

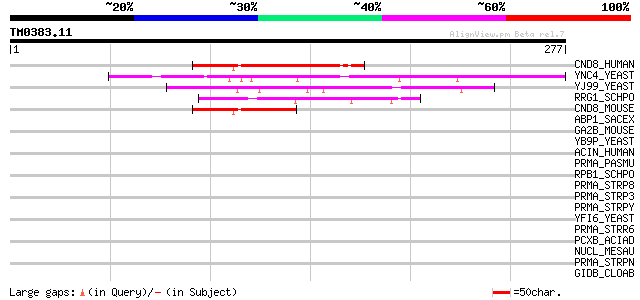
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0383.11
(277 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CND8_HUMAN (Q9H867) Protein C14orf138 64 4e-10
YNC4_YEAST (P53970) Hypothetical 27.7 kDa protein in UME3-HDA1 i... 60 4e-09
YJ99_YEAST (P47163) Hypothetical 39.0 kDa protein in ZMS1-MNS1 i... 49 2e-05
RRG1_SCHPO (P40389) Rapid response to glucose protein 1 47 5e-05
CND8_MOUSE (Q8C436) Protein C14orf138 homolog 47 7e-05
ABP1_SACEX (P38479) Actin binding protein 38 0.024
GA2B_MOUSE (O08795) Glucosidase II beta subunit precursor (Prote... 37 0.053
YB9P_YEAST (P38347) Hypothetical 48.0 kDa protein in MRPL37-RIF1... 36 0.12
ACIN_HUMAN (Q9UKV3) Apoptotic chromatin condensation inducer in ... 36 0.12
PRMA_PASMU (Q9CLW2) Ribosomal protein L11 methyltransferase (EC ... 35 0.15
RPB1_SCHPO (P36594) DNA-directed RNA polymerase II largest subun... 35 0.20
PRMA_STRP8 (Q8NZ98) Ribosomal protein L11 methyltransferase (EC ... 35 0.20
PRMA_STRP3 (Q8K5Q9) Ribosomal protein L11 methyltransferase (EC ... 35 0.20
PRMA_STRPY (Q99XW8) Ribosomal protein L11 methyltransferase (EC ... 34 0.34
YFI6_YEAST (P43597) Hypothetical 137.7 kDa protein in UGS1-FAB1 ... 34 0.45
PRMA_STRR6 (Q8DNP4) Ribosomal protein L11 methyltransferase (EC ... 34 0.45
PCXB_ACIAD (P20372) Protocatechuate 3,4-dioxygenase beta chain (... 34 0.45
NUCL_MESAU (P08199) Nucleolin (Protein C23) 34 0.45
PRMA_STRPN (Q97P62) Ribosomal protein L11 methyltransferase (EC ... 33 0.58
GIDB_CLOAB (Q97CW4) Methyltransferase gidB (EC 2.1.-.-) (Glucose... 33 0.58
>CND8_HUMAN (Q9H867) Protein C14orf138
Length = 144
Score = 63.9 bits (154), Expect = 4e-10
Identities = 40/89 (44%), Positives = 55/89 (60%), Gaps = 6/89 (6%)
Query: 92 GVTGAVMWDSGVVLGKFLE---HSVDLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVIL 148
G G V+WD+ +VL K+LE S D G L + ++ELGSG G VG +AA LG +V++
Sbjct: 36 GGVGCVVWDAAIVLSKYLETPEFSGD-GAHALSRRSVLELGSGTGAVGLMAATLGADVVV 94
Query: 149 TDLLDRLRLLRKNIETNMKHVSLRGSATA 177
TDL + LL+ NI N KH+ + GS A
Sbjct: 95 TDLEELQDLLKMNINMN-KHL-VTGSVQA 121
>YNC4_YEAST (P53970) Hypothetical 27.7 kDa protein in UME3-HDA1
intergenic region
Length = 246
Score = 60.5 bits (145), Expect = 4e-09
Identities = 70/249 (28%), Positives = 105/249 (42%), Gaps = 30/249 (12%)
Query: 50 GIQQPTLSKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKF- 108
G + +P + Q+ L G L L+ G G G V W +G +L ++
Sbjct: 7 GFGDLVVPRPKEHLGQTDLSFG----GKLLPALKICEDGGESGCGGKV-WIAGELLCEYI 61
Query: 109 LEHSVD--LGTLV---LQEKKIVELGSGCGLVGCIAALL-------GGEVILTDLLDRLR 156
LE SVD L V Q KK++ELGSG GLVG LL G +V +TD+ +
Sbjct: 62 LEKSVDHLLSKTVNGTKQFKKVLELGSGTGLVGLCVGLLEKNTFHDGTKVYVTDIDKLIP 121
