

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
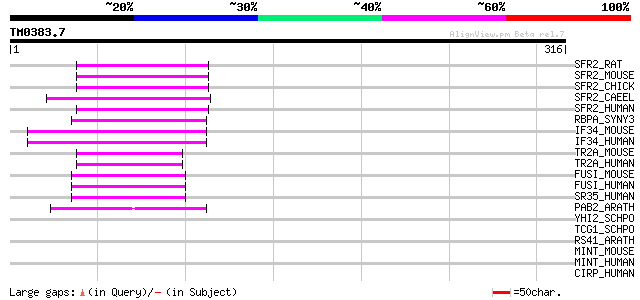
Query= TM0383.7
(316 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SFR2_RAT (Q6PDU1) Splicing factor, arginine/serine-rich 2 (Splic... 48 4e-05
SFR2_MOUSE (Q62093) Splicing factor, arginine/serine-rich 2 (Spl... 48 4e-05
SFR2_CHICK (P30352) Splicing factor, arginine/serine-rich 2 (Spl... 48 4e-05
SFR2_CAEEL (Q09511) Probable splicing factor, arginine/serine-ri... 47 6e-05
SFR2_HUMAN (Q01130) Splicing factor, arginine/serine-rich 2 (Spl... 45 3e-04
RBPA_SYNY3 (Q57014) Putative RNA-binding protein rbpA 44 5e-04
IF34_MOUSE (Q9Z1D1) Eukaryotic translation initiation factor 3 s... 44 5e-04
IF34_HUMAN (O75821) Eukaryotic translation initiation factor 3 s... 44 5e-04
TR2A_MOUSE (Q6PFR5) Transformer-2 protein homolog (TRA-2 alpha) 44 7e-04
TR2A_HUMAN (Q13595) Transformer-2 protein homolog (TRA-2 alpha) 44 7e-04
FUSI_MOUSE (Q9R0U0) FUS interacting serine-arginine rich protein... 44 7e-04
FUSI_HUMAN (O75494) FUS interacting serine-arginine rich protein... 44 7e-04
SR35_HUMAN (Q8WXF0) 35 kDa SR repressor protein (SRrp35) 43 9e-04
PAB2_ARATH (P42731) Polyadenylate-binding protein 2 (Poly(A)-bin... 43 9e-04
YHI2_SCHPO (O13620) Probable RNA-binding protein P22H7.02c 42 0.002
TCG1_SCHPO (Q9HEQ9) Single-stranded TG1-3 binding protein 42 0.002
RS41_ARATH (P92966) Arginine/serine-rich splicing factor RSP41 42 0.003
MINT_MOUSE (Q62504) Msx2-interacting protein (SMART/HDAC1 associ... 42 0.003
MINT_HUMAN (Q96T58) Msx2-interacting protein (SMART/HDAC1 associ... 42 0.003
CIRP_HUMAN (Q14011) Cold-inducible RNA-binding protein (Glycine-... 42 0.003
>SFR2_RAT (Q6PDU1) Splicing factor, arginine/serine-rich 2 (Splicing
factor SC35) (SC-35) (Splicing component, 35 kDa)
Length = 220
Score = 47.8 bits (112), Expect = 4e-05
Identities = 27/75 (36%), Positives = 43/75 (57%)
Query: 39 VDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNG 98
VD ++ S LRR+FE+YG + V++ R R FAF+RF ++DA +A+ +++G
Sbjct: 17 VDNLTYRTSPDTLRRVFEKYGRVGDVYIPRDRYTKESRGFAFVRFHDKRDAEDAMDAMDG 76
Query: 99 SWFFESFLMVNRARY 113
+ L V ARY
Sbjct: 77 AVLDGRELRVQMARY 91
>SFR2_MOUSE (Q62093) Splicing factor, arginine/serine-rich 2
(Splicing factor SC35) (SC-35) (Splicing component, 35
kDa) (PR264 protein)
Length = 220
Score = 47.8 bits (112), Expect = 4e-05
Identities = 27/75 (36%), Positives = 43/75 (57%)
Query: 39 VDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNG 98
VD ++ S LRR+FE+YG + V++ R R FAF+RF ++DA +A+ +++G
Sbjct: 17 VDNLTYRTSPDTLRRVFEKYGRVGDVYIPRDRYTKESRGFAFVRFHDKRDAEDAMDAMDG 76
Query: 99 SWFFESFLMVNRARY 113
+ L V ARY
Sbjct: 77 AVLDGRELRVQMARY 91
>SFR2_CHICK (P30352) Splicing factor, arginine/serine-rich 2
