

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0368c.9
(764 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
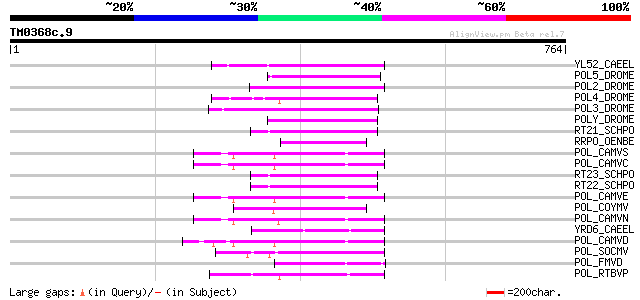
Score E
Sequences producing significant alignments: (bits) Value
YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III 110 1e-23
POL5_DROME (Q8I7P9) Retrovirus-related Pol polyprotein from tran... 101 8e-21
POL2_DROME (P20825) Retrovirus-related Pol polyprotein from tran... 101 8e-21
POL4_DROME (P10394) Retrovirus-related Pol polyprotein from tran... 100 2e-20
POL3_DROME (P04323) Retrovirus-related Pol polyprotein from tran... 96 4e-19
POLY_DROME (P10401) Retrovirus-related Pol polyprotein from tran... 91 9e-18
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 85 8e-16
RRPO_OENBE (P31843) RNA-directed DNA polymerase homolog (Reverse... 85 8e-16
POL_CAMVS (P03554) Enzymatic polyprotein [Contains: Aspartic pro... 83 3e-15
POL_CAMVC (P03555) Enzymatic polyprotein [Contains: Aspartic pro... 82 4e-15
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 81 9e-15
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 81 9e-15
POL_CAMVE (Q02964) Enzymatic polyprotein [Contains: Aspartic pro... 81 1e-14
POL_COYMV (P19199) Putative polyprotein [Contains: Coat protein;... 80 2e-14
POL_CAMVN (Q00962) Enzymatic polyprotein [Contains: Aspartic pro... 79 4e-14
YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II 78 1e-13
POL_CAMVD (P03556) Enzymatic polyprotein [Contains: Aspartic pro... 78 1e-13
POL_SOCMV (P15629) Enzymatic polyprotein [Contains: Aspartic pro... 70 3e-11
POL_FMVD (P09523) Enzymatic polyprotein [Contains: Aspartic prot... 69 4e-11
POL_RTBVP (P27502) Polyprotein (P194 protein) [Contains: Coat pr... 67 1e-10
>YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III
Length = 2186
Score = 110 bits (275), Expect = 1e-23
Identities = 68/238 (28%), Positives = 125/238 (51%), Gaps = 3/238 (1%)
Query: 279 EQRLSKLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQ 338
++++ ++E+ D+FA S ++ G + C + L G + + Q R + + I++
Sbjct: 903 DRKIWDVIEQFQDVFAISDDEL-GRNSGTECV-IELKEGAEPIRQKPRPIPLALKPEIRK 960
Query: 339 EVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLV 398
+ K+L + I E K P W + VV+VKK +G MC+DY +NK +++PLP+I+ +
Sbjct: 961 MIQKMLNQKVIRESKSP-WSSPVVLVKKKDGSIRMCIDYRKVNKVVKNNAHPLPNIEATL 1019
Query: 399 DGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLM 458
+ +L ++ D +G+ QI + K+ TAF + + +PFGL + A +Q M
Sbjct: 1020 QSLAGKKLYTVFDMIAGFWQIPLDEKSKEITAFAIGSELFEWNVLPFGLVISPALFQGTM 1079
Query: 459 DRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLNLKSSLLVYKE 516
+ + D +G VYV D+++ S H +++ EA R+RK M+L + KE
Sbjct: 1080 EEIIGDLLGVCAFVYVDDLLIASKDMEQHLQDVKEALTRIRKSGMKLRASKCHIAKKE 1137
>POL5_DROME (Q8I7P9) Retrovirus-related Pol polyprotein from
transposon opus [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1003
Score = 101 bits (251), Expect = 8e-21
Identities = 57/158 (36%), Positives = 84/158 (53%), Gaps = 4/158 (2%)
Query: 355 PTWLANVVMVKKANGK*P--MCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDA 412