Query: 157 LLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDP-------TPDFVVGSDVVYSENA 209
LL++NIE + ++ A EL WGE + D V+ +D VY E A
Sbjct: 122 LLKRNIELD----EVQYEVLARELWWGEPLSADFSPQEGAMQANNVDLVLAADCVYLEEA 177
Query: 210 VVDLVETLGQLSG-PNTTIFLAGELRNDAILEYFLEAAMNDFTIGRVDQTLWHPEYRSNR 268
L +TL L+ N + L + ++F +F + + Y R
Sbjct: 178 FPLLEKTLLDLTHCINPPVILMAYKKRRKADKHFFNKIKRNFDVLEITDFSKFEHYLKER 237
Query: 269 VVLYVLVKK 277
L+ L++K
Sbjct: 238 THLFQLIRK 246
>YJ99_YEAST (P47163) Hypothetical 39.0 kDa protein in ZMS1-MNS1
intergenic region
Length = 339
Score = 48.5 bits (114), Expect = 2e-05
Identities = 45/179 (25%), Positives = 80/179 (44%), Gaps = 19/179 (10%)
Query: 79 LSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHS--VDLGTLVLQEK-----KIVELGSG 131
++I ++P+ + A TG W++ + +G FL H +L + Q+ ++E+G+G
Sbjct: 117 ITIEETPNLISAASTTGFRTWEAALYMGDFLIHKPLQELAPVQGQDDGKKKLNVLEVGAG 176
Query: 132 CGLVGCIAALLGGEVI----LTDLLDRL--RLLRKNIETNMKHVSLRGSATATELTWGED 185
G+V + E + +TD L L++N E N + L WG D
Sbjct: 177 TGIVSLVILQKYHEFVNKMYVTDGDSNLVETQLKRNFELNNEVRENEPDIKLQRLWWGSD 236
Query: 186 PDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPN--TTIFLAGELRNDAILEYF 242
E I D VVG+DV Y + DL E L + + L+ +R+++ ++ F
Sbjct: 237 RVPEDI----DLVVGADVTYDPTILPDLCECLAECLALDRCKLCLLSATIRSESTVQLF 291
>RRG1_SCHPO (P40389) Rapid response to glucose protein 1
Length = 303
Score = 47.0 bits (110), Expect = 5e-05
Identities = 38/118 (32%), Positives = 57/118 (48%), Gaps = 12/118 (10%)
Query: 95 GAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCIAAL-LGGEVILTDLLD 153
G W S +L L DL + +ELG+G GLVG AA+ LG +V+ TDL D
Sbjct: 113 GFKTWGSAPLLSANLPKWEDLSNSI----NALELGAGTGLVGISAAIQLGWQVVCTDLPD 168
Query: 154 RLRLLRKNIETNMKHV-SLRGSATATELTWGEDPDRE-----LIDPTPDFVVGSDVVY 205
+ ++ N++ N + + GS + L W PD + LI P ++ SD +Y
Sbjct: 169 IVENMQYNVDYNSELIQQYAGSVSCHVLDWMNPPDDDNRPSWLIKPF-QRIIASDCIY 225
>CND8_MOUSE (Q8C436) Protein C14orf138 homolog
Length = 89
Score = 46.6 bits (109), Expect = 7e-05
Identities = 26/55 (47%), Positives = 34/55 (61%), Gaps = 4/55 (7%)
Query: 92 GVTGAVMWDSGVVLGKFLE---HSVDLGTLVLQEKKIVELGSGCGLVGCIAALLG 143
G G V+WD+ +VL K+LE S D G L + ++ELGSG G VG +AA LG
Sbjct: 36 GGVGCVVWDAAIVLSKYLETPGFSGD-GAHALSRRSVLELGSGTGAVGLMAATLG 89
>ABP1_SACEX (P38479) Actin binding protein
Length = 617
Score = 38.1 bits (87), Expect = 0.024
Identities = 20/43 (46%), Positives = 24/43 (55%)
Query: 2 ENRQEEEEEEEEEEDAPPPMVKLGSYGGEVRLVVPGEESAAEE 44
E +EEEEEEEEEE+AP P + + E P EE EE
Sbjct: 487 EGEEEEEEEEEEEEEAPAPSLPSRNAAPEPEPEQPQEEEEEEE 529
>GA2B_MOUSE (O08795) Glucosidase II beta subunit precursor (Protein
kinase C substrate, 60.1 kDa protein, heavy chain)
(PKCSH) (80K-H protein)
Length = 521
Score = 37.0 bits (84), Expect = 0.053
Identities = 15/20 (75%), Positives = 18/20 (90%)
