

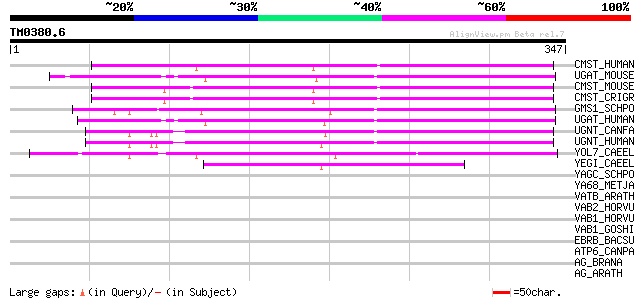
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0380.6
(347 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CMST_HUMAN (P78382) CMP-sialic acid transporter (CMP-Sia-Tr) (CM... 148 2e-35
UGAT_MOUSE (Q9R0M8) UDP-galactose translocator (UDP-galactose tr... 145 2e-34
CMST_MOUSE (Q61420) CMP-sialic acid transporter (CMP-Sia-Tr) (CM... 144 3e-34
CMST_CRIGR (O08520) CMP-sialic acid transporter (CMP-Sia-Tr) (CM... 144 4e-34
GMS1_SCHPO (P87041) UDP-galactose transporter (Golgi UDP-Gal tra... 143 7e-34
UGAT_HUMAN (P78381) UDP-galactose translocator (UDP-galactose tr... 141 2e-33
UGNT_CANFA (O77592) UDP-N-acetylglucosamine transporter (Golgi U... 132 1e-30
UGNT_HUMAN (Q9Y2D2) UDP-N-acetylglucosamine transporter (Golgi U... 131 2e-30
YOL7_CAEEL (Q02334) Hypothetical protein ZK370.7 in chromosome III 113 6e-25
YEGI_CAEEL (Q93890) Hypothetical protein M02B1.1 in chromosome IV 87 8e-17
YAGC_SCHPO (Q09875) Hypothetical protein C12G12.12 in chromosome I 37 0.096
YA68_METJA (Q58467) Hypothetical protein MJ1068 33 1.1
VATB_ARATH (P11574) Vacuolar ATP synthase subunit B (EC 3.6.3.14... 33 1.4
VAB2_HORVU (Q40079) Vacuolar ATP synthase subunit B isoform 2 (E... 32 1.8
VAB1_HORVU (Q40078) Vacuolar ATP synthase subunit B isoform 1 (E... 32 1.8
VAB1_GOSHI (Q43432) Vacuolar ATP synthase subunit B isoform 1 (E... 32 1.8
EBRB_BACSU (O31791) Multidrug resistance protein ebrB 32 2.4
ATP6_CANPA (Q03671) ATP synthase a chain precursor (EC 3.6.3.14)... 32 2.4
AG_BRANA (Q01540) Floral homeotic protein AGAMOUS 32 3.1
AG_ARATH (P17839) Floral homeotic protein AGAMOUS 32 3.1
>CMST_HUMAN (P78382) CMP-sialic acid transporter (CMP-Sia-Tr)
(CMP-SA-Tr) (Solute carrier family 35 member A1)
Length = 337
Score = 148 bits (373), Expect = 2e-35
Identities = 88/295 (29%), Positives = 158/295 (52%), Gaps = 7/295 (2%)
Query: 52 LTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDDNRLT 111
+T++ + I + +++ + K Y TTA + E +K +S+ L + G +
Sbjct: 20 MTLMAAVYTIALRYTRTSDKELYFSTTAVCITEVIKLLLSVGILAKETGSLGRFKASLRE 79
Query: 112 TTLD---EVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKKR 168
L E++ +P+ +Y V+N + + + +DA YQ+ I T + ++L +
Sbjct: 80 NVLGSPKELLKLSVPSLVYAVQNNMAFLALSNLDAAVYQVTYQLKIPCTALCTVLMLNRT 139
Query: 169 LSEIQWAAFILLTAGCTTAQ---LNSNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEAII 225
LS++QW + +L AG T Q + V Q P G+ L SGFAGVY E ++
Sbjct: 140 LSKLQWVSVFMLCAGVTLVQWKPAQATKVVVEQNPLLGFGAIAIAVLCSGFAGVYFEKVL 199
Query: 226 KKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSGI 285
