

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0404.11
(365 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
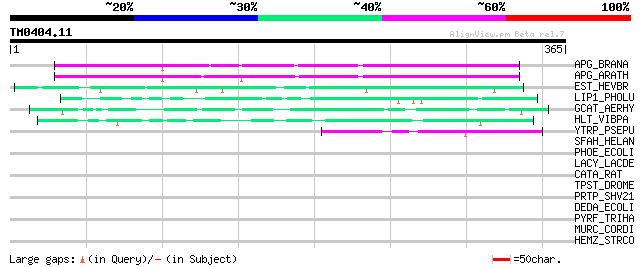
Score E
Sequences producing significant alignments: (bits) Value
APG_BRANA (P40603) Anter-specific proline-rich protein APG (Prot... 177 3e-44
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 172 1e-42
EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early nodule... 103 5e-22
LIP1_PHOLU (P40601) Lipase 1 precursor (EC 3.1.1.3) (Triacylglyc... 54 5e-07
GCAT_AERHY (P10480) Phosphatidylcholine-sterol acyltransferase p... 49 2e-05
HLT_VIBPA (Q99289) Thermolabile hemolysin precursor (TL) (Lecith... 47 8e-05
YTRP_PSEPU (P40604) Hypothetical 62.7 kDa protein in trpE-trpG i... 47 1e-04
SFAH_HELAN (P81098) Seed fatty acyl-ester hydrolase (EC 3.1.1.1)... 36 0.17
PHOE_ECOLI (P02932) Outer membrane pore protein E precursor 33 1.1
LACY_LACDE (P22733) Lactose permease (Lactose-proton symporter) ... 32 1.9
CATA_RAT (P04762) Catalase (EC 1.11.1.6) 32 1.9
TPST_DROME (Q9VYB7) Probable protein-tyrosine sulfotransferase (... 32 2.5
PRTP_SHV21 (P24911) Probable processing and transport protein 31 4.3
DEDA_ECOLI (P09548) DedA protein (DSG-1 protein) 31 4.3
PYRF_TRIHA (Q12709) Orotidine 5'-phosphate decarboxylase (EC 4.1... 31 5.6
MURC_CORDI (P61677) UDP-N-acetylmuramate--L-alanine ligase (EC 6... 30 9.6
HEMZ_STRCO (O50533) Probable ferrochelatase (EC 4.99.1.1) (Proto... 30 9.6
>APG_BRANA (P40603) Anter-specific proline-rich protein APG (Protein
CEX) (Fragment)
Length = 449
Score = 177 bits (450), Expect = 3e-44
Identities = 102/316 (32%), Positives = 159/316 (50%), Gaps = 17/316 (5%)
Query: 30 VPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGF 88
+P F FGDS+ D GNNN L + + +Y PYG+DFP G +GRFSNG+ D I++ LG
Sbjct: 123 IPAVFFFGDSIFDTGNNNNLDTKLKCNYRPYGMDFPMGVATGRFSNGRVASDYISKYLGV 182
Query: 89 DDFIPPYVSTS--------GDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQST 140
+ +P YV D+L GV+FAS AG +T + Q+ +Q
Sbjct: 183 KEIVPAYVDKKLQQNNELQQSDLLTGVSFASGGAGYLPQTSESWKVTTMLD-QLTYFQDY 241
Query: 141 VSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQA 200
++ ++G + + +SK + GSND + YF D + ++ +
Sbjct: 242 KKRMKKLVGKK-KTKKIVSKGAAIVVAGSNDLIYTYFGNGAQHLKND--VDSFTTMMADS 298
Query: 201 YTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVV 260
+ LY +GAR++ + G IGC+P+ QR + + C D+N A Q+FN+KL ++
Sbjct: 299 AASFVLQLYGYGARRIGVIGTPPIGCTPS---QRVKKKKICNEDLNYAAQLFNSKLVIIL 355
Query: 261 DQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTP-C 319
Q + LP++ ++Y + Y IF ++ SP YGF + CC +G G + C
Sbjct: 356 GQLSKTLPNSTIVYGDIYSIFSKMLESPEDYGFEEIKKPCCKIGLTKGGVFCKERTLKNM 415
Query: 320 ENRREYLFWDAFHPSE 335
N YLFWD HPS+
Sbjct: 416 SNASSYLFWDGLHPSQ 431