P W+ V K NG+ M VD+ LN D+YP+P I+ + N + + +D
Sbjct: 162 PIWI--VPKKPKPNGEKQYRMVVDFKRLNTVTIPDTYPIPDINATLASLGNAKYFTTLDL 219
Query: 413 YSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEV 472
SG+HQI M SD KTAF T Y + +PFGLKN A +QR++D + + +G+ V
Sbjct: 220 TSGFHQIHMKESDIPKTAFSTLNGKYEFLRLPFGLKNAPAIFQRMIDDILREHIGKVCYV 279
Query: 473 YVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLNLKSS 510
Y+ D+IV S H + L L K N+++NL+ S
Sbjct: 280 YIDDIIVFSEDYDTHWKNLRLVLASLSKANLQVNLEKS 317
>POL2_DROME (P20825) Retrovirus-related Pol polyprotein from
transposon 297 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1059
Score = 101 bits (251), Expect = 8e-21
Identities = 56/186 (30%), Positives = 91/186 (48%)
Query: 331 EKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYP 390
E E +Q+ +N+ L E PTW+ K + +DY LN+ D YP
Sbjct: 221 EVENQVQEMLNQGLIRESNSPYNSPTWVVPKKPDASGANKYRVVIDYRKLNEITIPDRYP 280
Query: 391 LPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNT 450
+P++D+++ + + +D G+HQI+M KTAF T +Y Y MPFGL+N
Sbjct: 281 IPNMDEILGKLGKCQYFTTIDLAKGFHQIEMDEESISKTAFSTKSGHYEYLRMPFGLRNA 340
Query: 451 GATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLNLKSS 510
AT+QR M+ + + ++ VY+ D+I+ S H + F +L N++L L
Sbjct: 341 PATFQRCMNNILRPLLNKHCLVYLDDIIIFSTSLTEHLNSIQLVFTKLADANLKLQLDKC 400
Query: 511 LLVYKE 516
+ KE
Sbjct: 401 EFLKKE 406
>POL4_DROME (P10394) Retrovirus-related Pol polyprotein from
transposon 412 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1237
Score = 99.8 bits (247), Expect = 2e-20
Identities = 64/234 (27%), Positives = 119/234 (50%), Gaps = 10/234 (4%)
Query: 279 EQRLSKLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQ 338
+ +L + E +D+FA + ++ N +L L + T+ R + E+ IQ
Sbjct: 276 KSQLENICSEYIDIFALESEPITV--NNLYKQQLRLKDDEPVYTKNYRSPHSQVEE-IQA 332
Query: 339 EVNKLLAAEFI*EIKYPTWLANVVMVKKANG------K*PMCVDYTDLNKACPKDSYPLP 392
+V KL+ + + E + + +++V K + K + +DY +NK D +PLP
Sbjct: 333 QVQKLIKDKIV-EPSVSQYNSPLLLVPKKSSPNSDKKKWRLVIDYRQINKKLLADKFPLP 391
Query: 393 SIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGA 452
ID ++D + S +D SG+HQI++ +D T+F T+ +Y + +PFGLK
Sbjct: 392 RIDDILDQLGRAKYFSCLDLMSGFHQIELDEGSRDITSFSTSNGSYRFTRLPFGLKIAPN 451
Query: 453 TYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLN 506
++QR+M F+ +Y+ D+IV E + + L E FG+ R++N++L+
Sbjct: 452 SFQRMMTIAFSGIEPSQAFLYMDDLIVIGCSEKHMLKNLTEVFGKCREYNLKLH 505
>POL3_DROME (P04323) Retrovirus-related Pol polyprotein from
transposon 17.6 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1058
Score = 95.9 bits (237), Expect = 4e-19
Identities = 66/236 (27%), Positives = 108/236 (44%), Gaps = 4/236 (1%)
Query: 274 LTESQEQRLSKLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTR--RRMGDE 331
L ++QRL LL++ D+ H+ N H + + L ++ + E
Sbjct: 165 LNNEEKQRLCALLQKYHDI--QYHEGDKLTFTNQTKHTINTKHNLPLYSKYSYPQAYEQE 222
Query: 332 KEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPL 391
E IQ +N+ + P W+ K + +DY LN+ D +P+
Sbjct: 223 VESQIQDMLNQGIIRTSNSPYNSPIWVVPKKQDASGKQKFRIVIDYRKLNEITVGDRHPI 282
