

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0383.5
(1806 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
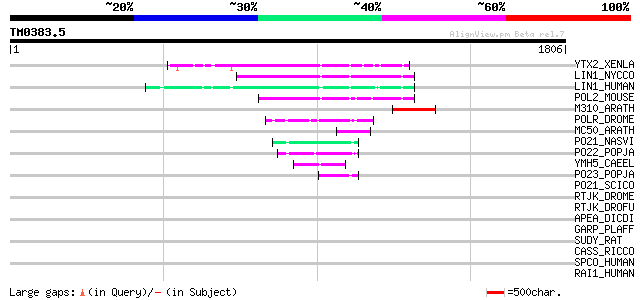
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 174 2e-42
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 160 3e-38
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 151 2e-35
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 149 7e-35
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 119 8e-26
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 83 8e-15
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 79 9e-14
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 68 3e-10
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 65 2e-09
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 53 9e-06
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 52 2e-05
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 44 0.005
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 40 0.045
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 39 0.13
APEA_DICDI (P51173) DNA-(apurinic or apyrimidinic site) lyase (E... 39 0.13
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 38 0.29
SUDY_RAT (P52847) DOPA/tyrosine sulfotransferase (EC 2.8.1.-) 36 1.1
CASS_RICCO (P59287) Casbene synthase, chloroplast precursor (EC ... 36 1.1
SPCO_HUMAN (Q01082) Spectrin beta chain, brain 1 (Spectrin, non-... 34 3.2
RAI1_HUMAN (Q7Z5J4) Retinoic acid induced protein 1 34 3.2
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein (ORF
2)
Length = 1308
Score = 174 bits (441), Expect = 2e-42
Identities = 202/833 (24%), Positives = 350/833 (41%), Gaps = 81/833 (9%)
Query: 514 WSSSLDVNIISLSSNHIQMEV--TTTIIDSKMV------------VTGIYGHPEAPQKWR 559
W+S +++L S+ Q EV T++I +++ + +Y P++ R
Sbjct: 64 WTSC---GVVTLFSDSFQPEVLSATSVIPGRLLHLRVRESGRTYNLMNVYAPTTGPERAR 120
Query: 560 TWELIKKL--TPDPEVPWLCLGDFNEVLKSSEKTGGNPINFVGAQAFQEAMDFCGL---- 613
+E + T D + + GDFN L + ++ + +E + L
Sbjct: 121 FFESLSAYMETIDSDEALIIGGDFNYTLDARDRNVPKKRDS-SESVLRELIAHFSLVDVW 179
Query: 614 RDMGFSGFQFTWSNMRPTPNTIQERLDRALVNECWTEHWPHTHVSHLDRFNSDHNPIYVI 673
R+ FT+ +R + Q R+DR ++ + + L F SDHN + +
Sbjct: 180 REQNPETVAFTYVRVRDG-HVSQSRIDRIYISSHLMSRAQSSTI-RLAPF-SDHNCVSLR 236
Query: 674 FLKAKRQKKKKIRLYRFEEMWLQEEDCRHVIHHSWPRGGRNITSK---------LGNVGA 724
A K + F L++E + +W RG R + +G V
Sbjct: 237 MSIAP--SLPKAAYWHFNNSLLEDEGFAKSVRDTW-RGWRAFQDEFATLNQWWDVGKVHL 293
Query: 725 NLT--EWAEATFGNIQKRIKSQLG-IIKRIQAAEQTEENLKKGKEAEKE--LENLLKLEE 779
L E+ ++ G I++ G ++ Q +E+ + + E++ L N+ + +
Sbjct: 294 KLLCQEYTKSVSGQRNAEIEALNGEVLDLEQRLSGSEDQALQCEYLERKEALRNMEQRQA 353
Query: 780 IKWSQRSRATWLAHGDRNTSFFHKKASQRKDRNSIARITDDRGRAWTEEEDIEEVLTKYF 839
RSR L DR + FF+ ++ +R I + + G + E I + ++
Sbjct: 354 RGAFVRSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFY 413
Query: 840 EDLFKAT--SPDDCEIVTNLVHGRITNEHKEIMEEPFTEEEVREALKKMHPTKAHGADGT 897
++LF SPD CE + + + ++ KE +E P T +E+ +AL+ M K+ G DG
Sbjct: 414 QNLFSPDPISPDACEELWDGLPV-VSERRKERLETPITLDELSQALRLMPHNKSPGLDGL 472
Query: 898 PALFFQKFWSIVGYDVTAQVLSILNDGADATHINQTLIALIPKIKKPSMAKDYRPISL*N 957
FFQ FW +G D + G + +++L+PK + K++RP+SL +
Sbjct: 473 TIEFFQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLS 532
Query: 958 VIFKLVTKTIANRLKSFLQDIVEETQSAFVPGRLITDNAIIAHECFHYMRRKTRGVKGVM 1017
+K+V K I+ RLKS L +++ QS VPGR I DN + + H+ RR +
Sbjct: 533 TDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTG---LSLA 589
Query: 1018 ALKTDMSKAYDRVEWNFLKSTLEQMGFTQRWTNLIMNCVSTVSFQVLLNGQPCRSFKPER 1077
L D KA+DRV+ +L TL+ F ++ + ++ V +N R