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 172 bits (436), Expect = 1e-42
Identities = 98/319 (30%), Positives = 161/319 (49%), Gaps = 19/319 (5%)
Query: 30 VPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGF 88
+P F FGDS+ D GNNN L + +++Y PYG+DF +GRFSNG D +A+ +G
Sbjct: 202 IPAVFFFGDSVFDTGNNNNLETKIKSNYRPYGMDFKFRVATGRFSNGMVASDYLAKYMGV 261
Query: 89 DDFIPPYVSTS--GDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVN 146
+ +P Y+ +D+L GV+FAS AG T + I Q+ +Q + +V
Sbjct: 262 KEIVPAYLDPKIQPNDLLTGVSFASGGAGYNPTTSEAANA-IPMLDQLTYFQDYIEKVNR 320
Query: 147 ILGNE---------DQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVL 197
++ E ++ +SK + + GSND + YF D Y ++
Sbjct: 321 LVRQEKSQYKLAGLEKTNQLISKGVAIVVGGSNDLIITYFGSGAQRLKNDI--DSYTTII 378
Query: 198 IQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLK 257
+ + LY +GAR++ + G +GC P+ QR + + C ++N A+Q+FN+KL
Sbjct: 379 ADSAASFVLQLYGYGARRIGVIGTPPLGCVPS---QRLKKKKICNEELNYASQLFNSKLL 435
Query: 258 SVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQT 317
++ Q + LP++ +Y++ Y I ++ +PA YGF T + CC G + C +
Sbjct: 436 LILGQLSKTLPNSTFVYMDIYTIISQMLETPAAYGFEETKKPCCKTGLLSAGALCKKSTS 495
Query: 318 P-CENRREYLFWDAFHPSE 335
C N YLFWD HP++
Sbjct: 496 KICPNTSSYLFWDGVHPTQ 514
>EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early
nodule-specific protein homolog) (Latex allergen Hev b
13)
Length = 391
Score = 103 bits (258), Expect = 5e-22
Identities = 94/364 (25%), Positives = 148/364 (39%), Gaps = 44/364 (12%)
Query: 4 NVNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYM---PY 60
N ++ ++ L+ +L L + P F FGDS D G A A Y PY
Sbjct: 8 NNPIITLSFLLCMLSL--AYASETCDFPAIFNFGDSNSDTGGK------AAAFYPLNPPY 59
Query: 61 GIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREET 120
G F +GR+S+G+ +D IAE ++ PY+S+ G + G +FA+A + I+ T
Sbjct: 60 GETFFHRSTGRYSDGRLIIDFIAESFNLP-YLSPYLSSLGSNFKHGADFATAGSTIKLPT 118
Query: 121 G--QQLGGRISFSGQVQNYQ--------STVSQVVNILGNEDQAANYLSKCIYSIGLGSN 170
GG F VQ Q + + I Y K +Y+ +G N
Sbjct: 119 TIIPAHGGFSPFYLDVQYSQFRQFIPRSQFIRETGGIFAELVPEEYYFEKALYTFDIGQN 178
Query: 171 DYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNE 230
D F + + + D L+ +++ ++ +Y+ GAR + G IGC
Sbjct: 179 DLTEG-----FLNLTVEEVNATVPD-LVNSFSANVKKIYDLGARTFWIHNTGPIGCLSFI 232
Query: 231 LAQ---RSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIAS 287
L +D C N+ Q FN+KLK +V Q LP A ++++ Y + + +
Sbjct: 233 LTYFPWAEKDSAGCAKAYNEVAQHFNHKLKEIVAQLRKDLPLATFVHVDIYSVKYSLFSE 292
Query: 288 PATYGFSNTNEGCCGV-GRNNGQITCLPMQT------------PCENRREYLFWDAFHPS 334
P +GF CCG G+ N +T T C + WD H +
Sbjct: 293 PEKHGFEFPLITCCGYGGKYNFSVTAPCGDTVTADDGTKIVVGSCACPSVRVNWDGAHYT 352
Query: 335 EAGN 338
EA N
Sbjct: 353 EAAN 356
>LIP1_PHOLU (P40601) Lipase 1 precursor (EC 3.1.1.3)
(Triacylglycerol lipase)
Length = 645