Query: 392 PSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTG 451
P++D+++ + +D G+HQI+M KTAF T +Y Y MPFGLKN
Sbjct: 283 PNMDEILGKLGRCNYFTTIDLAKGFHQIEMDPESVSKTAFSTKHGHYEYLRMPFGLKNAP 342
Query: 452 ATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLNL 507
AT+QR M+ + + ++ VY+ D+IV S H + L F +L K N++L L
Sbjct: 343 ATFQRCMNDILRPLLNKHCLVYLDDIIVFSTSLDEHLQSLGLVFEKLAKANLKLQL 398
>POLY_DROME (P10401) Retrovirus-related Pol polyprotein from
transposon gypsy [Contains: Reverse transcriptase (EC
2.7.7.49); Endonuclease]
Length = 1035
Score = 91.3 bits (225), Expect = 9e-18
Identities = 48/153 (31%), Positives = 80/153 (51%), Gaps = 1/153 (0%)
Query: 355 PTWLANVVMVKK-ANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAY 413
PTW+ + N + +D+ LN+ D YP+PSI ++ + + +D
Sbjct: 220 PTWVVDKKGTDAFGNPNKRLVIDFRKLNEKTIPDRYPMPSIPMILANLGKAKFFTTLDLK 279
Query: 414 SGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVY 473
SGYHQI + D++KT+F Y + +PFGL+N + +QR +D V +Q+G+ VY
Sbjct: 280 SGYHQIYLAEHDREKTSFSVNGGKYEFCRLPFGLRNASSIFQRALDDVLREQIGKICYVY 339
Query: 474 VGDMIVKSVLEANHHEELLEAFGRLRKHNMRLN 506
V D+I+ S E++H + L NMR++
Sbjct: 340 VDDVIIFSENESDHVRHIDTVLKCLIDANMRVS 372
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 84.7 bits (208), Expect = 8e-16
Identities = 48/175 (27%), Positives = 87/175 (49%), Gaps = 1/175 (0%)
Query: 332 KEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPL 391
K + + E+N+ L + I E K V+ V K G M VDY LNK + YPL
Sbjct: 424 KMQAMNDEINQGLKSGIIRESKAIN-ACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPL 482
Query: 392 PSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTG 451
P I++L+ + + + +D S YH I++ D+ K AF R + Y MP+G+
Sbjct: 483 PLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISTAP 542
Query: 452 ATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLN 506
A +Q ++ + + ++ Y+ D+++ S E+ H + + + +L+ N+ +N
Sbjct: 543 AHFQYFINTILGEAKESHVVCYMDDILIHSKSESEHVKHVKDVLQKLKNANLIIN 597
>RRPO_OENBE (P31843) RNA-directed DNA polymerase homolog (Reverse
transcriptase homolog)
Length = 142
Score = 84.7 bits (208), Expect = 8e-16
Identities = 39/119 (32%), Positives = 67/119 (55%)
Query: 373 MCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFM 432
MC+DY L K K+ YP+P +D L D + + +D SGY Q+++ D+ KT +
Sbjct: 8 MCIDYRALTKVTIKNKYPIPRVDDLFDRLAQATWFTKLDLRSGYWQVRIAKGDEPKTTCV 67
Query: 433 TARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEEL 491
T ++ ++ MPFGL N AT+ LM+ V + + + VY+ D++V ++ + HE +
Sbjct: 68 TRYGSFEFRVMPFGLTNALATFCNLMNNVLYEYLDHFVVVYLDDLVVYTIYSNSLHEHI 126
>POL_CAMVS (P03554) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 679
Score = 82.8 bits (203), Expect = 3e-15
Identities = 69/271 (25%), Positives = 125/271 (45%), Gaps = 18/271 (6%)
Query: 254 PIEETKELKFGDKVLKIGTRLTESQEQRLSKLLEENLDLFAWSHKDMSGIDPN----FIC 309
P+EE L G ++ + +T+ + Q++ +LLE+ + +DPN ++
Sbjct: 184 PLEEIAILSEGRRLSEEKLFITQQRMQKIEELLEKVCS--------ENPLDPNKTKQWMK 235
Query: 310 HRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVM---VKK 366
+ L K + + + +++ +LL + I K P ++ +K