Query: 2 ENRQEEEEEEEEEEDAPPPM 21
E +EEEEEEEEEE+APPP+
Sbjct: 316 EEPEEEEEEEEEEEEAPPPL 335
>YB9P_YEAST (P38347) Hypothetical 48.0 kDa protein in MRPL37-RIF1
intergenic region
Length = 419
Score = 35.8 bits (81), Expect = 0.12
Identities = 49/179 (27%), Positives = 76/179 (42%), Gaps = 29/179 (16%)
Query: 78 SLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVD---------LGTLVLQEKKIVEL 128
S SI SL A + G W S ++L + + +D L +++ K++EL
Sbjct: 202 STSIPLHEPSLTADNL-GWKTWGSSLILSQLVVDHLDYLHTTNVNMLANSDIKQIKVLEL 260
Query: 129 GSGCGLVGCIAAL----LGG----EVILTDLLDRLRLLRKNIETNMKHVSLRGSATATEL 180
G+G GLVG AL L G E+ +TDL + + L+KN+ N +L A L
Sbjct: 261 GAGTGLVGLSWALKWKELYGTENIEIFVTDLPEIVTNLKKNVSLN----NLGDFVQAEIL 316
Query: 181 TWGEDPDRELIDPTP-----DFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELR 234
W D ID D ++ +D +YS +V + + + T L LR
Sbjct: 317 DWTNPHD--FIDKFGHENEFDVILIADPIYSPQHPEWVVNMISKFLAASGTCHLEIPLR 373
>ACIN_HUMAN (Q9UKV3) Apoptotic chromatin condensation inducer in the
nucleus (Acinus)
Length = 1341
Score = 35.8 bits (81), Expect = 0.12
Identities = 38/135 (28%), Positives = 59/135 (43%), Gaps = 17/135 (12%)
Query: 2 ENRQEEEEEEEEEEDAPPPMVKLGSYGGEVRLVVPGEESAAEETMLLWGIQQPTLSKPNA 61
EN Q EEEEEE+E +A PP+ S EV L P E + T+ TL++ +
Sbjct: 931 ENGQREEEEEEKEPEAEPPVPPQVSV--EVALPPPAEHEVKKVTL------GDTLTRRSI 982
Query: 62 FVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVV----LGKFLEHSVDLGT 117
+S + + +D + + P G ++ S +V LG+ E GT
Sbjct: 983 SQQKSGVSITIDDPVRTAQVPSPPR-----GKISNIVHISNLVRPFTLGQLKELLGRTGT 1037
Query: 118 LVLQEKKIVELGSGC 132
LV + I ++ S C
Sbjct: 1038 LVEEAFWIDKIKSHC 1052
>PRMA_PASMU (Q9CLW2) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 293
Score = 35.4 bits (80), Expect = 0.15
Identities = 40/148 (27%), Positives = 62/148 (41%), Gaps = 20/148 (13%)
Query: 97 VMWDSGVVLGKFLEHSVDL-----GTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDL 151
VM D G+ G + L +L LQ+K +++ G G G++ A LG + +
Sbjct: 129 VMLDPGLAFGTGTHPTTALCLEWLDSLDLQDKTVIDFGCGSGILAIAALKLGAKSAVGID 188
Query: 152 LDRLRLL--RKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTPDFVVGSDVVYSENA 209
+D +L R N E N G A +L ED +L D VV + +
Sbjct: 189 IDPQAILASRNNAEQN-------GVADRLQLFLSEDKPADL---KADVVVANILA---GP 235
Query: 210 VVDLVETLGQLSGPNTTIFLAGELRNDA 237
+ +L + QL PN + L+G L A
Sbjct: 236 LKELYPVIRQLVKPNGVLGLSGILATQA 263
>RPB1_SCHPO (P36594) DNA-directed RNA polymerase II largest subunit
(EC 2.7.7.6) (RNA polymerase II subunit 1)
Length = 1752
Score = 35.0 bits (79), Expect = 0.20
Identities = 15/41 (36%), Positives = 25/41 (60%)
Query: 158 LRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTPDFV 198
L KN++T ++H +L +ATE+ + DP +I+ DFV
Sbjct: 1133 LAKNVQTQIEHTTLSTVTSATEIHYDPDPQDTVIEEDKDFV 1173