Score = 54.3 bits (129), Expect = 5e-07
Identities = 77/368 (20%), Positives = 130/368 (34%), Gaps = 105/368 (28%)
Query: 34 FIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIP 93
++FGDSL D GNN Y +D G + N D IA+ LG I
Sbjct: 29 YVFGDSLSDGGNNGR-----------YTVDGINGTESKLYN-----DFIAQQLG----IE 68
Query: 94 PYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQ 153
S G G N+A+ A + + N +T QV+ L +
Sbjct: 69 LVNSKKG-----GTNYAAGGA--------------TAVADLNNKHNTQDQVMGYLASHSN 109
Query: 154 AANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGA 213
A++ ++ IG ND P + +++ T+ A + Q+ L N GA
Sbjct: 110 RADHNGMYVHWIG--GNDVDAALRNP---ADAQKIITES-----AMAASSQVHALLNAGA 159
Query: 214 RKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNN------------------- 254
+++ + +G +P + G D+ + + N
Sbjct: 160 GLVIVPTVPDVGMTPKIMEFVLSKGGATSKDLAKIHAVVNGYPTIDKDTRLQVIHGVFKQ 219
Query: 255 ---------------KLKSVVDQFN-------------NQLPD-------ARVIYINAYG 279
K ++D +N NQL D ++ ++
Sbjct: 220 IGSDVSGGDAKKAEETTKQLIDGYNELSSNASKLVDNYNQLEDMALSQENGNIVRVDVNA 279
Query: 280 IFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNV 339
+ ++IA+P YGF NT C G N G +C T + + +LF D FHP+ +
Sbjct: 280 LLHEVIANPLRYGFLNTIGYACAQGVNAG--SCRSKDTGFDASKPFLFADDFHPTPEAHH 337
Query: 340 VIAQRAYS 347
+++Q S
Sbjct: 338 IVSQYTVS 345
>GCAT_AERHY (P10480) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43) (Glycerophospholipid-cholesterol
acyltransferase) (GCAT)
Length = 335
Score = 48.9 bits (115), Expect = 2e-05
Identities = 79/348 (22%), Positives = 136/348 (38%), Gaps = 66/348 (18%)
Query: 14 VMVLGLWGGWVGAAPQVPCY---FIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSG 70
V +LGL V AA P + +FGDSL D G + S R Y+P P G
Sbjct: 6 VCLLGLVALTVQAADSRPAFSRIVMFGDSLSDTGK---MYSKMRG-YLPSS---PPYYEG 58
Query: 71 RFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISF 130
RFSNG ++ + + G+ A+ A G +IS+
Sbjct: 59 RFSNGPVWLEQLT------------------NEFPGLTIANEAEGGPTAVAYN---KISW 97
Query: 131 SGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTT 190
+ + Q + +V L Q ++ + + +G+NDYL + + T
Sbjct: 98 NPKYQVINNLDYEVTQFL----QKDSFKPDDLVILWVGANDYL-----------AYGWNT 142
Query: 191 DQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQ 250
+Q A + A ++ + GA++++LF + +G +P+ +Q+ + + VS + NQ
Sbjct: 143 EQDAKRVRDAISDAANRMVLNGAKEILLFNLPDLGQNPSARSQKVVEAASHVSAYH--NQ 200
Query: 251 IFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQI 310
+ N + + P V F +++ P +G S+ C G G
Sbjct: 201 LLLNLARQLA-------PTGMVKLFEIDKQFAEMLRDPQNFGLSDQRNACYG-----GSY 248
Query: 311 TCLPMQTPCENRREYLFWDAFHPSE----AGNVVIAQRAYSAQSPSDA 354
P + + L AF+P E AGN ++AQ S + A
Sbjct: 249 VWKPFASRSASTDSQL--SAFNPQERLAIAGNPLLAQAVASPMAARSA 294
>HLT_VIBPA (Q99289) Thermolabile hemolysin precursor (TL)
(Lecithin-dependent haemolysin) (LDH) (Atypical