(Splicing factor SC35) (SC-35) (Splicing component, 35
kDa) (PR264 protein)
Length = 221
Score = 47.8 bits (112), Expect = 4e-05
Identities = 27/75 (36%), Positives = 43/75 (57%)
Query: 39 VDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNG 98
VD ++ S LRR+FE+YG + V++ R R FAF+RF ++DA +A+ +++G
Sbjct: 18 VDNLTYRTSPDTLRRVFEKYGRVGDVYIPRDRYTKESRGFAFVRFHDKRDAEDAMDAMDG 77
Query: 99 SWFFESFLMVNRARY 113
+ L V ARY
Sbjct: 78 AVLDGRELRVQMARY 92
>SFR2_CAEEL (Q09511) Probable splicing factor, arginine/serine-rich
2 (Splicing factor SC35) (SC-35) (Splicing component, 35
kDa)
Length = 196
Score = 47.0 bits (110), Expect = 6e-05
Identities = 29/93 (31%), Positives = 46/93 (49%)
Query: 22 GVDRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFI 81
G DR+ + ++ +D +S + +DLRR FERYG + V + R + + F F+
Sbjct: 6 GGDRRAAPDINGLTSLKIDNLSYQTTPNDLRRTFERYGDIGDVHIPRDKYSRQSKGFGFV 65
Query: 82 RFKYEQDACEAIRSLNGSWFFESFLMVNRARYD 114
RF +DA A+ +G L V A+YD
Sbjct: 66 RFYERRDAEHALDRTDGKLVDGRELRVTLAKYD 98
>SFR2_HUMAN (Q01130) Splicing factor, arginine/serine-rich 2
(Splicing factor SC35) (SC-35) (Splicing component, 35
kDa) (PR264 protein)
Length = 220
Score = 44.7 bits (104), Expect = 3e-04
Identities = 26/75 (34%), Positives = 42/75 (55%)
Query: 39 VDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNG 98
VD ++ S LRR+FE+Y + V++ R R FAF+RF ++DA +A+ +++G
Sbjct: 17 VDNLTYRTSPDTLRRVFEKYRRVGDVYIPRDRYTKESRGFAFVRFHDKRDAEDAMDAMDG 76
Query: 99 SWFFESFLMVNRARY 113
+ L V ARY
Sbjct: 77 AVLDGRELRVQMARY 91
>RBPA_SYNY3 (Q57014) Putative RNA-binding protein rbpA
Length = 101
Score = 43.9 bits (102), Expect = 5e-04
Identities = 25/77 (32%), Positives = 42/77 (54%)
Query: 36 TIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRS 95
+I+V +S +VS DL +F YG + V L R+ GR F F+ + + + AI +
Sbjct: 2 SIYVGNLSYDVSEADLTAVFAEYGSVKRVQLPTDRETGRMRGFGFVELEADAEETAAIEA 61
Query: 96 LNGSWFFESFLMVNRAR 112
L+G+ + L VN+A+
Sbjct: 62 LDGAEWMGRDLKVNKAK 78
>IF34_MOUSE (Q9Z1D1) Eukaryotic translation initiation factor 3
subunit 4 (eIF-3 delta) (eIF3 p44) (eIF-3 RNA-binding
subunit) (eIF3 p42) (Eif3p42) (eIF3g)
Length = 320
Score = 43.9 bits (102), Expect = 5e-04
Identities = 28/102 (27%), Positives = 46/102 (44%)
Query: 11 PVSRPEVVGSHGVDRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRR 70
P S + G Q R TI V +S++ DL+ +F +G + ++L + +
Sbjct: 215 PPSLRDGASRRGESMQPNRRADDNATIRVTNLSEDTRETDLQELFRPFGSISRIYLAKDK 274
Query: 71 KQGRKSRFAFIRFKYEQDACEAIRSLNGSWFFESFLMVNRAR 112
G+ FAFI F +DA AI ++G + L V A+
Sbjct: 275 TTGQSKGFAFISFHRREDAARAIAGVSGFGYDHLILNVEWAK 316
>IF34_HUMAN (O75821) Eukaryotic translation initiation factor 3
subunit 4 (eIF-3 delta) (eIF3 p44) (eIF-3 RNA-binding
