

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
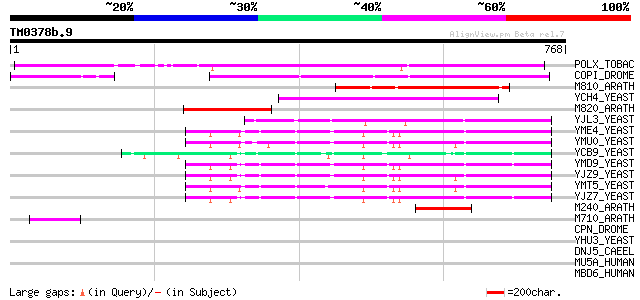
Query= TM0378b.9
(768 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 407 e-113
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 288 3e-77
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 225 3e-58
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 163 2e-39
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 121 8e-27
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 119 4e-26
YME4_YEAST (Q04711) Transposon Ty1 protein B 99 4e-20
YMU0_YEAST (Q04670) Transposon Ty1 protein B 98 7e-20
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 95 8e-19
YMD9_YEAST (Q03434) Transposon Ty1 protein B 92 7e-18
YJZ9_YEAST (P47100) Transposon Ty1 protein B 92 7e-18
YMT5_YEAST (Q04214) Transposon Ty1 protein B 91 9e-18
YJZ7_YEAST (P47098) Transposon Ty1 protein B 89 3e-17
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 79 6e-14
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 52 8e-06
CPN_DROME (Q02910) Calphotin 41 0.010
YHU3_YEAST (P38844) Hypothetical 33.4 kDa protein in RPL44B-RPC1... 38 0.12
DNJ5_CAEEL (Q09446) DnaJ homolog dnj-5 37 0.26
MU5A_HUMAN (P98088) Mucin 5AC (Mucin 5 subtype AC, tracheobronch... 36 0.44
MBD6_HUMAN (Q96DN6) Methyl-CpG binding domain protein 6 35 0.75
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 407 bits (1047), Expect = e-113
Identities = 268/750 (35%), Positives = 388/750 (51%), Gaps = 36/750 (4%)
Query: 7 GIQHRLTCPYVHQQNGRVEWKHRHITETGLALLAHAHMPLTFWGDAFETAAYLINQLPIP 66
GI+H T P Q NG E +R I E ++L A +P +FWG+A +TA YLIN+ P
Sbjct: 568 GIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACYLINRSPSV 627
Query: 67 TLDLSTPLQKLYDKKPDYSFLKCFGCLCYPHTRPYNTHKLAFRSAPCTFLGYSPMHKGYN 126
L P + +K+ YS LK FGC + H KL +S PC F+GY GY
Sbjct: 628 PLAFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYR 687
Query: 127 CLT-TEGKVIITRNVLFDETVFPFQFQTSKSSSDILLDSSLPTTSVIPLLPISNASPNVM 185
+ KVI +R+V+F E+ + +T+ S+ + + +P IP ++ N
Sbjct: 688 LWDPVKKKVIRSRDVVFRES----EVRTAADMSEKVKNGIIPNFVTIP------STSNNP 737
Query: 186 PHANSVMPTVATNGSAPVVQTTVSDFSQPA*DSGVVHASAQPTEPPVPPSSNVHSMTTRA 245
A S V+ G P + Q D GV PT+ S R
Sbjct: 738 TSAESTTDEVSEQGEQP--GEVIEQGEQL--DEGVEEVE-HPTQGEEQHQPLRRSERPRV 792
Query: 246 KTGIHRPKAYAASTALSSVPTSAK*ALSIPHWHQ---AMQEEYQALMNNNTWELVPLLHG 302
++ + Y + P S K LS P +Q AMQEE ++L N T++LV L G
Sbjct: 793 ESRRYPSTEYVLISD-DREPESLKEVLSHPEKNQLMKAMQEEMESLQKNGTYKLVELPKG 851
Query: 303 RDAIGCKWVFRTKYNSDGSINKHKPRLVAKGFHQTEGYDYNETFSPVVKPTTIRTVLSLA 362
+ + CKWVF+ K + D + ++K RLV KGF Q +G D++E FSPVVK T+IRT+LSLA
Sbjct: 852 KRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFDEIFSPVVKMTSIRTILSLA 911
Query: 363 LMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFS-KGSSHLVCKLKKALYGLKQAPRAWF 421
+ Q D AFL+G+L EE++M+QP GF G H+VCKL K+LYGLKQAPR W+
Sbjct: 912 ASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWY 971
Query: 422 DKLSSTLARFGFQAAKCDTSLFCK-FTKSETLYILVYVDDIIITGSSSTAIFSLIADLNA 480
K S + + D ++ K F+++ + +L+YVDD++I G I L DL+
Sbjct: 972 MKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSK 1031
Query: 481 VFPLKDLGSLHYFLGIEATHTKDG-GLILSQTKYINDLLIKAKMDSINPSPTPMTSGLKL 539
F +KDLG LG++ + L LSQ KYI +L + M + P TP+ LKL
Sbjct: 1032 SFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLERFNMKNAKPVSTPLAGHLKL 1091
Query: 540 S--------ANSGALFPKPLLYRSVVGALHYVTI-TRPEIAFPLNKVCQFMHSPLEPHWQ 590
S G + P Y S VG+L Y + TRP+IA + V +F+ +P + HW+
Sbjct: 1092 SKKMCPTTVEEKGNMAKVP--YSSAVGSLMYAMVCTRPDIAHAVGVVSRFLENPGKEHWE 1149
Query: 591 AVKRILRYLHGTATHGLYLRRPSSMSITAFADSDWGSDPDDRRSTSGMCIYIGGNLVSWA 650
AVK ILRYL GT T S + + D+D D D+R+S++G G +SW
Sbjct: 1150 AVKWILRYLRGT-TGDCLCFGGSDPILKGYTDADMAGDIDNRKSSTGYLFTFSGGAISWQ 1208
Query: 651 AKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSELRVPTSPSSLIYCDNLSVVHLVANP 710
