

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.8
(721 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
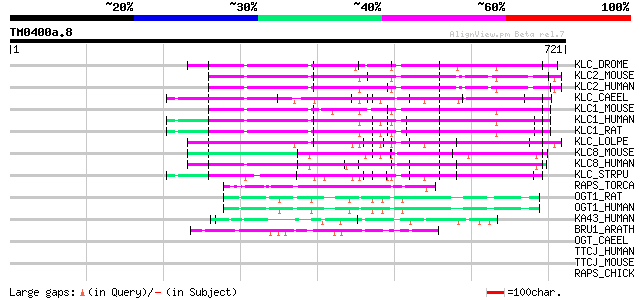
KLC_DROME (P46824) Kinesin light chain (KLC) 82 5e-15
KLC2_MOUSE (O88448) Kinesin light chain 2 (KLC 2) 80 2e-14
KLC2_HUMAN (Q9H0B6) Kinesin light chain 2 (KLC 2) 79 5e-14
KLC_CAEEL (P46822) Kinesin light chain (KLC) 77 1e-13
KLC1_MOUSE (O88447) Kinesin light chain 1 (KLC 1) 76 3e-13
KLC1_HUMAN (Q07866) Kinesin light chain 1 (KLC 1) 76 3e-13
KLC1_RAT (P37285) Kinesin light chain 1 (KLC 1) 75 8e-13
KLC_LOLPE (P46825) Kinesin light chain (KLC) 72 4e-12
KLC8_MOUSE (Q9DBS5) Kinesin-like protein 8 72 4e-12
KLC8_HUMAN (Q9NSK0) Kinesin-like protein 8 72 4e-12
KLC_STRPU (Q05090) Kinesin light chain (KLC) 67 1e-10
RAPS_TORCA (P09108) 43 kDa receptor-associated protein of the sy... 51 1e-05
OGT1_RAT (P56558) UDP-N-acetylglucosamine--peptide N-acetylgluco... 51 1e-05
OGT1_HUMAN (O15294) UDP-N-acetylglucosamine--peptide N-acetylglu... 51 1e-05
KA43_HUMAN (Q96AY4) TPR repeat containing protein KIAA1043 50 2e-05
BRU1_ARATH (Q6Q4D0) BRUSHY protein 1 (TONSOKU protein) (MGOUN pr... 45 9e-04
OGT_CAEEL (O18158) UDP-N-acetylglucosamine--peptide N-acetylgluc... 44 0.001
TTCJ_HUMAN (Q6DKK2) Tetratricopeptide repeat protein 19 (TPR rep... 42 0.006
TTCJ_MOUSE (Q8CC21) Tetratricopeptide repeat protein 19 (TPR rep... 41 0.010
RAPS_CHICK (O42393) 43 kDa receptor-associated protein of the sy... 41 0.010
>KLC_DROME (P46824) Kinesin light chain (KLC)
Length = 508
Score = 82.0 bits (201), Expect = 5e-15
Identities = 78/336 (23%), Positives = 141/336 (41%), Gaps = 43/336 (12%)
Query: 395 YDSKGDYEAALE--HYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKAL 452
Y S+G YE A+ L + ++ DVA++ + + Y +Y EA AL
Sbjct: 196 YASQGRYEVAVPLCKQALEDLERTSGHDHPDVATMLNILALVYRDQNKYKEAANLLNDAL 255
Query: 453 TVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL----RIFGKSKPGIPPEE 508
++ T GENHP VA+ LA LY K GK+KD++ C+ AL ++ GK P +
Sbjct: 256 SIRGKTLGENHPAVAATLNNLAVLYGKRGKYKDAEPLCKRALEIREKVLGKDHP-----D 310
Query: 509 IANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNY 567
+A L ++A + Q+ ++ K ++AL IY G +A + + Y G Y
Sbjct: 311 VAKQLNNLALLCQNQGKYDEVEKYYQRALDIYESKLGPDDPNVAKTKNNLAGCYLKQGRY 370
Query: 568 SDSYNIFKSAVAK-----FRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTI 622
+++ ++K + + F A K ++ +A + F+
Sbjct: 371 TEAEILYKQVLTRAHEREFGAIDSKNKPIWQVAEER-------------EEHKFDNRENT 417
Query: 623 LEKEYGQYH-------PDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPD 675
EYG +H P NL Y G + A + + + ++E A
Sbjct: 418 PYGEYGGWHKAAKVDSPTVTTTLKNLGALYRRQGMFEAAETLEDCAMRSKKEAYDLA--- 474
Query: 676 VDDEKRRLAELLKEAGRGRNRKSKRSLETLLDANSR 711
++ +L++LL + R++ K L+ + N++
Sbjct: 475 ---KQTKLSQLLTSNEKRRSKAIKEDLDFSEEKNAK 507
Score = 63.2 bits (152), Expect = 2e-09
Identities = 48/198 (24%), Positives = 86/198 (43%), Gaps = 7/198 (3%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y + G+Y A+P+ +++++ G DH +A + L Y +
Sbjct: 189 LHNLVIQYASQGRYEVAVPLCKQALEDLERTSGHDHPDVATMLNILAL--VYRDQNKYKE 246
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+G+ LGE P T +A + + ++ +AE LC+ ALEI
Sbjct: 247 AANLLNDALSIRGKTLGENHPAVAATLNNLAVLYGKRGKYKDAEPLCKRALEIREKVLGK 306
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSIGDAY 435
V + + + L+ ++G Y+ ++Y L + +VA +++ Y
Sbjct: 307 DHPDVAKQLNN--LALLCQNQGKYDEVEKYYQRALDIYESKLGPDDPNVAKTKNNLAGCY 364
Query: 436 LALARYDEAVFSYQKALT 453
L RY EA Y++ LT
Sbjct: 365 LKQGRYTEAEILYKQVLT 382
Score = 54.3 bits (129), Expect = 1e-06
Identities = 36/134 (26%), Positives = 58/134 (42%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + + K A+ + LN + L + EAA+L +
Sbjct: 194 IQYASQGRYEVAVPLCKQALEDLERTSGHDHPDVATMLNILALVYRDQNKYKEAANLLND 253
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A +I K G+ HP +NLA Y G+ DA + + + +RE+ LG +PDV
Sbjct: 254 ALSIRGKTLGENHPAVAATLNNLAVLYGKRGKYKDAEPLCKRALEIREKVLGKDHPDVAK 313
Query: 679 EKRRLAELLKEAGR 692
+ LA L + G+
Sbjct: 314 QLNNLALLCQNQGK 327
Score = 50.8 bits (120), Expect = 1e-05
Identities = 60/270 (22%), Positives = 114/270 (42%), Gaps = 15/270 (5%)
Query: 232 ALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDG 291
A+ L +AL+ E + G ++ L++LA +Y + +Y EA +L ++ I G
Sbjct: 205 AVPLCKQALEDLERTS-GHDHPDVATMLNILALVYRDQNKYKEAANLLNDALSIRGKTLG 263
Query: 292 QDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAH 351
++H A A L Y G +++ LEI+ +VLG+ P + +A
Sbjct: 264 ENHP-AVAATLNNLAVLYGKRGKYKDAEPLCKRALEIREKVLGKDHPDVAKQLNNLALLC 322
Query: 352 VQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA 411
++DE E+ Q AL+I+ +V ++ + + Y +G Y A Y
Sbjct: 323 QNQGKYDEVEKYYQRALDIYESKLGPDDPNVAKTKNN--LAGCYLKQGRYTEAEILY--- 377
Query: 412 SMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFS------YQKALTVFKSTKGENHPN 465
+ E++ ++DS + +E F Y + K+ K ++ P
Sbjct: 378 -KQVLTRAHEREFGAIDSKNKPIWQVAEEREEHKFDNRENTPYGEYGGWHKAAKVDS-PT 435
Query: 466 VASVYVRLADLYNKIGKFKDSKSYCENALR 495
V + L LY + G F+ +++ + A+R
Sbjct: 436 VTTTLKNLGALYRRQGMFEAAETLEDCAMR 465
>KLC2_MOUSE (O88448) Kinesin light chain 2 (KLC 2)
Length = 599
Score = 80.1 bits (196), Expect = 2e-14
Identities = 78/320 (24%), Positives = 142/320 (44%), Gaps = 35/320 (10%)
Query: 395 YDSKGDYEAALE--HYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKAL 452
Y S+G YE A+ L + ++ DVA++ + + Y +Y +A AL
Sbjct: 207 YASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKDAAHLLNDAL 266
Query: 453 TVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL----RIFGKSKPGIPPEE 508
+ + T G++HP VA+ LA LY K GK+K+++ C+ AL ++ GK P +
Sbjct: 267 AIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKFHP-----D 321
Query: 509 IANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNY 567
+A L ++A + Q+ E+ ++AL+IY G +A + + Y G Y
Sbjct: 322 VAKQLSNLALLCQNQGKAEEVEYYYRRALEIYATRLGPDDPNVAKTKNNLASCYLKQGKY 381
Query: 568 SDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEY 627
D+ ++K + + + EK+ FG + + E+ D + R + EY
Sbjct: 382 QDAETLYKEILTR---AHEKE---FGSVNGENKPIWMHAEEREESKDKRRDRRPM---EY 432
Query: 628 GQYH-------PDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEK 680
G ++ P L Y G+++ A + + R++ L A+ +
Sbjct: 433 GSWYKACKVDSPTVNTTLRTLGALYRPEGKLEAAHTLEDCASRSRKQGLDPAS------Q 486
Query: 681 RRLAELLKE-AGRGRNRKSK 699
++ ELLK+ +GRG R S+
Sbjct: 487 TKVVELLKDGSGRGHRRGSR 506
Score = 58.9 bits (141), Expect = 4e-08
Identities = 52/214 (24%), Positives = 90/214 (41%), Gaps = 12/214 (5%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y + G+Y A+P+ +++++ G DH +A + L Y ++
Sbjct: 200 LHNLVIQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILAL--VYRDQNKYKD 257
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+ + LG+ P T +A + + ++ EAE LC+ ALEI
Sbjct: 258 AAHLLNDALAIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGK 317
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSIGDAY 435
V + + L+ ++G E +Y L A + +VA +++ Y
Sbjct: 318 FHPDVAKQLSN--LALLCQNQGKAEEVEYYYRRALEIYATRLGPDDPNVAKTKNNLASCY 375
Query: 436 LALARYDEAVFSYQKALT-----VFKSTKGENHP 464
L +Y +A Y++ LT F S GEN P
Sbjct: 376 LKQGKYQDAETLYKEILTRAHEKEFGSVNGENKP 409
Score = 52.8 bits (125), Expect = 3e-06
Identities = 43/166 (25%), Positives = 68/166 (40%), Gaps = 8/166 (4%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + + K A+ + LN + L + +AA L +
Sbjct: 205 IQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKDAAHLLND 264
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A I EK G+ HP +NLA Y G+ +A + + + +RE+ LG +PDV
Sbjct: 265 ALAIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKFHPDVAK 324
Query: 679 EKRRLAELLKEAGRGRNRK--SKRSLETLL------DANSRLIKNN 716
+ LA L + G+ + +R+LE D N KNN
Sbjct: 325 QLSNLALLCQNQGKAEEVEYYYRRALEIYATRLGPDDPNVAKTKNN 370
>KLC2_HUMAN (Q9H0B6) Kinesin light chain 2 (KLC 2)
Length = 622
Score = 78.6 bits (192), Expect = 5e-14
Identities = 78/323 (24%), Positives = 143/323 (44%), Gaps = 34/323 (10%)
Query: 395 YDSKGDYEAALE--HYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKAL 452
Y S+G YE A+ L + ++ DVA++ + + Y +Y EA AL
Sbjct: 208 YASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKEAAHLLNDAL 267
Query: 453 TVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL----RIFGKSKPGIPPEE 508
+ + T G++HP VA+ LA LY K GK+K+++ C+ AL ++ GK P +