Sbjct: 590 FLSLDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGR 649
Query: 1078 GLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPRAPMISHLFFADDSILLAR- 1136
G+RQG PLS L+ L E F +++ + L G+ + + +ADD IL+A+
Sbjct: 650 GVRQGCPLSGQLYSLAIEPFLCLLRKR-----LTGLVLKEPDMRVVLSAYADDVILVAQD 704
Query: 1137 -ATKEEAMVLHDIIKGYEAFSGQRINFDKSE--LSTSRNV----PSTRVHELEQLLGVKA 1189
E A ++ Y A S RIN+ KS L S V P+ R E K
Sbjct: 705 LVDLERAQECQEV---YAAASSARINWSKSSGLLEGSLKVDFLPPAFRDISWES----KI 757
Query: 1190 VEKHGKYLGLPTIVGRSKTQVFSFVRERIWKKLKGWK--EKALSRSGREVLIKSIAQAIP 1247
++ G YL +Q F + E + +L WK K LS GR ++I + +
Sbjct: 758 IKYLGVYLSAEEY---PVSQNFIELEECVLTRLGKWKGFAKVLSMRGRALVINQLVASQI 814
Query: 1248 SYVMGLFLLPSTLCNQVERMINSFYWGGSADSRKLNWIRWDTLATPKNMGGLG 1300
Y + +++R + F W G +W+ + P GG G
Sbjct: 815 WYRLICLSPTQEFIAKIQRRLLDFLWIGK------HWVSAGVSSLPLKEGGQG 861
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 160 bits (405), Expect = 3e-38
Identities = 143/593 (24%), Positives = 269/593 (45%), Gaps = 28/593 (4%)
Query: 738 QKRIKSQLGIIKRIQAAEQTEENLKKGKEAEKELENLLKLEE---IKWSQRSRATWLAHG 794
++ + + +G +K+++ E + + KE K L ++E I+ +S++ +
Sbjct: 310 REEVNNLMGHLKQLEKEEHSNPKPSRRKEITKIRAELNEIENKRIIQQINKSKSWFFEKI 369
Query: 795 DRNTSFFHKKASQRKDRNSIARITDDRGRAWTEEEDIEEVLTKYFEDLF--KATSPDDCE 852
++ +++ ++ I+ I + T+ +I+++L +Y++ L+ K + + +
Sbjct: 370 NKIDKPLANLTRKKRVKSLISSIRNGNDEITTDPSEIQKILNEYYKKLYSHKYENLKEID 429
Query: 853 IVTNLVH-GRITNEHKEIMEEPFTEEEVREALKKMHPTKAHGADGTPALFFQKFWSIVGY 911
H R++ + E++ P + E+ ++ + K+ G DG + F+Q F +
Sbjct: 430 QYLEACHLPRLSQKEVEMLNRPISSSEIASTIQNLPKKKSPGPDGFTSEFYQTFKEELVP 489
Query: 912 DVTAQVLSILNDGADATHINQTLIALIPKI-KKPSMAKDYRPISL*NVIFKLVTKTIANR 970
+ +I +G + I LIPK K P+ ++YRPISL N+ K++ K + NR
Sbjct: 490 ILLNLFQNIEKEGILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNR 549
Query: 971 LKSFLQDIVEETQSAFVPGRLITDNAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRV 1030
++ ++ I+ Q F+PG N + ++ + K M L D KA+D +
Sbjct: 550 IQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQHINKLKN--KDHMILSIDAEKAFDNI 607
Query: 1031 EWNFLKSTLEQMGFTQRWTNLIMNCVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLF 1090
+ F+ TL+++G + LI S + ++LNG +SF G RQG PLSP LF
Sbjct: 608 QHPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLF 667
Query: 1091 ILCAEVFSSMIKAKVAAGVLHGIKITPRAPMISHLFFADDSILLARATKEEAMVLHDIIK 1150
+ EV + I+ + A + GI I +S FADD I+ T++ L ++IK
Sbjct: 668 NIVMEVLAIAIREEKA---IKGIHIGSEEIKLS--LFADDMIVYLENTRDSTTKLLEVIK 722
Query: 1151 GYEAFSGQRINFDKSE--LSTSRNVPSTRVHELEQLLGVKAVEKHGKYLG--LPTIVGRS 1206
Y SG +IN KS + T+ N V + + V K KYLG L V
Sbjct: 723 EYSNVSGYKINTHKSVAFIYTNNNQAEKTV---KDSIPFTVVPKKMKYLGVYLTKDVKDL 779
Query: 1207 KTQVFSFVRERIWKKLKGWKEKALSRSGREVLIKS--IAQAIPSYVMGLFLLPSTLCNQV 1264
+ + +R+ I + + WK S GR ++K + +AI ++ P + +
Sbjct: 780 YKENYETLRKEIAEDVNKWKNIPCSWLGRINIVKMSILPKAIYNFNAIPIKAPLSYFKDL 839
Query: 1265 ERMINSFYWGGSADSRKLNWIRWDTLATPKNMGGLGFRSFHSFNKALLAKQWW 1317
E++I F W ++K I L+ GG+ + K+++ K W
Sbjct: 840 EKIILHFIW-----NQKKPQIAKTLLSNKNKAGGITLPDLRLYYKSIVIKTAW 887
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 151 bits (381), Expect = 2e-35
Identities = 199/917 (21%), Positives = 373/917 (39%), Gaps = 78/917 (8%)
Query: 441 MISWNCRGLGTPRAVGALRKLITMERPDLVFLMETRKKAFEIQKTKMPSGLVHSIGVDCN 500
+++ N GL P L I + P + + ET + + K+ +I
Sbjct: 9 ILTLNVNGLNAPIKRHRLANWIKSQDPSVCCIQETHLTCRDTHRLKIKGW--RNI---YQ 63
Query: 501 GEGKSRGGGLACYWSSSLDVNIISLSSN---HIQMEVTTTIIDSKMVVTGIYGHPEAPQK 557
GK + G+A S D + + H M V +I ++ + IY P