+K Q VA S+TEAEY + E++WL+ L EL + ++YCD+ S + L N
Sbjct: 1209 SKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGL-HQKEYVVYCDSQSAIDLSKNS 1267
Query: 711 ILHTKTKHFALDLHFVRERVADKQVTVVNI 740
+ H +TKH + H++RE V D+ + V+ I
Sbjct: 1268 MYHARTKHIDVRYHWIREMVDDESLKVLKI 1297
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 288 bits (738), Expect = 3e-77
Identities = 166/478 (34%), Positives = 253/478 (52%), Gaps = 11/478 (2%)
Query: 277 WHQAMQEEYQALMNNNTWELVPLLHGRDAIGCKWVFRTKYNSDGSINKHKPRLVAKGFHQ 336
W +A+ E A NNTW + ++ + +WVF KYN G+ ++K RLVA+GF Q
Sbjct: 906 WEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQ 965
Query: 337 TEGYDYNETFSPVVKPTTIRTVLSLALMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFS 396
DY ETF+PV + ++ R +LSL + + + Q D AFLNG L EE++M+ P G S
Sbjct: 966 KYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGIS 1025
Query: 397 KGSSHLVCKLKKALYGLKQAPRAWFDKLSSTLARFGFQAAKCDTSLFC--KFTKSETLYI 454
S + VCKL KA+YGLKQA R WF+ L F + D ++ K +E +Y+
Sbjct: 1026 CNSDN-VCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYV 1084
Query: 455 LVYVDDIIITGSSSTAIFSLIADLNAVFPLKDLGSLHYFLGIEATHTKDGGLILSQTKYI 514
L+YVDD++I T + + L F + DL + +F+GI +D + LSQ+ Y+
Sbjct: 1085 LLYVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQED-KIYLSQSAYV 1143
Query: 515 NDLLIKAKMDSINPSPTPMTSGLKLS-ANSGALFPKPLLYRSVVGALHYVTI-TRPEIAF 572
+L K M++ N TP+ S + NS P RS++G L Y+ + TRP++
Sbjct: 1144 KKILSKFNMENCNAVSTPLPSKINYELLNSDEDCNTPC--RSLIGCLMYIMLCTRPDLTT 1201
Query: 573 PLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGLYLRRPSSM--SITAFADSDWGSDPD 630
+N + ++ WQ +KR+LRYL GT L ++ + I + DSDW
Sbjct: 1202 AVNILSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSEI 1261
Query: 631 DRRSTSGMCI-YIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSELRVP 689
DR+ST+G NL+ W K+Q VA SSTEAEY +L + E +WL+ LL+ + +
Sbjct: 1262 DRKSTTGYLFKMFDFNLICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFLLTSINIK 1321
Query: 690 TSPSSLIYCDNLSVVHLVANPILHTKTKHFALDLHFVRERVADKQVTVVNIPAPSQVS 747
IY DN + + NP H + KH + HF RE+V + + + IP +Q++
Sbjct: 1322 LENPIKIYEDNQGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTENQLA 1379
Score = 77.0 bits (188), Expect = 2e-13
Identities = 51/147 (34%), Positives = 73/147 (48%), Gaps = 6/147 (4%)
Query: 2 FLTQCGIQHRLTCPYVHQQNGRVEWKHRHITETGLALLAHAHMPLTFWGDAFETAAYLIN 61
F + GI + LT P+ Q NG E R ITE +++ A + +FWG+A TA YLIN
Sbjct: 563 FCVKKGISYHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLIN 622
Query: 62 QLPIPTL--DLSTPLQKLYDKKPDYSFLKCFGCLCYPHTRPYNTHKLAFRSAPCTFLGYS 119
++P L TP + ++KKP L+ FG Y H + K +S F+GY
Sbjct: 623 RIPSRALVDSSKTPYEMWHNKKPYLKHLRVFGATVYVHIK-NKQGKFDDKSFKSIFVGYE 681
Query: 120 PMHKGYNCL-TTEGKVIITRNVLFDET 145
P G+ K I+ R+V+ DET
Sbjct: 682 P--NGFKLWDAVNEKFIVARDVVVDET 706
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 225 bits (574), Expect = 3e-58
Identities = 116/241 (48%), Positives = 163/241 (67%), Gaps = 6/241 (2%)
Query: 452 LYILVYVDDIIITGSSSTAIFSLIADLNAVFPLKDLGSLHYFLGIEATHTKDGGLILSQT 511
+Y+L+YVDDI++TGSS+T + LI L++ F +KDLG +HYFLGI+ T GL LSQT
Sbjct: 1 MYLLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIK-THPSGLFLSQT 59
Query: 512 KYINDLLIKAKMDSINPSPTPMTSGLKLSAN-SGALFPKPLLYRSVVGALHYVTITRPEI 570
KY +L A M P TP+ LKL+++ S A +P P +RS+VGAL Y+T+TRP+I
Sbjct: 60 KYAEQILNNAGMLDCKPMSTPLP--LKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDI 117
Query: 571 AFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGLYLRRPSSMSITAFADSDWGSDPD 630
++ +N VCQ MH P + +KR+LRY+ GT HGLY+ + S +++ AF DSDW
Sbjct: 118 SYAVNIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTS 177
Query: 631 DRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSELRVPT 690
RRST+G C ++G N++SW+AK+Q V+RSSTE EYR+LAL E+ W S S R P+
Sbjct: 178 TRRSTTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW--SSASRSRDPS 235
Query: 691 S 691
+