K + ++ V+N +Y+ G+ + + D + KGFF+GY++ +IF ++ G+
Sbjct: 200 KSSDT-SLWVRNIQMYLSGIIVTLAGVYLSDGAEIKEKGFFYGYTYYVWFVIFLASVGGL 258
Query: 286 AVSMVMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLH 340
S+V+KY DNI+K +S + A++L+ + SV LFG ++L F LGT++V V+IYL+
Sbjct: 259 YTSVVVKYTDNIMKGFSAAAAIVLSTIASVMLFGLQITLTFALGTLLVCVSIYLY 313
>UGAT_MOUSE (Q9R0M8) UDP-galactose translocator (UDP-galactose
transporter) (UGT) (UDP-Gal-Tr) (Solute carrier family
35 member A2) (mUGT1)
Length = 390
Score = 145 bits (365), Expect = 2e-34
Identities = 103/329 (31%), Positives = 170/329 (51%), Gaps = 20/329 (6%)
Query: 26 VSDGETKVDSHRTQVQWKRKSMVTFALTILTSSQAILIV-WSKRAGKYEYSVTTANFMVE 84
VS G + S T +R ++ A+ ++ ++ IL + +++ + TTA M E
Sbjct: 18 VSSGALEPGS--TTAAHRRLKYISLAVLVVQNASLILSIRYARTLPGDRFFATTAVVMAE 75
Query: 85 TLKCAISLVALGRIWNKEGVTDDNRLTTTLDEVIVYP--------IPAALYLVKNLLQYY 136
LK L+ L K G + L L E ++ +P+ +Y ++N LQY
Sbjct: 76 VLKGLTCLLLL--FAQKRG--NVKHLVLFLHEAVLVQYVDTLKLAVPSLIYTLQNNLQYV 131
Query: 137 IFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLN----SN 192
+ + A +Q+ I++T + ++L + LS +QWA+ +LL G Q S
Sbjct: 132 AISNLPAATFQVTYQLKILTTALFSVLMLNRSLSRLQWASLLLLFTGVAIVQAQQAGGSG 191
Query: 193 SDHVLQTPFQGWVMAIAMALLSGFAGVYTEAIIKKRPSRNINVQNFWLYVFGMGFNAVAI 252
+ Q P G +A L SGFAGVY E I+K S ++ ++N L +FG V +
Sbjct: 192 PRPLDQNPGAGLAAVVASCLSSGFAGVYFEKILKGS-SGSVWLRNLQLGLFGTALGLVGL 250
Query: 253 LVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAV 312
+ AV ++GFF GY+ ++ N A G+ V++V+KYADNI+K ++TS++++L+ V
Sbjct: 251 WWAEGTAVASQGFFFGYTPAVWGVVLNQAFGGLLVAVVVKYADNILKGFATSLSIVLSTV 310
Query: 313 VSVFLFGFHLSLAFFLGTIVVSVAIYLHS 341
S+ LFGFHL F LG +V A+YL+S
Sbjct: 311 ASIRLFGFHLDPLFALGAGLVIGAVYLYS 339
>CMST_MOUSE (Q61420) CMP-sialic acid transporter (CMP-Sia-Tr)
(CMP-SA-Tr) (Solute carrier family 35 member A1)
Length = 336
Score = 144 bits (363), Expect = 3e-34
Identities = 86/296 (29%), Positives = 159/296 (53%), Gaps = 9/296 (3%)
Query: 52 LTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVAL----GRIWNKEGVTDD 107
+T++ ++ + + +++ + Y TTA + E +K IS+ L G + + +
Sbjct: 20 MTLVAAAYTVALRYTRTTAEELYFSTTAVCITEVIKLLISVGLLAKETGSLGRFKASLSE 79
Query: 108 NRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKK 167
N L + E+ +P+ +Y V+N + + + +DA YQ+ I T + ++L +
Sbjct: 80 NVLGSP-KELAKLSVPSLVYAVQNNMAFLALSNLDAAVYQVTYQLKIPCTALCTVLMLNR 138
Query: 168 RLSEIQWAAFILLTAGCTTAQ---LNSNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEAI 224
LS++QW + +L G T Q ++ V Q P G+ L SGFAGVY E +
Sbjct: 139 TLSKLQWISVFMLCGGVTLVQWKPAQASKVVVAQNPLLGFGAIAIAVLCSGFAGVYFEKV 198
Query: 225 IKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSG 284
