), Expect = 8e-14
Identities = 44/127 (34%), Positives = 71/127 (55%), Gaps = 13/127 (10%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E+ EV+E +K+ +E+ E +E EE E E+EEEE E+E+ +++E
Sbjct: 553 EESKEVQEESKEVQEDEE------------EVEEDEEEEEEEEEEEEEEEEEEEEEEEEE 600
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATED 516
+E EE E+ E + EE +D E D+ EE + E+ + ++ E E E +DEE E+
Sbjct: 601 EEEEEDEDEEDE-DDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEEEEEE 659
Query: 517 EEEESSE 523
EEEES +
Sbjct: 660 EEEESEK 666
Score = 72.8 bits (177), Expect = 2e-12
Identities = 51/162 (31%), Positives = 82/162 (50%), Gaps = 17/162 (10%)
Query: 350 NVRKDLAMYRESEYKNIQIYFEMYRESLLSFPSEQLHLRVTSLPHGFKEANEVEEATKDE 409
N +K +A E+E +Q + +E S+++ + +E E EE ++E
Sbjct: 526 NHKKKMAKIEEAE---LQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEE 582
Query: 410 EEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEA 469
EEE E +E EE E E+EEEE EDE+ DD E++ ++A ++ +A
Sbjct: 583 EEEE-------------EEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDA 629
Query: 470 TEDEEATEDVEATVDE-EEEATEVEEATDHEEVESEKESNKD 510
ED++ +D E DE E+E E EE + EE ESEK+ ++
Sbjct: 630 EEDDDEEDDDEEDDDEDEDEDEEDEEEEEEEEEESEKKIKRN 671
Score = 66.6 bits (161), Expect = 1e-10
Identities = 42/134 (31%), Positives = 72/134 (53%), Gaps = 9/134 (6%)
Query: 398 EANEVEEATKDEEEEASG-LSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+ ++VEE +E+ G + +V K+ ++E AE ++++ + E+ DK+
Sbjct: 496 DKSKVEEKNLSIQEQLIGTIGRVNVVPRRDNHKKKMAKIEEAELQKQKHVDKEE---DKK 552
Query: 457 DESEEASTEDGEATEDEEATED-----VEATVDEEEEATEVEEATDHEEVESEKESNKDE 511
+ES+E E E EDEE E+ E +EEEE E EE + EE E E E +DE
Sbjct: 553 EESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDE 612
Query: 512 EATEDEEEESSEDD 525
+ E++E+++ ED+
Sbjct: 613 DDAEEDEDDAEEDE 626
Score = 64.7 bits (156), Expect = 6e-10
Identities = 37/113 (32%), Positives = 59/113 (51%), Gaps = 1/113 (0%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTED-EDASDDK 455
KE E EE +++EEE + E +E EE E EDE+EE +D E+ DD
Sbjct: 563 KEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDA 622
Query: 456 EDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESN 508
E++ ++A +D E +DEE ++ E +E+EE E EE ++++ N
Sbjct: 623 EEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDEEEEEEEEEESEKKIKRNLRKN 675
Score = 43.5 bits (101), Expect = 0.001
Identities = 34/135 (25%), Positives = 65/135 (47%), Gaps = 8/135 (5%)
Query: 397 KEANEVEEAT-KDEEEEASGLSLHSVKSLTCEFKEATEEVEAAE--DEEEETTEDEDASD 453
KE ++E+ K EE+E K E K+ +E++ + ++E + E+++
Sbjct: 273 KEQEKIEKKKKKQEEKEKKKQEKERKKQEKKERKQKEKEMKKQKKIEKERKKKEEKEKKK 332
Query: 454 DKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEA 513
K D+ E + + + T +E + E V T+V +H+E E ++E +K+ E
Sbjct: 333 KKHDKENEETMQQPDQTSEE---TNNEIMVPLPSPLTDVTTPEEHKEGEHKEEEHKEGEH 389
Query: 514 TEDE--EEESSEDDY 526
E E EEE E+++
Sbjct: 390 KEGEHKEEEHKEEEH 404
Score = 40.0 bits (92), Expect = 0.015
Identities = 40/186 (21%), Positives = 80/186 (42%), Gaps = 11/186 (5%)
Query: 342 HKYGQEIKNVRKDLAMYRESEYKNIQIYFEMYRESLLSFPSEQLHLRVTSLPHGFKEANE 401
HK ++ K +KD ++ + K + + ++ SE + L T H K A E
Sbjct: 128 HKKDKKEKKEKKDKKEKKDKKEKKHKKEKKHKKDKKKKENSEVMSLYKTG-QHKPKNATE 186
Query: 402 VEEATKDEE-------EEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDD 454
E DEE GL L S ++ + ++ E T+D D +
Sbjct: 187 HGEENLDEEMVSEINNNAQGGLLLSSPYQYR---EQGGCGIISSVHETSNDTKDNDKENI 243
Query: 455 KEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEAT 514
ED+ E+ E+ T D++ + E + E+E+ + ++ + +E + +++ K +E
Sbjct: 244 SEDKKEDHQQEEMLKTLDKKERKQKEKEMKEQEKIEKKKKKQEEKEKKKQEKERKKQEKK 303
Query: 515 EDEEEE 520
E +++E
Sbjct: 304 ERKQKE 309
Score = 39.7 bits (91), Expect = 0.019
Identities = 30/134 (22%), Positives = 55/134 (40%), Gaps = 5/134 (3%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
KE + E+ K +E++ +K KE ++ E + +++ E+E+ +
Sbjct: 288 KEKKKQEKERKKQEKKERKQKEKEMKKQKKIEKERKKKEEKEKKKKKHDKENEETMQQPD 347
Query: 457 DESEEASTE-----DGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDE 511
SEE + E T+ E E EEE + +H+E E ++E +K E
Sbjct: 348 QTSEETNNEIMVPLPSPLTDVTTPEEHKEGEHKEEEHKEGEHKEGEHKEEEHKEEEHKKE 407
Query: 512 EATEDEEEESSEDD 525
E E + + D
Sbjct: 408 EHKSKEHKSKGKKD 421
Score = 39.7 bits (91), Expect = 0.019
Identities = 17/75 (22%), Positives = 44/75 (58%), Gaps = 3/75 (4%)
Query: 450 DASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNK 509
+ S+D +D +E +ED +++ E++ T+D++E + +E + E++E +K+ +
Sbjct: 230 ETSNDTKDNDKENISED---KKEDHQQEEMLKTLDKKERKQKEKEMKEQEKIEKKKKKQE 286
Query: 510 DEEATEDEEEESSED 524
++E + E+E ++
Sbjct: 287 EKEKKKQEKERKKQE 301
Score = 36.6 bits (83), Expect = 0.16
Identities = 24/125 (19%), Positives = 65/125 (51%), Gaps = 10/125 (8%)
Query: 400 NEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDES 459
N+ E ++D++E+ + +K+L K+ ++ E E+E+ + + ++KE +
Sbjct: 238 NDKENISEDKKEDHQQEEM--LKTLD---KKERKQKEKEMKEQEKIEKKKKKQEEKEKKK 292
Query: 460 EEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEE 519
+E + E E ++ +++ ++++ E E E+ + +K+ +K+ E T + +
Sbjct: 293 QEKERKKQEKKERKQKEKEM-----KKQKKIEKERKKKEEKEKKKKKHDKENEETMQQPD 347
Query: 520 ESSED 524
++SE+
Sbjct: 348 QTSEE 352
Score = 36.2 bits (82), Expect = 0.22
Identities = 28/133 (21%), Positives = 57/133 (42%), Gaps = 20/133 (15%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEET----------- 445
K+ ++E+ K +EE+ H KE E ++ + EET
Sbjct: 313 KKQKKIEKERKKKEEKEKKKKKHD--------KENEETMQQPDQTSEETNNEIMVPLPSP 364
Query: 446 -TEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESE 504
T+ + KE E +E ++GE E E E+ + ++EE E + ++ + +
Sbjct: 365 LTDVTTPEEHKEGEHKEEEHKEGEHKEGEHKEEEHKEEEHKKEEHKSKEHKSKGKKDKGK 424
Query: 505 KESNKDEEATEDE 517
K+ K ++A +++
Sbjct: 425 KDKGKHKKAKKEK 437
Score = 34.7 bits (78), Expect = 0.63
Identities = 23/115 (20%), Positives = 54/115 (46%), Gaps = 9/115 (7%)
Query: 420 SVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDV 479
S S+T TE + +D + T ++ D+K+D ++ A + E +
Sbjct: 41 SFDSITGRLLNETELEKNKDDNSKSETLLKEEKDEKDDVPTTSNDNLKNAHNNNEISSST 100
Query: 480 EAT----VDEEEEATEVEEATD-----HEEVESEKESNKDEEATEDEEEESSEDD 525
+ T V++++ V++ D H++ + EK+ KD++ +D++E+ + +
Sbjct: 101 DPTNIINVNDKDNENSVDKKKDKKEKKHKKDKKEKKEKKDKKEKKDKKEKKHKKE 155
Score = 34.7 bits (78), Expect = 0.63
Identities = 21/110 (19%), Positives = 52/110 (47%), Gaps = 1/110 (0%)
Query: 417 SLHSVKSLTCEFKEATEEVEAAED-EEEETTEDEDASDDKEDESEEASTEDGEATEDEEA 475
S+H + T + + + ED ++EE + D + K+ E E E E + ++
Sbjct: 227 SVHETSNDTKDNDKENISEDKKEDHQQEEMLKTLDKKERKQKEKEMKEQEKIEKKKKKQE 286
Query: 476 TEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
++ + E ++ + E +E++ +K+ K+ + E++E++ + D
Sbjct: 287 EKEKKKQEKERKKQEKKERKQKEKEMKKQKKIEKERKKKEEKEKKKKKHD 336
Score = 33.1 bits (74), Expect = 1.8
Identities = 38/166 (22%), Positives = 67/166 (39%), Gaps = 18/166 (10%)
Query: 340 EHHKYGQEIKNVRKDLAMYRESEYKNIQIYFEMYRESLLSFPSEQLHLRVTSLPHGFKEA 399
E K ++ K +K E+ + Q E E ++ PS L + P KE
Sbjct: 321 ERKKKEEKEKKKKKHDKENEETMQQPDQTSEETNNEIMVPLPSP---LTDVTTPEEHKEG 377
Query: 400 NEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDES 459
EE K+ E + E KE EE + E ++EE E S K+D+
Sbjct: 378 EHKEEEHKEGEHKEG------------EHKE--EEHKEEEHKKEEHKSKEHKSKGKKDKG 423
Query: 460 EEASTEDGEATEDEEATEDVEATV-DEEEEATEVEEATDHEEVESE 504
++ + +A +++ V+ + DE+++ E+ D E E +
Sbjct: 424 KKDKGKHKKAKKEKVKKHVVKNVIEDEDKDGVEIINLEDKEACEEQ 469
>YCF2_OENPI (P31568) Protein ycf2 (Fragment)
Length = 721
Score = 78.2 bits (191), Expect = 5e-14
Identities = 53/139 (38%), Positives = 70/139 (50%), Gaps = 10/139 (7%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTED-----EDA 451
+E EE + EEE G V+ E + EEVE EDEE E TE+ E+
Sbjct: 271 EEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEE 330
Query: 452 SDDKEDESEEASTEDGEATEDE-EATEDVEATVDEEEEATEVEEATDHEEVESEKE---- 506
+ EDE E + E+ E TE+E E TE+ +EE E TE E EEVE +E
Sbjct: 331 VEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEG 390
Query: 507 SNKDEEATEDEEEESSEDD 525
+ ++ E TEDEE E +E D
Sbjct: 391 TEEEVEGTEDEEVEGTEKD 409
Score = 77.8 bits (190), Expect = 6e-14