subunit) (eIF3 p42) (eIF3g)
Length = 320
Score = 43.9 bits (102), Expect = 5e-04
Identities = 28/102 (27%), Positives = 46/102 (44%)
Query: 11 PVSRPEVVGSHGVDRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRR 70
P S + G Q R TI V +S++ DL+ +F +G + ++L + +
Sbjct: 215 PPSLRDGASRRGESMQPNRRADDNATIRVTNLSEDTRETDLQELFRPFGSISRIYLAKDK 274
Query: 71 KQGRKSRFAFIRFKYEQDACEAIRSLNGSWFFESFLMVNRAR 112
G+ FAFI F +DA AI ++G + L V A+
Sbjct: 275 TTGQSKGFAFISFHRREDAARAIAGVSGFGYDHLILNVEWAK 316
>TR2A_MOUSE (Q6PFR5) Transformer-2 protein homolog (TRA-2 alpha)
Length = 281
Score = 43.5 bits (101), Expect = 7e-04
Identities = 24/60 (40%), Positives = 34/60 (56%)
Query: 39 VDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNG 98
V G+S + DLR +F RYG L GV + ++ GR FAF+ F+ D+ EA+ NG
Sbjct: 121 VFGLSLYTTERDLREVFSRYGPLSGVNVVYDQRTGRSRGFAFVYFERIDDSKEAMERANG 180
>TR2A_HUMAN (Q13595) Transformer-2 protein homolog (TRA-2 alpha)
Length = 282
Score = 43.5 bits (101), Expect = 7e-04
Identities = 24/60 (40%), Positives = 34/60 (56%)
Query: 39 VDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNG 98
V G+S + DLR +F RYG L GV + ++ GR FAF+ F+ D+ EA+ NG
Sbjct: 123 VFGLSLYTTERDLREVFSRYGPLSGVNVVYDQRTGRSRGFAFVYFERIDDSKEAMERANG 182
>FUSI_MOUSE (Q9R0U0) FUS interacting serine-arginine rich protein 1
(TLS-associated protein with Ser-Arg repeats)
(TLS-associated protein with SR repeats) (TASR)
(TLS-associated serine-arginine protein) (TLS-associated
SR protein) (Neural specific SR prot
Length = 262
Score = 43.5 bits (101), Expect = 7e-04
Identities = 22/65 (33%), Positives = 38/65 (57%)
Query: 36 TIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRS 95
++FV V+D+ DLRR F RYG + V++ R FA+++F+ +DA +A+ +
Sbjct: 11 SLFVRNVADDTRSEDLRREFGRYGPIVDVYVPLDFYTRRPRGFAYVQFEDVRDAEDALHN 70
Query: 96 LNGSW 100
L+ W
Sbjct: 71 LDRKW 75
>FUSI_HUMAN (O75494) FUS interacting serine-arginine rich protein 1
(TLS-associated protein with Ser-Arg repeats)
(TLS-associated protein with SR repeats) (TASR)
(TLS-associated serine-arginine protein) (TLS-associated
SR protein) (40 kDa SR-repressor pro
Length = 262
Score = 43.5 bits (101), Expect = 7e-04
Identities = 22/65 (33%), Positives = 38/65 (57%)
Query: 36 TIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRS 95
++FV V+D+ DLRR F RYG + V++ R FA+++F+ +DA +A+ +
Sbjct: 11 SLFVRNVADDTRSEDLRREFGRYGPIVDVYVPLDFYTRRPRGFAYVQFEDVRDAEDALHN 70
Query: 96 LNGSW 100
L+ W
Sbjct: 71 LDRKW 75
>SR35_HUMAN (Q8WXF0) 35 kDa SR repressor protein (SRrp35)
Length = 261
Score = 43.1 bits (100), Expect = 9e-04
Identities = 22/65 (33%), Positives = 37/65 (56%)
Query: 36 TIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRS 95
++F+ V+D DLRR F RYG + V++ R FA+++F+ +DA +A+ +