+K + ++ V+N +Y+ G+ + D + KGFF+GY++ +IF ++ G
Sbjct: 199 LKSSDT-SLWVRNIQMYLSGIVVTLAGTYLSDGAEIQEKGFFYGYTYYVWFVIFLASVGG 257
Query: 285 IAVSMVMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLH 340
+ S+V+KY DNI+K +S + A++L+ + SV LFG ++L+F LG ++V V+IYL+
Sbjct: 258 LYTSVVVKYTDNIMKGFSAAAAIVLSTIASVLLFGLQITLSFALGALLVCVSIYLY 313
>CMST_CRIGR (O08520) CMP-sialic acid transporter (CMP-Sia-Tr)
(CMP-SA-Tr) (Solute carrier family 35 member A1)
Length = 336
Score = 144 bits (362), Expect = 4e-34
Identities = 85/296 (28%), Positives = 160/296 (53%), Gaps = 9/296 (3%)
Query: 52 LTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVAL----GRIWNKEGVTDD 107
+T++ ++ + + +++ K Y TTA + E +K IS+ L G + + +
Sbjct: 20 MTLVAAAYTVALRYTRTTAKELYFSTTAVCVTEVIKLLISVGLLAKETGSLGRFKASLSE 79
Query: 108 NRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKK 167
N L + E++ +P+ +Y V+N + + + +DA YQ+ I T + ++L +
Sbjct: 80 NVLGSP-KELMKLSVPSLVYAVQNNMAFLALSNLDAAVYQVTYQLKIPCTALCTVLMLNR 138
Query: 168 RLSEIQWAAFILLTAGCTTAQ---LNSNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEAI 224
LS++QW + +L G Q + V Q+P G+ L SGFAGVY E +
Sbjct: 139 TLSKLQWVSVFMLCGGVILVQWKPAQATKVVVEQSPLLGFGAIAIAVLCSGFAGVYFEKV 198
Query: 225 IKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSG 284
+K + ++ V+N +Y+ G+ V + D + KGFF+GY++ +IF ++ G
Sbjct: 199 LKSSDT-SLWVRNIQMYLSGIVVTLVGTYLSDGAEIKEKGFFYGYTYYVWFVIFLASVGG 257
Query: 285 IAVSMVMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLH 340
+ S+V+KY DNI+K +S + A++L+ + SV LFG ++L+F +G ++V ++IYL+
Sbjct: 258 LYTSVVVKYTDNIMKGFSAAAAIVLSTIASVMLFGLQITLSFAMGALLVCISIYLY 313
>GMS1_SCHPO (P87041) UDP-galactose transporter (Golgi UDP-Gal
transporter)
Length = 353
Score = 143 bits (360), Expect = 7e-34
Identities = 97/318 (30%), Positives = 167/318 (52%), Gaps = 18/318 (5%)
Query: 40 VQWKRKSMVTFALTILTSSQAILIV---WSKRAGKYE---YSVTTANFMVETLKCAISLV 93
V+WK M AL +LT + LI+ +S+ Y+ Y +TA + E +K +
Sbjct: 8 VKWKGIPMKYIALVLLTVQNSALILTLNYSRIMPGYDDKRYFTSTAVLLNELIKLVVCF- 66
Query: 94 ALGRIWNKEGVTDDNRLTTTLDEVI-----VYPIPAALYLVKNLLQYYIFAYVDAPGYQI 148
++G ++ V + +L L ++ IPA LY +N LQY + A +Q+
Sbjct: 67 SVGYHQFRKNVGKEAKLRAFLPQIFGGDSWKLAIPAFLYTCQNNLQYVAAGNLTAASFQV 126
Query: 149 LKNFNIISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQL-NSNSDHVLQT----PFQG 203
I++T + ++L +RL ++W + LLT G QL N NSD + P G
Sbjct: 127 TYQLKILTTAIFSILLLHRRLGPMKWFSLFLLTGGIAIVQLQNLNSDDQMSAGPMNPVTG 186
Query: 204 WVMAIAMALLSGFAGVYTEAIIKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNK 263
+ + L+SG AGVY E ++K + ++ V+N L F + IL++D+ +