phospholipase) (Phospholipase A2) (Lysophospholipase)
Length = 418
Score = 47.0 bits (110), Expect = 8e-05
Identities = 64/334 (19%), Positives = 124/334 (36%), Gaps = 68/334 (20%)
Query: 19 LWGGWVGAAP-QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPS---GRFSN 74
+W P Q+ GDSL D GN + A FP S G FSN
Sbjct: 132 IWSNDAAMQPDQINKVVALGDSLSDTGN------IFNASQWR----FPNPNSWFLGHFSN 181
Query: 75 GKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQV 134
G + IA+ +P Y G G N A G+ G+Q+ ++++
Sbjct: 182 GFVWTEYIAKAKN----LPLYNWAVGG--AAGENQYIALTGV----GEQVSSYLTYAKLA 231
Query: 135 QNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYA 194
+NY+ + ++++ G ND++N Y+ YA
Sbjct: 232 KNYKP-------------------ANTLFTLEFGLNDFMN-------YNRGVPEVKADYA 265
Query: 195 DVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNN 254
+ LI+ L + GA+ +L + +P ++ + + + N+
Sbjct: 266 EALIR--------LTDAGAKNFMLMTLPDATKAPQFKYSTQEEIDKIRAKVLEMNEFIKA 317
Query: 255 KLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNG----QI 310
+ Q N + + + +F+ + ++P +GF N ++ C + R++
Sbjct: 318 QAMYYKAQGYN------ITLFDTHALFETLTSAPEEHGFVNASDPCLDINRSSSVDYMYT 371
Query: 311 TCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQR 344
L + +++FWD HP+ A + +A++
Sbjct: 372 HALRSECAASGAEKFVFWDVTHPTTATHRYVAEK 405
>YTRP_PSEPU (P40604) Hypothetical 62.7 kDa protein in trpE-trpG
intergenic region precursor
Length = 592
Score = 46.6 bits (109), Expect = 1e-04
Identities = 39/147 (26%), Positives = 64/147 (43%), Gaps = 13/147 (8%)
Query: 206 QTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNN 265
Q L GAR ++++ + +G +PN Q + +S + FN L S + Q
Sbjct: 180 QALQQGGARYIMVWLLPDLGQTPNFSGTPQQTPLSLLSGV------FNQSLLSQLGQI-- 231
Query: 266 QLPDARVIYINAYGIFQDIIASPATYGFSNTNE--GCCGVGRNNGQITCLPMQTPCENRR 323
DA +I +N + + +ASP+ +G + G C G + + +
Sbjct: 232 ---DAEIIPLNVPMLLSEALASPSQFGLATGQNLVGTCSSGEGCVENPVYGINGTTPDPT 288
Query: 324 EYLFWDAFHPSEAGNVVIAQRAYSAQS 350
+ LF D HP+ AG +IA AYS S
Sbjct: 289 KLLFNDLVHPTIAGQQLIADYAYSILS 315
>SFAH_HELAN (P81098) Seed fatty acyl-ester hydrolase (EC 3.1.1.1)
(Fragment)
Length = 18
Score = 35.8 bits (81), Expect = 0.17
Identities = 15/17 (88%), Positives = 15/17 (88%)
Query: 28 PQVPCYFIFGDSLVDNG 44
PQVP YFIFGDSL DNG
Sbjct: 2 PQVPXYFIFGDSLYDNG 18
>PHOE_ECOLI (P02932) Outer membrane pore protein E precursor
Length = 351
Score = 33.1 bits (74), Expect = 1.1
Identities = 41/171 (23%), Positives = 65/171 (37%), Gaps = 20/171 (11%)
Query: 125 GGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYST 184
G + SG N T Q + G +A + + Y NN ++ FYS
Sbjct: 199 GSDFAISGAYTNSDRTNEQNLQSRGTGKRAEAWATGLKYDA--------NNIYLATFYSE 250
Query: 185 SRQYT--TDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCV 242
+R+ T T +A+ Q + Q ++FG R +G + ++ +