Identities = 47/133 (35%), Positives = 68/133 (50%), Gaps = 4/133 (3%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E EE + EEE G V+ E + EEVE E+E E T E+ + ++D+E
Sbjct: 191 EEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEE 250
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKE----SNKDEE 512
E E +G E E E+VE T +E E E E T+ EEVE +E + ++ E
Sbjct: 251 VEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVE 310
Query: 513 ATEDEEEESSEDD 525
TEDEE E +E++
Sbjct: 311 GTEDEEVEGTEEE 323
Score = 76.3 bits (186), Expect = 2e-13
Identities = 57/142 (40%), Positives = 75/142 (52%), Gaps = 18/142 (12%)
Query: 401 EVEEATKDE-----EEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDK 455
E E T+DE EEE G V+ E + EEVE EDEE E TEDE+ +
Sbjct: 205 EEVEGTEDEEVEGTEEEVEGTE-EEVEGTEEEVEGTEEEVEGTEDEEVEGTEDEEVEGTE 263
Query: 456 E-----DESEEASTEDGEATEDE-EATED--VEATVDEEEEATEVEEATDHEEVESEKE- 506
E +E E + E+ E TE+E E TED VE T +E E E E T+ EEVE +E
Sbjct: 264 EEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEE 323
Query: 507 ---SNKDEEATEDEEEESSEDD 525
+ ++ E TEDEE E +E++
Sbjct: 324 VEGTEEEVEGTEDEEVEGTEEE 345
Score = 73.2 bits (178), Expect = 2e-12
Identities = 50/140 (35%), Positives = 70/140 (49%), Gaps = 18/140 (12%)
Query: 403 EEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE----DE 458
EE E+EE G V+ E + EEVE E+E E T E+ + ++D+E +E
Sbjct: 241 EEVEGTEDEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEE 300
Query: 459 SEEASTEDGEATEDEE----------ATEDVEATVDEEEEATEVEEATDHEEVESEKE-- 506
E + E+ E TEDEE E+VE T DEE E TE E EEVE +E
Sbjct: 301 EVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEV 360
Query: 507 --SNKDEEATEDEEEESSED 524
+ ++ E TE+E E + E+
Sbjct: 361 EGTEEEVEGTEEEVEGTEEE 380
Score = 72.0 bits (175), Expect = 4e-12
Identities = 55/149 (36%), Positives = 73/149 (48%), Gaps = 21/149 (14%)
Query: 389 VTSLPHGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTED 448
++ + HG E + EEE G T E E TEE +EE E TED
Sbjct: 162 LSDIVHGLLELEGALVGSSPTEEEVEG---------TEEEVEGTEEEVEGTEEEVEGTED 212
Query: 449 EDASDDKE-----DESEEASTEDGEATEDE-EATED--VEATVDEEEEATEVEEATDHEE 500
E+ +E +E E + E+ E TE+E E TED VE T DEE E TE E EE
Sbjct: 213 EEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEDEEVEGTEEEVEGTEEE 272
Query: 501 VESEKE----SNKDEEATEDEEEESSEDD 525
VE +E + ++ E TEDEE E +E++
Sbjct: 273 VEGTEEEVEGTEEEVEGTEDEEVEGTEEE 301
Score = 70.1 bits (170), Expect = 1e-11
Identities = 46/128 (35%), Positives = 62/128 (47%), Gaps = 2/128 (1%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E EE + EEE G V+ E E TEE +EE E TE+E ++E
Sbjct: 227 EEVEGTEEEVEGTEEEVEGTEDEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEE 286
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATED 516
E E +G E E E+VE T DEE E TE E EEVE ++ ++ E TE+
Sbjct: 287 VEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTED--EEVEGTEE 344
Query: 517 EEEESSED 524
E E + E+
Sbjct: 345 EVEGTEEE 352
Score = 66.6 bits (161), Expect = 1e-10
Identities = 47/120 (39%), Positives = 65/120 (54%), Gaps = 7/120 (5%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E EE + EEE G V+ E + EEVE EDEE E TE+E ++E
Sbjct: 293 EEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEE 352
Query: 457 ----DESEEASTEDGEATEDE-EATED-VEATVDEEEEATEVEEATDHEEVE-SEKESNK 509
+E E + E+ E TE+E E TE+ VE T +E E E E T+ EEVE +EK+S++
Sbjct: 353 VEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEDEEVEGTEKDSSQ 412
Score = 65.1 bits (157), Expect = 4e-10
Identities = 40/108 (37%), Positives = 56/108 (51%), Gaps = 10/108 (9%)
Query: 424 LTCEFKEATEEVEAAEDEEEETT----------EDEDASDDKEDESEEASTEDGEATEDE 473
L CE + EE+ EDE E E+E +DD+ED EA ED E++
Sbjct: 484 LDCEAEIPAEEIPEEEDELPEDALETEVAVWGVEEEGEADDEEDVLLEAQQEDELLEEED 543
Query: 474 EATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEES 521
E ++ E +DEEEE + EE HEE E E+E ++EE E +E +S
Sbjct: 544 EELDEEEDELDEEEEEPKEEEDELHEEEEEEEEEEEEEEEDELQENDS 591
Score = 53.9 bits (128), Expect = 1e-06
Identities = 39/88 (44%), Positives = 50/88 (56%), Gaps = 11/88 (12%)
Query: 440 DEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHE 499
+EE E TE+E ++E E E E+ E TEDEE VE T +EE E TE E E
Sbjct: 183 EEEVEGTEEEVEGTEEEVEGTE---EEVEGTEDEE----VEGT-EEEVEGTEEEVEGTEE 234
Query: 500 EVESEKESNKDEEATEDEEEESSEDDYI 527
EVE +E + E TEDEE E +ED+ +
Sbjct: 235 EVEGTEE---EVEGTEDEEVEGTEDEEV 259
Score = 51.6 bits (122), Expect = 5e-06
Identities = 40/114 (35%), Positives = 58/114 (50%), Gaps = 16/114 (14%)
Query: 381 PSEQLHLRVTSLPHGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAED 440
P+E++ LP ++A E E A EEE V EA +E E E+
Sbjct: 491 PAEEIPEEEDELP---EDALETEVAVWGVEEEGEADDEEDV------LLEAQQEDELLEE 541
Query: 441 EEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEE 494
E+EE E+ED D++E+E +E E+ E E+EE E+ E EEEE E++E
Sbjct: 542 EDEELDEEEDELDEEEEEPKE---EEDELHEEEEEEEEEE----EEEEEDELQE 588
Score = 42.7 bits (99), Expect = 0.002
Identities = 40/142 (28%), Positives = 60/142 (42%), Gaps = 23/142 (16%)
Query: 401 EVEEATKDEE-----EEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDK 455
E E T+DEE EE G V+ E + EEVE E+E E T E+ + +
Sbjct: 329 EEVEGTEDEEVEGTEEEVEGTE-EEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGT--- 384
Query: 456 EDESEEASTEDGEATEDE--EATEDVEATVDEEEEATEVEEAT-----------DHEEVE 502
+E E + E+ E TEDE E TE + D + + H++ E
Sbjct: 385 -EEEVEGTEEEVEGTEDEEVEGTEKDSSQFDNDRVTLLLRPKPRNPLDIQRLIYQHQKYE 443
Query: 503 SEKESNKDEEATEDEEEESSED 524
SE E + D++ ++ ED
Sbjct: 444 SELEEDDDDDEDVFAPQKMLED 465
Score = 37.7 bits (86), Expect = 0.074
Identities = 48/208 (23%), Positives = 72/208 (34%), Gaps = 80/208 (38%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDK- 455
+E EE + EEE G V+ E + EEVE EDEE E TE + + D
Sbjct: 358 EEVEGTEEEVEGTEEEVEGTE-EEVEGTEEEVEGTEEEVEGTEDEEVEGTEKDSSQFDND 416
Query: 456 --------------------------EDESEEASTEDGEATEDEEATEDV---------- 479
E E EE +D + ++ ED+
Sbjct: 417 RVTLLLRPKPRNPLDIQRLIYQHQKYESELEEDDDDDEDVFAPQKMLEDLFSELVWSPRI 476
Query: 480 ----------EATVDEE----------EEATEVE----------EATDHEEV-------- 501
EA + E E+A E E EA D E+V
Sbjct: 477 WHPWDFLLDCEAEIPAEEIPEEEDELPEDALETEVAVWGVEEEGEADDEEDVLLEAQQED 536
Query: 502 ----ESEKESNKDEEATEDEEEESSEDD 525
E ++E +++E+ ++EEEE E++
Sbjct: 537 ELLEEEDEELDEEEDELDEEEEEPKEEE 564
Score = 36.6 bits (83), Expect = 0.16
Identities = 28/115 (24%), Positives = 49/115 (42%), Gaps = 2/115 (1%)
Query: 346 QEIKNVRKDLAMYRESEYKNIQIYFEMYRESLLSFPSEQLHLRVTSLPHGFKEANEVEEA 405
+E++ +++ + E + + E E + E++ + +E EE
Sbjct: 300 EEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEE 359
Query: 406 TKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESE 460
+ EEE G V+ E + EEVE E EE E TEDE+ ++D S+
Sbjct: 360 VEGTEEEVEGTE-EEVEGTEEEVEGTEEEVEGTE-EEVEGTEDEEVEGTEKDSSQ 412
>MNN4_YEAST (P36044) MNN4 protein
Length = 1178
Score = 72.4 bits (176), Expect = 3e-12
Identities = 46/133 (34%), Positives = 67/133 (49%), Gaps = 4/133 (3%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
K+ E EE K EEEE K E K+ EE E + EEEE + E+ K+
Sbjct: 1042 KKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKQEEEEKKKK 1101
Query: 457 DESEEASTEDGE--ATEDEE--ATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEE 512
+E E+ E+GE EDEE ED E +EEEE + EE E E +K+ ++E+
Sbjct: 1102 EEEEKKKQEEGEKMKNEDEENKKNEDEEKKKNEEEEKKKQEEKNKKNEDEEKKKQEEEEK 1161
Query: 513 ATEDEEEESSEDD 525
+EEE+ +++
Sbjct: 1162 KKNEEEEKKKQEE 1174
Score = 48.5 bits (114), Expect = 4e-05
Identities = 28/93 (30%), Positives = 51/93 (54%), Gaps = 6/93 (6%)
Query: 434 EVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEA-TEV 492
E + E+EE++ E+E+ +E+E ++ E+ + E+EE + +EEE+ E
Sbjct: 1041 EKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKK-----EEEEKKKQEE 1095
Query: 493 EEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
EE EE E +K+ ++ EDEE + +ED+
Sbjct: 1096 EEKKKKEEEEKKKQEEGEKMKNEDEENKKNEDE 1128
Score = 43.9 bits (102), Expect = 0.001
Identities = 24/91 (26%), Positives = 48/91 (52%), Gaps = 6/91 (6%)
Query: 435 VEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEE 494
+E + E++ E+E+ +E+E ++ E+ + E+EE + EEEE + EE