Sbjct: 236 A 236
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 163 bits (412), Expect = 2e-39
Identities = 94/307 (30%), Positives = 170/307 (54%), Gaps = 3/307 (0%)
Query: 373 DFNNAFLNGELVEEVFMQQPPGF-SKGSSHLVCKLKKALYGLKQAPRAWFDKLSSTLARF 431
D + AFLN + E ++++QPPGF ++ + V +L +YGLKQAP W + +++TL +
Sbjct: 2 DVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKKI 61
Query: 432 GFQAAKCDTSLFCKFTKSETLYILVYVDDIIITGSSSTAIFSLIADLNAVFPLKDLGSLH 491
GF + + L+ + T +YI VYVDD+++ S + +L ++ +KDLG +
Sbjct: 62 GFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKVD 121
Query: 492 YFLGIEATHTKDGGLILSQTKYINDLLIKAKMDSINPSPTPMTSGLKLSANSGALFPKPL 551
FLG+ + +G + LS YI ++++++ + TP+ + L +
Sbjct: 122 KFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDIT 181
Query: 552 LYRSVVGALHYVTIT-RPEIAFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGLYLR 610
Y+S+VG L + T RP+I++P++ + +F+ P H ++ +R+LRYL+ T + L R
Sbjct: 182 PYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKYR 241
Query: 611 RPSSMSITAFADSDWGSDPDDRRSTSGMCIYIGGNLVSWAAKK-QTVVARSSTEAEYRSL 669
S +++T + D+ G+ D ST G + G V+W++KK + V+ STEAEY +
Sbjct: 242 SGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYITA 301
Query: 670 ALVITEI 676
+ + EI
Sbjct: 302 SETVMEI 308
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 121 bits (303), Expect = 8e-27
Identities = 64/125 (51%), Positives = 85/125 (67%), Gaps = 3/125 (2%)
Query: 241 MTTRAKTGIHR--PK-AYAASTALSSVPTSAK*ALSIPHWHQAMQEEYQALMNNNTWELV 297
M TR+K GI++ PK + +T + P S AL P W QAMQEE AL N TW LV
Sbjct: 1 MLTRSKAGINKLNPKYSLTITTTIKKEPKSVIFALKDPGWCQAMQEELDALSRNKTWILV 60
Query: 298 PLLHGRDAIGCKWVFRTKYNSDGSINKHKPRLVAKGFHQTEGYDYNETFSPVVKPTTIRT 357
P ++ +GCKWVF+TK +SDG++++ K RLVAKGFHQ EG + ET+SPVV+ TIRT
Sbjct: 61 PPPVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVRTATIRT 120
Query: 358 VLSLA 362
+L++A
Sbjct: 121 ILNVA 125
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 119 bits (297), Expect = 4e-26
Identities = 114/445 (25%), Positives = 198/445 (43%), Gaps = 28/445 (6%)
Query: 325 HKPRLVAKGFHQTEGYDYNETFSPVVKPTTIRTVLSLALMHQWPIRQFDFNNAFLNGELV 384
+K R+V +G Q+ Y+ + + I+ L +A ++ D N+AFL +L
Sbjct: 1337 YKARIVCRGDTQSPD-TYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLE 1395
Query: 385 EEVFMQQPPGFSKGSSHLVCKLKKALYGLKQAPRAWFDKLSSTLARFGFQAAKCDTSLFC 444
EE+++ P V KL KALYGLKQ+P+ W D L L G + L+
Sbjct: 1396 EEIYIPHPH-----DRRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLY- 1449
Query: 445 KFTKSETLYILVYVDDIIITGSSSTAIFSLIADLNAVFPLKDLGSL------HYFLGIEA 498
T+ + L I VYVDD +I S+ + I L + F LK G+L LG++
Sbjct: 1450 -QTEDKNLMIAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMDL 1508
Query: 499 THTKDGGLI-LSQTKYIN--DLLIKAKMDSINPSPTPMTSGLKLSANSGAL------FPK 549
+ K G I L+ +IN D ++ I S P S K+ L F +
Sbjct: 1509 VYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFRQ 1568
Query: 550 PLL-YRSVVGALHYVT-ITRPEIAFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGL 607
+L + ++G L+YV R +I F + KV + ++ P E + + +I++YL G+
Sbjct: 1569 GVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIGI 1628
Query: 608 YLRRP--SSMSITAFADSDWGSDPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAE 665
+ R + A D+ GS+ D +S G+ ++ G N+ + + K T SSTEAE
Sbjct: 1629 HYDRDCNKDKKVIAITDASVGSE-YDAQSRIGVILWYGMNIFNVYSNKSTNRCVSSTEAE 1687
Query: 666 YRSLALVITEIMWLRSLLSELRVPTSPSSLIYCDNLSVVHLVANPILHTKTKHFALDLHF 725
++ + L+ L EL + ++ D+ + + K K +
Sbjct: 1688 LHAIYEGYADSETLKVTLKELGEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTWIKTEI 1747
Query: 726 VRERVADKQVTVVNIPAPSQVSSTL 750
++E++ +K + ++ I ++ L
Sbjct: 1748 IKEKIKEKSIKLLKITGKGNIADLL 1772
Score = 33.5 bits (75), Expect = 2.2
Identities = 20/57 (35%), Positives = 26/57 (45%)
Query: 7 GIQHRLTCPYVHQQNGRVEWKHRHITETGLALLAHAHMPLTFWGDAFETAAYLINQL 63
GI H LT H NGR E R I LL +++ + FW A +A + N L