Sbjct: 64 ANGKQKKAGVAILVSDKTDFKPTKIKRDKEGHYIM-VKGSIQQEELTILNIYA-PNTGAP 121
Query: 558 WRTWELIKKLTPDPEVPWLCLGDFNEVLKSSEKTGGNPINFVGAQAFQEAMDFCGL---- 613
+++ L D + + +GDFN L + +++ IN Q A+ L
Sbjct: 122 RFIKQVLSDLQRDLDSHTIIMGDFNTPLSTLDRSTRQKIN-KDIQELNSALHQADLIDIY 180
Query: 614 RDMGFSGFQFTWSNMRPTPNTIQERLDRALVNECWTEHWPHTHVSHLDRFNSDHNPIYV- 672
R + ++T+ + P+ + D L ++ T + + SDH+ I +
Sbjct: 181 RTLHPKSTEYTFFS---APHHTYSKTDHILGSKTLLSKCKRTEI--ITNCLSDHSAIKLE 235
Query: 673 IFLKAKRQKKK---KIRLYRFEEMWLQEEDCRHVIHHSWPRGGRNITSKLGNVGANLTEW 729
+ +K Q K+ + W+ E + ++ T + NL +
Sbjct: 236 LRIKKLTQNHSTTWKLNNLLLNDYWVHNEMKAEIKKFFETNENKDTTYQ------NLWDT 289
Query: 730 AEATFG------NIQKR------IKSQLGIIKRIQAAEQTEENLKKGKEAEKELENLLKL 777
A+A N KR I + + +K ++ EQT + +E K L ++
Sbjct: 290 AKAVCRGKFIALNAHKRKQERSKIDTLISQLKELEKQEQTNSKASRRQEIIKIRAELKEI 349
Query: 778 EEIKWSQR---SRATWLAHGDRNTSFFHKKASQRKDRNSIARITDDRGRAWTEEEDIEEV 834
E K Q+ SR+ + ++ + +++++N I I +DRG T+ +I+
Sbjct: 350 ETQKTLQKINESRSWFFEKINKIDRPLARLIKKKREKNQIDTIKNDRGDITTDPTEIQTT 409
Query: 835 LTKYFEDLFKATSPDDCEIVTNLVHG----RITNEHKEIMEEPFTEEEVREALKKMHPTK 890
+ +Y++ L+ A ++ E + + R+ E E + P T E+ + + K
Sbjct: 410 IREYYKHLY-ANKLENLEEMDKFLDTYTLPRLNQEEVESLNRPITSSEIEAIINSLPNKK 468
Query: 891 AHGADGTPALFFQKFWSIVGYDVTAQVLSILNDGADATHINQTLIALIPKIKKPSMAKD- 949
+ G +G A F+Q++ + + SI +G + I LIPK + + K+
Sbjct: 469 SPGPEGFTAEFYQRYKEELVPFLLKLFQSIEKEGILPNSFYEASIILIPKPGRDTTKKEN 528
Query: 950 YRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQSAFVPGRLITDNAIIAHECFHYMRRK 1009
+RPISL N+ K++ K +AN+++ ++ ++ Q F+P N + ++ R
Sbjct: 529 FRPISLMNIDAKILNKILANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKSINIIQHINR- 587
Query: 1010 TRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGFTQRWTNLIMNCVSTVSFQVLLNGQP 1069
T+ M + D KA+D+++ F+ L ++G + +I + ++LNGQ
Sbjct: 588 TKDTNH-MIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQK 646
Query: 1070 CRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPRAPMISHLFFAD 1129
+ + G RQG PLSP L + EV + I+ + + GI++ +S FAD
Sbjct: 647 LEAPPLKTGTRQGCPLSPLLPNIVLEVLARAIRQEKE---IKGIQLGKEEVKLS--LFAD 701
Query: 1130 DSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSE---LSTSRNVPSTRVHELEQLLG 1186
D I+ A L +I + SG +IN KS+ + +R S + EL +
Sbjct: 702 DMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTNNRQTESQIMSELPFTIA 761
Query: 1187 VKAVEKHGKYLGLPTIVGRSKTQVFSFVRERIWKKLK----GWKEKALSRSGREVLIKS- 1241
K + KYLG+ + R +F + + ++K WK S GR ++K
Sbjct: 762 SKRI----KYLGIQ--LTRDVKDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRINIVKMA 815
Query: 1242 -IAQAIPSYVMGLFLLPSTLCNQVERMINSFYWGGSADSRKLNWIRWDTLATPKNMGGLG 1300
+ + I + LP T ++E+ F W ++K I TL+ GG+
Sbjct: 816 ILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIW-----NQKRAHIAKSTLSQKNKAGGIT 870
Query: 1301 FRSFHSFNKALLAKQWW 1317
F + KA + K W
Sbjct: 871 LPDFKLYYKATVTKTAW 887
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 149 bits (376), Expect = 7e-35
Identities = 129/519 (24%), Positives = 239/519 (45%), Gaps = 25/519 (4%)
Query: 809 KDRNSIARITDDRGRAWTEEEDIEEVLTKYFEDLF--KATSPDDCE-IVTNLVHGRITNE 865
+D+ I +I +++G T+ E+I+ + +++ L+ K + D+ + + ++ +
Sbjct: 411 RDKILINKIRNEKGDITTDPEEIQNTIRSFYKRLYSTKLENLDEMDKFLDRYQVPKLNQD 470
Query: 866 HKEIMEEPFTEEEVREALKKMHPTKAHGADGTPALFFQKFWSIVGYDVTAQVLSILNDGA 925
+ + P + +E+ + + K+ G DG A F+Q F + + I +G
Sbjct: 471 QVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGT 530
Query: 926 DATHINQTLIALIPKIKK-PSMAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQS 984
+ I LIPK +K P+ +++RPISL N+ K++ K +ANR++ ++ I+ Q