Sbjct: 187 FSAVLVACLISGLAGVYFEKVLKDT-NPSLWVRNVQLSFFSLFPCLFTILMKDYHNIAEN 245
Query: 264 GFFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHLS 323
GFF GY+ I L I A GI V++ + +ADNI+K +STS++++++++ SV+L F +S
Sbjct: 246 GFFFGYNSIVWLAILLQAGGGIIVALCVAFADNIMKNFSTSISIIISSLASVYLMDFKIS 305
Query: 324 LAFFLGTIVVSVAIYLHS 341
L F +G ++V A +L++
Sbjct: 306 LTFLIGVMLVIAATFLYT 323
>UGAT_HUMAN (P78381) UDP-galactose translocator (UDP-galactose
transporter) (UGT) (UDP-Gal-Tr) (Solute carrier family
35 member A2)
Length = 396
Score = 141 bits (356), Expect = 2e-33
Identities = 96/312 (30%), Positives = 162/312 (51%), Gaps = 18/312 (5%)
Query: 43 KRKSMVTFALTILTSSQAILIV-WSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNK 101
+R ++ A+ ++ ++ IL + +++ + TTA M E LK L+ L K
Sbjct: 33 RRLKYISLAVLVVQNASLILSIRYARTLPGDRFFATTAVVMAEVLKGLTCLLLL--FAQK 90
Query: 102 EGVTDDNRLTTTLDEVIVYP--------IPAALYLVKNLLQYYIFAYVDAPGYQILKNFN 153
G + L L E ++ +P+ +Y ++N LQY + + A +Q+
Sbjct: 91 RG--NVKHLVLFLHEAVLVQYVDTLKLAVPSLIYTLQNNLQYVAISNLPAATFQVTYQLK 148
Query: 154 IISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLNSNSDH----VLQTPFQGWVMAIA 209
I++T + ++L + LS +QWA+ +LL G Q + Q P G +A
Sbjct: 149 ILTTALFSVLMLNRSLSRLQWASLLLLFTGVAIVQAQQAGGGGPRPLDQNPGAGLAAVVA 208
Query: 210 MALLSGFAGVYTEAIIKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHGY 269
L SGFAGVY E I+K S ++ ++N L +FG V + + AV +GFF GY
Sbjct: 209 SCLSSGFAGVYFEKILKGS-SGSVWLRNLQLGLFGTALGLVGLWWAEGTAVATRGFFFGY 267
Query: 270 SFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLG 329
+ ++ N A G+ V++V+KYADNI+K ++TS++++L+ V S+ LFGFH+ F LG
Sbjct: 268 TPAVWGVVLNQAFGGLLVAVVVKYADNILKGFATSLSIVLSTVASIRLFGFHVDPLFALG 327
Query: 330 TIVVSVAIYLHS 341
+V A+YL+S
Sbjct: 328 AGLVIGAVYLYS 339
>UGNT_CANFA (O77592) UDP-N-acetylglucosamine transporter (Golgi
UDP-GlcNAc transporter) (Solute carrier family 35 member
A3)
Length = 326
Score = 132 bits (332), Expect = 1e-30
Identities = 92/314 (29%), Positives = 160/314 (50%), Gaps = 29/314 (9%)
Query: 48 VTFALTILTSSQAILIVWSKRAGKYE---YSVTTANFMVETLK---CAI--------SLV 93
++ + + ++ +L + R K E Y +TA + E LK C + SL
Sbjct: 8 LSLGILVFQTTSLVLTMRYSRTLKEEGPRYLSSTAVVVAELLKIMACILLVYKDSKCSLR 67
Query: 94 ALGRIWNKEGVTDDNRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFN 153
AL RI + E + E + IP+ +Y ++N L Y + +DA YQ+
Sbjct: 68 ALNRILHDE-------ILNKPMETLKLAIPSGIYTLQNNLLYVALSNLDAATYQVTYQLK 120
Query: 154 IISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLNSNSDHV------LQTPFQGWVMA 207
I++T + +L K+L QW + ++L G Q S+S + + F G +
Sbjct: 121 ILTTALFSVSMLSKKLGVYQWLSLVILMTGVAFVQWPSDSQELDSKELSAGSQFVGLMAV 180