>PRMA_STRP8 (Q8NZ98) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 317
Score = 35.0 bits (79), Expect = 0.20
Identities = 32/152 (21%), Positives = 63/152 (41%), Gaps = 23/152 (15%)
Query: 21 MVKLGSYGGEVRLVVPGEESAAEETMLLWGIQQPTLSKPNAFVSQSSLQLRLDSCGHSLS 80
+ +L S+G +V V + AEE W +P H L+
Sbjct: 82 LAELASFGLQVGQVTVDSQELAEED---WADNWKKYYEPARIT-------------HDLT 125
Query: 81 ILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQE-----KKIVELGSGCGLV 135
I+ S + A + D G+ G + + L++ + ++++G+G G++
Sbjct: 126 IVPSWTDYDASAGEKVIKLDPGMAFGTGTHPTTKMSLFALEQILRGGETVIDVGTGSGVL 185
Query: 136 GCIAALLGGEVILTDLLD--RLRLLRKNIETN 165
++LLG + I LD +R+ ++NI+ N
Sbjct: 186 SIASSLLGAKTIYAYDLDDVAVRVAQENIDLN 217
>PRMA_STRP3 (Q8K5Q9) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 317
Score = 35.0 bits (79), Expect = 0.20
Identities = 32/152 (21%), Positives = 63/152 (41%), Gaps = 23/152 (15%)
Query: 21 MVKLGSYGGEVRLVVPGEESAAEETMLLWGIQQPTLSKPNAFVSQSSLQLRLDSCGHSLS 80
+ +L S+G +V V + AEE W +P H L+
Sbjct: 82 LAELASFGLQVGQVTVDSQELAEED---WADNWKKYYEPARIT-------------HDLT 125
Query: 81 ILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQE-----KKIVELGSGCGLV 135
I+ S + A + D G+ G + + L++ + ++++G+G G++
Sbjct: 126 IVPSWTDYDASAGEKVIKLDPGMAFGTGTHPTTKMSLFALEQILRGGETVIDVGTGSGVL 185
Query: 136 GCIAALLGGEVILTDLLD--RLRLLRKNIETN 165
++LLG + I LD +R+ ++NI+ N
Sbjct: 186 SIASSLLGAKTIYAYDLDDVAVRVAQENIDLN 217
>PRMA_STRPY (Q99XW8) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 317
Score = 34.3 bits (77), Expect = 0.34
Identities = 32/152 (21%), Positives = 62/152 (40%), Gaps = 23/152 (15%)
Query: 21 MVKLGSYGGEVRLVVPGEESAAEETMLLWGIQQPTLSKPNAFVSQSSLQLRLDSCGHSLS 80
+ +L S+G +V V + AEE W +P H L+
Sbjct: 82 LAELASFGLQVGQVTVDSQELAEED---WADNWKKYYEPARIT-------------HDLT 125
Query: 81 ILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQE-----KKIVELGSGCGLV 135
I+ S + A + D G+ G + + L++ + ++++G+G G++
Sbjct: 126 IVPSWTDYDASAGEKVIKLDPGMAFGTGTHPTTKMSLFALEQILRGGETVIDVGTGSGVL 185
Query: 136 GCIAALLGGEVILTDLLD--RLRLLRKNIETN 165
++LLG + I LD +R+ + NI+ N
Sbjct: 186 SIASSLLGAKTIYAYDLDDVAVRVAQDNIDLN 217
>YFI6_YEAST (P43597) Hypothetical 137.7 kDa protein in UGS1-FAB1
intergenic region
Length = 1233
Score = 33.9 bits (76), Expect = 0.45
Identities = 20/44 (45%), Positives = 25/44 (56%)
Query: 1 MENRQEEEEEEEEEEDAPPPMVKLGSYGGEVRLVVPGEESAAEE 44
+E +EEEEEEEEEE++ VK + GE V E S EE
Sbjct: 463 VEVEKEEEEEEEEEENSTFSKVKKENVTGEQEAVRNNEVSGTEE 506
>PRMA_STRR6 (Q8DNP4) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 316
Score = 33.9 bits (76), Expect = 0.45
Identities = 23/96 (23%), Positives = 47/96 (48%), Gaps = 7/96 (7%)
Query: 77 HSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQE-----KKIVELGSG 131