Sbjct: 268 AIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKFHP-----D 322
Query: 509 IANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNY 567
+A L ++A + Q+ E+ ++AL+IY G +A + + Y G Y
Sbjct: 323 VAKQLSNLALLCQNQGKAEEVEYYYRRALEIYATRLGPDDPNVAKTKNNLASCYLKQGKY 382
Query: 568 SDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEY 627
D+ ++K + + + EK+ FG + E+ D ++ EY
Sbjct: 383 QDAETLYKEILTR---AHEKE---FGSVNGDNKPIWMHAEEREESKDKRRDSAPY--GEY 434
Query: 628 GQYH-------PDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEK 680
G ++ P +L Y G+++ A + + R++ L A+ +
Sbjct: 435 GSWYKACKVDSPTVNTTLRSLGALYRRQGKLEAAHTLEDCASRNRKQGLDPAS------Q 488
Query: 681 RRLAELLKE-AGRGRNRKSKRSL 702
++ ELLK+ +GR +R+S R +
Sbjct: 489 TKVVELLKDGSGRRGDRRSSRDM 511
Score = 58.9 bits (141), Expect = 4e-08
Identities = 56/241 (23%), Positives = 99/241 (40%), Gaps = 16/241 (6%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y + G+Y A+P+ +++++ G DH +A + L Y +
Sbjct: 201 LHNLVIQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILAL--VYRDQNKYKE 258
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+ + LG+ P T +A + + ++ EAE LC+ ALEI
Sbjct: 259 AAHLLNDALAIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGK 318
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSIGDAY 435
V + + L+ ++G E +Y L A + +VA +++ Y
Sbjct: 319 FHPDVAKQLSN--LALLCQNQGKAEEVEYYYRRALEIYATRLGPDDPNVAKTKNNLASCY 376
Query: 436 LALARYDEAVFSYQKALT-----VFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYC 490
L +Y +A Y++ LT F S G+N P +++ + K +DS Y
Sbjct: 377 LKQGKYQDAETLYKEILTRAHEKEFGSVNGDNKP----IWMHAEEREESKDKRRDSAPYG 432
Query: 491 E 491
E
Sbjct: 433 E 433
Score = 53.9 bits (128), Expect = 1e-06
Identities = 44/166 (26%), Positives = 68/166 (40%), Gaps = 8/166 (4%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + + K A+ + LN + L + EAA L +
Sbjct: 206 IQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKEAAHLLND 265
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A I EK G+ HP +NLA Y G+ +A + + + +RE+ LG +PDV
Sbjct: 266 ALAIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKFHPDVAK 325
Query: 679 EKRRLAELLKEAGRGRNRK--SKRSLETLL------DANSRLIKNN 716
+ LA L + G+ + +R+LE D N KNN
Sbjct: 326 QLSNLALLCQNQGKAEEVEYYYRRALEIYATRLGPDDPNVAKTKNN 371
>KLC_CAEEL (P46822) Kinesin light chain (KLC)
Length = 540
Score = 77.4 bits (189), Expect = 1e-13
Identities = 71/254 (27%), Positives = 115/254 (44%), Gaps = 25/254 (9%)
Query: 349 EAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLI-----YDSKGDYEA 403
E + A QF++ QMA +SA+V RL L Y S+G YE
Sbjct: 175 EEDMNASQFNQPTPANQMA----------ASANVGYEIPARLRTLHNLVIQYASQGRYEV 224
Query: 404 ALE--HYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGE 461
A+ L + ++ DVA++ + + Y +Y EA +AL++ + GE
Sbjct: 225 AVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKEAANLLNEALSIREKCLGE 284
Query: 462 NHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQ 521
+HP VA+ LA L+ K GKFKD++ C+ AL I K G ++A L ++A + Q
Sbjct: 285 SHPAVAATLNNLAVLFGKRGKFKDAEPLCKRALEIREKVL-GDDHPDVAKQLNNLALLCQ 343
Query: 522 SMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAK 580
+ E+ K K+AL+IY G +A + + Y G Y ++ ++K + +
Sbjct: 344 NQGKYEEVEKYYKRALEIYESKLGPDDPNVAKTKNNLSSAYLKQGKYKEAEELYKQILTR 403
Query: 581 F------RASGEKK 588
+ SGE K
Sbjct: 404 AHEREFGQISGENK 417
Score = 64.7 bits (156), Expect = 8e-10
Identities = 54/214 (25%), Positives = 91/214 (42%), Gaps = 12/214 (5%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y + G+Y A+P+ +++++ G DH +A + L Y +
Sbjct: 209 LHNLVIQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILAL--VYRDQNKYKE 266
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+ + LGE+ P T +A + +F +AE LC+ ALEI
Sbjct: 267 AANLLNEALSIREKCLGESHPAVAATLNNLAVLFGKRGKFKDAEPLCKRALEIREKVLGD 326
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSIGDAY 435
V + + + L+ ++G YE ++Y L + +VA +++ AY
Sbjct: 327 DHPDVAKQLNN--LALLCQNQGKYEEVEKYYKRALEIYESKLGPDDPNVAKTKNNLSSAY 384
Query: 436 LALARYDEAVFSYQKALT-----VFKSTKGENHP 464
L +Y EA Y++ LT F GEN P
Sbjct: 385 LKQGKYKEAEELYKQILTRAHEREFGQISGENKP 418
Score = 56.2 bits (134), Expect = 3e-07
Identities = 41/147 (27%), Positives = 65/147 (43%), Gaps = 2/147 (1%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + + K A+ + LN + L + EAA+L E
Sbjct: 214 IQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKEAANLLNE 273
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A +I EK G+ HP +NLA + G+ DA + + + +RE+ LG +PDV
Sbjct: 274 ALSIREKCLGESHPAVAATLNNLAVLFGKRGKFKDAEPLCKRALEIREKVLGDDHPDVAK 333
Query: 679 EKRRLAELLKEAGRGR--NRKSKRSLE 703
+ LA L + G+ + KR+LE
Sbjct: 334 QLNNLALLCQNQGKYEEVEKYYKRALE 360
Score = 53.5 bits (127), Expect = 2e-06
Identities = 62/247 (25%), Positives = 106/247 (42%), Gaps = 17/247 (6%)
Query: 204 TGLDNPDLGPFLLKQTRDMISSGENPQK-ALDLALRALKSFEMCADGKPSLELVMCLHVL 262
+G D+PD+ L ++ +N K A +L AL E C G+ + L+ L
Sbjct: 240 SGHDHPDVATML--NILALVYRDQNKYKEAANLLNEALSIREKCL-GESHPAVAATLNNL 296
Query: 263 ATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIENSILF 321
A ++ G++ +A P+ +R+++I G DH +AK + L G E +
Sbjct: 297 AVLFGKRGKFKDAEPLCKRALEIREKVLGDDHPDVAKQLNNLAL--LCQNQGKYEEVEKY 354
Query: 322 YTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMAL----EIHRGNGNG 377
Y LEI LG DP +T ++ A+++ ++ EAE L + L E G +G
Sbjct: 355 YKRALEIYESKLGPDDPNVAKTKNNLSSAYLKQGKYKEAEELYKQILTRAHEREFGQISG 414
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLA 437
+ + + A+ R KG+ A E A A +A V + ++G Y
Sbjct: 415 ENKPIWQIAEEREEN---KHKGEGATANEQ---AGWAKAAKVDSPTVTTTLKNLGALYRR 468
Query: 438 LARYDEA 444
+Y+ A
Sbjct: 469 QGKYEAA 475
Score = 52.0 bits (123), Expect = 5e-06
Identities = 45/178 (25%), Positives = 78/178 (43%), Gaps = 9/178 (5%)
Query: 520 YQSMNDLEKGLKLLKKAL----KIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFK 575
Y S E + L K+AL K GH +T+ I + ++Y Y ++ N+
Sbjct: 216 YASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNI---LALVYRDQNKYKEAANLLN 272
Query: 576 SAVA-KFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDT 634
A++ + + GE A+ LN + + +R +A L + A I EK G HPD
Sbjct: 273 EALSIREKCLGESHPAV-AATLNNLAVLFGKRGKFKDAEPLCKRALEIREKVLGDDHPDV 331
Query: 635 LGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGR 692
+NLA G+ ++ + + + + E KLG +P+V K L+ + G+
Sbjct: 332 AKQLNNLALLCQNQGKYEEVEKYYKRALEIYESKLGPDDPNVAKTKNNLSSAYLKQGK 389
Score = 50.4 bits (119), Expect = 2e-05
Identities = 45/205 (21%), Positives = 89/205 (42%), Gaps = 10/205 (4%)
Query: 472 RLADLYNKI------GKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMND 525
RL L+N + G+++ + C+ AL K+ G ++A L +A +Y+ N
Sbjct: 205 RLRTLHNLVIQYASQGRYEVAVPLCKQALEDLEKTS-GHDHPDVATMLNILALVYRDQNK 263
Query: 526 LEKGLKLLKKALKIYGHAPGQQS-TIAGIEAQMGVMYYMLGNYSDSYNIFKSAVA-KFRA 583
++ LL +AL I G+ +A + V++ G + D+ + K A+ + +
Sbjct: 264 YKEAANLLNEALSIREKCLGESHPAVAATLNNLAVLFGKRGKFKDAEPLCKRALEIREKV 323
Query: 584 SGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAG 643
G+ + LN + L C + E ++ A I E + G P+ +NL+
Sbjct: 324 LGDDHPDV-AKQLNNLALLCQNQGKYEEVEKYYKRALEIYESKLGPDDPNVAKTKNNLSS 382
Query: 644 TYDAMGRVDDAIEILEFVVGMREEK 668
Y G+ +A E+ + ++ E+
Sbjct: 383 AYLKQGKYKEAEELYKQILTRAHER 407
>KLC1_MOUSE (O88447) Kinesin light chain 1 (KLC 1)
Length = 537
Score = 76.3 bits (186), Expect = 3e-13
Identities = 79/323 (24%), Positives = 135/323 (41%), Gaps = 34/323 (10%)
Query: 395 YDSKGDYEAALEH--YVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKAL 452
Y S+G YE A+ L + ++ DVA++ + + Y +Y +A AL
Sbjct: 217 YASQGRYEVAVPSCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKDAANLLNDAL 276
Query: 453 TVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL----RIFGKSKPGIPPEE 508
+ + T G +HP VA+ LA LY K GK+K+++ C+ AL ++ GK P +
Sbjct: 277 AIREKTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHP-----D 331
Query: 509 IANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGH--APGQQSTIAGIEAQMGVMYYMLGN 566
+A L ++A + Q+ E+ ++AL IY P + +A + + Y G
Sbjct: 332 VAKQLNNLALLCQNQGKYEEVEYYYQRALGIYQTKLGPDRTPNVAKTKNNLASCYLKQGK 391
Query: 567 YSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKE 626
+ + ++K + + A FG ++ + E ++ E
Sbjct: 392 FKQAETLYKEILTR------AHEAEFGSVDDENKPIWMHAEEREECKGKQKDGSAF--GE 443
Query: 627 YGQYH-------PDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDE 679