Sbjct: 531 LPNSFYEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQV 590
Query: 985 AFVPGRLITDNAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGF 1044
F+PG N + HY+ + K M + D KA+D+++ F+ LE+ G
Sbjct: 591 GFIPGMQGWFNIRKSINVIHYINKLKD--KNHMIISLDAEKAFDKIQHPFMIKVLERSGI 648
Query: 1045 TQRWTNLIMNCVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAK 1104
+ N+I S + +NG+ + + G RQG PLSPYLF + EV + I+ +
Sbjct: 649 QGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQQ 708
Query: 1105 VAAGVLHGIKITPRAPMISHLFFADDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDK 1164
+ GI+I IS L ADD I+ K L ++I + G +IN +K
Sbjct: 709 KE---IKGIQIGKEEVKISLL--ADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNK 763
Query: 1165 S-ELSTSRNVPSTRVHELEQLLGVKAVEKHGKYLGLPTIVGRSK---TQVFSFVRERIWK 1220
S ++N + + E+ + V + KYLG+ T+ K + F +++ I +
Sbjct: 764 SMAFLYTKNKQAEK--EIRETTPFSIVTNNIKYLGV-TLTKEVKDLYDKNFKSLKKEIKE 820
Query: 1221 KLKGWKEKALSRSGREVLIKS--IAQAIPSYVMGLFLLPSTLCNQVERMINSFYWGGSAD 1278
L+ WK+ S GR ++K + +AI + +P+ N++E I F W
Sbjct: 821 DLRRWKDLPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVW----- 875
Query: 1279 SRKLNWIRWDTLATPKNMGGLGFRSFHSFNKALLAKQWW 1317
+ K I L + GG+ + +A++ K W
Sbjct: 876 NNKKPRIAKSLLKDKRTSGGITMPDLKLYYRAIVIKTAW 914
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 119 bits (298), Expect = 8e-26
Identities = 52/143 (36%), Positives = 89/143 (61%), Gaps = 1/143 (0%)
Query: 1245 AIPSYVMGLFLLPSTLCNQVERMINSFYWGGSADSRKLNWIRWDTLATPK-NMGGLGFRS 1303
A+P Y M F L LC ++ + F+W + RK++W+ W L K + GGLGFR
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 1304 FHSFNKALLAKQWWRLINRENSLMVQVIKARYYPKSTPLEANRGTNSSYLWKSVHAAREV 1363
FN+ALLAKQ +R+I++ ++L+ +++++RY+P S+ +E + GT SY W+S+ RE+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 1364 IQKGSIWKVGTGEKIHIWNDQWL 1386
+ +G + +G G +W D+W+
Sbjct: 122 LSRGLLRTIGDGIHTKVWLDRWI 144
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 82.8 bits (203), Expect = 8e-15
Identities = 88/354 (24%), Positives = 151/354 (41%), Gaps = 30/354 (8%)
Query: 832 EEVLTKYFEDLFKATSPDDCEIVTNLVHGRITNEHK-EIMEEPFTEEEVREALKKMHPTK 890
+E++ Y+ ++ SP C I +H E + TE+++R + ++ +
Sbjct: 307 QEIMVPYWREVMTQPSPSSCSGEV------IQMDHSLERVWSAITEQDLRAS--RVSLSS 358
Query: 891 AHGADG-TPALFFQKFWSIVGYDVTAQVLSILNDGADATH-INQTLIALIPKIKKPSMAK 948
+ G DG TP K V + ++++++ + H I IPK +
Sbjct: 359 SPGPDGITP-----KSAREVPSGIMLRIMNLILWCGNLPHSIRLARTVFIPKTVTAKRPQ 413
Query: 949 DYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQSAFVPGRLITDNAIIAHECFHYMRR 1008
D+RPIS+ +V+ + + +A RL S + + Q F+P DNA I +R
Sbjct: 414 DFRPISVPSVLVRQLNAILATRLNSSIN--WDPRQRGFLPTDGCADNATIVDLV---LRH 468
Query: 1009 KTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGFTQRWTNLIMNCVSTVSFQVLLNGQ 1068
+ + D+SKA+D + + TL G + + + + N + +G
Sbjct: 469 SHKHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQNTYEGGGTSLNGDGW 528
Query: 1069 PCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPRAPMISHLFFA 1128
F P RG++QGDPLSP LF L + + +++ A V G IT A FA
Sbjct: 529 SSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEIGAKV--GNAITNAAA------FA 580
Query: 1129 DDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVPSTRVHELE 1182
DD +L A T+ VL D + + G ++N DK + P + LE
Sbjct: 581 DDLVLFAE-TRMGLQVLLDKTLDFLSIVGLKLNADKCFTVGIKGQPKQKCTVLE 633
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 79.3 bits (194), Expect = 9e-14
Identities = 45/111 (40%), Positives = 59/111 (52%), Gaps = 1/111 (0%)
Query: 1064 LLNGQPCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPRAPMIS 1123
++NG P P RGLRQGDPLSPYLFILC EV S + + G L GI+++ +P I+