Sbjct: 714 GIHHILTSTQDHAANGRAERYIRTIITDATTLLRQSNLRVKFWEYAVTSATNIRNYL 770
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 99.0 bits (245), Expect = 4e-20
Identities = 119/543 (21%), Positives = 235/543 (42%), Gaps = 49/543 (9%)
Query: 244 RAKTGIHRPKAYAASTALSSVPTSAK*ALSIPH---------WHQAMQEEYQALMNNNTW 294
R+K IH A A ++ + T+ + +I + + +A +E L+ NTW
Sbjct: 781 RSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMNTW 840
Query: 295 ELVPLLHGRDAIGCKWVFRTKY----NSDGSINKHKPRLVAKG-FHQTEGYDYNETFSPV 349
+ + R I K V + + DG+ HK R VA+G + YD S
Sbjct: 841 D-TDKYYDRKEIDPKRVINSMFIFNRKRDGT---HKARFVARGDIQHPDTYDSGMQ-SNT 895
Query: 350 VKPTTIRTVLSLALMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFSKGSSHLVCKLKKA 409
V + T LSLAL + + I Q D ++A+L ++ EE++++ PP G + + +LKK+
Sbjct: 896 VHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHL--GMNDKLIRLKKS 953
Query: 410 LYGLKQAPRAWFDKLSSTLAR-FGFQAAKCDTSLFCKFTKSETLYILVYVDDIIITGSSS 468
LYGLKQ+ W++ + S L + G + + + +F K+ + I ++VDD+I+
Sbjct: 954 LYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF----KNSQVTICLFVDDMILFSKDL 1009
Query: 469 TAIFSLIADLNAVFPLK--DLG----SLHY-FLGIEATHTKDGGLILSQTKYINDLLIKA 521
A +I L + K +LG + Y LG+E + + + L + + + K
Sbjct: 1010 NANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKL 1069
Query: 522 KMDSINPS----PTPMTSGL-----KLSANSGALFPKPLLYRSVVGALHYVTIT-RPEIA 571
+ +NP P GL +L + K + ++G YV R ++
Sbjct: 1070 NV-PLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLL 1128
Query: 572 FPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGL--YLRRPS--SMSITAFADSDWGS 627
+ +N + Q + P +++++ T L + +P+ + A +D+ +G+
Sbjct: 1129 YYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGN 1188
Query: 628 DPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSELR 687
P +S G + G ++ + K ++ S+TEAE +++ + + L L+ EL
Sbjct: 1189 QP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELN 1247
Query: 688 VPTSPSSLIYCDNLSVVHLVANPILHTKTKHFALDLHFVRERVADKQVTVVNIPAPSQVS 747
L+ ++ +++N + + F +R+ V+ + V I ++
Sbjct: 1248 KKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIA 1307
Query: 748 STL 750
+
Sbjct: 1308 DVM 1310
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 98.2 bits (243), Expect = 7e-20
Identities = 117/547 (21%), Positives = 235/547 (42%), Gaps = 57/547 (10%)
Query: 244 RAKTGIHRPKAYAASTALSSVPTSAK*ALSIPH---------WHQAMQEEYQALMNNNTW 294
R+K IH A A ++ + T+ + +I + + QA +E L+ TW
Sbjct: 781 RSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIQAYHKEVNQLLKMKTW 840
Query: 295 ELVPLLHGRDAIGCKWVFRTKY----NSDGSINKHKPRLVAKGFHQTEGYDYNETFSPVV 350
+ + R I K V + + DG+ HK R VA+G Q + +T+ P +
Sbjct: 841 D-TDRYYDRKEIDPKRVINSMFIFNRKRDGT---HKARFVARGDIQ-----HPDTYDPGM 891
Query: 351 KPTTIR-----TVLSLALMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFSKGSSHLVCK 405
+ T+ T LSLAL + + I Q D ++A+L ++ EE++++ PP G + + +
Sbjct: 892 QSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHL--GMNDKLIR 949
Query: 406 LKKALYGLKQAPRAWFDKLSSTLAR-FGFQAAKCDTSLFCKFTKSETLYILVYVDDIIIT 464
LKK+LYGLKQ+ W++ + S L + G + + + +F K+ + I ++VDD+I+
Sbjct: 950 LKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF----KNSQVTICLFVDDMILF 1005
Query: 465 GSSSTAIFSLIADLNAVFPLK--DLG----SLHY-FLGIEATHTKDGGLILSQTKYINDL 517
A +I L + K +LG + Y LG+E + + + L + +
Sbjct: 1006 SKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEK 1065
Query: 518 LIKAKMDSINPS----PTPMTSGL-----KLSANSGALFPKPLLYRSVVGALHYVTIT-R 567
+ K + +NP P GL +L K + ++G YV R
Sbjct: 1066 IPKLNV-PLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFR 1124
Query: 568 PEIAFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGLYLRRPSSM----SITAFADS 623
++ + +N + Q + P + +++++ T L + + + +D+
Sbjct: 1125 FDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDA 1184
Query: 624 DWGSDPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLL 683
+G+ P +S G + G ++ + K ++ S+TEAE +++ + + L L+
