

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
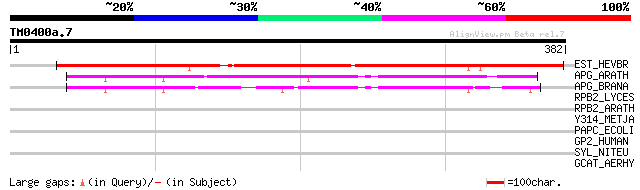
Query= TM0400a.7
(382 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early nodule... 308 1e-83
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 92 2e-18
APG_BRANA (P40603) Anter-specific proline-rich protein APG (Prot... 82 2e-15
RPB2_LYCES (Q42877) DNA-directed RNA polymerase II 135 kDa polyp... 35 0.32
RPB2_ARATH (P38420) DNA-directed RNA polymerase II 135 kDa polyp... 35 0.42
Y314_METJA (Q57762) Hypothetical protein MJ0314 32 2.7
PAPC_ECOLI (P07110) Outer membrane usher protein papC precursor 32 2.7
GP2_HUMAN (P55259) Pancreatic secretory granule membrane major g... 32 3.5
SYL_NITEU (Q820M7) Leucyl-tRNA synthetase (EC 6.1.1.4) (Leucine-... 30 7.8
GCAT_AERHY (P10480) Phosphatidylcholine-sterol acyltransferase p... 30 7.8
>EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early
nodule-specific protein homolog) (Latex allergen Hev b
13)
Length = 391
Score = 308 bits (789), Expect = 1e-83
Identities = 168/362 (46%), Positives = 224/362 (61%), Gaps = 21/362 (5%)
Query: 33 SSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEEL 92
+S C +PA++NFGDSNSDTG AAF L P G +FF +GR SDGRLIIDFI E
Sbjct: 26 ASETCDFPAIFNFGDSNSDTGGKAAAFYPLNPPYGETFFHRSTGRYSDGRLIIDFIAESF 85
Query: 93 EIPYLSAYLNSIGSNYRHGANFAAGGASIR------PVY-GFSPFYLGMQVAQFIQLQSH 145
+PYLS YL+S+GSN++HGA+FA G++I+ P + GFSPFYL +Q +QF Q
Sbjct: 86 NLPYLSPYLSSLGSNFKHGADFATAGSTIKLPTTIIPAHGGFSPFYLDVQYSQFRQFIPR 145
Query: 146 IENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRSIPEM 205
+QF T F +P F KALYT DIGQNDL G ++ + EEV ++P++
Sbjct: 146 -----SQF-IRETGGIFAELVPEEYYFEKALYTFDIGQNDLTEGFLNLTVEEVNATVPDL 199
Query: 206 MRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEF 265
+ +F+ +V+ +YD+GAR FWIHNTGPIGCL S I D+ GC +N++AQ F
Sbjct: 200 VNSFSANVKKIYDLGARTFWIHNTGPIGCL--SFILTYFPWAEKDSAGCAKAYNEVAQHF 257
Query: 266 NRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCC--GSYYGYRI- 322
N +LK+ V QLR++LP A F +VD+Y+ KY L S K GF PL CC G Y + +
Sbjct: 258 NHKLKEIVAQLRKDLPLATFVHVDIYSVKYSLFSEPEKHGFEFPLITCCGYGGKYNFSVT 317
Query: 323 -DCGKKAVV-NGT-VYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQ 379
CG +GT + C PS ++WDG HYT+AAN++ I G+ SDPPVP+
Sbjct: 318 APCGDTVTADDGTKIVVGSCACPSVRVNWDGAHYTEAANEYFFDQISTGAFSDPPVPLNM 377
Query: 380 AC 381
AC
Sbjct: 378 AC 379
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 92.4 bits (228), Expect = 2e-18
Identities = 89/335 (26%), Positives = 146/335 (43%), Gaps = 26/335 (7%)
Query: 40 PAVYNFGDSNSDTGVVYAAFAGLQS---PGGISF-FGNLSGRASDGRLIIDFITEELEIP 95
PAV+ FGDS DTG ++S P G+ F F +GR S+G + D++ + + +
Sbjct: 203 PAVFFFGDSVFDTGNNNNLETKIKSNYRPYGMDFKFRVATGRFSNGMVASDYLAKYMGVK 262
Query: 96 YL-SAYLNSI--GSNYRHGANFAAGGASIRPVYGFSPFYLGMQVAQFIQLQSHIENLLNQ 152
+ AYL+ ++ G +FA+GGA P + + M + Q Q +IE +
Sbjct: 263 EIVPAYLDPKIQPNDLLTGVSFASGGAGYNPTTSEAANAIPM-LDQLTYFQDYIEKVNRL 321
Query: 153 FSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRSIPE----MMRN 208
+++ + SK + + G NDL + + + I + +
Sbjct: 322 VRQEKSQYKLAGLEKTNQLISKGVAIVVGGSNDLIITYFGSGAQRLKNDIDSYTTIIADS 381