Query: 208 IAMALLSGFAGVYTEAIIKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFH 267
+ SGFAGVY E I+K+ +++ ++N L FG F + + + D + V GFF
Sbjct: 181 LTACFSSGFAGVYFEKILKET-KQSVWIRNIQLGFFGSIFGLMGVYIYDGELVSKNGFFQ 239
Query: 268 GYSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSVF-LFGFHLSLAF 326
GY+ +T +++ AL G+ ++ V+KYADNI+K ++TS++++L+ ++S F L F + F
Sbjct: 240 GYNRLTWIVVILQALGGLVIAAVIKYADNILKGFATSLSIILSTLISYFWLQDFVPTSVF 299
Query: 327 FLGTIVVSVAIYLH 340
FLG I+V A +L+
Sbjct: 300 FLGAILVITATFLY 313
>UGNT_HUMAN (Q9Y2D2) UDP-N-acetylglucosamine transporter (Golgi
UDP-GlcNAc transporter) (Solute carrier family 35 member
A3)
Length = 325
Score = 131 bits (330), Expect = 2e-30
Identities = 92/313 (29%), Positives = 160/313 (50%), Gaps = 28/313 (8%)
Query: 48 VTFALTILTSSQAILIVWSKRAGKYE---YSVTTANFMVETLK---CAI--------SLV 93
V+ + + ++ +L + R K E Y +TA + E LK C + SL
Sbjct: 8 VSLGILVFQTTSLVLTMRYSRTLKEEGPRYLSSTAVVVAELLKIMACILLVYKDSKCSLR 67
Query: 94 ALGRIWNKEGVTDDNRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFN 153
AL R+ + E + E + IP+ +Y ++N L Y + +DA YQ+
Sbjct: 68 ALNRVLHDE-------ILNKPMETLKLAIPSGIYTLQNNLLYVALSNLDAATYQVTYQLK 120
Query: 154 IISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLNSNS-----DHVLQTPFQGWVMAI 208
I++T + +L K+L QW + ++L G Q S+S + + F G + +
Sbjct: 121 ILTTALFSVSMLSKKLGVYQWLSLVILMTGVAFVQWPSDSQLDSKELSAGSQFVGLMAVL 180
Query: 209 AMALLSGFAGVYTEAIIKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHG 268
SGFAGVY E I+K+ +++ ++N L FG F + + + D + V GFF G
Sbjct: 181 TACFSSGFAGVYFEKILKET-KQSVWIRNIQLGFFGSIFGLMGVYIYDGELVSKNGFFQG 239
Query: 269 YSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSVF-LFGFHLSLAFF 327
Y+ +T +++ AL G+ ++ V+KYADNI+K ++TS++++L+ ++S F L F + FF
Sbjct: 240 YNRLTWIVVVLQALGGLVIAAVIKYADNILKGFATSLSIILSTLISYFWLQDFVPTSVFF 299
Query: 328 LGTIVVSVAIYLH 340
LG I+V A +L+
Sbjct: 300 LGAILVITATFLY 312
>YOL7_CAEEL (Q02334) Hypothetical protein ZK370.7 in chromosome III
Length = 355
Score = 113 bits (283), Expect = 6e-25
Identities = 88/349 (25%), Positives = 166/349 (47%), Gaps = 26/349 (7%)
Query: 13 RDSDVESVAVNNT-VSDGETKVDSHRTQVQWKRKSMVTFALTILTSSQAILIVWSKRAGK 71
+D D E + N+ V + S R +K V ++T + ++ + I +++
Sbjct: 11 QDEDKEKLLPNDKDVEKADESPSSSRPSFVFK--CYVIASMTFIWTAYTLTIKYTRSTVN 68
Query: 72 YE--YSVTTANFMVETLKCAISLVALGRIWNKEGVTDDNRLTTTLD--------EVIVYP 121
+ YS T+ E LK I+ ++ KE D + + + E+
Sbjct: 69 PDMMYSSTSVVLCAEILKLVITFA----MFYKECNFDSRQFSEQVSKYYINAPRELAKMS 124
Query: 122 IPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEIQWAAFILLT 181
+P+ Y ++N L + + +DA YQ+ ++ST + L ++ S +W A LL