Sbjct: 236 ASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEK 295
Query: 367 ANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDK 426
GK M V+Y +NKA D+Y LP+ D+L+ ++ S D SG+ Q+ + +
Sbjct: 296 RRGKKRMVVNYKAMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESR 355
Query: 427 DKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNM-EVYVGDMIVKSVLEA 485
TAF + +Y + +PFGLK + +QR MD F +V R VYV D++V S E
Sbjct: 356 PLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAF--RVFRKFCCVYVDDILVFSNNEE 413
Query: 486 NHHEELLEAFGRLRKHNMRLNLKSSLLVYKE 516
+H + + +H + L+ K + L K+
Sbjct: 414 DHLLHVAMILQKCNQHGIILSKKKAQLFKKK 444
>POL_CAMVC (P03555) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 679
Score = 82.4 bits (202), Expect = 4e-15
Identities = 69/271 (25%), Positives = 125/271 (45%), Gaps = 18/271 (6%)
Query: 254 PIEETKELKFGDKVLKIGTRLTESQEQRLSKLLEENLDLFAWSHKDMSGIDPN----FIC 309
P+EE L G ++ + +T+ + Q++ +LLE+ + +DPN ++
Sbjct: 184 PLEEIAILSEGRRLSEEKLFITQQRMQKIEELLEKVCS--------ENPLDPNKTKQWMK 235
Query: 310 HRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVM---VKK 366
+ L K + + + +++ +LL + I K P ++ +K
Sbjct: 236 ASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEK 295
Query: 367 ANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDK 426
GK M V+Y +NKA D+Y LP+ D+L+ ++ S D SG+ Q+ + +
Sbjct: 296 RRGKKRMVVNYKAMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESR 355
Query: 427 DKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNM-EVYVGDMIVKSVLEA 485
TAF + +Y + +PFGLK + +QR MD F +V R VYV D++V S E
Sbjct: 356 PLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAF--RVFRKFCCVYVDDILVFSNNEE 413
Query: 486 NHHEELLEAFGRLRKHNMRLNLKSSLLVYKE 516
+H + + +H + L+ K + L K+
Sbjct: 414 DHLLHVAMILQKCNQHGIILSKKKAQLFKKK 444
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 81.3 bits (199), Expect = 9e-15
Identities = 47/175 (26%), Positives = 87/175 (48%), Gaps = 1/175 (0%)
Query: 332 KEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPL 391
K + + E+N+ L + I E K V+ V K G M VDY LNK + YPL
Sbjct: 424 KMQAMNDEINQGLKSGIIRESKAIN-ACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPL 482
Query: 392 PSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTG 451
P I++L+ + + + +D S YH I++ D+ K AF R + Y MP+G+
Sbjct: 483 PLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISIAP 542
Query: 452 ATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLN 506
A +Q ++ + + ++ Y+ ++++ S E+ H + + + +L+ N+ +N
Sbjct: 543 AHFQYFINTILGEVKESHVVCYMDNILIHSKSESEHVKHVKDVLQKLKNANLIIN 597
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 81.3 bits (199), Expect = 9e-15
Identities = 47/175 (26%), Positives = 87/175 (48%), Gaps = 1/175 (0%)
Query: 332 KEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPL 391
K + + E+N+ L + I E K V+ V K G M VDY LNK + YPL
Sbjct: 424 KMQAMNDEINQGLKSGIIRESKAIN-ACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPL 482
Query: 392 PSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTG 451
P I++L+ + + + +D S YH I++ D+ K AF R + Y MP+G+