Sbjct: 1034 LEERKRREKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKK------EEEEKKKKEE 1087
Query: 495 ATDHEEVESEKESNKDEEATEDEEEESSEDD 525
++ E EK+ ++EE + EE E +++
Sbjct: 1088 EEKKKQEEEEKKKKEEEEKKKQEEGEKMKNE 1118
Score = 38.5 bits (88), Expect = 0.043
Identities = 22/77 (28%), Positives = 39/77 (50%)
Query: 449 EDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESN 508
ED + K E + + + E+++ E+ E EEEE + EE ++ E EK+
Sbjct: 1026 EDYAYAKLLEERKRREKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKK 1085
Query: 509 KDEEATEDEEEESSEDD 525
++EE + EEEE + +
Sbjct: 1086 EEEEKKKQEEEEKKKKE 1102
Score = 37.0 bits (84), Expect = 0.13
Identities = 19/78 (24%), Positives = 42/78 (53%)
Query: 448 DEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKES 507
D+D ED + E+ + E ++ E+ + +EEE+ + EE +E E +K+
Sbjct: 1018 DKDPIIVYEDYAYAKLLEERKRREKKKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKK 1077
Query: 508 NKDEEATEDEEEESSEDD 525
++E+ ++EEE+ +++
Sbjct: 1078 EEEEKKKKEEEEKKKQEE 1095
Score = 36.2 bits (82), Expect = 0.22
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 17/80 (21%)
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDD 454
G K NE EE K+E+EE K+ EE E + +EE+ ++ED
Sbjct: 1112 GEKMKNEDEENKKNEDEE----------------KKKNEE-EEKKKQEEKNKKNEDEEKK 1154
Query: 455 KEDESEEASTEDGEATEDEE 474
K++E E+ E+ E + EE
Sbjct: 1155 KQEEEEKKKNEEEEKKKQEE 1174
>NFL_HUMAN (P07196) Neurofilament triplet L protein (68 kDa
neurofilament protein) (Neurofilament light polypeptide)
(NF-L)
Length = 543
Score = 70.1 bits (170), Expect = 1e-11
Identities = 52/189 (27%), Positives = 91/189 (47%), Gaps = 10/189 (5%)
Query: 342 HKYGQEIKNVRKDLAMYRESEYKNI-------QIYFEMYRESLLSFPSEQLHLRVTSLPH 394
+K E++ + ++A Y + EY+++ I YR+ L + V S+
Sbjct: 352 NKLENELRTTKSEMARYLK-EYQDLLNVKMALDIEIAAYRKLLEGEETRLSFTSVGSITS 410
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDD 454
G+ ++++V + + S L S +S + +E + +E E ++ E+A D+
Sbjct: 411 GYSQSSQVFGRSAYGGLQTSSY-LMSTRSFPSYYTSHVQEEQTEVEETIEASKAEEAKDE 469
Query: 455 KEDESE-EASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEA 513
E E E +D E E+EEA E+ EA +E EEA E EE + EE E KE+ ++E+
Sbjct: 470 PPSEGEAEEEEKDKEEAEEEEAAEEEEAAKEESEEAKEEEEGGEGEEGEETKEAEEEEKK 529
Query: 514 TEDEEEESS 522
E EE +
Sbjct: 530 VEGAGEEQA 538
>RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase
regulator-interacting protein 1 (RPGR-interacting
protein 1)
Length = 1331
Score = 69.7 bits (169), Expect = 2e-11
Identities = 38/95 (40%), Positives = 55/95 (57%), Gaps = 3/95 (3%)
Query: 431 ATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEAT 490
A E + +E E+ E E E+ ++E + EE E+ E E+EE E+ E +EEEE
Sbjct: 895 APEGFQMSEAEKPEGEEKEEEGGEEEVKEEEVEEEEEEEEEEEEVKEEKE---EEEEEER 951
Query: 491 EVEEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
E EE + E+ E E+E K+EE E+EEEE E+D
Sbjct: 952 EEEEEKEEEKEEEEEEDEKEEEEEEEEEEEEEEED 986
Score = 62.8 bits (151), Expect = 2e-09
Identities = 35/94 (37%), Positives = 52/94 (55%)
Query: 430 EATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEA 489
E + EA + E EE E+ + KE+E EE E+ E E +E E+ E EEEE
Sbjct: 897 EGFQMSEAEKPEGEEKEEEGGEEEVKEEEVEEEEEEEEEEEEVKEEKEEEEEEEREEEEE 956
Query: 490 TEVEEATDHEEVESEKESNKDEEATEDEEEESSE 523
E E+ + EE E E+E ++EE E+EE+E+ +
Sbjct: 957 KEEEKEEEEEEDEKEEEEEEEEEEEEEEEDENKD 990
Score = 62.8 bits (151), Expect = 2e-09
Identities = 45/126 (35%), Positives = 63/126 (49%), Gaps = 22/126 (17%)
Query: 393 PHGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDAS 452
P GF + +E E+ +E+EE G E KE E E+EEEE E+E+
Sbjct: 896 PEGF-QMSEAEKPEGEEKEEEGGEE---------EVKE-----EEVEEEEEEEEEEEEVK 940
Query: 453 DDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEE 512
++KE+E EE E+ E E++E E EE+E E EE + EE E E E+ E
Sbjct: 941 EEKEEEEEEEREEEEEKEEEKEEEE-------EEDEKEEEEEEEEEEEEEEEDENKDVLE 993
Query: 513 ATEDEE 518
A+ EE
Sbjct: 994 ASFTEE 999
Score = 62.0 bits (149), Expect = 4e-09
Identities = 34/98 (34%), Positives = 51/98 (51%), Gaps = 10/98 (10%)
Query: 430 EATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEA 489
E E E E+ EE ++E+ +++E+E EE ++ + E+EE E EE
Sbjct: 905 EKPEGEEKEEEGGEEEVKEEEVEEEEEEEEEEEEVKEEKEEEEEE----------EREEE 954
Query: 490 TEVEEATDHEEVESEKESNKDEEATEDEEEESSEDDYI 527
E EE + EE E EKE ++EE E+EEEE D +
Sbjct: 955 EEKEEEKEEEEEEDEKEEEEEEEEEEEEEEEDENKDVL 992
Score = 60.8 bits (146), Expect = 8e-09
Identities = 33/99 (33%), Positives = 56/99 (56%), Gaps = 1/99 (1%)
Query: 428 FKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEE 487
F+ + E E++EEE E+E ++ E+E EE E+ E E++E E+ E +EE+
Sbjct: 899 FQMSEAEKPEGEEKEEEGGEEEVKEEEVEEEEEEEEEEE-EVKEEKEEEEEEEREEEEEK 957
Query: 488 EATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDDY 526
E + EE + E+ E E+E ++EE EDE ++ E +
Sbjct: 958 EEEKEEEEEEDEKEEEEEEEEEEEEEEEDENKDVLEASF 996
Score = 54.7 bits (130), Expect = 6e-07
Identities = 50/183 (27%), Positives = 78/183 (42%), Gaps = 8/183 (4%)
Query: 349 KNVRKDLAMYRESEYKNIQIYFEMYRESLLSFPSEQLHLRVTSLPHGF-KEANEVEEATK 407
K+++ D + E N I ++ +S P E + P G KE EE K
Sbjct: 864 KSIKGDFNLTDSGEKSNGSIKVQLDWKSHYLAP-EGFQMSEAEKPEGEEKEEEGGEEEVK 922
Query: 408 DEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDG 467
+EE E + + E +E EE E+E+EE E+E+ D+KE+E EE E+
Sbjct: 923 EEEVEEEEEEEEEEEEVKEEKEEEEEEEREEEEEKEEEKEEEEEEDEKEEEEEEEEEEEE 982
Query: 468 EATEDEEATEDVEATVDEEEEATEVEEATDHEEVESE----KESNKDEEATEDEEEESSE 523
E E++E + +EA+ EE ++ E+ E E K E +E E
Sbjct: 983 E--EEDENKDVLEASFTEEWVPFFSQDQIASTEIPIEAGQYPEKRKPPVIAEKKEREHQV 1040
Query: 524 DDY 526
Y
Sbjct: 1041 ASY 1043
>YCF2_OENVI (P31569) Protein ycf2 (Fragment)
Length = 630
Score = 68.6 bits (166), Expect = 4e-11
Identities = 53/147 (36%), Positives = 70/147 (47%), Gaps = 12/147 (8%)
Query: 389 VTSLPHGFKE-------ANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDE 441
++ + HG E ++ EE + EEE G V+ E + EEVE EDE
Sbjct: 162 LSDIVHGLLELEGALVGSSPTEEEVEGTEEEVEGTE-EEVEGTEEEVEGTEEEVEGTEDE 220
Query: 442 EEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEV 501
E E TE+E ++E E E E G E E E+VE T DEE E TE E EEV
Sbjct: 221 EVEGTEEEVEGTEEEVEGTEEEVE-GTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEV 279
Query: 502 ESEKES--NKDEEATEDEEE-ESSEDD 525
E +E +EE EEE E +E++
Sbjct: 280 EGTEEEVEGTEEEVEGTEEEVEGTEEE 306
Score = 67.8 bits (164), Expect = 7e-11
Identities = 48/131 (36%), Positives = 66/131 (49%), Gaps = 5/131 (3%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E EE + EEE G V+ E + EEVE E+E E T E+ + + E
Sbjct: 198 EEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGT---E 254
Query: 457 DESEEASTEDGEATEDE-EATEDVEATVDEEEEATEVEEATDHEEVE-SEKESNKDEEAT 514
+E E E+ E TE+E E TE+ +EE E TE E EEVE +E+E EE
Sbjct: 255 EEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEV 314
Query: 515 EDEEEESSEDD 525
E E++SS+ D
Sbjct: 315 EGTEKDSSQFD 325
Score = 63.5 bits (153), Expect = 1e-09
Identities = 47/132 (35%), Positives = 63/132 (47%), Gaps = 6/132 (4%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E EE + EEE G V+ E E TEE +EE E TE+E ++E
Sbjct: 191 EEVEGTEEEVEGTEEEVEGTE-EEVEGTEDEEVEGTEEEVEGTEEEVEGTEEEVEGTEEE 249
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKE----SNKDEE 512
E E E E E E E+VE T +EE E TE E EEVE +E + ++ E
Sbjct: 250 VEGTEEEVEGTEDEEVEGTEEEVEGT-EEEVEGTEEEVEGTEEEVEGTEEEVEGTEEEVE 308
Query: 513 ATEDEEEESSED 524
TE+E E + +D
Sbjct: 309 GTEEEVEGTEKD 320
Score = 62.8 bits (151), Expect = 2e-09
Identities = 37/107 (34%), Positives = 55/107 (50%), Gaps = 10/107 (9%)
Query: 424 LTCEFKEATEEVEAAEDEEEETT----------EDEDASDDKEDESEEASTEDGEATEDE 473
L CE + EE+ EDE E E+E +DD+ED EA ED E++
Sbjct: 395 LDCEAEIPAEEIPEEEDELPEDALETEVAVWGVEEEGEADDEEDVLLEAQQEDELLEEED 454
Query: 474 EATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEE 520
E ++ E +DEEEE + EE HEE E E+E ++E+ ++ + E
Sbjct: 455 EELDEEEDELDEEEEEPKEEEDELHEEEEEEEEEEEEEDELQENDSE 501
Score = 54.3 bits (129), Expect = 8e-07
Identities = 37/110 (33%), Positives = 51/110 (45%), Gaps = 2/110 (1%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E EE + EEE G V+ E + EEVE EDEE E TE+E ++E