Sbjct: 125 VPSFAYALQNNLDFVGLSNLDAGLYQVTTQLKVVSTAFFMMLFLGRKFSTRRWMAITLLM 184
Query: 182 AGCTTAQLNSNSDHVLQTPFQ-------GWVMAIAMALLSGFAGVYTEAIIKKRPSRNIN 234
G Q+N+ S T + G +A + +GFAGVY E ++K S
Sbjct: 185 FGVAFVQMNNVSASEANTKRETAENYIVGLSAVLATCVTAGFAGVYFEKMLKDGGSTPFW 244
Query: 235 VQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMKYA 294
++N +Y G+ ++A L DF + +KGFF GY+ ++ + G+ +S+VM+Y
Sbjct: 245 IRNMQMYSCGVISASIACLT-DFSRISDKGFFFGYTDKVWAVVILLGVGGLYISLVMRYL 303
Query: 295 DNIVKVYSTSVAMLLTAVVSVFLF-GFHLSLAFFLGTIVVSVAIYLHSA 342
DN+ K +++V+++L V+S+ +F + + F LGTI V +A+ L+++
Sbjct: 304 DNLYKSMASAVSIILVVVLSMLIFPDIFIGMYFVLGTICVVLAVLLYNS 352
>YEGI_CAEEL (Q93890) Hypothetical protein M02B1.1 in chromosome IV
Length = 311
Score = 86.7 bits (213), Expect = 8e-17
Identities = 50/165 (30%), Positives = 84/165 (50%), Gaps = 2/165 (1%)
Query: 122 IPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEIQWAAFILLT 181
IPA +Y+V+N L Y +++DA + I I + + IIL++ L+ QW A +L
Sbjct: 134 IPAMIYIVQNNLFYVAASHLDAATFMITSQLKIFTAAIFTVIILRRSLNRTQWFALAVLF 193
Query: 182 AGCTTAQLNSNS--DHVLQTPFQGWVMAIAMALLSGFAGVYTEAIIKKRPSRNINVQNFW 239
G + QL + ++PF G+V + LSGFAG+Y E I+K ++ ++N
Sbjct: 194 VGVSLVQLQGTKAKESSGESPFVGFVAVVVACCLSGFAGIYFEKILKGSAPVSLWMRNVQ 253
Query: 240 LYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSG 284
+ VF + + AI +QD V G +G+ I L + + + G
Sbjct: 254 MAVFSIPASFSAIYMQDSKTVNEYGLLYGFDSIVWLTVLWYGVGG 298
>YAGC_SCHPO (Q09875) Hypothetical protein C12G12.12 in chromosome I
Length = 324
Score = 36.6 bits (83), Expect = 0.096
Identities = 42/241 (17%), Positives = 93/241 (38%), Gaps = 21/241 (8%)
Query: 114 LDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEIQ 173
L + +PA + + + L Y A YQ+ + II + +LK+ + ++Q
Sbjct: 72 LKHKVFMALPAIMDICGSTLMNVGLLYTSASIYQMTRGSLIIFVALFATTLLKRTIGQLQ 131
Query: 174 WAAFILLTAGCTTAQLNSNSDHVLQTPFQGWVMAIAMALLSGF-------AGVYTEAIIK 226
W + + G + +S + P ++ I L+ F Y + I+
Sbjct: 132 WLSLSFVVLGVAIVGYSGSSSSIGSNP----ILGITAVLIGQFFLATQFTIEEYILSFIQ 187
Query: 227 KRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHG-YSFITVLMIFNHALSGI 285
PS + + +G+ F + +++ + +HG + + V+ FN +
Sbjct: 188 VDPSELVAYEG----TYGVFFVLLGMIISYYFIGSTTAGYHGWFDYSHVISRFNEVPALY 243
Query: 286 AVSMVMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLHSAGKM 345
+S V+ + V ++ L +A L +A G ++++A+ + S +
Sbjct: 244 VISGVILVSIAFFNVSGLAITKLHSATTRSL-----LDIARTFGIWIIAMAMGMESFHLL 298
Query: 346 Q 346
Q
Sbjct: 299 Q 299
>YA68_METJA (Q58467) Hypothetical protein MJ1068
Length = 507
Score = 33.1 bits (74), Expect = 1.1
Identities = 21/81 (25%), Positives = 38/81 (45%), Gaps = 3/81 (3%)