YG ++ P NL Y G+ +A E LE MR K G N
Sbjct: 444 YGGWYKACKVDSPTVTTTLKNLGALYRRQGKF-EAAETLE-EAAMRSRKQGLDNV----H 497
Query: 680 KRRLAELLKEAGRGRNRKSKRSL 702
K+R+AE+L + R+S+ SL
Sbjct: 498 KQRVAEVLNDPESMEKRRSRESL 520
Score = 55.8 bits (133), Expect = 4e-07
Identities = 55/243 (22%), Positives = 103/243 (41%), Gaps = 19/243 (7%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y + G+Y A+P +++++ G DH +A + L Y ++
Sbjct: 210 LHNLVIQYASQGRYEVAVPSCKQALEDLEKTSGHDHPDVATMLNILAL--VYRDQNKYKD 267
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+ + LG P T +A + + ++ EAE LC+ ALEI
Sbjct: 268 AANLLNDALAIREKTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGK 327
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSAN----GQEQDVASVDSSIGD 433
V + + + L+ ++G YE +E+Y ++ + + +VA +++
Sbjct: 328 DHPDVAKQLNN--LALLCQNQGKYE-EVEYYYQRALGIYQTKLGPDRTPNVAKTKNNLAS 384
Query: 434 AYLALARYDEAVFSYQKALT-----VFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKS 488
YL ++ +A Y++ LT F S EN P +++ + GK KD +
Sbjct: 385 CYLKQGKFKQAETLYKEILTRAHEAEFGSVDDENKP----IWMHAEEREECKGKQKDGSA 440
Query: 489 YCE 491
+ E
Sbjct: 441 FGE 443
Score = 51.6 bits (122), Expect = 7e-06
Identities = 35/134 (26%), Positives = 57/134 (42%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + K A+ + LN + L + +AA+L +
Sbjct: 215 IQYASQGRYEVAVPSCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKDAANLLND 274
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A I EK G+ HP +NLA Y G+ +A + + + +RE+ LG +PDV
Sbjct: 275 ALAIREKTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAK 334
Query: 679 EKRRLAELLKEAGR 692
+ LA L + G+
Sbjct: 335 QLNNLALLCQNQGK 348
Score = 38.5 bits (88), Expect = 0.063
Identities = 40/162 (24%), Positives = 69/162 (41%), Gaps = 6/162 (3%)
Query: 204 TGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLA 263
+G D+PD+ L ++ +N K L + G+ + L+ LA
Sbjct: 241 SGHDHPDVATML--NILALVYRDQNKYKDAANLLNDALAIREKTLGRDHPAVAATLNNLA 298
Query: 264 TIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIENSILFY 322
+Y G+Y EA P+ +R+++I G+DH +AK + L G E +Y
Sbjct: 299 VLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAKQLNNLAL--LCQNQGKYEEVEYYY 356
Query: 323 TAGLEIQGQVLG-ETDPRFGETCRYVAEAHVQALQFDEAERL 363
L I LG + P +T +A +++ +F +AE L
Sbjct: 357 QRALGIYQTKLGPDRTPNVAKTKNNLASCYLKQGKFKQAETL 398
>KLC1_HUMAN (Q07866) Kinesin light chain 1 (KLC 1)
Length = 569
Score = 76.3 bits (186), Expect = 3e-13
Identities = 79/322 (24%), Positives = 140/322 (42%), Gaps = 33/322 (10%)
Query: 395 YDSKGDYEAALE--HYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKAL 452
Y S+G YE A+ L + ++ DVA++ + + Y +Y +A AL
Sbjct: 219 YASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKDAANLLNDAL 278
Query: 453 TVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL----RIFGKSKPGIPPEE 508
+ + T G++HP VA+ LA LY K GK+K+++ C+ AL ++ GK P +
Sbjct: 279 AIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHP-----D 333
Query: 509 IANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNY 567
+A L ++A + Q+ E+ ++AL+IY G +A + + Y G +
Sbjct: 334 VAKQLNNLALLCQNQGKYEEVEYYYQRALEIYQTKLGPDDPNVAKTKNNLASCYLKQGKF 393
Query: 568 SDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEY 627
+ ++K + + + E++ FG ++ + E ++ + EY
Sbjct: 394 KQAETLYKEILTR---AHERE---FGSVDDENKPIWMHAEEREECKGKQKDGTSF--GEY 445
Query: 628 GQYH-------PDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEK 680
G ++ P NL Y G+ +A E LE MR K G N K
Sbjct: 446 GGWYKACKVDSPTVTTTLKNLGALYRRQGKF-EAAETLE-EAAMRSRKQGLDNV----HK 499
Query: 681 RRLAELLKEAGRGRNRKSKRSL 702
+R+AE+L + R+S+ SL
Sbjct: 500 QRVAEVLNDPENMEKRRSRESL 521
Score = 60.1 bits (144), Expect = 2e-08
Identities = 56/241 (23%), Positives = 101/241 (41%), Gaps = 16/241 (6%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y + G+Y A+P+ +++++ G DH +A + L Y ++
Sbjct: 212 LHNLVIQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILAL--VYRDQNKYKD 269
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+ + LG+ P T +A + + ++ EAE LC+ ALEI
Sbjct: 270 AANLLNDALAIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGK 329
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSIGDAY 435
V + + + L+ ++G YE +Y L + +VA +++ Y
Sbjct: 330 DHPDVAKQLNN--LALLCQNQGKYEEVEYYYQRALEIYQTKLGPDDPNVAKTKNNLASCY 387
Query: 436 LALARYDEAVFSYQKALT-----VFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYC 490
L ++ +A Y++ LT F S EN P +++ + GK KD S+
Sbjct: 388 LKQGKFKQAETLYKEILTRAHEREFGSVDDENKP----IWMHAEEREECKGKQKDGTSFG 443
Query: 491 E 491
E
Sbjct: 444 E 444
Score = 53.1 bits (126), Expect = 2e-06
Identities = 35/134 (26%), Positives = 58/134 (43%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + + K A+ + LN + L + +AA+L +
Sbjct: 217 IQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKDAANLLND 276
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A I EK G+ HP +NLA Y G+ +A + + + +RE+ LG +PDV
Sbjct: 277 ALAIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAK 336
Query: 679 EKRRLAELLKEAGR 692
+ LA L + G+
Sbjct: 337 QLNNLALLCQNQGK 350
Score = 49.3 bits (116), Expect = 4e-05
Identities = 51/218 (23%), Positives = 87/218 (39%), Gaps = 13/218 (5%)
Query: 472 RLADLYNKI------GKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMND 525
RL L+N + G+++ + C+ AL K+ G ++A L +A +Y+ N
Sbjct: 208 RLRTLHNLVIQYASQGRYEVAVPLCKQALEDLEKTS-GHDHPDVATMLNILALVYRDQNK 266
Query: 526 LEKGLKLLKKALKIYGHAPGQQS-TIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRAS 584
+ LL AL I G+ +A + V+Y G Y ++ + K A+
Sbjct: 267 YKDAANLLNDALAIREKTLGKDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKV 326
Query: 585 GEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGT 644
K LN + L C + E ++ A I + + G P+ +NLA
Sbjct: 327 LGKDHPDVAKQLNNLALLCQNQGKYEEVEYYYQRALEIYQTKLGPDDPNVAKTKNNLASC 386
Query: 645 YDAMGRVDDAIEIL-EFVVGMREEKLGTANPDVDDEKR 681
Y G+ A + E + E + G+ VDDE +
Sbjct: 387 YLKQGKFKQAETLYKEILTRAHEREFGS----VDDENK 420
Score = 48.9 bits (115), Expect = 5e-05
Identities = 70/313 (22%), Positives = 117/313 (37%), Gaps = 48/313 (15%)
Query: 204 TGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLA 263
+G D+PD+ L ++ +N K L + GK + L+ LA
Sbjct: 243 SGHDHPDVATML--NILALVYRDQNKYKDAANLLNDALAIREKTLGKDHPAVAATLNNLA 300
Query: 264 TIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIENSILFY 322
+Y G+Y EA P+ +R+++I G+DH +AK + L G E +Y
Sbjct: 301 VLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAKQLNNLAL--LCQNQGKYEEVEYYY 358
Query: 323 TAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASV 382
LEI LG DP +T +A +++ +F +AE L + L
Sbjct: 359 QRALEIYQTKLGPDDPNVAKTKNNLASCYLKQGKFKQAETLYKEIL-------------- 404
Query: 383 EESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYD 442
A R G + D ++ A G+++D S G
Sbjct: 405 -TRAHEREFGSVDDENKPI------WMHAEEREECKGKQKDGTSFGEYGG---------- 447
Query: 443 EAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKP 502
+ KA V + P V + L LY + GKF+ +++ E A+R +
Sbjct: 448 -----WYKACKV-------DSPTVTTTLKNLGALYRRQGKFEAAETLEEAAMRSRKQGLD 495
Query: 503 GIPPEEIANGLID 515
+ + +A L D
Sbjct: 496 NVHKQRVAEVLND 508
>KLC1_RAT (P37285) Kinesin light chain 1 (KLC 1)
Length = 556
Score = 74.7 bits (182), Expect = 8e-13
Identities = 78/322 (24%), Positives = 139/322 (42%), Gaps = 33/322 (10%)
Query: 395 YDSKGDYEAALE--HYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKAL 452
Y S+G YE A+ L + ++ DVA++ + + Y +Y +A AL
Sbjct: 219 YASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKDAANLLNDAL 278
Query: 453 TVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL----RIFGKSKPGIPPEE 508
+ + T G +HP VA+ LA LY K GK+K+++ C+ AL ++ GK P +
Sbjct: 279 AIREKTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHP-----D 333
Query: 509 IANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNY 567
+A L ++A + Q+ E+ ++AL+IY G +A + + Y G +
Sbjct: 334 VAKQLNNLALLCQNQGKYEEVEYYYQRALEIYQTKLGPDDPNVAKTKNNLASCYLKQGKF 393
Query: 568 SDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEY 627
+ ++K + + + E++ FG ++ + E ++ + EY
Sbjct: 394 KQAETLYKEILTR---AHERE---FGSVDDENKPIWMHAEEREECKGKQKDGSSF--GEY 445
Query: 628 GQYH-------PDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEK 680
G ++ P NL Y G+ +A E LE +R K G N K
Sbjct: 446 GGWYKACKVDSPTVTTTLKNLGALYRRQGKF-EAAETLE-EAALRSRKQGLDNV----HK 499