Sbjct: 11 SLFIRNVADATRPEDLRREFGRYGPIVDVYIPLDFYTRRPRGFAYVQFEDVRDAEDALYN 70
Query: 96 LNGSW 100
LN W
Sbjct: 71 LNRKW 75
>PAB2_ARATH (P42731) Polyadenylate-binding protein 2
(Poly(A)-binding protein 2) (PABP 2)
Length = 629
Score = 43.1 bits (100), Expect = 9e-04
Identities = 23/89 (25%), Positives = 42/89 (46%), Gaps = 1/89 (1%)
Query: 24 DRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRF 83
+R K ++V ++++ + DL+ F YG + + + +G+ F F+ F
Sbjct: 204 ERDSTANKTKFTNVYVKNLAESTTDDDLKNAFGEYGKITSAVVMKDG-EGKSKGFGFVNF 262
Query: 84 KYEQDACEAIRSLNGSWFFESFLMVNRAR 112
+ DA A+ SLNG F + V RA+
Sbjct: 263 ENADDAARAVESLNGHKFDDKEWYVGRAQ 291
>YHI2_SCHPO (O13620) Probable RNA-binding protein P22H7.02c
Length = 833
Score = 42.0 bits (97), Expect = 0.002
Identities = 26/59 (44%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query: 69 RRKQGRKSRFAFIRFKYEQDACEAIRSLNGSWFFESFLMVNRARYDYTNRRNVTNRHSK 127
+ K+G RFAFI FK E+DA +AIR LN S+ S + V+RA DY + +SK
Sbjct: 39 KTKEGVSRRFAFIGFKNEEDADKAIRYLNKSYVETSRIEVHRA-LDYRSANEKLRPYSK 96
>TCG1_SCHPO (Q9HEQ9) Single-stranded TG1-3 binding protein
Length = 348
Score = 42.0 bits (97), Expect = 0.002
Identities = 25/82 (30%), Positives = 45/82 (54%), Gaps = 4/82 (4%)
Query: 35 FTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRR-KQGRK---SRFAFIRFKYEQDAC 90
F +FV +S + ++R +FE G + V + RR ++G + S AF+ F ++D
Sbjct: 44 FRVFVGRLSTSTKKSEIRSLFETVGTVRKVTIPFRRVRRGTRLVPSGIAFVTFNNQEDVD 103
Query: 91 EAIRSLNGSWFFESFLMVNRAR 112
+AI +LNG + ++V +AR
Sbjct: 104 KAIETLNGKTLDDREIVVQKAR 125
Score = 36.2 bits (82), Expect = 0.11
Identities = 27/109 (24%), Positives = 48/109 (43%), Gaps = 18/109 (16%)
Query: 36 TIFVDGVSDNVSYHDLRRMFERYG--------------VLHGVFLQRRRKQGRKSRFAFI 81
+I+V G+S ++ L+ MF+ Y ++ + L+ +++GR F F+
Sbjct: 206 SIYVSGLSVTLTNEGLKEMFDAYNPTRARIAVRSLPPYIIRRIKLRGEQRRGRG--FGFV 263
Query: 82 RFKYEQDACEAIRSLNGSWFFESFLMVNRA--RYDYTNRRNVTNRHSKV 128
F +D AI +NG + L+V A R D N N N + +
Sbjct: 264 SFANAEDQSRAIEEMNGKQVGDLTLVVKSAVFREDKQNDENEKNENEPI 312
>RS41_ARATH (P92966) Arginine/serine-rich splicing factor RSP41
Length = 356
Score = 41.6 bits (96), Expect = 0.003
Identities = 29/92 (31%), Positives = 44/92 (47%), Gaps = 10/92 (10%)
Query: 29 RTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQD 88
R K +F I D + N DL R FE YG + V ++R FAFI+++ ++D
Sbjct: 93 RPSKTLFVINFD--AQNTRTRDLERHFEPYGKIVNVRIRRN--------FAFIQYEAQED 142
Query: 89 ACEAIRSLNGSWFFESFLMVNRARYDYTNRRN 120
A A+ + N S + + V A D +R N
Sbjct: 143 ATRALDATNSSKLMDKVISVEYAVKDDDSRGN 174
Score = 32.3 bits (72), Expect = 1.6
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 11/67 (16%)
Query: 50 DLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNGSWFFESFLMVN 109
DL R+F +YG + V + K+ FAF+ + E+DA +AIR+L+ FE