Sbjct: 220 EEVEGTEEEVEGTEEEVEGTE-EEVEGTEEEVEGTEEEVEGTEDEEVEGTEEEVEGTEEE 278
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKE 506
E E E G E E E+VE T +E E E E T+ + + + +
Sbjct: 279 VEGTEEEVE-GTEEEVEGTEEEVEGTEEEVEGTEEEVEGTEKDSSQFDND 327
Score = 52.4 bits (124), Expect = 3e-06
Identities = 40/95 (42%), Positives = 53/95 (55%), Gaps = 13/95 (13%)
Query: 433 EEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEV 492
EEVE E+E E T E+ + ++ E E + E+ E TEDEE VE T +EE E TE
Sbjct: 184 EEVEGTEEEVEGTEEEVEGTE----EEVEGTEEEVEGTEDEE----VEGT-EEEVEGTEE 234
Query: 493 EEATDHEEVESEKESNKDEEATEDEEEESSEDDYI 527
E EEVE +E + E TE EE E +ED+ +
Sbjct: 235 EVEGTEEEVEGTEE---EVEGTE-EEVEGTEDEEV 265
Score = 47.4 bits (111), Expect = 9e-05
Identities = 40/124 (32%), Positives = 59/124 (47%), Gaps = 24/124 (19%)
Query: 381 PSEQLHLRVTSLPHGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAED 440
P+E++ LP ++A E E A EEE V EA +E E E+
Sbjct: 402 PAEEIPEEEDELP---EDALETEVAVWGVEEEGEADDEEDV------LLEAQQEDELLEE 452
Query: 441 EEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEE 500
E+EE E+ED D++E+E +E E++E E+ EEEE E EE + +E
Sbjct: 453 EDEELDEEEDELDEEEEEPKE---------EEDELHEE------EEEEEEEEEEEDELQE 497
Query: 501 VESE 504
+SE
Sbjct: 498 NDSE 501
Score = 34.3 bits (77), Expect = 0.82
Identities = 46/207 (22%), Positives = 71/207 (34%), Gaps = 79/207 (38%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDK- 455
+E EE + EEE G V+ E + EEVE E+E E T +D D+
Sbjct: 270 EEVEGTEEEVEGTEEEVEGTE-EEVEGTEEEVEGTEEEVEGTEEEVEGTEKDSSQFDNDR 328
Query: 456 -------------------------EDESEEASTEDGEATEDEEATEDV----------- 479
E E EE +D + ++ ED+
Sbjct: 329 VTLLLRPKPRNPLDIQRLIYQHQKYESELEEDDDDDEDVFAPQKMLEDLFSELVWSPRIW 388
Query: 480 ---------EATVDEE----------EEATEVE----------EATDHEEV--------- 501
EA + E E+A E E EA D E+V
Sbjct: 389 HPWDFLLDCEAEIPAEEIPEEEDELPEDALETEVAVWGVEEEGEADDEEDVLLEAQQEDE 448
Query: 502 ---ESEKESNKDEEATEDEEEESSEDD 525
E ++E +++E+ ++EEEE E++
Sbjct: 449 LLEEEDEELDEEEDELDEEEEEPKEEE 475
>NFL_MOUSE (P08551) Neurofilament triplet L protein (68 kDa
neurofilament protein) (Neurofilament light polypeptide)
(NF-L)
Length = 542
Score = 66.6 bits (161), Expect = 1e-10
Identities = 51/187 (27%), Positives = 91/187 (48%), Gaps = 13/187 (6%)
Query: 342 HKYGQEIKNVRKDLAMYRESEYKNI-------QIYFEMYRESLLSFPSEQLHLRVTSLPH 394
+K E+++ + ++A Y + EY+++ I YR+ L + V S+
Sbjct: 352 NKLENELRSTKSEMARYLK-EYQDLLNVKMALDIEIAAYRKLLEGEETRLSFTSVGSITS 410
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDD 454
G+ ++++V + ++S L S +S + +E + +E E T+ E+A D+
Sbjct: 411 GYSQSSQVFGRSAYSGLQSSSY-LMSARSFPAYYTSHVQEEQTEVEETIEATKAEEAKDE 469
Query: 455 KEDESE-EASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEE---VESEKESNKD 510
E E E ++ E E+EE E+ EA DE E+ E EE + EE ESE+E K+
Sbjct: 470 PPSEGEAEEEEKEKEEGEEEEGAEEEEAAKDESEDTKEEEEGGEGEEEDTKESEEEEKKE 529
Query: 511 EEATEDE 517
E A E++
Sbjct: 530 ESAGEEQ 536
Score = 30.8 bits (68), Expect = 9.0
Identities = 38/182 (20%), Positives = 70/182 (37%), Gaps = 20/182 (10%)
Query: 333 NAEGKLLEHHKYGQEIKNVRKDLAMYRESEYKNIQIYFEMYRESLLSFPSEQLHLRVTSL 392
+AEG+L+E K E R +L +S I +++ E + + Q+ S+
Sbjct: 186 DAEGRLMERRKGADEAALARAELEKRIDSLMDEIAFLKKVHEEEIAELQA-QIQYAQISV 244
Query: 393 PHGFKEANEVEEATKDEEEEASGLSLHSVKS-----------LTCEFKEATEEVEAAEDE 441
++ A KD + L+ ++++ LT + T+ V AA+DE
Sbjct: 245 EMDVSSKPDLSAALKDIRAQYEKLAAKNMQNAEEWFKSRFTVLTESAAKNTDAVRAAKDE 304
Query: 442 EEE--------TTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVE 493
E T E E E ++ + + D A +D ++ E +T+ E
Sbjct: 305 VSESRRLLKAKTLEIEACRGMNEALEKQLQELEDKQNADISAMQDTINKLENELRSTKSE 364
Query: 494 EA 495
A
Sbjct: 365 MA 366
>CNG4_BOVIN (Q28181) 240 kDa protein of rod photoreceptor
CNG-channel [Contains: Glutamic acid-rich protein
(GARP); Cyclic-nucleotide-gated cation channel 4 (CNG
channel 4) (CNG-4) (Cyclic nucleotide-gated cation
channel modulatory subunit)]
Length = 1394
Score = 66.2 bits (160), Expect = 2e-10
Identities = 44/132 (33%), Positives = 63/132 (47%), Gaps = 15/132 (11%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E ++E +DEEEE + +E EE E+EE E E+E+ +KE
Sbjct: 348 RELPRIQEEKEDEEEEKE------------DGEEEEEEGREKEEEEGEEKEEEEGR-EKE 394
Query: 457 DESEEASTEDGEATEDEEA--TEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEAT 514
+E E E+G E+EE ED E EEEE EE E+ E E ++ E
Sbjct: 395 EEEGEKKEEEGREKEEEEGGEKEDEEGREKEEEEGRGKEEEEGGEKEEEEGRGKEEVEGR 454
Query: 515 EDEEEESSEDDY 526
E+EE+E E D+
Sbjct: 455 EEEEDEEEEQDH 466
Score = 63.2 bits (152), Expect = 2e-09
Identities = 49/152 (32%), Positives = 71/152 (46%), Gaps = 7/152 (4%)
Query: 376 SLLSFPSEQLHLRVTSL-PHGFK-EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATE 433
S+L E+ HL + + PH + E E +T EA+ K + +E
Sbjct: 293 SILPGEQEESHLILEEVDPHWEEDEHQEGSTSTSPRTSEAAPADEEKGKVVEQTPRELPR 352
Query: 434 EVEAAEDEEEE----TTEDEDASDDKEDESEEASTEDGEATEDEEA-TEDVEATVDEEEE 488
E EDEEEE E+E+ + +E+E EE E+G E+EE ++ E EEEE
Sbjct: 353 IQEEKEDEEEEKEDGEEEEEEGREKEEEEGEEKEEEEGREKEEEEGEKKEEEGREKEEEE 412
Query: 489 ATEVEEATDHEEVESEKESNKDEEATEDEEEE 520
E E+ E+ E E ++EE E EEEE
Sbjct: 413 GGEKEDEEGREKEEEEGRGKEEEEGGEKEEEE 444
Score = 57.0 bits (136), Expect = 1e-07
Identities = 47/147 (31%), Positives = 64/147 (42%), Gaps = 19/147 (12%)
Query: 392 LPHGFKEANEVEEATKD-EEEEASGLSLHSVKSLTCEFKEATE-EVEAAEDEEEE--TTE 447
LP +E + EE +D EEEE G + E +E E E E E +EEE E
Sbjct: 350 LPRIQEEKEDEEEEKEDGEEEEEEGREKEEEEGEEKEEEEGREKEEEEGEKKEEEGREKE 409
Query: 448 DEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKES 507
+E+ + +++E E E+G E+EE E EEEE EE EE E E+E
Sbjct: 410 EEEGGEKEDEEGREKEEEEGRGKEEEEGGE------KEEEEGRGKEEVEGREEEEDEEEE 463
Query: 508 NKDE---------EATEDEEEESSEDD 525
++ ED EES D
Sbjct: 464 QDHSVLLDSYLVPQSEEDRSEESETQD 490
Score = 53.9 bits (128), Expect = 1e-06
Identities = 38/130 (29%), Positives = 61/130 (46%), Gaps = 7/130 (5%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
KE E EE E+EE G + E +E ++ E ++EEE +++ + +E
Sbjct: 364 KEDGEEEEEEGREKEEEEGEEKEEEEGREKEEEEGEKKEEEGREKEEEEGGEKEDEEGRE 423
Query: 457 DESEEA---STEDGEATEDEE--ATEDVEATVDEEEEATEVEEAT--DHEEVESEKESNK 509
E EE E+G E+EE E+VE +EE+E E + + D V +E
Sbjct: 424 KEEEEGRGKEEEEGGEKEEEEGRGKEEVEGREEEEDEEEEQDHSVLLDSYLVPQSEEDRS 483
Query: 510 DEEATEDEEE 519
+E T+D+ E
Sbjct: 484 EESETQDQSE 493
Score = 48.1 bits (113), Expect = 5e-05
Identities = 49/164 (29%), Positives = 67/164 (39%), Gaps = 21/164 (12%)
Query: 381 PSEQLHLRVTSLPHGFKEA-------NEVEEATKDEEEEASGLSLHSVKSLTCEFKEATE 433
P Q ++ +SLP A + +E A G S +TC+ + T
Sbjct: 235 PEPQPTIQASSLPPPQDSARLMAWILHRLEMALPQPVIRGKGGEQESDAPVTCDVQ--TI 292
Query: 434 EVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATE-----DEEATEDVEATVDE--- 485
+ E EE +E +EDE +E ST T DEE + VE T E
Sbjct: 293 SILPGEQEESHLILEEVDPHWEEDEHQEGSTSTSPRTSEAAPADEEKGKVVEQTPRELPR 352
Query: 486 --EEEATEVEEATDHEEVESEKESNKDEEATEDEEEE--SSEDD 525
EE+ E EE D EE E E ++EE E EEEE E++
Sbjct: 353 IQEEKEDEEEEKEDGEEEEEEGREKEEEEGEEKEEEEGREKEEE 396
Score = 45.8 bits (107), Expect = 3e-04
Identities = 33/128 (25%), Positives = 52/128 (39%), Gaps = 6/128 (4%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E + EE +++EEE G E +E EE ++EEE ++E+ KE
Sbjct: 396 EEGEKKEEEGREKEEEEGGEKEDE------EGREKEEEEGRGKEEEEGGEKEEEEGRGKE 449
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATED 516
+ ED E +D D EE+ +E E D EV + + A
Sbjct: 450 EVEGREEEEDEEEEQDHSVLLDSYLVPQSEEDRSEESETQDQSEVGGAQAQGEVGGAQAL 509
Query: 517 EEEESSED 524
EE ++D
Sbjct: 510 SEESETQD 517
Score = 41.2 bits (95), Expect = 0.007
Identities = 42/170 (24%), Positives = 66/170 (38%), Gaps = 47/170 (27%)
Query: 401 EVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDA--------S 452
E EE + EEEE G E EEVE E+EE+E E + +
Sbjct: 417 EDEEGREKEEEEGRGKEEEEGGEKEEEEGRGKEEVEGREEEEDEEEEQDHSVLLDSYLVP 476
Query: 453 DDKEDESEEASTED---------------GEATEDEEATEDVEATVDEEEEATEV----- 492
+ED SEE+ T+D +A +E T+D ++ V ++ +EV
Sbjct: 477 QSEEDRSEESETQDQSEVGGAQAQGEVGGAQALSEESETQD-QSEVGGAQDQSEVGGAQA 535
Query: 493 -----------------EEATDHEEVESEK-ESNKDEEATEDEEEESSED 524
+++T H+E++ E + ATE+ E ED
Sbjct: 536 QGEVGGAQEQDGVGGAQDQSTSHQELQEEALADSSGVPATEEHPELQVED 585