Query: 681 RRLAELLKEAGRGRNRKSKRSL 702
+R+AE+L + R+S+ SL
Sbjct: 500 QRVAEVLNDPENVEKRRSRESL 521
Score = 60.1 bits (144), Expect = 2e-08
Identities = 56/241 (23%), Positives = 100/241 (41%), Gaps = 16/241 (6%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y + G+Y A+P+ +++++ G DH +A + L Y ++
Sbjct: 212 LHNLVIQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILAL--VYRDQNKYKD 269
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+ + LG P T +A + + ++ EAE LC+ ALEI
Sbjct: 270 AANLLNDALAIREKTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGK 329
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSIGDAY 435
V + + + L+ ++G YE +Y L + +VA +++ Y
Sbjct: 330 DHPDVAKQLNN--LALLCQNQGKYEEVEYYYQRALEIYQTKLGPDDPNVAKTKNNLASCY 387
Query: 436 LALARYDEAVFSYQKALT-----VFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYC 490
L ++ +A Y++ LT F S EN P +++ + GK KD S+
Sbjct: 388 LKQGKFKQAETLYKEILTRAHEREFGSVDDENKP----IWMHAEEREECKGKQKDGSSFG 443
Query: 491 E 491
E
Sbjct: 444 E 444
Score = 53.1 bits (126), Expect = 2e-06
Identities = 35/134 (26%), Positives = 58/134 (43%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + + K A+ + LN + L + +AA+L +
Sbjct: 217 IQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKDAANLLND 276
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A I EK G+ HP +NLA Y G+ +A + + + +RE+ LG +PDV
Sbjct: 277 ALAIREKTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAK 336
Query: 679 EKRRLAELLKEAGR 692
+ LA L + G+
Sbjct: 337 QLNNLALLCQNQGK 350
Score = 49.3 bits (116), Expect = 4e-05
Identities = 51/218 (23%), Positives = 87/218 (39%), Gaps = 13/218 (5%)
Query: 472 RLADLYNKI------GKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMND 525
RL L+N + G+++ + C+ AL K+ G ++A L +A +Y+ N
Sbjct: 208 RLRTLHNLVIQYASQGRYEVAVPLCKQALEDLEKTS-GHDHPDVATMLNILALVYRDQNK 266
Query: 526 LEKGLKLLKKALKIYGHAPGQQS-TIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRAS 584
+ LL AL I G+ +A + V+Y G Y ++ + K A+
Sbjct: 267 YKDAANLLNDALAIREKTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKV 326
Query: 585 GEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGT 644
K LN + L C + E ++ A I + + G P+ +NLA
Sbjct: 327 LGKDHPDVAKQLNNLALLCQNQGKYEEVEYYYQRALEIYQTKLGPDDPNVAKTKNNLASC 386
Query: 645 YDAMGRVDDAIEIL-EFVVGMREEKLGTANPDVDDEKR 681
Y G+ A + E + E + G+ VDDE +
Sbjct: 387 YLKQGKFKQAETLYKEILTRAHEREFGS----VDDENK 420
Score = 48.9 bits (115), Expect = 5e-05
Identities = 71/313 (22%), Positives = 119/313 (37%), Gaps = 48/313 (15%)
Query: 204 TGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLA 263
+G D+PD+ L ++ +N K L + G+ + L+ LA
Sbjct: 243 SGHDHPDVATML--NILALVYRDQNKYKDAANLLNDALAIREKTLGRDHPAVAATLNNLA 300
Query: 264 TIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIENSILFY 322
+Y G+Y EA P+ +R+++I G+DH +AK + L G E +Y
Sbjct: 301 VLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAKQLNNLAL--LCQNQGKYEEVEYYY 358
Query: 323 TAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASV 382
LEI LG DP +T +A +++ +F +AE L + L
Sbjct: 359 QRALEIYQTKLGPDDPNVAKTKNNLASCYLKQGKFKQAETLYKEIL-------------- 404
Query: 383 EESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYD 442
A R G + D ++ A G+++D SS G+
Sbjct: 405 -TRAHEREFGSVDDENKPI------WMHAEEREECKGKQKD----GSSFGE--------- 444
Query: 443 EAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKP 502
+ KA V + P V + L LY + GKF+ +++ E ALR +
Sbjct: 445 --YGGWYKACKV-------DSPTVTTTLKNLGALYRRQGKFEAAETLEEAALRSRKQGLD 495
Query: 503 GIPPEEIANGLID 515
+ + +A L D
Sbjct: 496 NVHKQRVAEVLND 508
>KLC_LOLPE (P46825) Kinesin light chain (KLC)
Length = 571
Score = 72.4 bits (176), Expect = 4e-12
Identities = 52/193 (26%), Positives = 91/193 (46%), Gaps = 12/193 (6%)
Query: 395 YDSKGDYEAALE--HYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKAL 452
Y S+G YE A+ L + ++ DVA++ + + Y +Y EA AL
Sbjct: 230 YASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQGKYKEAANLLNDAL 289
Query: 453 TVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL----RIFGKSKPGIPPEE 508
+ + T G +HP VA+ LA LY K GK+KD++ C+ AL ++ GK P +
Sbjct: 290 GIREKTLGPDHPAVAATLNNLAVLYGKRGKYKDAEPLCKRALVIREKVLGKDHP-----D 344
Query: 509 IANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNY 567
+A L ++A + Q+ E+ + ++AL+IY G +A + + Y G Y
Sbjct: 345 VAKQLNNLALLCQNQGKYEEVERYYQRALEIYQKELGPDDPNVAKTKNNLASAYLKQGKY 404
Query: 568 SDSYNIFKSAVAK 580
+ ++K + +
Sbjct: 405 KQAEILYKEVLTR 417
Score = 57.8 bits (138), Expect = 1e-07
Identities = 52/235 (22%), Positives = 97/235 (41%), Gaps = 16/235 (6%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y + G+Y A+P+ +++++ G DH +A + L Y G +
Sbjct: 223 LHNLVIQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILAL--VYRDQGKYKE 280
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+ + LG P T +A + + ++ +AE LC+ AL I
Sbjct: 281 AANLLNDALGIREKTLGPDHPAVAATLNNLAVLYGKRGKYKDAEPLCKRALVIREKVLGK 340
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSIGDAY 435
V + + + L+ ++G YE +Y L + +VA +++ AY
Sbjct: 341 DHPDVAKQLNN--LALLCQNQGKYEEVERYYQRALEIYQKELGPDDPNVAKTKNNLASAY 398
Query: 436 LALARYDEAVFSYQKALT-----VFKSTKGENHPNVASVYVRLADLYNKIGKFKD 485
L +Y +A Y++ LT F +N P ++++ + K+KD
Sbjct: 399 LKQGKYKQAEILYKEVLTRAHEKEFGKVDDDNKP----IWMQAEEREENKAKYKD 449
Score = 57.0 bits (136), Expect = 2e-07
Identities = 63/270 (23%), Positives = 115/270 (42%), Gaps = 15/270 (5%)
Query: 232 ALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDG 291
A+ L +AL+ E + G ++ L++LA +Y + G+Y EA +L ++ I G
Sbjct: 239 AVPLCKQALEDLEKTS-GHDHPDVATMLNILALVYRDQGKYKEAANLLNDALGIREKTLG 297
Query: 292 QDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAH 351
DH A A L Y G +++ L I+ +VLG+ P + +A
Sbjct: 298 PDHP-AVAATLNNLAVLYGKRGKYKDAEPLCKRALVIREKVLGKDHPDVAKQLNNLALLC 356
Query: 352 VQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA 411
+++E ER Q ALEI++ +V ++ + + Y +G Y+ A Y
Sbjct: 357 QNQGKYEEVERYYQRALEIYQKELGPDDPNVAKTKNN--LASAYLKQGKYKQAEILY--- 411
Query: 412 SMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKAL------TVFKSTKGENHPN 465
+ E++ VD ++ +E Y+ + K+ K ++ P
Sbjct: 412 -KEVLTRAHEKEFGKVDDDNKPIWMQAEEREENKAKYKDGAPQPDYGSWLKAVKVDS-PT 469
Query: 466 VASVYVRLADLYNKIGKFKDSKSYCENALR 495
V + L LY + GK++ +++ E ALR
Sbjct: 470 VTTTLKNLGALYRRQGKYEAAETLEECALR 499
Score = 54.3 bits (129), Expect = 1e-06
Identities = 46/166 (27%), Positives = 68/166 (40%), Gaps = 8/166 (4%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + + K A+ + LN + L + EAA+L +
Sbjct: 228 IQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQGKYKEAANLLND 287
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A I EK G HP +NLA Y G+ DA + + + +RE+ LG +PDV
Sbjct: 288 ALGIREKTLGPDHPAVAATLNNLAVLYGKRGKYKDAEPLCKRALVIREKVLGKDHPDVAK 347
Query: 679 EKRRLAELLKEAGRGR--NRKSKRSLETLL------DANSRLIKNN 716
+ LA L + G+ R +R+LE D N KNN
Sbjct: 348 QLNNLALLCQNQGKYEEVERYYQRALEIYQKELGPDDPNVAKTKNN 393
Score = 53.1 bits (126), Expect = 2e-06
Identities = 53/213 (24%), Positives = 86/213 (39%), Gaps = 11/213 (5%)
Query: 472 RLADLYNKI------GKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMND 525
RL L+N + G+++ + C+ AL K+ G ++A L +A +Y+
Sbjct: 219 RLRTLHNLVIQYASQGRYEVAVPLCKQALEDLEKTS-GHDHPDVATMLNILALVYRDQGK 277
Query: 526 LEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRAS 584
++ LL AL I G +A + V+Y G Y D+ + K A+
Sbjct: 278 YKEAANLLNDALGIREKTLGPDHPAVAATLNNLAVLYGKRGKYKDAEPLCKRALVIREKV 337
Query: 585 GEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGT 644
K LN + L C + E ++ A I +KE G P+ +NLA
Sbjct: 338 LGKDHPDVAKQLNNLALLCQNQGKYEEVERYYQRALEIYQKELGPDDPNVAKTKNNLASA 397
Query: 645 YDAMGRVDDAIEIL--EFVVGMREEKLGTANPD 675
Y G+ A EIL E + E++ G + D
Sbjct: 398 YLKQGKYKQA-EILYKEVLTRAHEKEFGKVDDD 429
Score = 50.4 bits (119), Expect = 2e-05
Identities = 42/177 (23%), Positives = 74/177 (41%), Gaps = 7/177 (3%)
Query: 520 YQSMNDLEKGLKLLKKAL----KIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFK 575
Y S E + L K+AL K GH +T+ I + ++Y G Y ++ N+
Sbjct: 230 YASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNI---LALVYRDQGKYKEAANLLN 286
Query: 576 SAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTL 635
A+ + LN + + +R +A L + A I EK G+ HPD
Sbjct: 287 DALGIREKTLGPDHPAVAATLNNLAVLYGKRGKYKDAEPLCKRALVIREKVLGKDHPDVA 346