Sbjct: 1185 SYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLV 1243
Query: 684 SELRVPTSPSSLIYCDNLSVVHLVANPILHTKTKHFALDLHFVRERVADKQVTVVNIPAP 743
EL L+ ++ +++N + + F +R+ V+ + V I
Sbjct: 1244 QELNKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETK 1303
Query: 744 SQVSSTL 750
++ +
Sbjct: 1304 KNIADVM 1310
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 94.7 bits (234), Expect = 8e-19
Identities = 140/653 (21%), Positives = 253/653 (38%), Gaps = 89/653 (13%)
Query: 155 KSSSDILLDSSLPTTSVIPLLPISNASPNV-------MPHANSVMPTVATNG--SAPVVQ 205
KSS ++ DS L +PL +++ SP +PH +S + G + V+
Sbjct: 1132 KSSKNVTADSILDD---LPLPDLTHQSPTDTSDVSKDIPHIHSRQTNSSLGGMDDSNVLT 1188
Query: 206 TTVSDFSQPA*DSGVVHASAQPTEPP-----VPPSSNVHSMTTRAKTGIHRPKAYAASTA 260
TT S + + S PP S A G+ K +
Sbjct: 1189 TTKSKKRSLEDNETEIEVSRDTWNNKNMRSLEPPRSKKRINLIAAIKGVKSIKPVRTTLR 1248
Query: 261 LSSVPTSAK*ALSIPHWHQAMQEEYQALMNNNTWELVPLLHGRD-----AIGCKWVFRTK 315
T K + +A +E L+ NTW+ D I ++F K
Sbjct: 1249 YDEAITYNKDNKEKDRYVEAYHKEISQLLKMNTWDTNKYYDRNDIDPKKVINSMFIFNKK 1308
Query: 316 YNSDGSINKHKPRLVAKG-FHQTEGYDYNETFSPVVKPTTIRTVLSLALMHQWPIRQFDF 374
DG+ HK R VA+G + YD ++ S V + T LS+AL + + I Q D
Sbjct: 1309 --RDGT---HKARFVARGDIQHPDTYD-SDMQSNTVHHYALMTSLSIALDNDYYITQLDI 1362
Query: 375 NNAFLNGELVEEVFMQQPPGFSKGSSHLVCKLKKALYGLKQAPRAWFDKLSSTLARFGFQ 434
++A+L ++ EE++++ PP G + + +L+K+LYGLKQ+ W++ + S L
Sbjct: 1363 SSAYLYADIKEELYIRPPPHL--GLNDKLLRLRKSLYGLKQSGANWYETIKSYLINC--- 1417
Query: 435 AAKCD----TSLFCKFTKSETLYILVYVDDIIITGSSSTAIFSLIADLNAVFPLK--DLG 488
CD C F K+ + I ++VDD+I+ A +I L + K +LG
Sbjct: 1418 ---CDMQEVRGWSCVF-KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLG 1473
Query: 489 ----SLHY-FLGIEATHTKDGGLILSQTKYINDLLIKAKMDSINPSPTPMTSGLKLSANS 543
+ Y LG+E + + + L K + + K+ +N P L+
Sbjct: 1474 ESDNEIQYDILGLEIKYQRSKYMKLGMEKSLTE-----KLPKLNVPLNPKGKKLRAPGQP 1528
Query: 544 GALFPKPLL-------------------------YRSVVGALHYVTITRPEIAFPLNKVC 578
G + L Y+ L+Y+ I FP +V
Sbjct: 1529 GHYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVL 1588
Query: 579 QFMHSPLEPHWQAVKRILRYLHGTATHGLYLRRPSSMSITAFADSDWGSDPDDRRSTSGM 638
+ ++ W + L + T +P + + A +D+ +G+ P +S G
Sbjct: 1589 DMTYELIQFMWDTRDKQLIWHKNKPT------KPDN-KLVAISDASYGNQP-YYKSQIGN 1640
Query: 639 CIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSELRVPTSPSSLIYC 698
+ G ++ + K ++ S+TEAE +++ I + L L+ EL L+
Sbjct: 1641 IFLLNGKVIGGKSTKASLTCTSTTEAEIHAVSEAIPLLNNLSHLVQELNKKPIIKGLL-T 1699
Query: 699 DNLSVVHLVANPILHT-KTKHFALDLHFVRERVADKQVTVVNIPAPSQVSSTL 750
D+ S + ++ + + + F +R+ V+ + V I ++ +
Sbjct: 1700 DSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVM 1752
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 91.7 bits (226), Expect = 7e-18
Identities = 118/545 (21%), Positives = 235/545 (42%), Gaps = 53/545 (9%)
Query: 244 RAKTGIHRPKAYAASTALSSVPTSAK*ALSIPH---------WHQAMQEEYQALMNNNTW 294
R+K IH A A ++ + T+ + +I + + +A +E L+ TW
Sbjct: 781 RSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTW 840
Query: 295 ELVPLLHGRD-----AIGCKWVFRTKYNSDGSINKHKPRLVAKG-FHQTEGYDYNETFSP 348
+ ++ I ++F K DG+ HK R VA+G + YD S
Sbjct: 841 DTDEYYDRKEIDPKRVINSMFIFNKK--RDGT---HKARFVARGDIQHPDTYDSGMQ-SN 894
Query: 349 VVKPTTIRTVLSLALMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFSKGSSHLVCKLKK 408
V + T LSLAL + + I Q D ++A+L ++ EE++++ PP G + + +LKK
Sbjct: 895 TVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHL--GMNDKLIRLKK 952
Query: 409 ALYGLKQAPRAWFDKLSSTLAR-FGFQAAKCDTSLFCKFTKSETLYILVYVDDIIITGSS 467
+LYGLKQ+ W++ + S L + G + + + +F K+ + I ++VDD+I+
Sbjct: 953 SLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF----KNSQVTICLFVDDMILFSKD 1008
Query: 468 STAIFSLIADLNAVFPLK--DLG----SLHY-FLGIEATHTKDGGLILSQTKYINDLLIK 520
A +I L + K +LG + Y LG+E + + + L + + + K
Sbjct: 1009 LNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPK 1068
Query: 521 AKMDSINPS----PTPMTSGL-----KLSANSGALFPKPLLYRSVVGALHYVTIT-RPEI 570