Score = 32.0 bits (71), Expect = 4.1
Identities = 17/39 (43%), Positives = 22/39 (55%)
Query: 420 SVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDE 458
S + L E E EE E E+E EE E E+A +KE+E
Sbjct: 1356 SEQILLVEVPEKQEEKEKKEEETEEKEEGEEARKEKEEE 1394
>NUCL_RAT (P13383) Nucleolin (Protein C23)
Length = 712
Score = 65.9 bits (159), Expect = 3e-10
Identities = 42/129 (32%), Positives = 65/129 (49%), Gaps = 4/129 (3%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKED 457
E E E+ + ++E+E VK + + +A A+EDE+EE +DED DD E+
Sbjct: 148 EDEEDEDDSDEDEDEEDEFEPPVVKGV--KPAKAAPAAPASEDEDEEDDDDEDDDDDDEE 205
Query: 458 ESEEASTEDG--EATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATE 515
E EE +E+ E T + + + + E EE + +E E E E +DEE E
Sbjct: 206 EEEEDDSEEEVMEITPAKGKKTPAKVVPVKAKSVAEEEEDDEDDEDEEEDEDEEDEEDDE 265
Query: 516 DEEEESSED 524
DE+EE E+
Sbjct: 266 DEDEEEEEE 274
Score = 55.1 bits (131), Expect = 4e-07
Identities = 36/122 (29%), Positives = 52/122 (42%), Gaps = 35/122 (28%)
Query: 439 EDEEEETTEDEDASDDKEDE-----------------------SEEASTEDGEATEDEEA 475
+ +E+E EDED SD+ EDE SE+ ED + +D++
Sbjct: 143 DSDEDEDEEDEDDSDEDEDEEDEFEPPVVKGVKPAKAAPAAPASEDEDEEDDDDEDDDDD 202
Query: 476 TEDVEATVDEEEEATEVEEATDHEE------------VESEKESNKDEEATEDEEEESSE 523
E+ E D EEE E+ A + E E++ DE+ EDE+EE E
Sbjct: 203 DEEEEEEDDSEEEVMEITPAKGKKTPAKVVPVKAKSVAEEEEDDEDDEDEEEDEDEEDEE 262
Query: 524 DD 525
DD
Sbjct: 263 DD 264
Score = 47.8 bits (112), Expect = 7e-05
Identities = 32/106 (30%), Positives = 55/106 (51%), Gaps = 7/106 (6%)
Query: 400 NEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDES 459
+E EE D EEE ++ K + + A E+E++E EDE+ +D+EDE
Sbjct: 203 DEEEEEEDDSEEEVMEITPAKGKKTPAKVVPVKAKSVAEEEEDDEDDEDEEEDEDEEDE- 261
Query: 460 EEASTEDGEATEDEEATEDVEATVDE-EEEATEVEEATDHEEVESE 504
ED E ++EE E V+A + ++E T+ +EA + ++ + E
Sbjct: 262 -----EDDEDEDEEEEEEPVKAAPGKRKKEMTKQKEAPEAKKQKIE 302
Score = 46.2 bits (108), Expect = 2e-04
Identities = 33/102 (32%), Positives = 46/102 (44%), Gaps = 19/102 (18%)
Query: 433 EEVEAAEDEEEETTEDEDASDDK--------------EDESEEASTEDGEATEDEEATED 478
EE E +D EEE E A K E+E ++ ED E EDEE ED
Sbjct: 204 EEEEEEDDSEEEVMEITPAKGKKTPAKVVPVKAKSVAEEEEDDEDDEDEEEDEDEEDEED 263
Query: 479 VEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEE 520
DE+E+ E EE + +KE K +EA E ++++
Sbjct: 264 -----DEDEDEEEEEEPVKAAPGKRKKEMTKQKEAPEAKKQK 300
>NFL_RAT (P19527) Neurofilament triplet L protein (68 kDa
neurofilament protein) (Neurofilament light polypeptide)
(NF-L)
Length = 541
Score = 65.9 bits (159), Expect = 3e-10
Identities = 49/192 (25%), Positives = 94/192 (48%), Gaps = 12/192 (6%)
Query: 342 HKYGQEIKNVRKDLAMYRESEYKNI-------QIYFEMYRESLLSFPSEQLHLRVTSLPH 394
+K E+++ + ++A Y + EY+++ I YR+ L + V S+
Sbjct: 352 NKLENELRSTKSEMARYLK-EYQDLLNVKMALDIEIAAYRKLLEGEETRLSFTSVGSITS 410
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDD 454
G+ ++++V + ++S L S ++ + +E ++ +E E T+ E+A D+
Sbjct: 411 GYSQSSQVFGRSAYSGLQSSSY-LMSARAFPAYYTSHVQEEQSEVEETIEATKAEEAKDE 469
Query: 455 KEDESE-EASTEDGEATEDEEATEDVEATVDEEEEATEVE--EATDHEEVESEKESNKDE 511
E E E ++ E E+EE E+ EA DE E+A E E E + + ESE+E K+E
Sbjct: 470 PPSEGEAEEEEKEKEEGEEEEGAEEEEAAKDESEDAKEEEGGEGEEEDTKESEEEEKKEE 529
Query: 512 EATEDEEEESSE 523
A E++ + +
Sbjct: 530 SAGEEQAAKKKD 541
>NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3
Length = 802
Score = 65.5 bits (158), Expect = 3e-10
Identities = 39/128 (30%), Positives = 67/128 (51%), Gaps = 4/128 (3%)
Query: 404 EATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEE-EETTEDEDASDDKEDESEEA 462
E + D + L +S EF EE EA +EE EE E ED +D++E++ EE
Sbjct: 17 ELSVDSNSNENELMNNSSADDGIEFDAPEEEREAEREEENEEQHELEDVNDEEEEDKEEK 76
Query: 463 STEDGEAT---EDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEE 519
E+GE E+EE + D++++ E EE + ++ + + + + DEE E+EEE
Sbjct: 77 GEENGEVINTEEEEEEEHQQKGGNDDDDDDNEEEEEEEEDDDDDDDDDDDDEEEEEEEEE 136
Query: 520 ESSEDDYI 527
E +++ +
Sbjct: 137 EGNDNSSV 144
Score = 62.0 bits (149), Expect = 4e-09
Identities = 39/126 (30%), Positives = 65/126 (50%), Gaps = 19/126 (15%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E +E+E+ +EEE+ E E EV E+EEEE + + +DD +
Sbjct: 57 EEQHELEDVNDEEEEDKE------------EKGEENGEVINTEEEEEEEHQQKGGNDDDD 104
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEV--ESEKESNKDEEAT 514
D++EE E+ + +D++ +D DEEEE E EE D+ V +S E +DEE
Sbjct: 105 DDNEEEEEEEEDDDDDDDDDDD-----DEEEEEEEEEEGNDNSSVGSDSAAEDGEDEEDK 159
Query: 515 EDEEEE 520
+D+ ++
Sbjct: 160 KDKTKD 165
Score = 61.6 bits (148), Expect = 5e-09
Identities = 43/137 (31%), Positives = 66/137 (47%), Gaps = 23/137 (16%)
Query: 396 FKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEE-------VEAAEDEEEETTED 448
F E EA ++EE E H ++ + E +E EE V E+EEEE +
Sbjct: 41 FDAPEEEREAEREEENEEQ----HELEDVNDEEEEDKEEKGEENGEVINTEEEEEEEHQQ 96
Query: 449 EDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESN 508
+ +DD +D++EE E+ + +D++ +D DEEEE E EE D+ V S+
Sbjct: 97 KGGNDDDDDDNEEEEEEEEDDDDDDDDDDD-----DEEEEEEEEEEGNDNSSVGSDS--- 148
Query: 509 KDEEATEDEEEESSEDD 525
A ED E+E + D
Sbjct: 149 ----AAEDGEDEEDKKD 161
Score = 61.2 bits (147), Expect = 6e-09
Identities = 36/127 (28%), Positives = 68/127 (53%), Gaps = 5/127 (3%)
Query: 394 HGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEA---TEEVEAAEDEEEETTEDED 450
H ++ N+ EE K+E+ E +G +++ + E ++ ++ + E+EEEE +D+D
Sbjct: 60 HELEDVNDEEEEDKEEKGEENGEVINTEEEEEEEHQQKGGNDDDDDDNEEEEEEEEDDDD 119
Query: 451 ASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKD 510
DD +D+ EE E+ E ++ D A E+EE + ++ T +EVE +E+ +
Sbjct: 120 DDDDDDDDEEEEEEEEEEGNDNSSVGSDSAAEDGEDEE--DKKDKTKDKEVELRRETLEK 177
Query: 511 EEATEDE 517
E+ DE
Sbjct: 178 EQKDVDE 184
Score = 38.1 bits (87), Expect = 0.057
Identities = 26/93 (27%), Positives = 45/93 (47%), Gaps = 17/93 (18%)
Query: 448 DEDASDDKEDE-----SEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDH---- 498
DE+ + D +D S ++++ + E + A + +E EEE E EE +
Sbjct: 3 DENHNSDVQDIPSPELSVDSNSNENELMNNSSADDGIEFDAPEEEREAEREEENEEQHEL 62
Query: 499 EEVESEKESNKDEEA--------TEDEEEESSE 523
E+V E+E +K+E+ TE+EEEE +
Sbjct: 63 EDVNDEEEEDKEEKGEENGEVINTEEEEEEEHQ 95
>MP62_LYTPI (P91753) Mitotic apparatus protein p62
Length = 411
Score = 65.5 bits (158), Expect = 3e-10
Identities = 42/138 (30%), Positives = 68/138 (48%), Gaps = 11/138 (7%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEE--------EETTEDE 449
E +E E+ ++EEEE + K + + A D++ + E+
Sbjct: 183 ELDEDEDDDEEEEEEEEEIQTAKGKKRPAPSAKGPAKKLAKVDKDGTSKRKVPNGSVENG 242
Query: 450 DASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEAT---DHEEVESEKE 506
A DD ED+ E+ + G+ E+EEAT E DEEEE E +E D ++ + ++E
Sbjct: 243 HAIDDDEDDEEDEDYKVGDEEEEEEATSGEEEEEDEEEEEEEDDEEMALGDDDDEDDDEE 302
Query: 507 SNKDEEATEDEEEESSED 524
++DEE +DE+EE ED
Sbjct: 303 DDEDEEGMDDEDEEEEED 320
Score = 53.1 bits (126), Expect = 2e-06
Identities = 46/185 (24%), Positives = 68/185 (35%), Gaps = 68/185 (36%)
Query: 409 EEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTE--------------------- 447
+E+E G ++ ++ +E E EDEEEE T
Sbjct: 120 QEDEEFGKDFEGAEAYEVGDEDLEDEDEGEEDEEEEETPKKGSPKRIVKKIAAVKGRMKG 179
Query: 448 -----DEDASDDKEDESEE--------------------------------------AST 464
DED DD+E+E EE S
Sbjct: 180 KGDELDEDEDDDEEEEEEEEEIQTAKGKKRPAPSAKGPAKKLAKVDKDGTSKRKVPNGSV 239
Query: 465 EDGEATEDEEATEDVE----ATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEE 520
E+G A +D+E E+ E +EEEEAT EE + EE E E++ + +D+E++
Sbjct: 240 ENGHAIDDDEDDEEDEDYKVGDEEEEEEATSGEEEEEDEEEEEEEDDEEMALGDDDDEDD 299
Query: 521 SSEDD 525
EDD
Sbjct: 300 DEEDD 304
Score = 48.9 bits (115), Expect = 3e-05
Identities = 38/139 (27%), Positives = 64/139 (45%), Gaps = 7/139 (5%)
Query: 392 LPHGFKEANEVEEATKDEEEEAS---GLSLHSVKSLTCEFKEATEEVEAAEDEEEETTED 448
+P+G E + +D+EE+ G ++ + E +E EE E ED+EE D
Sbjct: 234 VPNGSVENGHAIDDDEDDEEDEDYKVGDEEEEEEATSGEEEEEDEEEEEEEDDEEMALGD 293
Query: 449 EDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESN 508
+D DD E++ E+ E+G EDEE ED + V ++A T + + ++N
Sbjct: 294 DDDEDDDEEDDED---EEGMDDEDEEEEED-SSPVKPAKKAKGKVNGTAKPKGTPKSQAN 349