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRIN 72
Query: 1124 HLFFADDSILLARATKEEAMVLHDIIKGYEAFSGQRINFDKSELSTSRNVP 1174
HL FADD+ AR A + F G +N S L ++P
Sbjct: 73 HLLFADDT-SSARWIPLAAQIWPIFFLSMRLFQGNPVNHPMSNLYFLGSLP 122
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 67.8 bits (164), Expect = 3e-10
Identities = 70/281 (24%), Positives = 113/281 (39%), Gaps = 20/281 (7%)
Query: 855 TNLVHGRITNEHKEIMEEPFTEEEVREALKKMHPTKAHGADGTPALFFQKFWSIVGYDVT 914
T + R+ E K + P + +E++E + A G DG W+ + +
Sbjct: 301 TRHLRSRMQGEIKNLWR-PISNDEIKEV--EACKRTAAGPDGMTTTA----WNSIDECIK 353
Query: 915 AQVLSILNDGADATHINQTLIALIPKIKKPSMAKDYRPISL*NVIFKLVTKTIANRLKSF 974
+ I+ G + LIPK +RP+S+ +V + + +ANR+
Sbjct: 354 SLFNMIMYHGQCPRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE- 412
Query: 975 LQDIVEETQSAFVPGRLITDNAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRVEWNF 1034
+++ Q AF+ + +N + R K +KG+ D+ KA+D VE
Sbjct: 413 -HGLLDTRQRAFIVADGVAENTSLLSAMIKEARMK---IKGLYIAILDVKKAFDSVEHRS 468
Query: 1035 LKSTLEQMGFTQRWTNLIMNCVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLFILCA 1094
+ L + N IM ++ + R +P RG+RQGDPLSP LF
Sbjct: 469 ILDALRRKKLPLEMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCVM 528
Query: 1095 EVFSSMIKAKVAAGVLHGIKITPRAPMISHLFFADDSILLA 1135
+ + G L G A I L FADD +LLA
Sbjct: 529 DAVLRRLPEN--TGFLMG------AEKIGALVFADDLVLLA 561
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 64.7 bits (156), Expect = 2e-09
Identities = 69/265 (26%), Positives = 127/265 (47%), Gaps = 22/265 (8%)
Query: 873 PFTEEEVREALKKMHPTKAHGADGTPALFFQKFWSIVGYDVTAQVLSI-LNDGADATHIN 931
P EE++ A+K P+ A G+DG +I + + + L G T
Sbjct: 5 PIAREEIQCAIKGWKPS-APGSDGLTVQ------AITRTRLPRNFVQLHLLRGHVPTPWT 57
Query: 932 QTLIALIPKIKKPSMAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQSAFVPGRL 991
LIPK ++RPI++ + + +L+ + +A RL++ ++ + A + G L
Sbjct: 58 AMRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVELHPAQKGYARIDGTL 117
Query: 992 ITDNAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGFTQRWTNL 1051
+ N+++ + + RR+ R V++L D+ KA+D V + + L+++G + +N
Sbjct: 118 V--NSLLL-DTYISSRREQRKTYNVVSL--DVRKAFDTVSHSSICRALQRLGIDEGTSNY 172
Query: 1052 IMNCVSTVSFQVLLN-GQPCRSFKPERGLRQGDPLSPYLF-ILCAEVFSSMIKAKVAAGV 1109
I +S + + + G R RG++QGDPLSP+LF + E+ S+ G
Sbjct: 173 ITGSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQSTPGIGGT 232
Query: 1110 LHGIKITPRAPMISHLFFADDSILL 1134
+ KI P+++ FADD +LL
Sbjct: 233 IGEEKI----PVLA---FADDLLLL 250
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 52.8 bits (125), Expect = 9e-06
Identities = 43/172 (25%), Positives = 74/172 (43%), Gaps = 4/172 (2%)
Query: 922 NDGADATHINQTLIALIPKIKKPSMAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEE 981
+D A +I IPK PS +YRPISL + +++ + I +R++S ++
Sbjct: 657 SDSAIPNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSP 716
Query: 982 TQSAFVPGRLITDNAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQ 1041
Q F+ R + + + +H + + K + L D +KA+D+V L L
Sbjct: 717 HQHGFLNFRSCPSSLVRSISLYHSI---LKNEKSLDILFFDFAKAFDKVSHPILLKKLAL 773
Query: 1042 MGFTQRWTNLIMNCVSTVSFQVLLNGQPCRSFKP-ERGLRQGDPLSPYLFIL 1092
G + + + +F V +N + P G+ QG P LFIL
Sbjct: 774 FGLDKLTCSWFKEFLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFIL 825
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 51.6 bits (122), Expect = 2e-05
Identities = 34/131 (25%), Positives = 60/131 (44%), Gaps = 6/131 (4%)
Query: 1004 HYMRRKTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGFTQRWTNLIMNCVSTVSFQV 1063
HY++ + K + D+ KA+D V + + G + IM+ ++ +