+ +NP P GL +L + K + ++G YV R ++
Sbjct: 1069 LNV-PLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDL 1127
Query: 571 AFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGL--YLRRPS--SMSITAFADSDWG 626
+ +N + Q + P +++++ T L + +P+ + A +D+ +G
Sbjct: 1128 LYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYG 1187
Query: 627 SDPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSEL 686
+ P +S G + G ++ + K ++ S+TEAE +++ + + L L+ EL
Sbjct: 1188 NQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQEL 1246
Query: 687 RVPTSPSSLIYCDNLSVVHLVANPILHT-KTKHFALDLHFVRERVADKQVTVVNIPAPSQ 745
L+ D+ S + ++ + + + F +R+ V+ + V I
Sbjct: 1247 NKKPIIKGLL-TDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKN 1305
Query: 746 VSSTL 750
++ +
Sbjct: 1306 IADVM 1310
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 91.7 bits (226), Expect = 7e-18
Identities = 113/544 (20%), Positives = 232/544 (41%), Gaps = 51/544 (9%)
Query: 244 RAKTGIHRPKAYAASTALSSVPTSAK*ALSIPH---------WHQAMQEEYQALMNNNTW 294
R+K IH A A ++ + T+ + +I + + +A +E L+ TW
Sbjct: 1208 RSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTW 1267
Query: 295 ELVPLLHGRD-----AIGCKWVFRTKYNSDGSINKHKPRLVAKG-FHQTEGYDYNETFSP 348
+ ++ I ++F K DG+ HK R VA+G + YD S
Sbjct: 1268 DTDEYYDRKEIDPKRVINSMFIFNKK--RDGT---HKARFVARGDIQHPDTYDSGMQ-SN 1321
Query: 349 VVKPTTIRTVLSLALMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFSKGSSHLVCKLKK 408
V + T LSLAL + + I Q D ++A+L ++ EE++++ PP G + + +LKK
Sbjct: 1322 TVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHL--GMNDKLIRLKK 1379
Query: 409 ALYGLKQAPRAWFDKLSSTL-ARFGFQAAKCDTSLFCKFTKSETLYILVYVDDIIITGSS 467
+LYGLKQ+ W++ + S L + G + + + +F K+ + I ++VDD+++ +
Sbjct: 1380 SLYGLKQSGANWYETIKSYLIQQCGMEEVRGWSCVF----KNSQVTICLFVDDMVLFSKN 1435
Query: 468 STAIFSLIADLNAVFPLK--DLG----SLHY-FLGIEATHTKDGGLILSQTKYINDLLIK 520
+ +I L + K +LG + Y LG+E + + + L + + + K
Sbjct: 1436 LNSNKRIIEKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPK 1495
Query: 521 AKMDSINPS----PTPMTSGL-----KLSANSGALFPKPLLYRSVVGALHYVTIT-RPEI 570
+ +NP P GL +L K + ++G YV R ++
Sbjct: 1496 LNV-PLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDL 1554
Query: 571 AFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGLYLRRPSSM----SITAFADSDWG 626
+ +N + Q + P + +++++ T L + + + +D+ +G
Sbjct: 1555 LYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASYG 1614
Query: 627 SDPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSEL 686
+ P +S G + G ++ + K ++ S+TEAE +++ + + L L+ EL
Sbjct: 1615 NQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQEL 1673
Query: 687 RVPTSPSSLIYCDNLSVVHLVANPILHTKTKHFALDLHFVRERVADKQVTVVNIPAPSQV 746
L+ ++ +++N + + F +R+ V+ + V I +
Sbjct: 1674 DKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNI 1733
Query: 747 SSTL 750
+ +
Sbjct: 1734 ADVM 1737
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 91.3 bits (225), Expect = 9e-18
Identities = 115/543 (21%), Positives = 231/543 (42%), Gaps = 49/543 (9%)
Query: 244 RAKTGIHRPKAYAASTALSSVPTSAK*ALSIPH---------WHQAMQEEYQALMNNNTW 294
R+K IH A A ++ + T+ + +I + + +A +E L+ TW
Sbjct: 781 RSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTW 840
Query: 295 ELVPLLHGRDAIGCKWVFRTKY----NSDGSINKHKPRLVAKG-FHQTEGYDYNETFSPV 349
+ + R I K V + + DG+ HK R VA+G + YD S
Sbjct: 841 D-TDKYYDRKEIDPKRVINSMFIFNRKRDGT---HKARFVARGDIQHPDTYDSGMQ-SNT 895
Query: 350 VKPTTIRTVLSLALMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFSKGSSHLVCKLKKA 409
V + T LSLAL + + I Q D ++A+L ++ EE++++ PP G + + +LKK+
Sbjct: 896 VHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHL--GMNDKLIRLKKS 953
Query: 410 LYGLKQAPRAWFDKLSSTLAR-FGFQAAKCDTSLFCKFTKSETLYILVYVDDIIITGSSS 468
LYGLKQ+ W++ + S L + G + + C F S+ + I ++VDD+++ +
Sbjct: 954 LYGLKQSGANWYETIKSYLIKQCGMEEVR---GWSCVFENSQ-VTICLFVDDMVLFSKNL 1009