Query: 509 KDEEATEDEEEESSEDDYI 527
K + + E + D I
Sbjct: 350 KGMKEKKTYSLEDMKQDLI 368
>A32B_HUMAN (Q92688) Acidic leucine-rich nuclear phosphoprotein 32
family member B (PHAPI2 protein) (Silver-stainable
protein SSP29) (Acidic protein rich in leucines)
Length = 251
Score = 65.5 bits (158), Expect = 3e-10
Identities = 38/99 (38%), Positives = 55/99 (55%), Gaps = 2/99 (2%)
Query: 427 EFKEATEEVEAAEDEEE-ETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDE 485
E ++ EV+ ++EEE E EDE+ DD++ E EE ED E ED E ED + +E
Sbjct: 154 EAPDSDAEVDGVDEEEEDEEGEDEEDEDDEDGEEEEFDEEDDE-DEDVEGDEDDDEVSEE 212
Query: 486 EEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSED 524
EEE EE D +E E E+E K E+ + ++E +D
Sbjct: 213 EEEFGLDEEDEDEDEDEEEEEGGKGEKRKRETDDEGEDD 251
Score = 58.5 bits (140), Expect = 4e-08
Identities = 32/96 (33%), Positives = 52/96 (53%), Gaps = 3/96 (3%)
Query: 433 EEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEA---TEDEEATEDVEATVDEEEEA 489
E+ EA + + E DE+ D++ ++ E+ EDGE E+++ EDVE D++E +
Sbjct: 151 EDQEAPDSDAEVDGVDEEEEDEEGEDEEDEDDEDGEEEEFDEEDDEDEDVEGDEDDDEVS 210
Query: 490 TEVEEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
E EE EE E E E ++EE + E+ + DD
Sbjct: 211 EEEEEFGLDEEDEDEDEDEEEEEGGKGEKRKRETDD 246
Score = 57.8 bits (138), Expect = 7e-08
Identities = 34/87 (39%), Positives = 48/87 (55%), Gaps = 3/87 (3%)
Query: 440 DEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATE-VEEATDH 498
D E++ D DA D DE EE E+GE EDE+ + E DEE++ E VE D
Sbjct: 149 DREDQEAPDSDAEVDGVDEEEE--DEEGEDEEDEDDEDGEEEEFDEEDDEDEDVEGDEDD 206
Query: 499 EEVESEKESNKDEEATEDEEEESSEDD 525
+EV E+E +E EDE+E+ E++
Sbjct: 207 DEVSEEEEEFGLDEEDEDEDEDEEEEE 233
Score = 45.8 bits (107), Expect = 3e-04
Identities = 29/62 (46%), Positives = 37/62 (58%), Gaps = 3/62 (4%)
Query: 466 DGEATEDEEATE-DVEAT-VDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSE 523
DG ED+EA + D E VDEEEE E E+ D ++ + E+E DEE EDE+ E E
Sbjct: 146 DGYDREDQEAPDSDAEVDGVDEEEEDEEGEDEEDEDDEDGEEE-EFDEEDDEDEDVEGDE 204
Query: 524 DD 525
DD
Sbjct: 205 DD 206
>CENB_SHEEP (P49451) Major centromere autoantigen B (Centromere
protein B) (CENP-B) (Fragment)
Length = 239
Score = 65.1 bits (157), Expect = 4e-10
Identities = 43/130 (33%), Positives = 65/130 (49%), Gaps = 1/130 (0%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
K E EE ++EEEE G + E +EA E E E+EE E D D +++E
Sbjct: 43 KSEGEEEEEEEEEEEEEEGEGEEEEEEDGEEEEEAGEGEELGEEEEVEEEGDVDTVEEEE 102
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDH-EEVESEKESNKDEEATE 515
+E EE+S+E EA + + + + E + T H E E + ES+ +EE +
Sbjct: 103 EEEEESSSEGLEAEDWAQGVVEAGGSFGGYGAQEEAQCPTLHFLEGEEDSESDSEEEEED 162
Query: 516 DEEEESSEDD 525
D+E+E EDD
Sbjct: 163 DDEDEDDEDD 172
Score = 60.1 bits (144), Expect = 1e-08
Identities = 37/79 (46%), Positives = 48/79 (59%), Gaps = 6/79 (7%)
Query: 445 TTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESE 504
TT + +++E+E EE E+GE E+EE ED E EEEEA E EE + EEVE E
Sbjct: 39 TTALKSEGEEEEEEEEEEEEEEGEGEEEEE--EDGE----EEEEAGEGEELGEEEEVEEE 92
Query: 505 KESNKDEEATEDEEEESSE 523
+ + EE E+EEE SSE
Sbjct: 93 GDVDTVEEEEEEEEESSSE 111
Score = 46.2 bits (108), Expect = 2e-04
Identities = 35/125 (28%), Positives = 57/125 (45%), Gaps = 10/125 (8%)
Query: 401 EVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESE 460
E EE EEEE +G + E + + VE E+EEEE++ + ++D
Sbjct: 64 EEEEEEDGEEEEEAGEGEELGEEEEVEEEGDVDTVEEEEEEEEESSSEGLEAEDWAQGVV 123
Query: 461 EASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEE 520
EA G EEA ++ EE++ E +SE+E D+E +DE++E
Sbjct: 124 EAGGSFGGYGAQEEAQCPTLHFLEGEEDS----------ESDSEEEEEDDDEDEDDEDDE 173
Query: 521 SSEDD 525
+D+
Sbjct: 174 EEDDE 178
Score = 38.1 bits (87), Expect = 0.057
Identities = 20/46 (43%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query: 425 TCEFKEATEEVEA-AEDEEEETTEDEDASDDKEDESEEASTEDGEA 469
T F E E+ E+ +E+EEE+ EDED DD+E++ E GEA
Sbjct: 142 TLHFLEGEEDSESDSEEEEEDDDEDEDDEDDEEEDDEVPVPSFGEA 187
Score = 33.5 bits (75), Expect = 1.4
Identities = 17/43 (39%), Positives = 24/43 (55%)
Query: 485 EEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDDYI 527
EEEE E EE + E E E+E + +EE E EE E++ +
Sbjct: 47 EEEEEEEEEEEEEEGEGEEEEEEDGEEEEEAGEGEELGEEEEV 89
Score = 33.5 bits (75), Expect = 1.4
Identities = 17/50 (34%), Positives = 28/50 (56%)
Query: 476 TEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
T +++ +EEEE E EE + E E E+E ++EE + EE E++
Sbjct: 39 TTALKSEGEEEEEEEEEEEEEEGEGEEEEEEDGEEEEEAGEGEELGEEEE 88
>CENB_MOUSE (P27790) Major centromere autoantigen B (Centromere
protein B) (CENP-B)
Length = 599
Score = 65.1 bits (157), Expect = 4e-10
Identities = 42/133 (31%), Positives = 62/133 (46%), Gaps = 30/133 (22%)
Query: 423 SLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEAT 482
++T FK EE E E+EEEE E+E +++E+E EE E GE E+ E + E
Sbjct: 396 TITTSFKSEGEEEEEEEEEEEEEEEEEGEGEEEEEEEEEGEEEGGEGEEEGEEEVEEEGE 455
Query: 483 VD-----EEEEATEVEEATDHEE-------------------------VESEKESNKDEE 512
VD EEE ++E EA D + +E ++S+ D +
Sbjct: 456 VDDSDEEEEESSSEGLEAEDWAQGVVEASGGFGGYSVQEEAQFPTLHFLEGGEDSDSDSD 515
Query: 513 ATEDEEEESSEDD 525
ED+EEE ED+
Sbjct: 516 EEEDDEEEDEEDE 528
Score = 65.1 bits (157), Expect = 4e-10
Identities = 44/137 (32%), Positives = 68/137 (49%), Gaps = 13/137 (9%)
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDD 454
G +E E EE ++EEEE G E +E EE E+E EE E+E DD
Sbjct: 405 GEEEEEEEEEEEEEEEEEGEGEEEEE------EEEEGEEEGGEGEEEGEEEVEEEGEVDD 458
Query: 455 KEDESEEASTEDGEATEDEEATEDVEA-----TVDEEEEATEVE--EATDHEEVESEKES 507
++E EE+S+E EA + + + +V EE + + E + + +S++E
Sbjct: 459 SDEEEEESSSEGLEAEDWAQGVVEASGGFGGYSVQEEAQFPTLHFLEGGEDSDSDSDEEE 518
Query: 508 NKDEEATEDEEEESSED 524
+ +EE EDE+EE ED
Sbjct: 519 DDEEEDEEDEDEEDDED 535
Score = 46.2 bits (108), Expect = 2e-04
Identities = 42/137 (30%), Positives = 64/137 (46%), Gaps = 13/137 (9%)
Query: 396 FKEANEVEEATKDEEEEASGLSLHSVKSL--TCEFKEATEEVEAAEDEEE-------ETT 446
+++A ++ E ++ SGL L V++L +A E + A E T
Sbjct: 337 YRQAMLLKAMAALEGQDPSGLQLGLVEALHFVAAAWQAVEPSDIATCFREAGFGGGLNAT 396
Query: 447 EDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKE 506
+ E+E EE E+ E E+ E E+ E + EEE E EE + EEVE E E
Sbjct: 397 ITTSFKSEGEEEEEEEEEEEEEEEEEGEGEEEEEEEEEGEEEGGEGEEEGE-EEVEEEGE 455
Query: 507 SNKDEEATEDEEEESSE 523
+++ E+EEE SSE
Sbjct: 456 V---DDSDEEEEESSSE 469
Score = 33.1 bits (74), Expect = 1.8
Identities = 26/90 (28%), Positives = 41/90 (44%), Gaps = 8/90 (8%)
Query: 397 KEANEVEEATKDEEEEAS-GLSLHSVKSLTCEFKEATEEVEAAEDEEEETT------EDE 449
+E EV+++ ++EEE +S GL E E+ + T ED
Sbjct: 451 EEEGEVDDSDEEEEESSSEGLEAEDWAQGVVEASGGFGGYSVQEEAQFPTLHFLEGGEDS 510
Query: 450 DASDDKEDESEEASTEDGEATEDEEATEDV 479
D SD E+E +E E+ E ED+E ++V
Sbjct: 511 D-SDSDEEEDDEEEDEEDEDEEDDEDGDEV 539
>NFM_HUMAN (P07197) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M) (Neurofilament 3)
Length = 915
Score = 64.7 bits (156), Expect = 6e-10
Identities = 48/149 (32%), Positives = 71/149 (47%), Gaps = 5/149 (3%)
Query: 375 ESLLSFPSEQLHLRVTSLPHGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEE 434
E L+ +E+L S+ KEA E +E + EEE VK+ E KE E
Sbjct: 469 EEALTAITEEL---AASMKEEKKEAAEEKEEEPEAEEEEVAAKKSPVKATAPEVKEEEGE 525
Query: 435 VEAAEDEEEETTEDEDASDDKEDE--SEEASTEDGEATEDEEATEDVEATVDEEEEATEV 492
E E +EEE EDE A D+ +E SE+ + + E E EE + EA +E E E
Sbjct: 526 KEEEEGQEEEEEEDEGAKSDQAEEGGSEKEGSSEKEEGEQEEGETEAEAEGEEAEAKEEK 585
Query: 493 EEATDHEEVESEKESNKDEEATEDEEEES 521
+ EEV +++E D + + E+ +S
Sbjct: 586 KVEEKSEEVATKEELVADAKVEKPEKAKS 614
Score = 57.8 bits (138), Expect = 7e-08
Identities = 37/126 (29%), Positives = 60/126 (47%), Gaps = 4/126 (3%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKED 457
E +VE+ + EE + ++ S+ E KEA EE E EE E E+E A+
Sbjct: 457 EETKVEDEKSEMEEALTAITEELAASMKEEKKEAAEEKE----EEPEAEEEEVAAKKSPV 512
Query: 458 ESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDE 517
++ ++ E ++EE ++ E DE ++ + EE +E SEKE + EE +
Sbjct: 513 KATAPEVKEEEGEKEEEEGQEEEEEEDEGAKSDQAEEGGSEKEGSSEKEEGEQEEGETEA 572
Query: 518 EEESSE 523
E E E
Sbjct: 573 EAEGEE 578
Score = 53.1 bits (126), Expect = 2e-06