Sbjct: 251 TRKMTPITGGFAN-KTQNFEAVAQYQFDFGLRP----SLGYVLSKGKDIEGIGDEDLVNY 305
Query: 243 SDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGF 293
D+ A FN + + VD NQL + IN DI+A TY F
Sbjct: 306 IDVG-ATYYFNKNMSAFVDYKINQLDSDNKLNINN----DDIVAVGMTYQF 351
>LACY_LACDE (P22733) Lactose permease (Lactose-proton symporter)
(Lactose transport protein)
Length = 627
Score = 32.3 bits (72), Expect = 1.9
Identities = 26/108 (24%), Positives = 46/108 (42%), Gaps = 12/108 (11%)
Query: 136 NYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSND------YLNNYFMPQFYSTSRQYT 189
N ++T+ QV +LG DQ +L+ + GLG N Y +Y + S YT
Sbjct: 226 NEKTTLKQVFKVLGQNDQLL-WLAFAYWFYGLGINTLNALQLYYFSYILGDARGYSLLYT 284
Query: 190 TDQYADVLIQAYTEQLQTLYN-----FGARKMVLFGIGQIGCSPNELA 232
+ + ++ ++ L +N + ++L GIG + LA
Sbjct: 285 INTFVGLISASFFPSLAKKFNRNRLFYACIAVMLLGIGVFSVASGSLA 332
>CATA_RAT (P04762) Catalase (EC 1.11.1.6)
Length = 526
Score = 32.3 bits (72), Expect = 1.9
Identities = 49/221 (22%), Positives = 88/221 (39%), Gaps = 37/221 (16%)
Query: 63 DFPGGPSGRFSNGKTTVDVIAEL--LGFDDF-IPPYVSTSGDDILRGVNFASAAAGIREE 119
D+P P G+ + + AE+ + FD +PP + S D +L+G FA R
Sbjct: 306 DYPLIPVGKLVLNRNPANYFAEVEQMAFDPSNMPPGIEPSPDKMLQGRLFAYPDTH-RHR 364
Query: 120 TGQ---QLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNY 176
G Q+ + +V NYQ ++ N+ A NY Y N++
Sbjct: 365 LGPNYLQIPVNCPYRARVANYQRDGPMCMH--DNQGGAPNY--------------YPNSF 408
Query: 177 FMPQFYSTSRQYTTDQYADV-----LIQAYTEQLQTLY----NFGARKMVLFGIGQIGCS 227
P+ ++ ++ + ADV + Q++T Y N RK + I
Sbjct: 409 SAPEQQGSALEHHSQCSADVKRFNSANEDNVTQVRTFYTKVLNEEERKRLCENIANHLKD 468
Query: 228 PNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLP 268
QR V + D + + ++++++DQ+N+Q P
Sbjct: 469 AQLFIQRK-----AVKNFTDVHPDYGARVQALLDQYNSQKP 504
>TPST_DROME (Q9VYB7) Probable protein-tyrosine sulfotransferase (EC
2.8.2.20) (Tyrosylprotein sulfotransferase) (TPST)
Length = 499
Score = 32.0 bits (71), Expect = 2.5
Identities = 28/133 (21%), Positives = 55/133 (41%), Gaps = 12/133 (9%)
Query: 75 GKTTVDVIAELLGFDDFIPPYVSTSG-DDILRGVNFASAAAGIREETGQQLGGRISFSGQ 133
G+ DV+ ++ I P +S G D ++ A +++ T + R+
Sbjct: 305 GQIPGDVVRDMAD----IAPMLSVLGYDPYANPPDYGKPDAWVQDNTSKLKANRML---- 356
Query: 134 VQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQY 193
++S QV+ + +ED N + + +N Y N +P+ +S RQ+ Q+
Sbjct: 357 ---WESKAKQVLQMSSSEDDNTNTIINNSNNKDNNNNQYTINKIIPEQHSRQRQHVQQQH 413
Query: 194 ADVLIQAYTEQLQ 206
Q + +Q Q
Sbjct: 414 LQQQQQQHLQQQQ 426
>PRTP_SHV21 (P24911) Probable processing and transport protein
Length = 679
Score = 31.2 bits (69), Expect = 4.3
Identities = 21/81 (25%), Positives = 37/81 (44%), Gaps = 11/81 (13%)
Query: 264 NNQLPDARVIYINAYGIFQDIIA-----SPATYGFSNTNEGCCGVGRNNGQITCLPMQTP 318
+NQ+ DA + +N + IF++I A S Y S ++ C V + L +
Sbjct: 248 DNQIRDASLSKLNQHTIFENISAPVLELSNLIYWSSGAHKKCANVENTSEMAKLLSYEAK 307