Query: 469 TAIFSLIADLNAVFPLK--DLG----SLHY-FLGIEATHTKDGGLILSQTKYINDLLIKA 521
+ +I L + K +LG + Y LG+E + + + L + + + K
Sbjct: 1010 NSNKRIIDKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKL 1069
Query: 522 KMDSINPS----PTPMTSGL-----KLSANSGALFPKPLLYRSVVGALHYVTIT-RPEIA 571
+ +NP P GL +L K + ++G YV R ++
Sbjct: 1070 NV-PLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLL 1128
Query: 572 FPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGLYLRRPSSM----SITAFADSDWGS 627
+ +N + Q + P + +++++ T L + + + +D+ +G+
Sbjct: 1129 YYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASYGN 1188
Query: 628 DPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSELR 687
P +S G + G ++ + K ++ S+TEAE +++ + + L L+ EL
Sbjct: 1189 QP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELD 1247
Query: 688 VPTSPSSLIYCDNLSVVHLVANPILHTKTKHFALDLHFVRERVADKQVTVVNIPAPSQVS 747
L+ ++ +++N + + F +R+ V+ + V I ++
Sbjct: 1248 KKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIA 1307
Query: 748 STL 750
+
Sbjct: 1308 DVM 1310
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 89.4 bits (220), Expect = 3e-17
Identities = 117/545 (21%), Positives = 234/545 (42%), Gaps = 53/545 (9%)
Query: 244 RAKTGIHRPKAYAASTALSSVPTSAK*ALSIPH---------WHQAMQEEYQALMNNNTW 294
R+K IH A A ++ + T+ + +I + + +A +E L+ TW
Sbjct: 1208 RSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTW 1267
Query: 295 ELVPLLHGRD-----AIGCKWVFRTKYNSDGSINKHKPRLVAKG-FHQTEGYDYNETFSP 348
+ ++ I ++F K DG+ HK R VA+G + YD S
Sbjct: 1268 DTDEYYDRKEIDPKRVINSMFIFNKK--RDGT---HKARFVARGDIQHPDTYDTGMQ-SN 1321
Query: 349 VVKPTTIRTVLSLALMHQWPIRQFDFNNAFLNGELVEEVFMQQPPGFSKGSSHLVCKLKK 408
V + T LSLAL + + I Q D ++A+L ++ EE++++ PP G + + +LKK
Sbjct: 1322 TVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHL--GMNDKLIRLKK 1379
Query: 409 ALYGLKQAPRAWFDKLSSTLAR-FGFQAAKCDTSLFCKFTKSETLYILVYVDDIIITGSS 467
+ YGLKQ+ W++ + S L + G + + + +F K+ + I ++VDD+I+
Sbjct: 1380 SHYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF----KNSQVTICLFVDDMILFSKD 1435
Query: 468 STAIFSLIADLNAVFPLK--DLG----SLHY-FLGIEATHTKDGGLILSQTKYINDLLIK 520
A +I L + K +LG + Y LG+E + + + L + + + K
Sbjct: 1436 LNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPK 1495
Query: 521 AKMDSINPS----PTPMTSGL-----KLSANSGALFPKPLLYRSVVGALHYVTIT-RPEI 570
+ +NP P GL +L + K + ++G YV R ++
Sbjct: 1496 LNV-PLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDL 1554
Query: 571 AFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGL--YLRRPS--SMSITAFADSDWG 626
+ +N + Q + P +++++ T L + +P+ + A +D+ +G
Sbjct: 1555 LYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYG 1614
Query: 627 SDPDDRRSTSGMCIYIGGNLVSWAAKKQTVVARSSTEAEYRSLALVITEIMWLRSLLSEL 686
+ P +S G + G ++ + K ++ S+TEAE +++ + + L L+ EL
Sbjct: 1615 NQP-YYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQEL 1673
Query: 687 RVPTSPSSLIYCDNLSVVHLVANPILHT-KTKHFALDLHFVRERVADKQVTVVNIPAPSQ 745
L+ D+ S + ++ + + + F +R+ V+ + V I
Sbjct: 1674 NKKPIIKGLL-TDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKN 1732
Query: 746 VSSTL 750
++ +
Sbjct: 1733 IADVM 1737
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 78.6 bits (192), Expect = 6e-14
Identities = 35/78 (44%), Positives = 49/78 (61%)
Query: 562 YVTITRPEIAFPLNKVCQFMHSPLEPHWQAVKRILRYLHGTATHGLYLRRPSSMSITAFA 621
Y+TITRP++ F +N++ QF + QAV ++L Y+ GT GL+ S + + AFA
Sbjct: 2 YLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAFA 61
Query: 622 DSDWGSDPDDRRSTSGMC 639
DSDW S PD RRS +G C
Sbjct: 62 DSDWASCPDTRRSVTGFC 79
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 51.6 bits (122), Expect = 8e-06
Identities = 24/70 (34%), Positives = 38/70 (54%)
Query: 28 HRHITETGLALLAHAHMPLTFWGDAFETAAYLINQLPIPTLDLSTPLQKLYDKKPDYSFL 87
+R I E ++L +P TF DA TA ++IN+ P ++ P + + P YS+L
Sbjct: 2 NRTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSYL 61
Query: 88 KCFGCLCYPH 97
+ FGC+ Y H
Sbjct: 62 RRFGCVAYIH 71