Identities = 40/133 (30%), Positives = 70/133 (52%), Gaps = 14/133 (10%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAA-EDEEEETTEDEDASDDK 455
K E+ E TK E+E++ ++LT TEE+ A+ ++E++E E+++ +
Sbjct: 450 KFVEEIIEETKVEDEKSE-----MEEALTA----ITEELAASMKEEKKEAAEEKEEEPEA 500
Query: 456 EDESEEASTEDGEAT----EDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDE 511
E+E A +AT ++EE ++ E +EEEE E ++ EE SEKE + ++
Sbjct: 501 EEEEVAAKKSPVKATAPEVKEEEGEKEEEEGQEEEEEEDEGAKSDQAEEGGSEKEGSSEK 560
Query: 512 EATEDEEEESSED 524
E E EE E+ +
Sbjct: 561 EEGEQEEGETEAE 573
Score = 50.8 bits (120), Expect = 8e-06
Identities = 39/132 (29%), Positives = 63/132 (47%), Gaps = 9/132 (6%)
Query: 394 HGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASD 453
H F E E +DE+ E + L KE E+ EAAE++EEE +E+
Sbjct: 449 HKFVEEIIEETKVEDEKSEMEEALTAITEELAASMKE--EKKEAAEEKEEEPEAEEEEVA 506
Query: 454 DKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEA 513
K+ + + E ++EE ++ E +EEEE E ++ EE SEKE + ++E
Sbjct: 507 AKKSPVKATAPE----VKEEEGEKEEEEGQEEEEEEDEGAKSDQAEEGGSEKEGSSEKE- 561
Query: 514 TEDEEEESSEDD 525
+ E+E E +
Sbjct: 562 --EGEQEEGETE 571
Score = 35.0 bits (79), Expect = 0.48
Identities = 27/103 (26%), Positives = 45/103 (43%), Gaps = 6/103 (5%)
Query: 427 EFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTED------GEATEDEEATEDVE 480
E ++ EV E +EEE E ++A +++ E +E +D E+ EEA +V
Sbjct: 687 EEAKSKAEVGKGEQKEEEEKEVKEAPKEEKVEKKEEKPKDVPEKKKAESPVKEEAVAEVV 746
Query: 481 ATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSE 523
+ E E + + ++ EKE K EEE S +
Sbjct: 747 TITKSVKVHLEKETKEEGKPLQQEKEKEKAGGEGGSEEEGSDK 789
Score = 33.9 bits (76), Expect = 1.1
Identities = 35/139 (25%), Positives = 59/139 (42%), Gaps = 12/139 (8%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEF-KEATEEVEAAEDEEEET-------TED 448
K+ E ++A +EEA + KS+ KE EE + + E+E+ +E+
Sbjct: 725 KDVPEKKKAESPVKEEAVAEVVTITKSVKVHLEKETKEEGKPLQQEKEKEKAGGEGGSEE 784
Query: 449 EDASDDKEDESEEASTEDGEATEDEEATEDV-EATVDEEEEATEVEEATDHEEVESEKES 507
E + + +E +GE EE ++ E EEE V D + +K
Sbjct: 785 EGSDKGAKGSRKEDIAVNGEVEGKEEVEQETKEKGSGREEEKGVVTNGLDLSPADEKKGG 844
Query: 508 NKDEE---ATEDEEEESSE 523
+K EE T+ E+ +SE
Sbjct: 845 DKSEEKVVVTKTVEKITSE 863
Score = 32.0 bits (71), Expect = 4.1
Identities = 25/100 (25%), Positives = 50/100 (50%), Gaps = 9/100 (9%)
Query: 429 KEATEEVEAAEDEEEETTEDEDASDDKEDESEEA--STEDGEATEDEEATEDV------E 480
++A V + EE ++ + + KE+E +E + ++ + + EE +DV E
Sbjct: 675 EKAKSPVPKSPVEEAKSKAEVGKGEQKEEEEKEVKEAPKEEKVEKKEEKPKDVPEKKKAE 734
Query: 481 ATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEE 520
+ V +EE EV T +V EKE+ ++ + + E+E+
Sbjct: 735 SPV-KEEAVAEVVTITKSVKVHLEKETKEEGKPLQQEKEK 773
Score = 31.2 bits (69), Expect = 6.9
Identities = 19/54 (35%), Positives = 31/54 (57%), Gaps = 5/54 (9%)
Query: 479 VEATVDE---EEEATEVEEATDH--EEVESEKESNKDEEATEDEEEESSEDDYI 527
VE ++E E+E +E+EEA EE+ + + K E A E EEE +E++ +
Sbjct: 452 VEEIIEETKVEDEKSEMEEALTAITEELAASMKEEKKEAAEEKEEEPEAEEEEV 505
>NUCL_HUMAN (P19338) Nucleolin (Protein C23)
Length = 706
Score = 64.3 bits (155), Expect = 7e-10
Identities = 42/128 (32%), Positives = 66/128 (50%), Gaps = 13/128 (10%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTED-EDASDDKE 456
++ E EE +DE+E+ + ++K A A+EDE++E ED ED DD+E
Sbjct: 151 DSEEDEEDDEDEDEDEDEIEPAAMK--------AAAAAPASEDEDDEDDEDDEDDDDDEE 202
Query: 457 DESEEASTEDGEATEDEEA----TEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEE 512
D+SEE + E A + A + DE+EE + +E D +E + + + DEE
Sbjct: 203 DDSEEEAMETTPAKGKKAAKVVPVKAKNVAEDEDEEEDDEDEDDDDDEDDEDDDDEDDEE 262
Query: 513 ATEDEEEE 520
E+EEEE
Sbjct: 263 EEEEEEEE 270
Score = 60.8 bits (146), Expect = 8e-09
Identities = 36/122 (29%), Positives = 66/122 (53%), Gaps = 18/122 (14%)
Query: 422 KSLTCEFKEATEEVEAAEDEEEETTEDED----------------ASDDKEDESEEASTE 465
K+ E + E+ ++ EDEE++ EDED AS+D++DE +E +
Sbjct: 137 KNAKKEDSDEEEDDDSEEDEEDDEDEDEDEDEIEPAAMKAAAAAPASEDEDDEDDEDDED 196
Query: 466 DGEATEDEEATEDVEATVDEEEEATEVE--EATDHEEVESEKESNKDEEATEDEEEESSE 523
D + ED+ E +E T + ++A +V +A + E E E+E ++DE+ +DE++E +
Sbjct: 197 DDDDEEDDSEEEAMETTPAKGKKAAKVVPVKAKNVAEDEDEEEDDEDEDDDDDEDDEDDD 256
Query: 524 DD 525
D+
Sbjct: 257 DE 258
Score = 53.1 bits (126), Expect = 2e-06
Identities = 42/142 (29%), Positives = 67/142 (46%), Gaps = 19/142 (13%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDE------- 449
+E +E E+ +DE E A+ + + + E E E+ E +D+EE+ +E+E
Sbjct: 156 EEDDEDEDEDEDEIEPAAMKAAAAAPASEDEDDEDDEDDEDDDDDEEDDSEEEAMETTPA 215
Query: 450 -----------DASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDH 498
A + EDE EE ED E +D+E ED + DEEEE E EE
Sbjct: 216 KGKKAAKVVPVKAKNVAEDEDEEEDDED-EDDDDDEDDEDDDDEDDEEEEEEEEEEPVKE 274
Query: 499 EEVESEKESNKDEEATEDEEEE 520
+ +KE K + A E ++++
Sbjct: 275 APGKRKKEMAKQKAAPEAKKQK 296
Score = 48.1 bits (113), Expect = 5e-05
Identities = 34/126 (26%), Positives = 53/126 (41%), Gaps = 29/126 (23%)
Query: 429 KEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDE------------EAT 476
K A + A++ + ED D +D + E +E ED + EDE A+
Sbjct: 124 KGAAIPAKGAKNGKNAKKEDSDEEEDDDSEEDEEDDEDEDEDEDEIEPAAMKAAAAAPAS 183
Query: 477 EDVEATVDEEEEATEVEEATDHEEVESE-----------------KESNKDEEATEDEEE 519
ED + DE++E + +E D EE E K +DE+ ED+E+
Sbjct: 184 EDEDDEDDEDDEDDDDDEEDDSEEEAMETTPAKGKKAAKVVPVKAKNVAEDEDEEEDDED 243
Query: 520 ESSEDD 525
E +DD
Sbjct: 244 EDDDDD 249
Score = 47.4 bits (111), Expect = 9e-05
Identities = 36/122 (29%), Positives = 62/122 (50%), Gaps = 12/122 (9%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEV-----EAAEDEEEETTEDEDAS 452
E +E +E D+EE+ S ++++ + K+A + V AEDE+EE +DED
Sbjct: 189 EDDEDDEDDDDDEEDDS--EEEAMETTPAKGKKAAKVVPVKAKNVAEDEDEE-EDDEDED 245
Query: 453 DDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEE 512
DD +++ E+ ED E E+EE E V+ + ++ E A E++K+ + E
Sbjct: 246 DDDDEDDEDDDDEDDEEEEEEEEEEPVKEAPGKRKK----EMAKQKAAPEAKKQKVEGTE 301
Query: 513 AT 514
T
Sbjct: 302 PT 303
Score = 33.9 bits (76), Expect = 1.1
Identities = 15/70 (21%), Positives = 36/70 (51%)
Query: 427 EFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEE 486
E +E +E E +D+E++ +D++ +++E+E EE ++ +E + A ++
Sbjct: 235 EDEEEDDEDEDDDDDEDDEDDDDEDDEEEEEEEEEEPVKEAPGKRKKEMAKQKAAPEAKK 294
Query: 487 EEATEVEEAT 496
++ E T
Sbjct: 295 QKVEGTEPTT 304
>SEC7_YEAST (P11075) Protein transport protein SEC7
Length = 2009
Score = 63.5 bits (153), Expect = 1e-09
Identities = 39/132 (29%), Positives = 65/132 (48%), Gaps = 5/132 (3%)
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDD 454
G + A E + + +G+ T + + E + EDE+E+ EDED D+
Sbjct: 53 GPENAGSTAETKETSNDATNGMKTPEETEDTNDKRHDDEGEDGDEDEDEDEDEDEDNGDE 112
Query: 455 KEDE----SEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKD 510
+++ S E S+EDGE +E E E++ E+EE+ E + T + E S ++
Sbjct: 113 DDEDVDSSSSETSSEDGEDSESVSG-ESTESSSGEDEESDESDGNTSNSSSGDESGSEEE 171
Query: 511 EEATEDEEEESS 522
EE E+EEEE +
Sbjct: 172 EEEEEEEEEEEN 183
Score = 62.8 bits (151), Expect = 2e-09
Identities = 39/138 (28%), Positives = 65/138 (46%), Gaps = 9/138 (6%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAA-----EDEEEETTEDEDA 451
KE N ++ K E S + + EE E +DE E+ EDED
Sbjct: 42 KETNGEDQKCKGPENAGSTAETKETSNDATNGMKTPEETEDTNDKRHDDEGEDGDEDEDE 101
Query: 452 SDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESE----KES 507
+D+++++ + ED +++ E ++ED E + E+TE D E ES+ S
Sbjct: 102 DEDEDEDNGDEDDEDVDSSSSETSSEDGEDSESVSGESTESSSGEDEESDESDGNTSNSS 161
Query: 508 NKDEEATEDEEEESSEDD 525
+ DE +E+EEEE E++
Sbjct: 162 SGDESGSEEEEEEEEEEE 179
Score = 54.3 bits (129), Expect = 8e-07
Identities = 39/137 (28%), Positives = 61/137 (44%), Gaps = 8/137 (5%)
Query: 390 TSLPHGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDE 449
+S+ K N ++E K+ E T E KE + + EET E
Sbjct: 25 SSVNPSVKPQNAIKEEAKETNGEDQKCKGPENAGSTAETKETSNDATNGMKTPEET---E 81
Query: 450 DASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVE-SEKESN 508
D +D + D+ E DG+ EDE+ ED + +++E+ T E+ E SE S
Sbjct: 82 DTNDKRHDDEGE----DGDEDEDEDEDEDEDNGDEDDEDVDSSSSETSSEDGEDSESVSG 137
Query: 509 KDEEATEDEEEESSEDD 525
+ E++ E+EES E D
Sbjct: 138 ESTESSSGEDEESDESD 154
Score = 40.4 bits (93), Expect = 0.011