Query: 209 FTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQ 268
V LY GAR + T P+GC+P+ + KK + C N +Q FN +
Sbjct: 382 AASFVLQLYGYGARRIGVIGTPPLGCVPSQRL----KKKKI----CNEELNYASQLFNSK 433
Query: 269 LKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYYGYRIDCGKKA 328
L + QL + LP + F Y+D+YT +++ + GF + CC + KK+
Sbjct: 434 LLLILGQLSKTLPNSTFVYMDIYTIISQMLETPAAYGFEETKKPCCKTGLLSAGALCKKS 493
Query: 329 VVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAK 363
C N S ++ WDGVH TQ A K + K
Sbjct: 494 T------SKICPNTSSYLFWDGVHPTQRAYKTINK 522
>APG_BRANA (P40603) Anter-specific proline-rich protein APG (Protein
CEX) (Fragment)
Length = 449
Score = 82.0 bits (201), Expect = 2e-15
Identities = 97/352 (27%), Positives = 150/352 (42%), Gaps = 55/352 (15%)
Query: 40 PAVYNFGDSNSDTGVVYAAFAGLQS---PGGISF-FGNLSGRASDGRLIIDFITEELEIP 95
PAV+ FGDS DTG L+ P G+ F G +GR S+GR+ D+I++ L +
Sbjct: 124 PAVFFFGDSIFDTGNNNNLDTKLKCNYRPYGMDFPMGVATGRFSNGRVASDYISKYLGVK 183
Query: 96 YL-SAYLNSI--------GSNYRHGANFAAGGASIRPVYGFSPFYLGMQVAQFIQLQSHI 146
+ AY++ S+ G +FA+GGA P S + + + Q Q +
Sbjct: 184 EIVPAYVDKKLQQNNELQQSDLLTGVSFASGGAGYLPQTSES-WKVTTMLDQLTYFQDYK 242
Query: 147 ENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDL-----GFGLMHTSEEEVLRS 201
+ + +T+ + SK + G NDL G G H + + S
Sbjct: 243 KRMKKLVGKKKTK----------KIVSKGAAIVVAGSNDLIYTYFGNGAQHLKND--VDS 290
Query: 202 IPEMMRNFTYD-VQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNK 260
MM + V LY GAR + T PIGC P+ + KK + C N
Sbjct: 291 FTTMMADSAASFVLQLYGYGARRIGVIGTPPIGCTPSQRV----KKKKI----CNEDLNY 342
Query: 261 IAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCC--GSYY 318
AQ FN +L + QL + LP + Y D+Y+ +++ + GF + CC G
Sbjct: 343 AAQLFNSKLVIILGQLSKTLPNSTIVYGDIYSIFSKMLESPEDYGFEEIKKPCCKIGLTK 402
Query: 319 GYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAA-----NKWVAKHI 365
G + C ++ + N N S ++ WDG+H +Q A K V K+I
Sbjct: 403 G-GVFCKERTLKN-------MSNASSYLFWDGLHPSQRAYEISNRKLVKKYI 446
>RPB2_LYCES (Q42877) DNA-directed RNA polymerase II 135 kDa
polypeptide (EC 2.7.7.6) (RNA polymerase II subunit 2)
Length = 1191
Score = 35.0 bits (79), Expect = 0.32
Identities = 21/84 (25%), Positives = 40/84 (47%), Gaps = 2/84 (2%)
Query: 142 LQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRS 201
+Q+ ++ ++++ + P + DF++ +Y I+ GQ L +M S+ E
Sbjct: 50 IQNTMQEIVDESADIEIRPESQHNPGHQSDFAETIYKINFGQIYLSKPMMTESDGETATL 109
Query: 202 IPE--MMRNFTYDVQVLYDVGARV 223
P+ +RN TY + DV RV
Sbjct: 110 FPKAARLRNLTYSAPLYVDVTKRV 133
>RPB2_ARATH (P38420) DNA-directed RNA polymerase II 135 kDa
polypeptide (EC 2.7.7.6) (RNA polymerase II subunit 2)
Length = 1188
Score = 34.7 bits (78), Expect = 0.42
Identities = 21/84 (25%), Positives = 39/84 (46%), Gaps = 2/84 (2%)
Query: 142 LQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRS 201
+Q+ ++ ++++ + P + DF++ +Y I GQ L +M S+ E
Sbjct: 51 IQNTMQEIVDESADIEIRPESQHNPGHQSDFAETIYKISFGQIYLSKPMMTESDGETATL 110
Query: 202 IPE--MMRNFTYDVQVLYDVGARV 223
P+ +RN TY + DV RV
Sbjct: 111 FPKAARLRNLTYSAPLYVDVTKRV 134
>Y314_METJA (Q57762) Hypothetical protein MJ0314
Length = 263
Score = 32.0 bits (71), Expect = 2.7