Query: 319 CENRREYL------FWDAFHP 333
+N R+Y+ F+D + P
Sbjct: 308 MQNYRKYICKNSTHFFDKYKP 328
>DEDA_ECOLI (P09548) DedA protein (DSG-1 protein)
Length = 219
Score = 31.2 bits (69), Expect = 4.3
Identities = 19/49 (38%), Positives = 25/49 (50%), Gaps = 6/49 (12%)
Query: 2 DLNVNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALR 50
DLNV+MMVV ML+ + VG A +FG+ L N N+ R
Sbjct: 68 DLNVHMMVVLMLIAAI------VGDAVNYTIGRLFGEKLFSNPNSKIFR 110
>PYRF_TRIHA (Q12709) Orotidine 5'-phosphate decarboxylase (EC
4.1.1.23) (OMP decarboxylase) (OMPDCase) (OMPdecase)
(Uridine 5'-monophosphate synthase) (UMP synthase)
Length = 379
Score = 30.8 bits (68), Expect = 5.6
Identities = 20/85 (23%), Positives = 37/85 (43%), Gaps = 4/85 (4%)
Query: 70 GRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIR---EETGQQLGG 126
G F G+ T + DF+ ++S + +F G + E+ QQ G
Sbjct: 261 GNFMTGEYTQACVEAAREHKDFVMGFISQEALNTQADDDFIHMTPGCQLPPEDEDQQTNG 320
Query: 127 RISFSGQVQNYQSTVSQVVNILGNE 151
++ GQ Q Y +T +++ I G++
Sbjct: 321 KVGGDGQGQQY-NTAHKIIGIAGSD 344
>MURC_CORDI (P61677) UDP-N-acetylmuramate--L-alanine ligase (EC
6.3.2.8) (UDP-N-acetylmuramoyl-L-alanine synthetase)
Length = 477
Score = 30.0 bits (66), Expect = 9.6
Identities = 37/167 (22%), Positives = 68/167 (40%), Gaps = 25/167 (14%)
Query: 65 PGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVS-----TSGDDILRGVNFASAAAGIR-- 117
P G R + G TVDVI + G + + T+G ++ R S +G+R
Sbjct: 267 PAGTRARITVGDDTVDVIMRIPGHHMVLNACAALTAGVTAGAELERLAAGISDFSGVRRR 326
Query: 118 -EETGQQLGGRISFSGQVQNYQSTVSQVVNIL--GNEDQAANYLSKCIYSIGLGSNDYLN 174
E G+ GG + +Y ++V +L + Q A + + + +
Sbjct: 327 FEFHGRVSGGAFHDAEVYDDYAHHPTEVTAVLKAARQQQEARGIGRVVVA---------- 376
Query: 175 NYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGI 221
F P YS + ++ ++A+ L + + L FGAR+ + G+
Sbjct: 377 --FQPHLYSRTMEFAA-EFAEAL--SLADHAVILDIFGAREQPVEGV 418
>HEMZ_STRCO (O50533) Probable ferrochelatase (EC 4.99.1.1)
(Protoheme ferro-lyase) (Heme synthetase)
Length = 375
Score = 30.0 bits (66), Expect = 9.6
Identities = 13/33 (39%), Positives = 17/33 (51%)
Query: 33 YFIFGDSLVDNGNNNALRSLARADYMPYGIDFP 65
YF+FG N N AL R D+ +G+D P
Sbjct: 52 YFLFGGVSPINDQNRALLDALRKDFAEHGLDLP 84
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,958,860
Number of Sequences: 164201
Number of extensions: 1954926
Number of successful extensions: 4435
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 4412
Number of HSP's gapped (non-prelim): 19
length of query: 365
length of database: 59,974,054
effective HSP length: 112
effective length of query: 253
effective length of database: 41,583,542
effective search space: 10520636126
effective search space used: 10520636126
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0404.11