>CPN_DROME (Q02910) Calphotin
Length = 865
Score = 41.2 bits (95), Expect = 0.010
Identities = 29/103 (28%), Positives = 48/103 (46%), Gaps = 6/103 (5%)
Query: 166 LPTTSVIPLLPISNASPNVMPHANSVMPTVATNGSAPVVQTTVSDFSQPA*DSGVVHASA 225
+P SV+ P++ A P V+ + P VA ++P V PA ++ VV +
Sbjct: 227 IPEVSVVATKPLAAAEPVVVAPPATETPVVAPAAASPHVSVA------PAVETAVVAPVS 280
Query: 226 QPTEPPVPPSSNVHSMTTRAKTGIHRPKAYAASTALSSVPTSA 268
TEPPV ++ + T A + AA+T +++ PT A
Sbjct: 281 ASTEPPVAAATLTTAPETPALAPVVAESQVAANTVVATPPTPA 323
>YHU3_YEAST (P38844) Hypothetical 33.4 kDa protein in RPL44B-RPC10
intergenic region precursor
Length = 325
Score = 37.7 bits (86), Expect = 0.12
Identities = 34/128 (26%), Positives = 58/128 (44%), Gaps = 1/128 (0%)
Query: 136 ITRNVLFDETVFPFQFQTSKSSSDILLDSSLPTTSVIPLLPISNASPNVMPHANSVMPTV 195
+T + F + PF Q S SSS SS T+S L ++++S + A +
Sbjct: 154 LTSSSAFVSSTAPFSSQLSSSSSSETSSSSFSTSSSSAPLSLTSSSSSSSSFATIITLAP 213
Query: 196 ATNGSAPVVQTTVSDFSQPA*DSGVVHASAQPTEPPVPPSSNVHSMTTRAKTGIHRPKAY 255
+++ S T S S A +S ++ T + PSS+V + T+RA T + +
Sbjct: 214 SSSKSGNSQLTLASSSSTSAVESSQTGSTIARTTSTLVPSSSVDT-TSRATTSMPLESSS 272
Query: 256 AASTALSS 263
S ++SS
Sbjct: 273 TQSISVSS 280
>DNJ5_CAEEL (Q09446) DnaJ homolog dnj-5
Length = 915
Score = 36.6 bits (83), Expect = 0.26
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Query: 118 YSPMHKGYNCLTTEGKVIITRNVLFDETVFPFQFQTSKSSSDILLDSSLPTTSVIPLLPI 177
YSP H+ + + ++ + L FPF Q SS++IL +L S P LP
Sbjct: 71 YSPRHESWGLPSNANTTLLGSSSLTSSNWFPFTDQKETSSNEILSSLNLAAGSSSPALP- 129
Query: 178 SNASPNVMPH 187
++P+ +PH
Sbjct: 130 --STPSRVPH 137
>MU5A_HUMAN (P98088) Mucin 5AC (Mucin 5 subtype AC,
tracheobronchial) (Tracheobronchial mucin) (TBM) (Major
airway glycoprotein) (Fragment)
Length = 1233
Score = 35.8 bits (81), Expect = 0.44
Identities = 35/117 (29%), Positives = 46/117 (38%), Gaps = 12/117 (10%)
Query: 153 TSKSSSDILLDSSLPTTSVIPLLPISNAS-PNVMPHANSVMPTVATNGSAPVVQTTVSDF 211
TS S S +PTTS S S P P S +PT +T SAP TT +
Sbjct: 26 TSMPSGPGTTPSPVPTTSTTSAPTTSTTSGPGTTP---SPVPTTSTT-SAPTTSTTSAST 81
Query: 212 SQPA*DSGVVHASAQPTEPPVPPSSNVHSMTTRAKTGIHRPKAYAASTALSSVPTSA 268
+ G T PVP +S + TT + T+LS VPT++
Sbjct: 82 ASTTSGPGT-------TPSPVPTTSTTSAPTTSTTSASTASTTSGPGTSLSPVPTTS 131
Score = 33.5 bits (75), Expect = 2.2
Identities = 29/121 (23%), Positives = 50/121 (40%), Gaps = 12/121 (9%)
Query: 153 TSKSSSDILLDSSLPTTSVIPLLPISNASPNVMPHANSVMPTVATNGSAPVVQTTVSDFS 212
TS +S S +PTTS + ++P + S T + G+ P T S S
Sbjct: 50 TSTTSGPGTTPSPVPTTS-------TTSAPTTSTTSASTASTTSGPGTTPSPVPTTSTTS 102
Query: 213 QPA*DSGVVHASAQPTEP-----PVPPSSNVHSMTTRAKTGIHRPKAYAASTALSSVPTS 267
P + ++ + P PVP +S + TT +G + +T+ +S PT+
Sbjct: 103 APTTSTTSASTASTTSGPGTSLSPVPTTSTTSAPTTSTTSGPGTTPSPVPTTSTTSAPTT 162
Query: 268 A 268
+
Sbjct: 163 S 163
>MBD6_HUMAN (Q96DN6) Methyl-CpG binding domain protein 6
Length = 1003
Score = 35.0 bits (79), Expect = 0.75
Identities = 29/106 (27%), Positives = 45/106 (42%), Gaps = 12/106 (11%)
Query: 158 SDILLDSSLPTTSVIPLLPISNASPNVMPHANSVMPTVATNGSAPVVQTTVSDFSQPA*D 217
SD L LP ++ +P P + P V + + G AP V+ + P
Sbjct: 264 SDALTPPPLPPSNNLPAHPGPASQPPVSSATMHLPLVLGPLGGAPTVE---GPGAPPFLA 320
Query: 218 SGVVHASAQPTEPPVPPSSNVHSMTTRAKTGIHRPKAYAASTALSS 263
S ++ A+A+ PP+PP S + RP+A A S + SS
Sbjct: 321 SSLLSAAAKAQHPPLPPPSTLQG---------RRPRAQAPSASHSS 357
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 89,853,713
Number of Sequences: 164201
Number of extensions: 3783646
Number of successful extensions: 9335
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 9216
Number of HSP's gapped (non-prelim): 101
length of query: 768
length of database: 59,974,054
effective HSP length: 118
effective length of query: 650
effective length of database: 40,598,336
effective search space: 26388918400
effective search space used: 26388918400
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0378b.9