Sbjct: 483 PLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISIAP 542
Query: 452 ATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLN 506
A +Q ++ + + ++ Y+ ++++ S E+ H + + + +L+ N+ +N
Sbjct: 543 AHFQYFINTILGEVKESHVVCYMDNILIHSKSESEHVKHVKDVLQKLKNANLIIN 597
>POL_CAMVE (Q02964) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 679
Score = 80.9 bits (198), Expect = 1e-14
Identities = 68/271 (25%), Positives = 125/271 (46%), Gaps = 18/271 (6%)
Query: 254 PIEETKELKFGDKVLKIGTRLTESQEQRLSKLLEENLDLFAWSHKDMSGIDPN----FIC 309
P++E L G ++ + +T+ + Q++ +LLE+ + +DPN ++
Sbjct: 184 PLKEIAILSEGRRLSEEKLFITQQRMQKIEELLEKVCS--------ENPLDPNKTKQWMK 235
Query: 310 HRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVM---VKK 366
+ L K + + + +++ +LL + I K P ++ +K
Sbjct: 236 ASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEK 295
Query: 367 ANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDK 426
GK M V+Y +NKA D+Y LP+ D+L+ ++ S D SG+ Q+ + +
Sbjct: 296 RRGKKRMVVNYKAMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESR 355
Query: 427 DKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNM-EVYVGDMIVKSVLEA 485
TAF + +Y + +PFGLK + +QR MD F +V R VYV D++V S E
Sbjct: 356 PLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAF--RVFRKFCCVYVDDILVFSNNEE 413
Query: 486 NHHEELLEAFGRLRKHNMRLNLKSSLLVYKE 516
+H + + +H + L+ K + L K+
Sbjct: 414 DHLLHVAMILQKCNQHGIILSKKKAQLFKKK 444
>POL_COYMV (P19199) Putative polyprotein [Contains: Coat protein;
Protease (EC 3.4.23.-); Reverse transcriptase (EC
2.7.7.49); Ribonuclease H (EC 3.1.26.4)]
Length = 1886
Score = 80.5 bits (197), Expect = 2e-14
Identities = 55/193 (28%), Positives = 91/193 (46%), Gaps = 11/193 (5%)
Query: 309 CHRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*--EIKYPTWLANV----- 361
C ++P IK++ + + + E+ + +++N LL + I E K+ + V
Sbjct: 1391 CKLNIINPDIKIMGRPIKHVTPGDEEAMTRQINLLLQMKVIRPSESKHRSTAFIVRSGTE 1450
Query: 362 ---VMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQ 418
+ K+ GK M +Y LN+ D Y LP I+ ++ +++ S D SG+ Q
Sbjct: 1451 IDPITGKEKKGKERMVFNYKLLNENTESDQYSLPGINTIISKVGRSKIYSKFDLKSGFWQ 1510
Query: 419 IKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMI 478
+ M TAF+ Y + MPFGLKN A +QR MD VF + + VY+ D++
Sbjct: 1511 VAMEEESVPWTAFLAGNKLYEWLVMPFGLKNAPAIFQRKMDNVFKG-TEKFIAVYIDDIL 1569
Query: 479 VKSVLEANHHEEL 491
V S H + L
Sbjct: 1570 VFSETAEQHSQHL 1582
>POL_CAMVN (Q00962) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 680
Score = 79.0 bits (193), Expect = 4e-14
Identities = 68/271 (25%), Positives = 123/271 (45%), Gaps = 18/271 (6%)
Query: 254 PIEETKELKFGDKVLKIGTRLTESQEQRLSKLLEENLDLFAWSHKDMSGIDPN----FIC 309
P+EE L G ++ + +T+ + Q+ +LLE+ + +DPN ++
Sbjct: 185 PLEEIAILSEGRRLSEEKLFITQQRMQKTEELLEKVCS--------ENPLDPNKTKQWMK 236
Query: 310 HRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKAN- 368
+ L K + + + +++ +LL + I K P ++ +A
Sbjct: 237 ASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEN 296
Query: 369 --GK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDK 426
G M V+Y +NKA D+Y LP+ D+L+ ++ S D SG+ Q+ + +