Query: 240 LYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVK 299
L + MG+ + +V F ++ Y+ T ++ LS S++ Y N+
Sbjct: 133 LIIMAMGYYFLDSIVAFFSNILTGFQLQNYASSTRVV---RILSVFIFSLIFIYLFNVHN 189
Query: 300 VYSTSVAMLLTAVVSVFLFGF 320
Y SV+ LL AVV + ++G+
Sbjct: 190 AYVPSVSYLLMAVVMIIIYGY 210
>VATB_ARATH (P11574) Vacuolar ATP synthase subunit B (EC 3.6.3.14)
(V-ATPase B subunit) (Vacuolar proton pump B subunit)
(V-ATPase 57 kDa subunit)
Length = 486
Score = 32.7 bits (73), Expect = 1.4
Identities = 34/115 (29%), Positives = 49/115 (42%), Gaps = 25/115 (21%)
Query: 1 MEYRKIKDE-------DKVRDSDVESVAVNNTVSDGETKVDSHRTQVQWKRKSMVTFALT 53
MEYR + DKV+ + + VN + DG T+ R QV
Sbjct: 17 MEYRTVSGVAGPLVILDKVKGPKYQEI-VNIRLGDGSTR----RGQV------------L 59
Query: 54 ILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDDN 108
+ +A++ V+ +G + TT F E LK +SL LGRI+N G DN
Sbjct: 60 EVDGEKAVVQVFEGTSG-IDNKFTTVQFTGEVLKTPVSLDMLGRIFNGSGKPIDN 113
>VAB2_HORVU (Q40079) Vacuolar ATP synthase subunit B isoform 2 (EC
3.6.3.14) (V-ATPase B subunit 2) (Vacuolar proton pump B
subunit 2)
Length = 483
Score = 32.3 bits (72), Expect = 1.8
Identities = 34/115 (29%), Positives = 49/115 (42%), Gaps = 25/115 (21%)
Query: 1 MEYRKIKDE-------DKVRDSDVESVAVNNTVSDGETKVDSHRTQVQWKRKSMVTFALT 53
MEYR + DKV+ + + VN + DG T+ R QV
Sbjct: 14 MEYRTVSGVAGPLVILDKVKGPKYQEI-VNIRLGDGTTR----RGQV------------L 56
Query: 54 ILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDDN 108
+ +A++ V+ +G + TT F E LK +SL LGRI+N G DN
Sbjct: 57 EVDGEKAVVQVFEGTSG-IDNKYTTVQFTGEVLKTPVSLDMLGRIFNGSGKPIDN 110
>VAB1_HORVU (Q40078) Vacuolar ATP synthase subunit B isoform 1 (EC
3.6.3.14) (V-ATPase B subunit 1) (Vacuolar proton pump B
subunit 1)
Length = 488
Score = 32.3 bits (72), Expect = 1.8
Identities = 34/115 (29%), Positives = 49/115 (42%), Gaps = 25/115 (21%)
Query: 1 MEYRKIKDE-------DKVRDSDVESVAVNNTVSDGETKVDSHRTQVQWKRKSMVTFALT 53
MEYR + DKV+ + + VN + DG T+ R QV
Sbjct: 19 MEYRTVSGVAGPLVILDKVKGPKYQEI-VNIRLGDGTTR----RGQV------------L 61
Query: 54 ILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDDN 108
+ +A++ V+ +G + TT F E LK +SL LGRI+N G DN
Sbjct: 62 EVDGEKAVVQVFEGTSG-IDNKYTTVQFTGEVLKTPVSLDMLGRIFNGSGKPIDN 115
>VAB1_GOSHI (Q43432) Vacuolar ATP synthase subunit B isoform 1 (EC
3.6.3.14) (V-ATPase B subunit 1) (Vacuolar proton pump B
subunit 1)
Length = 488
Score = 32.3 bits (72), Expect = 1.8
Identities = 34/115 (29%), Positives = 49/115 (42%), Gaps = 25/115 (21%)
Query: 1 MEYRKIKDE-------DKVRDSDVESVAVNNTVSDGETKVDSHRTQVQWKRKSMVTFALT 53
MEYR + DKV+ + + VN + DG T+ R QV