Query: 636 GVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGR 692
+NLA G+ ++ + + + +++LG +P+V K LA + G+
Sbjct: 347 KQLNNLALLCQNQGKYEEVERYYQRALEIYQKELGPDDPNVAKTKNNLASAYLKQGK 403
Score = 45.1 bits (105), Expect = 7e-04
Identities = 51/213 (23%), Positives = 89/213 (40%), Gaps = 19/213 (8%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
L+ LA +Y G+Y +A P+ +R++ I G+DH +AK + L G E
Sbjct: 307 LNNLAVLYGKRGKYKDAEPLCKRALVIREKVLGKDHPDVAKQLNNLAL--LCQNQGKYEE 364
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMAL----EIHRG 373
+Y LEI + LG DP +T +A A+++ ++ +AE L + L E G
Sbjct: 365 VERYYQRALEIYQKELGPDDPNVAKTKNNLASAYLKQGKYKQAEILYKEVLTRAHEKEFG 424
Query: 374 NGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGD 433
+ + + A+ R ++K Y+ S + V + ++G
Sbjct: 425 KVDDDNKPIWMQAEER-----EENKAKYKDGAPQPDYGSWLKAVKVDSPTVTTTLKNLGA 479
Query: 434 AYLALARYDE-------AVFSYQKALTVFKSTK 459
Y +Y+ A+ S + AL V + TK
Sbjct: 480 LYRRQGKYEAAETLEECALRSRKSALEVVRQTK 512
>KLC8_MOUSE (Q9DBS5) Kinesin-like protein 8
Length = 619
Score = 72.4 bits (176), Expect = 4e-12
Identities = 58/211 (27%), Positives = 102/211 (47%), Gaps = 11/211 (5%)
Query: 375 GNGSSASVEESAD-----RRLMGLI--YDSKGDYEAALE--HYVLASMAMSANGQEQDVA 425
G G++A+ + + R L L+ Y ++G YE A+ L + ++ DVA
Sbjct: 194 GQGAAAAQQGGYEIPARLRTLHNLVIQYAAQGRYEVAVPLCKQALEDLERTSGRGHPDVA 253
Query: 426 SVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKD 485
++ + + Y +Y EA AL++ +ST G +HP VA+ LA LY K GK+K+
Sbjct: 254 TMLNILALVYRDQNKYKEAAHLLNDALSIRESTLGRDHPAVAATLNNLAVLYGKRGKYKE 313
Query: 486 SKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPG 545
++ C+ AL I K G ++A L ++A + Q+ E + ++AL IY G
Sbjct: 314 AEPLCQRALEIREKVL-GTDHPDVAKQLNNLALLCQNQGKYEAVERYYQRALAIYESQLG 372
Query: 546 QQS-TIAGIEAQMGVMYYMLGNYSDSYNIFK 575
+ +A + + Y G YS++ ++K
Sbjct: 373 PDNPNVARTKNNLASCYLKQGKYSEAEALYK 403
Score = 57.4 bits (137), Expect = 1e-07
Identities = 55/237 (23%), Positives = 93/237 (39%), Gaps = 45/237 (18%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y G+Y A+P+ +++++ G+ H +A + L Y +
Sbjct: 214 LHNLVIQYAAQGRYEVAVPLCKQALEDLERTSGRGHPDVATMLNILAL--VYRDQNKYKE 271
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+ LG P T +A + + ++ EAE LCQ ALEI
Sbjct: 272 AAHLLNDALSIRESTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCQRALEIREKVLGT 331
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLA 437
V + + + L+ ++G YEA +E Y
Sbjct: 332 DHPDVAKQLNN--LALLCQNQGKYEA-VERY----------------------------- 359
Query: 438 LARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL 494
YQ+AL +++S G ++PNVA LA Y K GK+ ++++ + L
Sbjct: 360 ----------YQRALAIYESQLGPDNPNVARTKNNLASCYLKQGKYSEAEALYKEIL 406
Score = 55.5 bits (132), Expect = 5e-07
Identities = 36/134 (26%), Positives = 59/134 (43%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + + K A+ + + LN + L + EAA L +
Sbjct: 219 IQYAAQGRYEVAVPLCKQALEDLERTSGRGHPDVATMLNILALVYRDQNKYKEAAHLLND 278
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A +I E G+ HP +NLA Y G+ +A + + + +RE+ LGT +PDV
Sbjct: 279 ALSIRESTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCQRALEIREKVLGTDHPDVAK 338
Query: 679 EKRRLAELLKEAGR 692
+ LA L + G+
Sbjct: 339 QLNNLALLCQNQGK 352
Score = 48.1 bits (113), Expect = 8e-05
Identities = 61/271 (22%), Positives = 108/271 (39%), Gaps = 15/271 (5%)
Query: 232 ALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDG 291
A+ L +AL+ E + G+ ++ L++LA +Y + +Y EA +L ++ I G
Sbjct: 230 AVPLCKQALEDLERTS-GRGHPDVATMLNILALVYRDQNKYKEAAHLLNDALSIRESTLG 288
Query: 292 QDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAH 351
+DH A A L Y G + + LEI+ +VLG P + +A
Sbjct: 289 RDHP-AVAATLNNLAVLYGKRGKYKEAEPLCQRALEIREKVLGTDHPDVAKQLNNLALLC 347
Query: 352 VQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA 411
+++ ER Q AL I+ + +V + + + Y +G Y A Y
Sbjct: 348 QNQGKYEAVERYYQRALAIYESQLGPDNPNVARTKNN--LASCYLKQGKYSEAEALY--- 402
Query: 412 SMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKG-------ENHP 464
+ Q+ SVD ++ +E S + + + G + P
Sbjct: 403 -KEILTCAHVQEFGSVDDDHKPIWMHAEEREEMSRSRPRDSSAPYAEYGGWYKACRVSSP 461
Query: 465 NVASVYVRLADLYNKIGKFKDSKSYCENALR 495
V + L LY + GK + +++ E ALR
Sbjct: 462 TVNTTLKNLGALYRRQGKLEAAETLEECALR 492
Score = 48.1 bits (113), Expect = 8e-05
Identities = 57/249 (22%), Positives = 102/249 (40%), Gaps = 38/249 (15%)
Query: 472 RLADLYNKI------GKFKDSKSYCENAL----RIFGKSKPGIPPEEIANGLIDVAAIYQ 521
RL L+N + G+++ + C+ AL R G+ P ++A L +A +Y+
Sbjct: 210 RLRTLHNLVIQYAAQGRYEVAVPLCKQALEDLERTSGRGHP-----DVATMLNILALVYR 264
Query: 522 SMNDLEKGLKLLKKALKIYGHAPGQQS-TIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAK 580
N ++ LL AL I G+ +A + V+Y G Y ++ + + A+
Sbjct: 265 DQNKYKEAAHLLNDALSIRESTLGRDHPAVAATLNNLAVLYGKRGKYKEAEPLCQRAL-- 322
Query: 581 FRASGEKKSALFGI-------ALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPD 633
E + + G LN + L C + ++ A I E + G +P+
Sbjct: 323 -----EIREKVLGTDHPDVAKQLNNLALLCQNQGKYEAVERYYQRALAIYESQLGPDNPN 377
Query: 634 TLGVYSNLAGTYDAMGRVDDAIEIL-EFVVGMREEKLGTANPDVDDEKRRL---AELLKE 689
+NLA Y G+ +A + E + ++ G+ VDD+ + + AE +E
Sbjct: 378 VARTKNNLASCYLKQGKYSEAEALYKEILTCAHVQEFGS----VDDDHKPIWMHAEEREE 433
Query: 690 AGRGRNRKS 698
R R R S
Sbjct: 434 MSRSRPRDS 442
>KLC8_HUMAN (Q9NSK0) Kinesin-like protein 8
Length = 625
Score = 72.4 bits (176), Expect = 4e-12
Identities = 57/216 (26%), Positives = 103/216 (47%), Gaps = 11/216 (5%)
Query: 375 GNGSSASVEESAD-----RRLMGLI--YDSKGDYEAALE--HYVLASMAMSANGQEQDVA 425
G G++A+ + + R L L+ Y ++G YE A+ L + ++ DVA
Sbjct: 200 GQGATAAQQGGYEIPARLRTLHNLVIQYAAQGRYEVAVPLCKQALEDLERTSGRGHPDVA 259
Query: 426 SVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKD 485
++ + + Y +Y EA AL++ +ST G +HP VA+ LA LY K GK+K+
Sbjct: 260 TMLNILALVYRDQNKYKEAAHLLNDALSIRESTLGPDHPAVAATLNNLAVLYGKRGKYKE 319
Query: 486 SKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIY-GHAP 544
++ C+ AL I K G ++A L ++A + Q+ E + ++AL IY G
Sbjct: 320 AEPLCQRALEIREKVL-GTNHPDVAKQLNNLALLCQNQGKYEAVERYYQRALAIYEGQLG 378
Query: 545 GQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAK 580
+A + + Y G Y+++ ++K + +
Sbjct: 379 PDNPNVARTKNNLASCYLKQGKYAEAETLYKEILTR 414
Score = 57.4 bits (137), Expect = 1e-07
Identities = 49/198 (24%), Positives = 83/198 (41%), Gaps = 7/198 (3%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIEN 317
LH L Y G+Y A+P+ +++++ G+ H +A + L Y +
Sbjct: 220 LHNLVIQYAAQGRYEVAVPLCKQALEDLERTSGRGHPDVATMLNILAL--VYRDQNKYKE 277
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
+ L I+ LG P T +A + + ++ EAE LCQ ALEI
Sbjct: 278 AAHLLNDALSIRESTLGPDHPAVAATLNNLAVLYGKRGKYKEAEPLCQRALEIREKVLGT 337
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSIGDAY 435
+ V + + + L+ ++G YEA +Y LA +VA +++ Y
Sbjct: 338 NHPDVAKQLNN--LALLCQNQGKYEAVERYYQRALAIYEGQLGPDNPNVARTKNNLASCY 395
Query: 436 LALARYDEAVFSYQKALT 453
L +Y EA Y++ LT
Sbjct: 396 LKQGKYAEAETLYKEILT 413
Score = 54.7 bits (130), Expect = 8e-07
Identities = 36/134 (26%), Positives = 58/134 (42%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y G Y + + K A+ + + LN + L + EAA L +
Sbjct: 225 IQYAAQGRYEVAVPLCKQALEDLERTSGRGHPDVATMLNILALVYRDQNKYKEAAHLLND 284
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A +I E G HP +NLA Y G+ +A + + + +RE+ LGT +PDV
Sbjct: 285 ALSIRESTLGPDHPAVAATLNNLAVLYGKRGKYKEAEPLCQRALEIREKVLGTNHPDVAK 344
Query: 679 EKRRLAELLKEAGR 692
+ LA L + G+
Sbjct: 345 QLNNLALLCQNQGK 358
Score = 53.5 bits (127), Expect = 2e-06
Identities = 64/272 (23%), Positives = 112/272 (40%), Gaps = 17/272 (6%)
Query: 232 ALDLALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDG 291
A+ L +AL+ E + G+ ++ L++LA +Y + +Y EA +L ++ I G
Sbjct: 236 AVPLCKQALEDLERTS-GRGHPDVATMLNILALVYRDQNKYKEAAHLLNDALSIRESTLG 294
Query: 292 QDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAH 351
DH A A L Y G + + LEI+ +VLG P + +A
Sbjct: 295 PDHP-AVAATLNNLAVLYGKRGKYKEAEPLCQRALEIREKVLGTNHPDVAKQLNNLALLC 353
Query: 352 VQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA 411
+++ ER Q AL I+ G + +V + + + Y +G Y A Y
Sbjct: 354 QNQGKYEAVERYYQRALAIYEGQLGPDNPNVARTKNN--LASCYLKQGKYAEAETLY--- 408
Query: 412 SMAMSANGQEQDVASVDSSIGDAY--------LALARYDEAVFSYQKALTVFKSTKGENH 463
+ Q+ SVD + ++ +R+ E Y + +K+ K +
Sbjct: 409 -KEILTRAHVQEFGSVDDDHKPIWMHAEEREEMSKSRHHEGGTPYAEYGGWYKACK-VSS 466