Sbjct: 297 GRGNKRMVVNYKAMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESR 356
Query: 427 DKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNM-EVYVGDMIVKSVLEA 485
TAF + +Y + +PFGLK + +QR MD F +V R VYV D++V S E
Sbjct: 357 PLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAF--RVFRKFCCVYVDDIVVFSNNEE 414
Query: 486 NHHEELLEAFGRLRKHNMRLNLKSSLLVYKE 516
+H + + +H + L+ K + L K+
Sbjct: 415 DHLLHVAMILQKCNQHGIILSKKKAQLFKKK 445
>YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II
Length = 1268
Score = 77.8 bits (190), Expect = 1e-13
Identities = 49/186 (26%), Positives = 91/186 (48%), Gaps = 6/186 (3%)
Query: 334 KVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKK-ANGK*PMCVDY--TDLNKACPKDSYP 390
+ ++ E+N+L I I Y W A +V++KK GK +C D+ + LN A + +P
Sbjct: 454 EAVETELNRLQEMGVIVPITYAKWAAPIVVIKKKGTGKIRVCADFKCSGLNAALKDEFHP 513
Query: 391 LPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNT 450
LP+ + + + S +D Y Q+++ + T R + Y M FGLK
Sbjct: 514 LPTSEDIFSRLKGT-VYSQIDLKDAYLQVELDEEAQKLAVINTHRGIFKYLRMTFGLKPA 572
Query: 451 GATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLNLKSS 510
A++Q++MD++ + G + VY D+I+ + H + L E F R +++ R++ +
Sbjct: 573 PASFQKIMDKMVSGLTG--VAVYWDDIIISASSIEEHEKILRELFERFKEYGFRVSAEKC 630
Query: 511 LLVYKE 516
K+
Sbjct: 631 AFAQKQ 636
>POL_CAMVD (P03556) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 674
Score = 77.8 bits (190), Expect = 1e-13
Identities = 74/290 (25%), Positives = 130/290 (44%), Gaps = 24/290 (8%)
Query: 239 ENLDSRGEGRVNRPTPIEETKELKFGDKVLKIGTRLTESQ----EQRLSKLLEENLDLFA 294
E++ R + + P I K +L G RL+E + +QR+ K+ EE L+
Sbjct: 162 ESMKKRSKTQQPEPVNISTNKIA-----ILSEGRRLSEEKLFITQQRMQKI-EELLEKVC 215
Query: 295 WSHKDMSGIDPN----FICHRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI* 350
+ +DPN ++ + L K + + + +++ +LL + I
Sbjct: 216 SENP----LDPNKTKQWMKASIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIK 271
Query: 351 EIKYPTWLANVVM---VKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELL 407
K P ++ +K GK M V+Y +NKA D+Y P+ D+L+ ++
Sbjct: 272 PSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNPPNKDELLTLIRGKKIF 331
Query: 408 SLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVG 467
S D SG+ Q+ + + TAF + +Y + +PFGLK + +QR MD F +V
Sbjct: 332 SSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAF--RVF 389
Query: 468 RNM-EVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLNLKSSLLVYKE 516
R VYV D++V S E +H + + +H + L+ K + L K+
Sbjct: 390 RKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILSKKKAQLFKKK 439
>POL_SOCMV (P15629) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 692
Score = 69.7 bits (169), Expect = 3e-11
Identities = 61/243 (25%), Positives = 114/243 (46%), Gaps = 15/243 (6%)
Query: 284 KLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTRR---RMGDEKEKVIQQEV 340
K +EE L+ H + N + + L ++ + T R + D +E ++E
Sbjct: 166 KTIEEQLEEVCSEHPLDETKNKNGLLIEIRLKDPLQEINVTNRIPYTIRDVQE--FKEEC 223
Query: 341 NKLLAAEFI*EIKYPT-----WLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSID 395
LL I E + P ++ N +K+ GK M ++Y +N+A DSY LP D