Identities = 36/144 (25%), Positives = 65/144 (45%), Gaps = 16/144 (11%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEA---AEDEEEETTEDEDASDD 454
E N V A K + E +S + S + + + + A +E ED++ + E+ ++ +
Sbjct: 3 EQNSVVNAEKGDGEISSNVETASSVNPSVKPQNAIKEEAKETNGEDQKCKGPENAGSTAE 62
Query: 455 KEDESEEAST-----EDGEATED-------EEATEDVEATVDEEEE-ATEVEEATDHEEV 501
++ S +A+ E+ E T D E+ ED + DE+E+ E +E D
Sbjct: 63 TKETSNDATNGMKTPEETEDTNDKRHDDEGEDGDEDEDEDEDEDEDNGDEDDEDVDSSSS 122
Query: 502 ESEKESNKDEEATEDEEEESSEDD 525
E+ E +D E+ E ESS +
Sbjct: 123 ETSSEDGEDSESVSGESTESSSGE 146
Score = 37.4 bits (85), Expect = 0.097
Identities = 24/101 (23%), Positives = 44/101 (42%), Gaps = 2/101 (1%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEAT--EEVEAAEDEEEETTEDEDASDDK 455
E + +E DE++E S S E E+ E E++ E+EE+ E + + +
Sbjct: 101 EDEDEDEDNGDEDDEDVDSSSSETSSEDGEDSESVSGESTESSSGEDEESDESDGNTSNS 160
Query: 456 EDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEAT 496
E S E+ E E+EE E+ +++ ++T
Sbjct: 161 SSGDESGSEEEEEEEEEEEEEENAGEPAIAHQDSVPTNDST 201
Score = 32.3 bits (72), Expect = 3.1
Identities = 26/124 (20%), Positives = 50/124 (39%), Gaps = 14/124 (11%)
Query: 404 EATKDEEEEASGLSLHSVKSLTCEFKEATEE--------------VEAAEDEEEETTEDE 449
E + ++ E++ +S S +S + E +E+ E E E+EEEE E+E
Sbjct: 123 ETSSEDGEDSESVSGESTESSSGEDEESDESDGNTSNSSSGDESGSEEEEEEEEEEEEEE 182
Query: 450 DASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNK 509
+A + + T D A ++ +T + T E + ++K+ K
Sbjct: 183 NAGEPAIAHQDSVPTNDSTAPRSTHTRNISLSSNGSNTNSTIILVKTTLETILNDKDIKK 242
Query: 510 DEEA 513
+ A
Sbjct: 243 NSNA 246
>NFM_BOVIN (O77788) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M) (Fragment)
Length = 810
Score = 63.5 bits (153), Expect = 1e-09
Identities = 46/145 (31%), Positives = 66/145 (44%), Gaps = 14/145 (9%)
Query: 394 HGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDE---- 449
H F E E +DE+ E + L KE +E EA E EE+E E+E
Sbjct: 335 HKFVEEIIEETKVEDEKSEMEEALTAITEELAVSVKEEVKEEEAEEKEEKEEAEEEVVAA 394
Query: 450 -------DASDDKEDESEEASTEDGEATEDEE---ATEDVEATVDEEEEATEVEEATDHE 499
A + KE+E E+ E E E+EE ++ E E+E ++E EE E
Sbjct: 395 KKSPVKATAPELKEEEGEKEEEEGQEEEEEEEEAAKSDQAEEGGSEKEGSSEKEEGEQEE 454
Query: 500 EVESEKESNKDEEATEDEEEESSED 524
E E+E E +E A E +EE+ E+
Sbjct: 455 EGETEAEGEGEEAAAEAKEEKKMEE 479
Score = 60.1 bits (144), Expect = 1e-08
Identities = 50/150 (33%), Positives = 75/150 (49%), Gaps = 10/150 (6%)
Query: 375 ESLLSFPSEQLHLRVTSLPHGFKEANEVEEATKDE-EEEASGLSLHSVKSLTCEFKEATE 433
E L+ +E+L S+ KE E+ K+E EEE VK+ E KE
Sbjct: 355 EEALTAITEEL---AVSVKEEVKEEEAEEKEEKEEAEEEVVAAKKSPVKATAPELKEEEG 411
Query: 434 EVEAAEDEEEETTEDEDASDDKEDE---SEEASTEDGEATEDEEATEDVEATVDEEEEAT 490
E E E +EEE E+E A D+ +E +E S+E E ++EE + EA + EE A
Sbjct: 412 EKEEEEGQEEEEEEEEAAKSDQAEEGGSEKEGSSEKEEGEQEEEG--ETEAEGEGEEAAA 469
Query: 491 EVEEATDHEEVESEKESNKDEEATEDEEEE 520
E +E EE ++E+ + K+E A E + E+
Sbjct: 470 EAKEEKKMEE-KAEEVAPKEELAAEAKVEK 498
Score = 44.7 bits (104), Expect = 6e-04
Identities = 34/97 (35%), Positives = 52/97 (53%), Gaps = 7/97 (7%)
Query: 429 KEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEE 488
+E E E +E EE T E+ + ++E +E E+ E E EEA E+V A +
Sbjct: 343 EETKVEDEKSEMEEALTAITEELAVSVKEEVKEEEAEEKE--EKEEAEEEVVAAKKSPVK 400
Query: 489 ATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
AT A + +E E EKE + +E E+EEEE+++ D
Sbjct: 401 AT----APELKEEEGEKEEEEGQEE-EEEEEEAAKSD 432
Score = 40.0 bits (92), Expect = 0.015
Identities = 28/97 (28%), Positives = 48/97 (48%), Gaps = 7/97 (7%)
Query: 430 EATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDV--EATVDEEE 487
E E+ E E+E++E E KE+++E+ + + E ++A V E+ V EE
Sbjct: 593 EKGEQKEKVEEEKKEAKESP-----KEEKAEKKEEKPKDVPEKKKAESPVKAESPVKEEV 647
Query: 488 EATEVEEATDHEEVESEKESNKDEEATEDEEEESSED 524
A V+ + + E E EK K++E + EE E+
Sbjct: 648 PAKPVKVSPEKEAKEEEKPQEKEKEKEKVEEVGGKEE 684
Score = 39.3 bits (90), Expect = 0.025
Identities = 36/125 (28%), Positives = 54/125 (42%), Gaps = 21/125 (16%)
Query: 416 LSLHSVKSLTCEFKEATEEVEAAEDEEE----ETTEDEDASDDKEDESEEASTEDGEATE 471
L H S+ K +VEA + + + E +E +D++ E EEA T A
Sbjct: 307 LYTHRQPSIAISSKIQKTKVEAPKLKVQHKFVEEIIEETKVEDEKSEMEEALT----AIT 362
Query: 472 DEEATEDVEATVDEEEEATEVEEATDHEEVES-------------EKESNKDEEATEDEE 518
+E A E +EE E E +E + E V + E+E K+EE ++EE
Sbjct: 363 EELAVSVKEEVKEEEAEEKEEKEEAEEEVVAAKKSPVKATAPELKEEEGEKEEEEGQEEE 422
Query: 519 EESSE 523
EE E
Sbjct: 423 EEEEE 427
Score = 38.9 bits (89), Expect = 0.033
Identities = 29/116 (25%), Positives = 49/116 (42%), Gaps = 25/116 (21%)
Query: 398 EANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKED 457
E E E ++EEEE + A D+ EE +++ S +KE+
Sbjct: 412 EKEEEEGQEEEEEEEEA----------------------AKSDQAEEGGSEKEGSSEKEE 449
Query: 458 ESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEA 513
+E E+GE + E E +E++ + EE EE+ +E + K E+A
Sbjct: 450 GEQE---EEGETEAEGEGEEAAAEAKEEKKMEEKAEEVAPKEELAAEAKVEKPEKA 502
Score = 38.5 bits (88), Expect = 0.043
Identities = 26/105 (24%), Positives = 51/105 (47%), Gaps = 7/105 (6%)
Query: 421 VKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVE 480
V+ + + + E+ E E EEE E +++ +++ E +E +D + E+ E
Sbjct: 581 VEEVKPKAEAGAEKGEQKEKVEEEKKEAKESPKEEKAEKKEEKPKDVPEKKKAESPVKAE 640
Query: 481 ATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDD 525
+ V EE A V+ V EKE+ ++E+ E E+E+ ++
Sbjct: 641 SPVKEEVPAKPVK-------VSPEKEAKEEEKPQEKEKEKEKVEE 678
Score = 36.2 bits (82), Expect = 0.22
Identities = 29/129 (22%), Positives = 53/129 (40%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
++A + EE KD E+ S +S E A + E E +E + ++ +KE
Sbjct: 615 EKAEKKEEKPKDVPEKKKAESPVKAESPVKEEVPAKPVKVSPEKEAKEEEKPQEKEKEKE 674
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATED 516
E E+G E + + V+ +EE E +E E E +N + + D
Sbjct: 675 KVEEVGGKEEGGLKESRKEDIAINGEVEGKEEEQETKEKGSGGEEEKGVVTNGLDVSPGD 734
Query: 517 EEEESSEDD 525
E++ + +
Sbjct: 735 EKKGGDKSE 743
Score = 36.2 bits (82), Expect = 0.22
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query: 473 EEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESSEDDYI 527
EE E+ + DE+ E E A E S KE K+EEA E EE+E +E++ +
Sbjct: 339 EEIIEETKVE-DEKSEMEEALTAITEELAVSVKEEVKEEEAEEKEEKEEAEEEVV 392
Score = 31.2 bits (69), Expect = 6.9
Identities = 36/141 (25%), Positives = 56/141 (39%), Gaps = 23/141 (16%)
Query: 397 KEANEVE-----EATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDA 451
KEA E E E K++ EE G +K E EVE E+E+E +
Sbjct: 658 KEAKEEEKPQEKEKEKEKVEEVGGKEEGGLKESRKEDIAINGEVEGKEEEQETKEKGSGG 717
Query: 452 SDDKE------DESEEASTEDGEATEDE-EATEDVEATVDE---------EEEATEVEEA 495
++K D S + G+ +E++ T+ VE E + T ++
Sbjct: 718 EEEKGVVTNGLDVSPGDEKKGGDKSEEKVVVTKMVEKITSEGGDGATKYITKSVTVTQKV 777
Query: 496 TDHEEVESEK--ESNKDEEAT 514
+HEE EK + K E+ T
Sbjct: 778 EEHEETFEEKLVSTKKVEKVT 798
Score = 30.8 bits (68), Expect = 9.0
Identities = 34/127 (26%), Positives = 56/127 (43%), Gaps = 10/127 (7%)
Query: 403 EEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEE--ETTEDEDASDDKEDES- 459
E K E G V+ E KE+ +E +A + EE+ + E + A + ES
Sbjct: 583 EVKPKAEAGAEKGEQKEKVEEEKKEAKESPKEEKAEKKEEKPKDVPEKKKAESPVKAESP 642
Query: 460 --EEASTEDGEATEDEEATEDV--EATVDEEEEATEVEEATDHEEVESEKES---NKDEE 512
EE + + + ++EA E+ + E+E+ EV + ES KE N + E
Sbjct: 643 VKEEVPAKPVKVSPEKEAKEEEKPQEKEKEKEKVEEVGGKEEGGLKESRKEDIAINGEVE 702
Query: 513 ATEDEEE 519
E+E+E
Sbjct: 703 GKEEEQE 709
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,982,794
Number of Sequences: 164201
Number of extensions: 2967253
Number of successful extensions: 65494
Number of sequences better than 10.0: 2209
Number of HSP's better than 10.0 without gapping: 1100
Number of HSP's successfully gapped in prelim test: 1183
Number of HSP's that attempted gapping in prelim test: 20566
Number of HSP's gapped (non-prelim): 16145
length of query: 527
length of database: 59,974,054
effective HSP length: 115
effective length of query: 412
effective length of database: 41,090,939
effective search space: 16929466868
effective search space used: 16929466868
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0476b.10