Query: 464 PNVASVYVRLADLYNKIGKFKDSKSYCENALR 495
P V + L LY + GK + +++ E ALR
Sbjct: 467 PTVNTTLRNLGALYRRQGKLEAAETLEECALR 498
Score = 49.3 bits (116), Expect = 4e-05
Identities = 62/267 (23%), Positives = 107/267 (39%), Gaps = 50/267 (18%)
Query: 435 YLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL 494
Y A RY+ AV ++AL + T G HP+VA++ LA +Y K+K++ +AL
Sbjct: 227 YAAQGRYEVAVPLCKQALEDLERTSGRGHPDVATMLNILALVYRDQNKYKEAAHLLNDAL 286
Query: 495 RIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIE 554
I +S G +A L ++A +Y ++ L ++AL+I G
Sbjct: 287 SI-RESTLGPDHPAVAATLNNLAVLYGKRGKYKEAEPLCQRALEIREKVLGT-------- 337
Query: 555 AQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAAD 614
N+ D VAK LN + L C +
Sbjct: 338 -----------NHPD--------VAK--------------QLNNLALLCQNQGKYEAVER 364
Query: 615 LFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEIL-EFVVGMREEKLGTAN 673
++ A I E + G +P+ +NLA Y G+ +A + E + ++ G+
Sbjct: 365 YYQRALAIYEGQLGPDNPNVARTKNNLASCYLKQGKYAEAETLYKEILTRAHVQEFGS-- 422
Query: 674 PDVDDEKRRL---AELLKEAGRGRNRK 697
VDD+ + + AE +E + R+ +
Sbjct: 423 --VDDDHKPIWMHAEEREEMSKSRHHE 447
Score = 46.2 bits (108), Expect = 3e-04
Identities = 39/174 (22%), Positives = 71/174 (40%), Gaps = 1/174 (0%)
Query: 520 YQSMNDLEKGLKLLKKALKIYGHAPGQ-QSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAV 578
Y + E + L K+AL+ G+ +A + + ++Y Y ++ ++ A+
Sbjct: 227 YAAQGRYEVAVPLCKQALEDLERTSGRGHPDVATMLNILALVYRDQNKYKEAAHLLNDAL 286
Query: 579 AKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVY 638
+ ++ LN + + +R EA L + A I EK G HPD
Sbjct: 287 SIRESTLGPDHPAVAATLNNLAVLYGKRGKYKEAEPLCQRALEIREKVLGTNHPDVAKQL 346
Query: 639 SNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGR 692
+NLA G+ + + + + E +LG NP+V K LA + G+
Sbjct: 347 NNLALLCQNQGKYEAVERYYQRALAIYEGQLGPDNPNVARTKNNLASCYLKQGK 400
>KLC_STRPU (Q05090) Kinesin light chain (KLC)
Length = 686
Score = 67.4 bits (163), Expect = 1e-10
Identities = 57/220 (25%), Positives = 100/220 (44%), Gaps = 17/220 (7%)
Query: 373 GNGNGSSASVEESADRRLMGLI-----YDSKGDYEAALE--HYVLASMAMSANGQEQDVA 425
G+G+ S+A+ RL L Y S+ YE A+ L + ++ DVA
Sbjct: 198 GSGSVSAAAGGYEIPARLRTLHNLVIQYASQSRYEVAVPLCKQALEDLEKTSGHDHPDVA 257
Query: 426 SVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKD 485
++ + + Y +Y EA AL + + T G +HP VA+ LA LY K GK+K+
Sbjct: 258 TMLNILALVYRDQNKYKEAGNLLHDALAIREKTLGPDHPAVAATLNNLAVLYGKRGKYKE 317
Query: 486 SKSYCENAL----RIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYG 541
++ C+ AL ++ GK P ++A L ++A + Q+ E+ ++AL+IY
Sbjct: 318 AEPLCKRALEIREKVLGKDHP-----DVAKQLNNLALLCQNQGKYEEVEWYYQRALEIYE 372
Query: 542 HAPG-QQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAK 580
G +A + + Y G Y + ++K + +
Sbjct: 373 KKLGPDDPNVAKTKNNLAAAYLKQGKYKAAETLYKQVLTR 412
Score = 53.5 bits (127), Expect = 2e-06
Identities = 57/245 (23%), Positives = 102/245 (41%), Gaps = 27/245 (11%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQL----GDTYAMMG 313
LH L Y + +Y A+P+ +++++ G DH +A + L + Y G
Sbjct: 218 LHNLVIQYASQSRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKEAG 277
Query: 314 NIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRG 373
N+ + L I+ + LG P T +A + + ++ EAE LC+ ALEI
Sbjct: 278 NLLHD------ALAIREKTLGPDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREK 331
Query: 374 NGNGSSASVEESADRRLMGLIYDSKGDYEAALEHY--VLASMAMSANGQEQDVASVDSSI 431
V + + + L+ ++G YE +Y L + +VA +++
Sbjct: 332 VLGKDHPDVAKQLNN--LALLCQNQGKYEEVEWYYQRALEIYEKKLGPDDPNVAKTKNNL 389
Query: 432 GDAYLALARYDEAVFSYQKALT-------VFKSTKGENHPNVASVYVRLADLYNKIGKFK 484
AYL +Y A Y++ LT + +N P ++++ + K GKFK
Sbjct: 390 AAAYLKQGKYKAAETLYKQVLTRAHEREFGLSADDKDNKP----IWMQAEEREEK-GKFK 444
Query: 485 DSKSY 489
D+ Y
Sbjct: 445 DNAPY 449
Score = 51.2 bits (121), Expect = 9e-06
Identities = 45/177 (25%), Positives = 75/177 (41%), Gaps = 7/177 (3%)
Query: 520 YQSMNDLEKGLKLLKKAL----KIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFK 575
Y S + E + L K+AL K GH +T+ I + ++Y Y ++ N+
Sbjct: 225 YASQSRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNI---LALVYRDQNKYKEAGNLLH 281
Query: 576 SAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTL 635
A+A + LN + + +R EA L + A I EK G+ HPD
Sbjct: 282 DALAIREKTLGPDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVA 341
Query: 636 GVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGR 692
+NLA G+ ++ + + + E+KLG +P+V K LA + G+
Sbjct: 342 KQLNNLALLCQNQGKYEEVEWYYQRALEIYEKKLGPDDPNVAKTKNNLAAAYLKQGK 398
Score = 50.8 bits (120), Expect = 1e-05
Identities = 61/269 (22%), Positives = 109/269 (39%), Gaps = 17/269 (6%)
Query: 204 TGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSLELVMCLHVLA 263
+G D+PD+ L ++ +N K L + G + L+ LA
Sbjct: 249 SGHDHPDVATML--NILALVYRDQNKYKEAGNLLHDALAIREKTLGPDHPAVAATLNNLA 306
Query: 264 TIYCNLGQYNEAIPILERSIDIPVLEDGQDHA-LAKFAGCMQLGDTYAMMGNIENSILFY 322
+Y G+Y EA P+ +R+++I G+DH +AK + L G E +Y
Sbjct: 307 VLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAKQLNNLAL--LCQNQGKYEEVEWYY 364
Query: 323 TAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASV 382
LEI + LG DP +T +A A+++ ++ AE L + L G SA
Sbjct: 365 QRALEIYEKKLGPDDPNVAKTKNNLAAAYLKQGKYKAAETLYKQVLTRAHEREFGLSADD 424
Query: 383 EESADRRLMGLIYDSKGDYEAALEH-----YVLASMAMSANGQEQDVASVDSSIGDAYLA 437
+++ + + KG ++ + + A+ S + V + ++G Y
Sbjct: 425 KDNKPIWMQAEEREEKGKFKDNAPYGDYGGWHKAAKVDSRSRSSPTVTTTLKNLGALYRR 484
Query: 438 LARYDE-------AVFSYQKALTVFKSTK 459
+YD A+ S + AL + + TK
Sbjct: 485 QGKYDAAEILEECAMKSRRNALDMVRETK 513
Score = 49.7 bits (117), Expect = 3e-05
Identities = 34/134 (25%), Positives = 55/134 (40%)
Query: 559 VMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEE 618
+ Y Y + + K A+ + LN + L + EA +L +
Sbjct: 223 IQYASQSRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKEAGNLLHD 282
Query: 619 ARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDD 678
A I EK G HP +NLA Y G+ +A + + + +RE+ LG +PDV
Sbjct: 283 ALAIREKTLGPDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAK 342
Query: 679 EKRRLAELLKEAGR 692
+ LA L + G+
Sbjct: 343 QLNNLALLCQNQGK 356
Score = 49.7 bits (117), Expect = 3e-05
Identities = 51/216 (23%), Positives = 84/216 (38%), Gaps = 8/216 (3%)
Query: 472 RLADLYNKIGKFKDSKSY------CENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMND 525
RL L+N + ++ Y C+ AL K+ G ++A L +A +Y+ N
Sbjct: 214 RLRTLHNLVIQYASQSRYEVAVPLCKQALEDLEKTS-GHDHPDVATMLNILALVYRDQNK 272
Query: 526 LEKGLKLLKKALKIYGHAPG-QQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRAS 584
++ LL AL I G +A + V+Y G Y ++ + K A+
Sbjct: 273 YKEAGNLLHDALAIREKTLGPDHPAVAATLNNLAVLYGKRGKYKEAEPLCKRALEIREKV 332
Query: 585 GEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGT 644
K LN + L C + E ++ A I EK+ G P+ +NLA
Sbjct: 333 LGKDHPDVAKQLNNLALLCQNQGKYEEVEWYYQRALEIYEKKLGPDDPNVAKTKNNLAAA 392
Query: 645 YDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEK 680
Y G+ A + + V+ E+ + D D K
Sbjct: 393 YLKQGKYKAAETLYKQVLTRAHEREFGLSADDKDNK 428
>RAPS_TORCA (P09108) 43 kDa receptor-associated protein of the
synapse (RAPSYN) (Acetylcholine receptor-associated 43
kDa protein) (43 kDa postsynaptic protein)
Length = 411
Score = 50.8 bits (120), Expect = 1e-05
Identities = 69/281 (24%), Positives = 119/281 (41%), Gaps = 25/281 (8%)
Query: 278 ILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETD 337
++ERS ++P G+ AL GC L ++ MG E+ + F A E Q +G+ +
Sbjct: 31 VVERSTELP----GRFRAL----GC--LITAHSEMGKYEDMLRFAVAQSEAARQ-MGDPE 79
Query: 338 PRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRL-MGLIYD 396
R E +A H + +F EA C+ L G ++ + L MG +
Sbjct: 80 -RVTEAYLNLARGHEKLCEFSEAVAYCRTCL-----GAEGGPLRLQFNGQVCLSMGNAFL 133
Query: 397 SKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVF-SYQKALTVF 455
++ ALE + A N + V S+G Y+ L Y++A+F + A V
Sbjct: 134 GLSAFQKALECFEKALRYAHGNDDKMLECRVCCSLGAFYVQLKDYEKALFFPCKSAELVA 193
Query: 456 KSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLID 515
+G + A +A Y K+G+ D+ CE +++I P + + L+
Sbjct: 194 DYGRGWSLKYKAMSRYHMAAAYRKLGRMDDAMECCEESMKI-ALQHQDRPLQALC--LLC 250
Query: 516 VAAIYQSMNDLEKGLKLLKKALKI---YGHAPGQQSTIAGI 553
A I++ +D+ K L + +L I G+ GQ + I
Sbjct: 251 FADIHRHRSDIGKALPRYESSLNIMTEIGNRLGQAHVLLNI 291
>OGT1_RAT (P56558) UDP-N-acetylglucosamine--peptide
N-acetylglucosaminyltransferase 110 kDa subunit (EC