Identities = 38/136 (27%), Positives = 58/136 (41%), Gaps = 22/136 (16%)
Query: 190 LMHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNL 249
+M S E + R ++RN D++ + I NT I P+ S+ Y G L
Sbjct: 62 IMKCSHETIRR----ILRNNNIDIRK----SSESLIIKNTKKINLNPSESLAYI--LGVL 111
Query: 250 DANGCVIPHNKIAQEFNRQLK----DQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQG 305
+ +G V NK + +LK D + + +RNL F Y++ Y K+E N Q
Sbjct: 112 NGDGSV---NKQESNYVIELKVTDKDFIEEFKRNLENIGFKYINEYVRKFE---NKKDQY 165
Query: 306 FVNPLEVCCGSYYGYR 321
V G YY Y+
Sbjct: 166 VVRVRSK--GFYYWYK 179
>PAPC_ECOLI (P07110) Outer membrane usher protein papC precursor
Length = 836
Score = 32.0 bits (71), Expect = 2.7
Identities = 29/103 (28%), Positives = 45/103 (43%), Gaps = 13/103 (12%)
Query: 22 IQIPSGNGSYSSSSKCS---YPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRA 78
I +P G G+ S S S Y + + D+ +D Y+ AGL S GG++
Sbjct: 570 ISVPLGTGTASYSGSMSNDRYVNMAGYTDTFNDGLDSYSLNAGLNSGGGLT--------- 620
Query: 79 SDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASI 121
R I + + + LSA + S+ Y A+GGA+I
Sbjct: 621 -SQRQINAYYSHRSPLANLSANIASLQKGYTSFGVSASGGATI 662
>GP2_HUMAN (P55259) Pancreatic secretory granule membrane major
glycoprotein GP2 precursor (Pancreatic zymogen granule
membrane protein GP-2) (ZAP75)
Length = 527
Score = 31.6 bits (70), Expect = 3.5
Identities = 15/43 (34%), Positives = 24/43 (54%), Gaps = 3/43 (6%)
Query: 299 SNASKQGFVNPLEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKN 341
++A ++G+ NP+E S YG +DCG V +PC+N
Sbjct: 19 ASAVQRGYGNPIEA---SSYGLDLDCGAPGTPEAHVCFDPCQN 58
>SYL_NITEU (Q820M7) Leucyl-tRNA synthetase (EC 6.1.1.4)
(Leucine--tRNA ligase) (LeuRS)
Length = 869
Score = 30.4 bits (67), Expect = 7.8
Identities = 21/86 (24%), Positives = 38/86 (43%), Gaps = 5/86 (5%)
Query: 276 LRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYYGYRIDCGKKAVVNGTVY 335
+++ +P AK+TY ++ + +L S + L C YY + + + G Y
Sbjct: 91 IQKGVPPAKWTYDNIAYMRSQLQSLGFAIDWQRELATCDPQYYRWNQWLFLRMLEKGIAY 150
Query: 336 GNPCKNPSQHISWDGVHYTQAANKWV 361
+Q ++WD V T AN+ V
Sbjct: 151 -----QKTQVVNWDPVDQTVLANEQV 171
>GCAT_AERHY (P10480) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43)
(Glycerophospholipid-cholesterol acyltransferase)
(GCAT)
Length = 335
Score = 30.4 bits (67), Expect = 7.8
Identities = 18/60 (30%), Positives = 33/60 (55%), Gaps = 4/60 (6%)
Query: 32 SSSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEE 91
++ S+ ++ + FGDS SDTG +Y+ G P ++ GR S+G + ++ +T E
Sbjct: 18 AADSRPAFSRIVMFGDSLSDTGKMYSKMRG-YLPSSPPYY---EGRFSNGPVWLEQLTNE 73
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.138 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,087,072
Number of Sequences: 164201
Number of extensions: 2184951
Number of successful extensions: 4574
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 4559
Number of HSP's gapped (non-prelim): 10
length of query: 382
length of database: 59,974,054
effective HSP length: 112
effective length of query: 270
effective length of database: 41,583,542
effective search space: 11227556340
effective search space used: 11227556340
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0400a.7