Sbjct: 11 HYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMSTITDAYTNI 70
Query: 1064 LLNGQPCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPRAPMIS 1123
++ G+ G++QGDPLSP LF + + + + + G +TP A I+
Sbjct: 71 VVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQP-----GASMTP-ACKIA 124
Query: 1124 HLFFADDSILL 1134
L FADD +LL
Sbjct: 125 SLAFADDLLLL 135
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 43.5 bits (101), Expect = 0.005
Identities = 87/380 (22%), Positives = 161/380 (41%), Gaps = 36/380 (9%)
Query: 808 RKDRNSIARIT-DDRGRAWTEEEDIEEVLTKYFEDLFKATSPDDCEIVTNLVHGRITNEH 866
+KD ++ AR+ D+ + T+ + + Y++D+F AT TN ++ H
Sbjct: 97 KKDISAAARVVLDENDKIATKIPPVRHMFD-YWKDVF-ATGGGSA--ATN-INRAPPAPH 151
Query: 867 KEIMEEPFTEEEVREALKKMHPTKAHGADG-TPALFFQKFWSIVGYDVTAQVLSI-LNDG 924
E + +P + E++ A + K G DG TP + W+ + + +I + G
Sbjct: 152 METLWDPVSLIEIKSA--RASNEKGAGPDGVTP-----RSWNALDDRYKRLLYNIFVFYG 204
Query: 925 ADATHINQTLIALIPKIKKPSMAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQS 984
+ I + PKI+ +RP+S+ +VI + K +A R S +E Q+
Sbjct: 205 RVPSPIKGSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILARRFVSCYT--YDERQT 262
Query: 985 AFVPGRLITDNAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGF 1044
A++P + N + +R + + + D+ KA++ V + L + + G
Sbjct: 263 AYLPIDGVCINVSMLTAIIAEAKRLRKELHIAIL---DLVKAFNSVYHSALIDAITEAGC 319
Query: 1045 TQRWTNLIMNCVSTVSFQVLLNGQPCRSFKPERGLRQGDPLSPYLFILCAEVFSSMIKAK 1104
+ I + + V ++ G+ C G+ QGDPLS LF L E K
Sbjct: 320 PPGVVDYIADMYNNVITEMQFEGK-CELASILAGVYQGDPLSGPLFTLAYE--------K 370
Query: 1105 VAAGVLHGIKITPRAPMISHLFFADDSILLARATKEEAMVLHDIIKGYEAFS--GQRINF 1162
+ + + ++ ++DD +LLA + H++ K E + G RIN
Sbjct: 371 ALRALNNEGRFDIADVRVNASAYSDDGLLLAMTV---IGLQHNLDKFGETLAKIGLRINS 427
Query: 1163 DKSELSTSRNVPSTRVHELE 1182
KS+ T VPS R +++
Sbjct: 428 RKSK--TVSLVPSGREKKMK 445
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 40.4 bits (93), Expect = 0.045
Identities = 143/711 (20%), Positives = 256/711 (35%), Gaps = 114/711 (16%)
Query: 439 MKMISWNCRGLGTPRAVGALRKLITMERPDLVFLMETRKKAFEIQKTKMPSGLVHSIGVD 498
+K+ WN RGL R LR ++ D++ ET + Q+ +P L++
Sbjct: 6 LKIGLWNARGL--TRGSEELRIFLSDHDIDVMLTTETHMRVG--QRIYLPGYLMYHAH-- 59
Query: 499 CNGEGKSRGGGLACYWSSSLDVNIISLSSN-------HIQMEVTTTIIDSKMVVTGIYGH 551
+ G SRGG S + +S+N H+Q V T + V +Y
Sbjct: 60 -HPSGNSRGGSAVIIKSRLCHSPLTPISTNDRQIARVHLQTSVGT------VTVAAVYLP 112
Query: 552 PEAPQKWRTWELIKKLTPDPEVPWLCLGDFNEVLKSSEKTGGNPINFVGAQAFQEAMDFC 611
P ++W + K + ++ GD+N + GNP + + QE +
Sbjct: 113 PA--ERWIVDDF-KSMFAALGNKFIAGGDYN----AKHAWWGNPRSCPRGKMLQEVIAHG 165
Query: 612 GLRDMGFSGFQFTWSNMRPTPNTIQERLDRALVNECWTEHWPHTHVSHLDRFNSDHNPIY 671
+ + F N TP+ + + V L +SDH PI
Sbjct: 166 QYQVLATGEPTFYSYNPLLTPSALDFFITCGY-------GMGRLDVQTLQELSSDHLPIL 218
Query: 672 VIFLKAKRQKKKKIRLYRFEEMWLQEEDCRHVIHHSWPRGGRNITSKLGNVGANLTEWAE 731
+ +K +++RL + H++ + +L V + E +
Sbjct: 219 AVLHATPLKKPQRVRL---------------LAHNADINIFKTHLEQLSEVNMQILEAVD 263
Query: 732 ATFGNIQKRIKSQLGIIKRIQAAEQTEENLKKGKEAEKELE---NLLKLEEIKWSQRSRA 788
N S+L ++ A E EA + L+ ++L L +K +R R
Sbjct: 264 --IDNATSLFMSKLSEAAQLAAPRNRHE-----VEAFRPLQLPSSILALLRLK--RRVRK 314
Query: 789 TWLAHGDRNTSFFHK-------KASQRKDRNSIARITDDRGRAWTEEEDIEEVLTKYFED 841
+ GD H KA R+ + I D+ G + + + TK F+
Sbjct: 315 EYARTGDPRMQQIHSRLANCLHKALARRKQAQIDTFLDNLGADASTNYSLWRI-TKRFKA 373
Query: 842 L-------------FKATSPDDCEIVTNLVHGRIT----------NEHKEIMEEPF---- 874
+ TS + E+ N + R T + +E +E PF
Sbjct: 374 QPTPKSAIKNPSGGWCRTSLEKTEVFANNLEQRFTPYNYAPESLCRQVEEYLESPFQMSL 433
Query: 875 -----TEEEVREALKKMHPTKAHGADGTPALFFQKFWSIVGYDVTAQVLSILNDGADATH 929