Sbjct: 224 EDLLKKGLIRESQSPHSAPAFYVENHNEIKR--GKRRMVINYKKMNEATIGDSYKLPRKD 281
Query: 396 KLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAF-MTARANYSYQTMPFGLKNTGATY 454
+++ + S +DA SGY+Q+++H + K TAF + +Y + + FGLK + Y
Sbjct: 282 FILEKIKGSLWFSSLDAKSGYYQLRLHENTKPLTAFSCPPQKHYEWNVLSFGLKQAPSIY 341
Query: 455 QRLMDRVFADQVGRNMEVYVGDMIV-KSVLEANHHEELLEAFGRLRKHNMRLNLKSSLLV 513
QR MD+ + Y+ D+++ + H ++ R+++ + ++ K S L+
Sbjct: 342 QRFMDQSLKG-LEHICLAYIDDILIFTKGSKEQHVNDVRIVLQRIKEKGIIISKKKSKLI 400
Query: 514 YKE 516
+E
Sbjct: 401 QQE 403
>POL_FMVD (P09523) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 666
Score = 69.3 bits (168), Expect = 4e-11
Identities = 43/153 (28%), Positives = 79/153 (51%), Gaps = 2/153 (1%)
Query: 365 KKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSS 424
++ GK M V+Y +N+A DS+ LP++ +L+ + S D SG+ Q+ +
Sbjct: 287 ERRRGKKRMVVNYKAINQATIGDSHNLPNMQELLTLLRGKSIFSSFDCKSGFWQVVLDEE 346
Query: 425 DKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLE 484
+ TAF + ++ ++ +PFGLK + +QR M + + VYV D+IV S E
Sbjct: 347 SQKLTAFTCPQGHFQWKVVPFGLKQAPSIFQRHMQTAL-NGADKFCMVYVDDIIVFSNSE 405
Query: 485 ANHHEELLEAFGRLRKHNMRLNLKSSLLVYKEE 517
+H+ + + K+ + L+ K + L +KE+
Sbjct: 406 LDHYNHVYAVLKIVEKYGIILSKKKANL-FKEK 437
>POL_RTBVP (P27502) Polyprotein (P194 protein) [Contains: Coat
protein; Protease (EC 3.4.23.-); Reverse transcriptase
(EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4)]
Length = 1675
Score = 67.4 bits (163), Expect = 1e-10
Identities = 55/246 (22%), Positives = 106/246 (42%), Gaps = 9/246 (3%)
Query: 276 ESQEQRLSKLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKV 335
E Q L +++E L+ + D++ ++C ++P I + T +KE V
Sbjct: 1141 EIGNQSLITMVKE-LEALGFIGDDITKNRTTWVCDFKIINPDINITCATIPYTPADKE-V 1198
Query: 336 IQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANG-----K*PMCVDYTDLNKACPKDSYP 390
++++ +LL + I + + + + K + +Y LN D +
Sbjct: 1199 FEKQIKELLDNKLIKKADPTCRHRTAAFIVRNHSEEVAQKPRIVYNYKRLNDNMHTDPFN 1258
Query: 391 LPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNT 450
+P +++ + S D +G+H +K+ KD T F + Y++ PFG+ N
Sbjct: 1259 IPHKISMINLIQKANIFSKFDLKAGFHHMKLKDDFKDWTTFTCSEGLYTWNVCPFGIANA 1318
Query: 451 GATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLNLKSS 510
+QR M F D + +Y+ D+++ S E H E L F R+++ L+ K S
Sbjct: 1319 PCAFQRFMQESFGDL--KFALLYIDDILIASNNEKEHIEHLKIFFNRVKEVGCVLSKKKS 1376
Query: 511 LLVYKE 516
+ KE
Sbjct: 1377 KMFLKE 1382
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.337 0.145 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 76,800,684
Number of Sequences: 164201
Number of extensions: 3017094
Number of successful extensions: 11866
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 11827
Number of HSP's gapped (non-prelim): 54
length of query: 764
length of database: 59,974,054
effective HSP length: 118
effective length of query: 646
effective length of database: 40,598,336
effective search space: 26226525056
effective search space used: 26226525056
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0368c.9