H L+I+ S + A + D G+ G + + L++ + ++++G+G
Sbjct: 121 HDLTIVPSWTDYEATAGEKIIKLDPGMAFGTGTHPTTKMSLFALEQVLRGGETVLDVGTG 180
Query: 132 CGLVGCIAALLGGEVILTDLLD--RLRLLRKNIETN 165
G++ ++LLG + I LD +R+ ++NIE N
Sbjct: 181 SGVLSIASSLLGAKEIFAYDLDDVAVRVAQENIELN 216
>PCXB_ACIAD (P20372) Protocatechuate 3,4-dioxygenase beta chain (EC
1.13.11.3) (3,4-PCD)
Length = 241
Score = 33.9 bits (76), Expect = 0.45
Identities = 20/67 (29%), Positives = 35/67 (51%), Gaps = 3/67 (4%)
Query: 178 TELTWGEDPDRELIDPTPDFVVG--SDVVYS-ENAVVDLVETLGQLSGPNTTIFLAGELR 234
+++ WG R D P + G + V+ S +NA++ + ETL +++ P+ + G
Sbjct: 2 SQIIWGAYAQRNTEDHPPAYAPGYKTSVLRSPKNALISIAETLSEVTAPHFSADKFGPKD 61
Query: 235 NDAILEY 241
ND IL Y
Sbjct: 62 NDLILNY 68
>NUCL_MESAU (P08199) Nucleolin (Protein C23)
Length = 713
Score = 33.9 bits (76), Expect = 0.45
Identities = 17/43 (39%), Positives = 25/43 (57%), Gaps = 2/43 (4%)
Query: 2 ENRQEEEEEEEEEEDAPPPMVKLGSYGGEVRLVVPGEESAAEE 44
E+ +E+EE+EEE++ PP+VK G+V P E EE
Sbjct: 153 EDDSDEDEEDEEEDEFEPPVVK--GKQGKVAAAAPASEDEDEE 193
>PRMA_STRPN (Q97P62) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 316
Score = 33.5 bits (75), Expect = 0.58
Identities = 23/96 (23%), Positives = 47/96 (48%), Gaps = 7/96 (7%)
Query: 77 HSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTLVLQE-----KKIVELGSG 131
H L+I+ S + A + D G+ G + + L++ + ++++G+G
Sbjct: 121 HDLTIVPSWTDYEATAGEMIIKLDPGMAFGTGTHPTTKMSLFALEQVLRGGETVLDVGTG 180
Query: 132 CGLVGCIAALLGGEVILTDLLD--RLRLLRKNIETN 165
G++ ++LLG + I LD +R+ ++NIE N
Sbjct: 181 SGVLSIASSLLGAKEIFAYDLDDVAVRVAQENIELN 216
>GIDB_CLOAB (Q97CW4) Methyltransferase gidB (EC 2.1.-.-) (Glucose
inhibited division protein B)
Length = 239
Score = 33.5 bits (75), Expect = 0.58
Identities = 29/95 (30%), Positives = 49/95 (51%), Gaps = 10/95 (10%)
Query: 119 VLQEKKIVELGSGCGLVGCIAALLGGE---VILTDLLDRLRLLRKNI-ETNMKHVS-LRG 173
+++ K+I+++G+G G G A++ + V+L L R+ L + I E N+K++S + G
Sbjct: 68 IVKAKRIIDVGTGAGFPGLPIAIMNDDIEVVLLDSLQKRVNFLNEVIKELNLKNISTVHG 127
Query: 174 SATATELTWGEDPD-RELIDPTPDFVVGSDVVYSE 207
A +G D D RE D V + V SE
Sbjct: 128 RAE----DFGVDKDYREKFDVAVSRAVANMAVLSE 158
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.135 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,583,198
Number of Sequences: 164201
Number of extensions: 1430821
Number of successful extensions: 11624
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 10816
Number of HSP's gapped (non-prelim): 432
length of query: 277
length of database: 59,974,054
effective HSP length: 109
effective length of query: 168
effective length of database: 42,076,145
effective search space: 7068792360
effective search space used: 7068792360
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0383.11