Sbjct: 17 DLERLFRKYGKVERVDM--------KAGFAFVYMEDERDAEDAIRALDR---FEYGRTGR 65
Query: 110 RARYDYT 116
R R ++T
Sbjct: 66 RLRVEWT 72
>MINT_MOUSE (Q62504) Msx2-interacting protein (SMART/HDAC1
associated repressor protein)
Length = 3644
Score = 41.6 bits (96), Expect = 0.003
Identities = 21/86 (24%), Positives = 45/86 (51%), Gaps = 3/86 (3%)
Query: 32 KKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACE 91
K T+F+ + +YHDLR +F+R+G + + + +K ++AF+++ C+
Sbjct: 436 KATRTLFIGNLEKTTTYHDLRNIFQRFGEIVDIDI---KKVNGVPQYAFLQYCDIASVCK 492
Query: 92 AIRSLNGSWFFESFLMVNRARYDYTN 117
AI+ ++G + + L + + TN
Sbjct: 493 AIKKMDGEYLGNNRLKLGFGKSMPTN 518
Score = 31.2 bits (69), Expect = 3.5
Identities = 23/84 (27%), Positives = 39/84 (46%), Gaps = 11/84 (13%)
Query: 37 IFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSL 96
+++DG+S NVS L R F RYG + V R + A + + +DA A++
Sbjct: 520 VWLDGLSSNVSDQYLTRHFCRYGPVVKVVFDRLK------GMALVLYSEIEDAQAAVKET 573
Query: 97 NGSWFFESFLMVNRARYDYTNRRN 120
G + N+ + D+ NR +
Sbjct: 574 KG-----RKIGGNKIKVDFANRES 592
>MINT_HUMAN (Q96T58) Msx2-interacting protein (SMART/HDAC1
associated repressor protein)
Length = 3664
Score = 41.6 bits (96), Expect = 0.003
Identities = 21/86 (24%), Positives = 45/86 (51%), Gaps = 3/86 (3%)
Query: 32 KKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACE 91
K T+F+ + +YHDLR +F+R+G + + + +K ++AF+++ C+
Sbjct: 435 KATRTLFIGNLEKTTTYHDLRNIFQRFGEIVDIDI---KKVNGVPQYAFLQYCDIASVCK 491
Query: 92 AIRSLNGSWFFESFLMVNRARYDYTN 117
AI+ ++G + + L + + TN
Sbjct: 492 AIKKMDGEYLGNNRLKLGFGKSMPTN 517
>CIRP_HUMAN (Q14011) Cold-inducible RNA-binding protein
(Glycine-rich RNA-binding protein CIRP) (A18 hnRNP)
Length = 172
Score = 41.6 bits (96), Expect = 0.003
Identities = 23/82 (28%), Positives = 43/82 (52%)
Query: 37 IFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSL 96
+FV G+S + + L ++F +YG + V + + R+ R F F+ F+ DA +A+ ++
Sbjct: 8 LFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAM 67
Query: 97 NGSWFFESFLMVNRARYDYTNR 118
NG + V++A NR
Sbjct: 68 NGKSVDGRQIRVDQAGKSSDNR 89
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,314,717
Number of Sequences: 164201
Number of extensions: 1439781
Number of successful extensions: 4624
Number of sequences better than 10.0: 219
Number of HSP's better than 10.0 without gapping: 128
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 4335
Number of HSP's gapped (non-prelim): 352
length of query: 316
length of database: 59,974,054
effective HSP length: 110
effective length of query: 206
effective length of database: 41,911,944
effective search space: 8633860464
effective search space used: 8633860464
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0383.7