Sbjct: 19 MEYRTVSGVAGPLVILDKVKGPKYQEI-VNIRLGDGTTR----RGQV------------L 61
Query: 54 ILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDDN 108
+ +A++ V+ +G + TT F E LK +SL LGRI+N G DN
Sbjct: 62 EVDGEKAVVQVFEGTSG-IDNKYTTVQFTGEVLKTPVSLDMLGRIFNGSGKPIDN 115
>EBRB_BACSU (O31791) Multidrug resistance protein ebrB
Length = 117
Score = 32.0 bits (71), Expect = 2.4
Identities = 31/105 (29%), Positives = 54/105 (50%), Gaps = 9/105 (8%)
Query: 244 GMGFNAVAILVQDFDAVMNK---GFFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKV 300
G+ + A+AI+ + F + M K GF + V++ F A + ++ S+ K D +
Sbjct: 3 GLLYLALAIVSEVFGSTMLKLSEGFTQAWPIAGVIVGFLSAFTFLSFSL--KTID-LSSA 59
Query: 301 YST--SVAMLLTAVVSVFLFGFHLSLAFFLG-TIVVSVAIYLHSA 342
Y+T V LTA+V LFG +SL G T+V++ + L+ +
Sbjct: 60 YATWSGVGTALTAIVGFLLFGETISLKGVFGLTLVIAGVVVLNQS 104
>ATP6_CANPA (Q03671) ATP synthase a chain precursor (EC 3.6.3.14)
(ATPase protein 6) (ATP synthase subunit 6)
Length = 246
Score = 32.0 bits (71), Expect = 2.4
Identities = 30/103 (29%), Positives = 46/103 (44%), Gaps = 27/103 (26%)
Query: 246 GFNAVAILVQDFDAVMNK-----GFFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKV 300
G + +AI +D ++N G GY F + IFN L +SM+
Sbjct: 57 GVSVIAI----YDTILNLVNGQIGRKGGYYFPLIFTIFNFILIANLISMI---------P 103
Query: 301 YSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLHSAG 343
YS +++ L AVVS SL ++G +V + +YLH G
Sbjct: 104 YSFAISAQLVAVVS-------FSLTLWIGNVV--LGLYLHGWG 137
>AG_BRANA (Q01540) Floral homeotic protein AGAMOUS
Length = 252
Score = 31.6 bits (70), Expect = 3.1
Identities = 16/42 (38%), Positives = 24/42 (57%)
Query: 50 FALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAIS 91
+ L++L ++ LIV+S R YEYS + +E K AIS
Sbjct: 49 YELSVLCDAEVALIVFSSRGRLYEYSNNSVKGTIERYKKAIS 90
>AG_ARATH (P17839) Floral homeotic protein AGAMOUS
Length = 252
Score = 31.6 bits (70), Expect = 3.1
Identities = 16/42 (38%), Positives = 24/42 (57%)
Query: 50 FALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAIS 91
+ L++L ++ LIV+S R YEYS + +E K AIS
Sbjct: 49 YELSVLCDAEVALIVFSSRGRLYEYSNNSVKGTIERYKKAIS 90
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,057,305
Number of Sequences: 164201
Number of extensions: 1354831
Number of successful extensions: 5273
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 5233
Number of HSP's gapped (non-prelim): 29
length of query: 347
length of database: 59,974,054
effective HSP length: 111
effective length of query: 236
effective length of database: 41,747,743
effective search space: 9852467348
effective search space used: 9852467348
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0380.6