T EEV+ + K+ KA G D L + ++ + I N D +
Sbjct: 434 PLSAVTLEEVKNLIAKLPLKKAPGED----LLDNRTIRLLPDQALQFLALIFNSVLDVGY 489
Query: 930 I-----NQTLIALIPKIKKPSMAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQS 984
+ ++I + K P+ YRP SL + K++ + I NRL + +D+ +
Sbjct: 490 FPKAWKSASIIMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLT-CKDVTKAIPK 548
Query: 985 AFVPGRLITDNAIIAHECFHYMRRKTRGVKGVMALKTDMSKAYDRVEWNFLKSTLEQMGF 1044
RL H ++ + + D+ +A+DRV L +++ F
Sbjct: 549 FQFGFRLQHGTPEQLHRVVNFALEAMENKEYAVGAFLDIQQAFDRVWHPGLLYKAKRL-F 607
Query: 1045 TQRWTNLIMNCVSTVSFQVLLNGQPCRSFKP-ERGLRQGDPLSPYLFILCA 1094
+ ++ + + +F V ++G S KP G+ QG L P L+ + A
Sbjct: 608 PPQLYLVVKSFLEERTFHVSVDGYK-SSIKPIAAGVPQGSVLGPTLYSVFA 657
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 38.9 bits (89), Expect = 0.13
Identities = 71/320 (22%), Positives = 122/320 (37%), Gaps = 40/320 (12%)
Query: 833 EVLTKYFEDLFKATS--PDDCEIVTNLVHGRITNEHKEIMEEPFTEEEVREALKKMHPTK 890
EV + E FKA + P+ ++ + T + +P T EEV+E + K+ P K
Sbjct: 397 EVFANHLEQRFKALAFAPESHSLM--VAESLQTPFQMALPADPVTLEEVKELVSKLKPKK 454
Query: 891 AHGADGTPALFFQKFWSIVGYDVTAQVLSILNDGADATHI-----NQTLIALIPKIKKPS 945
A G D L + ++ ++ I N + ++I ++ K+P
Sbjct: 455 APGED----LLDNRTIRLLPDQALLYLVLIFNSILRVGYFPKARPTASIIMILKPGKQPL 510
Query: 946 MAKDYRPISL*NVIFKLVTKTIANRLKSFLQDIVEETQSAFVPGRLITDNAIIAHECFHY 1005
YRP SL + K++ + I NR+ + +++ RL H ++
Sbjct: 511 DVDSYRPTSLLPSLGKMLERLILNRILT-SEEVTRAIPKFQFGFRLQHGTPEQLHRVVNF 569
Query: 1006 MRRKTRGVKGVMALKTDMSKAYDRVEWN-----FLKSTLEQMGFTQRWTNLIMNCVSTVS 1060
+ + D+ +A+DRV W+ KS L F LI +
Sbjct: 570 ALEALEKKEYAGSCFLDIQQAFDRV-WHPGLLYKAKSLLSPQLF-----QLIKSFWEGRK 623
Query: 1061 FQVLLNGQPCRSFKP--ERGLRQGDPLSPYLFILCAEVFSSMIKAKVAAGVLHGIKITPR 1118
F V +G CRS E G+ QG L P L+ + + M G+ G +
Sbjct: 624 FSVTADG--CRSSVKFIEAGVPQGSVLGPTLYSIFT---ADMPNQNAVTGLAEGEVLIAT 678
Query: 1119 APMISHLFFADDSILLARAT 1138
+ADD +L ++T
Sbjct: 679 --------YADDIAVLTKST 690
>APEA_DICDI (P51173) DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (Class II
apurinic/apyrimidinic(AP)-endonuclease)
Length = 361
Score = 38.9 bits (89), Expect = 0.13
Identities = 59/268 (22%), Positives = 99/268 (36%), Gaps = 38/268 (14%)
Query: 434 VPPRAMKMISWNCRGLGTPRAVGALRKLITMERPDLVFLMETRKKAFEIQKTKMPSGL-V 492
V MK+ISWN G + + G + + E PD++ L ET+ I+K +MP G
Sbjct: 100 VEENQMKIISWNVAGFKSVLSKG-FTECVEKENPDVLCLQETKINPSNIKKDQMPKGYEY 158
Query: 493 HSIGVDCNGEGKSRGGGLACYWSSSLDVNIISLSSNHIQMEVTTT------IIDSKMVVT 546
H I D G G G+ + I ++ + + V T I+++ +
Sbjct: 159 HFIEADQKGH---HGTGVLTKKKPNAITFGIGIAKHDNEGRVITLEYVQFYIVNTYIPNA 215
Query: 547 GIYGHPEAPQKWRTWEL-----IKKLTPDPEVPWLCLGDFN----EVLKSSEKTGGNPIN 597
G G + + W++ ++KL + W GD N E+ + KT
Sbjct: 216 GTRGLQRLDYRIKEWDVDFQAYLEKLNATKPIIW--CGDLNVAHTEIDLKNPKTNKKSAG 273
Query: 598 FVGAQAFQEAMDFCGLRDMGF-----------SGFQFTWSNM-RPTPNTIQERLDRALVN 645
F +E F + G+ G WS + + RLD +V+
Sbjct: 274 F----TIEERTSFSNFLEKGYVDSYRHFNPGKEGSYTFWSYLGGGRSKNVGWRLDYFVVS 329
Query: 646 ECWTEHWPHTHVSHLDRFNSDHNPIYVI 673
+ + + SDH PI V+
Sbjct: 330 KRLMDSIKISPFHRTSVMGSDHCPIGVV 357
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 37.7 bits (86), Expect = 0.29
Identities = 37/160 (23%), Positives = 65/160 (40%), Gaps = 19/160 (11%)
Query: 737 IQKRIKSQLGIIKRIQAAEQTEENLKKGKEAEKELENLLKLEEIKWSQRSRATWLAHGDR 796
I+K+ K Q K+ Q E+ ++ K+ K+ EKE++ K+E+ + + +
Sbjct: 278 IEKKKKKQEEKEKKKQEKERKKQEKKERKQKEKEMKKQKKIEKERKKKEEKE-------- 329
Query: 797 NTSFFHKKASQRKDRNSIARITDDRGRAWTEEEDIEEVLTKYFEDLFKATSPDDCEIVTN 856
KK K+ + D T EE E++ L T+P++ + +