2.4.1.-) (O-GlcNAc transferase p110 subunit)
Length = 1036
Score = 50.8 bits (120), Expect = 1e-05
Identities = 98/462 (21%), Positives = 173/462 (37%), Gaps = 110/462 (23%)
Query: 279 LERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDP 338
L+RS L Q+ LA+ LG+ Y G ++ +I Y L ++ + D
Sbjct: 61 LDRSAHFSTLAIKQNPLLAE--AYSNLGNVYKERGQLQEAIEHYRHALRLKPDFI---DG 115
Query: 339 RFGETCRYVA--------EAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRL 390
VA +A+V ALQ++ + L GN + +EE+ L
Sbjct: 116 YINLAAALVAAGDMEGAVQAYVSALQYNPDLYCVRSDL----GNLLKALGRLEEAKACYL 171
Query: 391 ---------------MGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAY 435
+G +++++G+ A+ H+ + ++D + DAY
Sbjct: 172 KAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHF-------------EKAVTLDPNFLDAY 218
Query: 436 LALAR-------YDEAVFSYQKALTVFKSTKGENHPNVASVY-----VRLA-DLYNKIGK 482
+ L +D AV +Y +AL++ H N+A VY + LA D Y + +
Sbjct: 219 INLGNVLKEARIFDRAVAAYLRALSL-SPNHAVVHGNLACVYYEQGLIDLAIDTYRRAIE 277
Query: 483 ----FKDSKSYCENALRIFGKSKPGIPPEEI------------ANGLIDVAAIYQSMNDL 526
F D+ NAL K K + E A+ L ++A I + ++
Sbjct: 278 LQPHFPDAYCNLANAL----KEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNI 333
Query: 527 EKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGE 586
E+ ++L +KAL+++ S +A + Q G + L +Y ++ I
Sbjct: 334 EEAVRLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRI------------- 380
Query: 587 KKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYD 646
S F A + MG + + A + A Q +P +SNLA +
Sbjct: 381 --SPTFADAYSNMGNTLKEMQDVQGALQCYTRAI--------QINPAFADAHSNLASIHK 430
Query: 647 AMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLK 688
G + +AI + ++ PD D LA L+
Sbjct: 431 DSGNIPEAIASYRTALKLK--------PDFPDAYCNLAHCLQ 464
>OGT1_HUMAN (O15294) UDP-N-acetylglucosamine--peptide
N-acetylglucosaminyltransferase 110 kDa subunit (EC
2.4.1.-) (O-GlcNAc transferase p110 subunit)
Length = 1036
Score = 50.8 bits (120), Expect = 1e-05
Identities = 98/462 (21%), Positives = 173/462 (37%), Gaps = 110/462 (23%)
Query: 279 LERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDP 338
L+RS L Q+ LA+ LG+ Y G ++ +I Y L ++ + D
Sbjct: 61 LDRSAHFSTLAIKQNPLLAE--AYSNLGNVYKERGQLQEAIEHYRHALRLKPDFI---DG 115
Query: 339 RFGETCRYVA--------EAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRL 390
VA +A+V ALQ++ + L GN + +EE+ L
Sbjct: 116 YINLAAALVAAGDMEGAVQAYVSALQYNPDLYCVRSDL----GNLLKALGRLEEAKACYL 171
Query: 391 ---------------MGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAY 435
+G +++++G+ A+ H+ + ++D + DAY
Sbjct: 172 KAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHF-------------EKAVTLDPNFLDAY 218
Query: 436 LALAR-------YDEAVFSYQKALTVFKSTKGENHPNVASVY-----VRLA-DLYNKIGK 482
+ L +D AV +Y +AL++ H N+A VY + LA D Y + +
Sbjct: 219 INLGNVLKEARIFDRAVAAYLRALSL-SPNHAVVHGNLACVYYEQGLIDLAIDTYRRAIE 277
Query: 483 ----FKDSKSYCENALRIFGKSKPGIPPEEI------------ANGLIDVAAIYQSMNDL 526
F D+ NAL K K + E A+ L ++A I + ++
Sbjct: 278 LQPHFPDAYCNLANAL----KEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNI 333
Query: 527 EKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGE 586
E+ ++L +KAL+++ S +A + Q G + L +Y ++ I
Sbjct: 334 EEAVRLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRI------------- 380
Query: 587 KKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYD 646
S F A + MG + + A + A Q +P +SNLA +
Sbjct: 381 --SPTFADAYSNMGNTLKEMQDVQGALQCYTRAI--------QINPAFADAHSNLASIHK 430
Query: 647 AMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLK 688
G + +AI + ++ PD D LA L+
Sbjct: 431 DSGNIPEAIASYRTALKLK--------PDFPDAYCNLAHCLQ 464
>KA43_HUMAN (Q96AY4) TPR repeat containing protein KIAA1043
Length = 1642
Score = 50.1 bits (118), Expect = 2e-05
Identities = 80/391 (20%), Positives = 151/391 (38%), Gaps = 46/391 (11%)
Query: 268 NLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLE 327
N+ EAI E+ + + G + L + LGD Y +G+ E +I +Y L
Sbjct: 7 NMNVMEEAIGYFEQQLAMLQQLSGNESVLDRGRAYGNLGDCYEALGDYEEAIKYYEQYLS 66
Query: 328 IQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESAD 387
+ + D + R + H +A + L + G + +
Sbjct: 67 VAQSLNRMQDQ--AKAYRGLGNGHRAMGSLQQALVCFEKRLVVAHELGEAFNKAQAYGE- 123
Query: 388 RRLMGLIYDSKGDYEAA---LEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEA 444
+G ++ G+YE A LE + + M E D A +G Y + YD A
Sbjct: 124 ---LGSLHSQLGNYEQAISCLERQLNIARDMKDRALESDAA---CGLGGVYQQMGEYDTA 177
Query: 445 VFSYQKALTVFKSTKGENHPNV-ASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPG 503
+ +Q L + + T N+P Y L Y +G F+ + Y E L I +
Sbjct: 178 LQYHQLDLQIAEET---NNPTCQGRAYGNLGLTYESLGTFERAVVYQEQHLSIAAQM--- 231
Query: 504 IPPEEIANGLIDVAAI---YQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVM 560
++A + +++ + ++ + + + L++ L++ G++ A I +G+
Sbjct: 232 ---NDLAAKTVSYSSLGRTHHALQNYSQAVMYLQEGLRL-AEQLGRREDEAKIRHGLGLS 287
Query: 561 YYMLGNYSDSYNIFKSAVAKF-------RASGEKKSALFGIALNQMGLACVQRYAI---- 609
+ GN ++ + A A F + S + K +LF L +QR +
Sbjct: 288 LWASGNLEEAQHQLYRASALFETIRHEAQLSTDYKLSLFD--LQTSSYQALQRVLVSLGH 345
Query: 610 -NEAADLFEEART------ILEKEYGQYHPD 633
+EA + E RT ++E++ GQ D
Sbjct: 346 HDEALAVAERGRTRAFADLLVERQTGQQDSD 376
Score = 47.4 bits (111), Expect = 1e-04
Identities = 51/194 (26%), Positives = 76/194 (38%), Gaps = 55/194 (28%)
Query: 262 LATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILF 321
L +++ LG Y +AI LER ++I D +D AL A C LG Y MG + ++ +
Sbjct: 124 LGSLHSQLGNYEQAISCLERQLNI--ARDMKDRALESDAAC-GLGGVYQQMGEYDTALQY 180
Query: 322 YTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSAS 381
+ L+I + T CQ R GN
Sbjct: 181 HQLDLQIAEETNNPT---------------------------CQ-----GRAYGN----- 203
Query: 382 VEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVD---SSIGDAYLAL 438
+GL Y+S G +E A+ V +S Q D+A+ SS+G + AL
Sbjct: 204 ---------LGLTYESLGTFERAV---VYQEQHLSIAAQMNDLAAKTVSYSSLGRTHHAL 251
Query: 439 ARYDEAVFSYQKAL 452
Y +AV Q+ L
Sbjct: 252 QNYSQAVMYLQEGL 265
>BRU1_ARATH (Q6Q4D0) BRUSHY protein 1 (TONSOKU protein) (MGOUN
protein 3)
Length = 1311
Score = 44.7 bits (104), Expect = 9e-04
Identities = 75/347 (21%), Positives = 144/347 (40%), Gaps = 59/347 (17%)
Query: 236 ALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLE-DGQDH 294
A RA + E D + E + + I N G+Y +A+ DI V G+D
Sbjct: 9 AKRAYRKAEEVGDRR---EQARWANNVGDILKNHGEYVDALKWFRIDYDISVKYLPGKD- 64
Query: 295 ALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETD-----PRFGETCRYV-- 347
C LG+ Y + N E ++++ L++ + + + G T +
Sbjct: 65 ---LLPTCQSLGEIYLRLENFEEALIYQKKHLQLAEEANDTVEKQRACTQLGRTYHEMFL 121
Query: 348 -AEAHVQALQ-----FDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDY 401
+E +A+Q F +A L Q+ E + SS +EE + +++ G
Sbjct: 122 KSEDDCEAIQSAKKYFKKAMELAQILKE--KPPPGESSGFLEEYIN------AHNNIGML 173
Query: 402 EAALEHYVLASMAMSANGQ---EQDVASVDS-------SIGDAYLALARYDEAVFSYQKA 451
+ L++ A + Q E++V D+ ++G+ ++AL +DEA +
Sbjct: 174 DLDLDNPEAARTILKKGLQICDEEEVREYDAARSRLHHNLGNVFMALRSWDEAKKHIEMD 233
Query: 452 LTVFKSTKGENH-PNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIA 510
+ + NH A Y+ LA+L+NK K+ D AL +GK+ +A
Sbjct: 234 INICHKI---NHVQGEAKGYINLAELHNKTQKYID-------ALLCYGKA------SSLA 277
Query: 511 NGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQM 557
+ D +A+ + +E K++KK++K+ ++ + + A+M
Sbjct: 278 KSMQDESAL---VEQIEHNTKIVKKSMKVMEELREEELMLKKLSAEM 321
Score = 32.0 bits (71), Expect = 5.9
Identities = 43/206 (20%), Positives = 82/206 (38%), Gaps = 29/206 (14%)
Query: 383 EESADRRL-------MGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAY 435
EE DRR +G I + G+Y AL+ + + +D+ S+G+ Y
Sbjct: 17 EEVGDRREQARWANNVGDILKNHGEYVDALKWFRIDYDISVKYLPGKDLLPTCQSLGEIY 76
Query: 436 LALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKI--------GKFKDSK 487
L L ++EA+ +K L + + A +L Y+++ + +K
Sbjct: 77 LRLENFEEALIYQKKHLQLAEEANDTVEKQRAC--TQLGRTYHEMFLKSEDDCEAIQSAK 134
Query: 488 SYCENALRIFGKSKPGIPP-------EEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIY 540
Y + A+ + K PP EE N ++ + +++ E +LKK L+I
Sbjct: 135 KYFKKAMELAQILKEKPPPGESSGFLEEYINAHNNIGMLDLDLDNPEAARTILKKGLQIC 194
Query: 541 GHAPGQQSTIAGIEAQMGVMYYMLGN 566
+ + +A +++ LGN
Sbjct: 195 -----DEEEVREYDAARSRLHHNLGN 215
>OGT_CAEEL (O18158) UDP-N-acetylglucosamine--peptide
N-acetylglucosaminyltransferase (EC 2.4.1.-) (O-GlcNAc)
(OGT)
Length = 1151
Score = 44.3 bits (103), Expect = 0.001
Identities = 65/294 (22%), Positives = 112/294 (37%), Gaps = 68/294 (23%)
Query: 391 MGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALAR-------YDE 443
+G +++S+G+ A+ H+ + ++D + DAY+ L +D
Sbjct: 301 LGCVFNSQGEIWLAIHHF-------------EKAVTLDPNFLDAYINLGNVLKEARIFDR 347
Query: 444 AVFSYQKALTVFKSTKGENHPNVASVY-----VRLA-DLYNKI----GKFKDSKSYCENA 493
AV +Y +AL + H N+A VY + LA D Y K F D+ NA
Sbjct: 348 AVSAYLRALNL-SGNHAVVHGNLACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANA 406
Query: 494 LRIFGKSKPGIPPEEI-----------ANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGH 542
L+ G + E++ A+ ++A I + +E +L KAL+IY
Sbjct: 407 LKEKGSV---VEAEQMYMKALELCPTHADSQNNLANIKREQGKIEDATRLYLKALEIYPE 463
Query: 543 APGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLA 602
S +A I Q G + + +Y ++ I + + G L +MG +
Sbjct: 464 FAAAHSNLASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMGN--------TLKEMGDS 515
Query: 603 CVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIE 656
N A Q +P +SNLA + G + +AI+
Sbjct: 516 SAAIACYNRAI---------------QINPAFADAHSNLASIHKDAGNMAEAIQ 554
Score = 41.2 bits (95), Expect = 0.010
Identities = 75/397 (18%), Positives = 143/397 (35%), Gaps = 76/397 (19%)
Query: 335 ETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLI 394
++DP T ++ + Q +++ + +A+++ N A + +G
Sbjct: 153 QSDPNNLPTLLLLSAINFQTKNLEKSMQYSMLAIKV-----NNQCAEAYSN-----LGNY 202
Query: 395 YDSKGDYEAALEHYVLA---------------SMAMSANGQEQ-------------DVAS 426
Y KG + ALE+Y LA + +S EQ D+
Sbjct: 203 YKEKGQLQDALENYKLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAYFNALQINPDLYC 262
Query: 427 VDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDS 486
V S +G+ A+ R +EA Y KA+ E P A + L ++N G+ +
Sbjct: 263 VRSDLGNLLKAMGRLEEAKVCYLKAI--------ETQPQFAVAWSNLGCVFNSQGEIWLA 314
Query: 487 KSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQ 546
+ E A+ + + I++ + + ++ + +AL + G+
Sbjct: 315 IHHFEKAVTL---------DPNFLDAYINLGNVLKEARIFDRAVSAYLRALNLSGNHAVV 365
Query: 547 QSTIAGIEAQMGVMYYMLGNYSDSYNI---FKSAVAKF-RASGEKKSALFGIALNQMGLA 602
+A + + G++ + Y + ++ F A A EK S + + L
Sbjct: 366 HGNLACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEAEQMYMKALE 425
Query: 603 CVQRYA--------INEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDA 654
+A I E+A + K + +P+ +SNLA G+++DA
Sbjct: 426 LCPTHADSQNNLANIKREQGKIEDATRLYLKAL-EIYPEFAAAHSNLASILQQQGKLNDA 484
Query: 655 IEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAG 691
IL + +R P D + LKE G
Sbjct: 485 --ILHYKEAIR------IAPTFADAYSNMGNTLKEMG 513
Score = 32.0 bits (71), Expect = 5.9
Identities = 37/174 (21%), Positives = 67/174 (38%), Gaps = 32/174 (18%)
Query: 481 GKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIY 540
G + +++ YC +F +P L+ ++AI +LEK ++ A+K+
Sbjct: 139 GNYVEAEKYCN---LVFQSDPNNLPT------LLLLSAINFQTKNLEKSMQYSMLAIKVN 189
Query: 541 GHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMG 600
Q Y LGNY + A+ ++ + + K +N +
Sbjct: 190 N--------------QCAEAYSNLGNYYKEKGQLQDALENYKLAVKLKPEFIDAYIN-LA 234
Query: 601 LACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDA 654
A V + +A + A Q +PD V S+L AMGR+++A
Sbjct: 235 AALVSGGDLEQAVTAYFNAL--------QINPDLYCVRSDLGNLLKAMGRLEEA 280
>TTCJ_HUMAN (Q6DKK2) Tetratricopeptide repeat protein 19 (TPR repeat
protein 19)
Length = 380
Score = 42.0 bits (97), Expect = 0.006
Identities = 62/281 (22%), Positives = 111/281 (39%), Gaps = 33/281 (11%)
Query: 280 ERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPR 339
++ D EDG D A A+ ++ M E + L L + Q TD +
Sbjct: 77 QQGADGAAAEDGADEAEAEIIQLLKRAKLSIMKDEPEEAELILHDALRLAYQ----TDNK 132
Query: 340 FGETCRYVAEAHVQAL--QFDEAERLCQMALEIHRGNG--NGSSASVEESADRRLMGLIY 395
T Y A++ + Q + AE+L + + G G +A +E S + IY
Sbjct: 133 KAITYTYDLMANLAFIRGQLENAEQLFKATMSYLLGGGMKQEDNAIIEISLK---LASIY 189
Query: 396 DSKGDYEAALEHYVLASMAMSANGQEQ-----DVASVDSS------IGDAYLALARY--- 441
++ E A+ Y + + + D+ SV+ +G A ARY
Sbjct: 190 AAQNRQEFAVAGYEFCISTLEEKIEREKELAEDIMSVEEKANTHLLLGMCLDACARYLLF 249
Query: 442 ----DEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIF 497
+A Y+KAL + + +GE HP + LA + G+F ++ Y + A +
Sbjct: 250 SKQPSQAQRMYEKALQISEEIQGERHPQTIVLMSDLATTLDAQGRFDEAYIYMQRASDLA 309
Query: 498 GKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALK 538
+ I E+ L ++AA+ + ++ ++ALK
Sbjct: 310 RQ----INHPELHMVLSNLAAVLMHRERYTQAKEIYQEALK 346
Score = 42.0 bits (97), Expect = 0.006
Identities = 32/127 (25%), Positives = 56/127 (43%), Gaps = 9/127 (7%)
Query: 530 LKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKS 589
+++ K IY Q+ +AG E + + + K + EK +
Sbjct: 179 IEISLKLASIYAAQNRQEFAVAGYE-------FCISTLEEKIEREKELAEDIMSVEEKAN 231
Query: 590 A--LFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDA 647
L G+ L+ + ++A ++E+A I E+ G+ HP T+ + S+LA T DA
Sbjct: 232 THLLLGMCLDACARYLLFSKQPSQAQRMYEKALQISEEIQGERHPQTIVLMSDLATTLDA 291
Query: 648 MGRVDDA 654
GR D+A
Sbjct: 292 QGRFDEA 298
>TTCJ_MOUSE (Q8CC21) Tetratricopeptide repeat protein 19 (TPR repeat
protein 19)
Length = 365
Score = 41.2 bits (95), Expect = 0.010
Identities = 21/64 (32%), Positives = 36/64 (55%)
Query: 591 LFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGR 650
L G+ L+ + +++A ++E+A I ++ G+ HP T+ + S+LA T DA G
Sbjct: 225 LLGMCLDSCARYLLFSKQLSQAQRMYEKALQICQEIQGERHPQTIVLMSDLATTLDAQGH 284
Query: 651 VDDA 654
DDA
Sbjct: 285 FDDA 288
Score = 37.7 bits (86), Expect = 0.11
Identities = 63/271 (23%), Positives = 107/271 (39%), Gaps = 32/271 (11%)
Query: 289 EDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVA 348
ED D A A+ ++ M E + L L + E+D R T Y
Sbjct: 77 EDASDEAEAEIIQLLKQAKLSIMKDEPEAAELILHDALRLAY----ESDNRKAITYTYDL 132
Query: 349 EAHVQAL--QFDEAERLCQMALEIHRGNG--NGSSASVEESADRRLMGLIYDSKGDYEAA 404
A++ + Q + AE+L + + G G +A +E S + IY ++ E A
Sbjct: 133 MANLAFIRGQLENAEQLFKATMSYLLGGGMKQEDNAIIEISLK---LANIYAAQNKQEFA 189
Query: 405 LEHYVLASMAMSAN-GQEQDVAS--VDSSIGDAYLAL-------ARY-------DEAVFS 447
L Y + +E+++A + + YL L ARY +A
Sbjct: 190 LAGYEFCISTLEGKIEREKELAEDIMSEETANTYLLLGMCLDSCARYLLFSKQLSQAQRM 249
Query: 448 YQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPE 507
Y+KAL + + +GE HP + LA + G F D+ Y + A + + I
Sbjct: 250 YEKALQICQEIQGERHPQTIVLMSDLATTLDAQGHFDDAYIYMQRASDLARE----INHP 305
Query: 508 EIANGLIDVAAIYQSMNDLEKGLKLLKKALK 538
E+ L ++AAI + ++ ++ALK
Sbjct: 306 ELHMVLSNLAAILIHRERYTQAKEIYQEALK 336
>RAPS_CHICK (O42393) 43 kDa receptor-associated protein of the
synapse (RAPsyn) (Acetylcholine receptor-associated 43
kDa protein) (43 kDa postsynaptic protein)
Length = 411
Score = 41.2 bits (95), Expect = 0.010
Identities = 63/274 (22%), Positives = 115/274 (40%), Gaps = 20/274 (7%)
Query: 287 VLEDGQDHA-LAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRF-GETC 344
VLE D A + GC L +A MG ++ + F ++ ++ DP + E
Sbjct: 31 VLEKSADPAGRFRVLGC--LITAHAEMGRYKDMLKFAVVQIDTAREL---EDPNYLTEGY 85
Query: 345 RYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRL-MGLIYDSKGDYEA 403
+A ++ + +F + C+ L N G++ S++ + L MG + ++
Sbjct: 86 LNLARSNEKLCEFQKTISYCKTCL-----NMQGTTVSLQLNGQVSLSMGNAFLGLSIFQK 140
Query: 404 ALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKS-TKGEN 462
ALE + A N + V S+G+ Y + Y++A+F KA + G +
Sbjct: 141 ALECFEKALRYAHNNDDKMLECRVCCSLGNFYAQIKDYEKALFFPCKAAELVNDYGAGWS 200
Query: 463 HPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQS 522
A +A Y K+G+ D+ CE +++I + G P + A L+ A I+ S
Sbjct: 201 LKYRAMSQYHMAVAYRKLGRLADAMDCCEESMKI--ALQHGDRPLQ-ALCLLCFADIHLS 257
Query: 523 MNDLEKGLKLLKKALKI---YGHAPGQQSTIAGI 553
D++ A+ I G+ GQ + G+
Sbjct: 258 RRDVQTAFPRYDSAMSIMTEIGNRLGQIQVLLGV 291
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.132 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 81,306,497
Number of Sequences: 164201
Number of extensions: 3433899
Number of successful extensions: 8801
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 77
Number of HSP's that attempted gapping in prelim test: 8468
Number of HSP's gapped (non-prelim): 287
length of query: 721
length of database: 59,974,054
effective HSP length: 118
effective length of query: 603
effective length of database: 40,598,336
effective search space: 24480796608
effective search space used: 24480796608
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0400a.8