Sbjct: 330 -----KKKKKHDKENEETMQQPDQ-----TSEETNNEIMVPLPSPLTDVTTPEEHKEGEH 379
Query: 857 LVHGRITNEHKEIMEEPFTEEEVREALKKMHPTKAHGADG 896
EHKE E E + E K+ H +K H + G
Sbjct: 380 KEEEHKEGEHKE-GEHKEEEHKEEEHKKEEHKSKEHKSKG 418
>SUDY_RAT (P52847) DOPA/tyrosine sulfotransferase (EC 2.8.1.-)
Length = 299
Score = 35.8 bits (81), Expect = 1.1
Identities = 23/96 (23%), Positives = 44/96 (44%), Gaps = 3/96 (3%)
Query: 614 RDMGFSGFQFT-WSNMRPTPNTIQERLDRALVNECWTEHWPHTHVSHLDRFNSDHNPIYV 672
+D+ S + F +N++P P T +E L++ L W HV H +++
Sbjct: 134 KDVAVSYYHFDLMNNIQPLPGTWEEYLEKFLAGNVAYGSW-FDHVKSWWEKREGHPILFL 192
Query: 673 IFLKAKRQKKKKIR-LYRFEEMWLQEEDCRHVIHHS 707
+ K+ KK+I+ + F + L E ++HH+
Sbjct: 193 YYEDLKKNPKKEIKKIANFLDKTLDEHTLERIVHHT 228
>CASS_RICCO (P59287) Casbene synthase, chloroplast precursor (EC
4.2.3.8)
Length = 601
Score = 35.8 bits (81), Expect = 1.1
Identities = 26/122 (21%), Positives = 52/122 (42%), Gaps = 6/122 (4%)
Query: 822 GRAWTEEEDIEEVLTKYFEDLFKATSPDDCEIVTNLVHGRITNEHKEIMEEPFTEEEVRE 881
G ++ E DIEE+L+K F +C++ T + R+ +H M + ++
Sbjct: 127 GVSYHFENDIEELLSKIFNSQPDLVDEKECDLYTAAIVFRVFRQHGFKMSSD-VFSKFKD 185
Query: 882 ALKKMHPTKAHGADGTPALFFQKFWSIVGYDVTAQVLSILNDGADATHINQTLIALIPKI 941
+ K + A G +LF S+ G D+ + + D ++ + + L P +
Sbjct: 186 SDGKFKESLRGDAKGMLSLFEASHLSVHGEDILEEAFAFTKD-----YLQSSAVELFPNL 240
Query: 942 KK 943
K+
Sbjct: 241 KR 242
>SPCO_HUMAN (Q01082) Spectrin beta chain, brain 1 (Spectrin,
non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin
beta chain)
Length = 2364
Score = 34.3 bits (77), Expect = 3.2
Identities = 43/185 (23%), Positives = 80/185 (43%), Gaps = 16/185 (8%)
Query: 717 SKLGNVGANLTEWAEATFGNIQKRIKSQLGIIKRIQAAEQ---TEENLKKGKEAEKELEN 773
SK+ + A L + AE G + +R + + + + EQ E + E ++ E+
Sbjct: 1674 SKVDKLYAGLKDLAEERRGKLDERHRL-FQLNREVDDLEQWIAEREVVAGSHELGQDYEH 1732
Query: 774 LLKLEEIKWSQRSRATWLAHGDRNTSFFHKKA----SQRKDRNSIARITDDRGRAWTEEE 829
+ L+E ++ + +R T +R + H S D +IA D AW +
Sbjct: 1733 VTMLQE-RFREFARDTGNIGQERVDTVNHLADELINSGHSDAATIAEWKDGLNEAWADLL 1791
Query: 830 DIEEVLTKYFEDLFKATS--PDDCEIVTNLVHGRITNEHKEIMEEPFTEEEVREALKKMH 887
++ + T+ ++ D EI GRI ++HK++ EE ++ E L++MH
Sbjct: 1792 ELIDTRTQILAASYELHKFYHDAKEIF-----GRIQDKHKKLPEELGRDQNTVETLQRMH 1846
Query: 888 PTKAH 892
T H
Sbjct: 1847 TTFEH 1851
>RAI1_HUMAN (Q7Z5J4) Retinoic acid induced protein 1
Length = 1906
Score = 34.3 bits (77), Expect = 3.2
Identities = 35/153 (22%), Positives = 59/153 (37%), Gaps = 18/153 (11%)
Query: 374 RNTGTKVEEASKIRPQNNNTMPY*DPEEERLRDEHSYSHGNRGLPCVSESQIGSGE-DCG 432
R + +V SK +P++ +T P++ + + S+ + LP S S+ +GE DC
Sbjct: 536 RGSPARVNSNSKAKPESVSTCSVTSPDDMSTKSDDSFQSLHGSLPLDSFSKFVAGERDCP 595
Query: 433 AVPPRAMKMISWNCRGLGTPRAVGALRKLITMERPDLVFLMETRKKAFEIQ--------- 483
+ A+ LG A+G E P LV ++ K F ++
Sbjct: 596 RLLLSALAQEDLASEILGLQEAIGEKADKAWAEAPSLV--KDSSKPPFSLENHSACLDSV 653
Query: 484 ------KTKMPSGLVHSIGVDCNGEGKSRGGGL 510
+ P L S+ +D G K GL
Sbjct: 654 AKSAWPRPGEPEALPDSLQLDKGGNAKDFSPGL 686
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.333 0.144 0.484
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 216,004,271
Number of Sequences: 164201
Number of extensions: 9311384
Number of successful extensions: 24131
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 24073
Number of HSP's gapped (non-prelim): 53
length of query: 1806
length of database: 59,974,054
effective HSP length: 125
effective length of query: 1681
effective length of database: 39,448,929
effective search space: 66313649649
effective search space used: 66313649649
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0383.5


