

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0367.1
(481 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
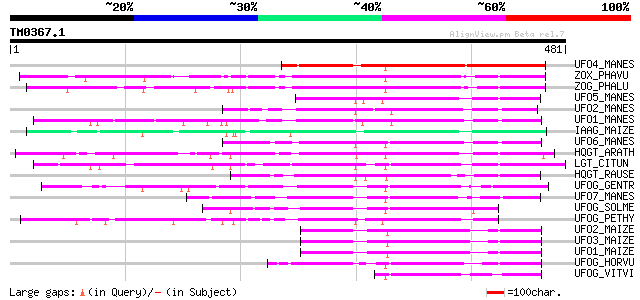
Score E
Sequences producing significant alignments: (bits) Value
UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC 2.4.1... 199 1e-50
ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40) (Ze... 175 3e-43
ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203) (... 168 2e-41
UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC 2.4.1... 153 1e-36
UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC 2.4.1... 152 2e-36
UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC 2.4.1... 144 4e-34
IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (E... 144 5e-34
UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC 2.4.1... 142 1e-33
HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (E... 133 1e-30
LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.21... 130 8e-30
HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.2... 129 2e-29
UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 115 2e-25
UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC 2.4.1... 109 1e-23
UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 104 6e-22
UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 103 8e-22
UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 103 1e-21
UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 101 5e-21
UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 100 8e-21
UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 98 4e-20
UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 77 8e-14
>UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
4) (Fragment)
Length = 241
Score = 199 bits (506), Expect = 1e-50
Identities = 102/235 (43%), Positives = 154/235 (65%), Gaps = 14/235 (5%)
Query: 236 EKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFV 295
+KAERG + SV +T ELL WL +P SV+Y C GS+ ++ QL E+ G+E+++ F+
Sbjct: 7 DKAERGDKASVDNT-ELLKWLDLWEPGSVIYACLGSISGLTSWQLAELGLGLESTNQPFI 65
Query: 296 WVVPAEKRGKEEEEEEEKEKW-LPEGFEEK-----GMILRGWAPQLLILGHPSVGAFLTH 349
WV+ +E E+ E EKW L EG+EE+ +RGW+PQ+LIL HP++GAF TH
Sbjct: 66 WVI------REGEKSEGLEKWILEEGYEERKRKREDFWIRGWSPQVLILSHPAIGAFFTH 119
Query: 350 CGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVV 409
CG NS LE +SAGVPI+ P+ +QFY EKL+ EV G+GV VG E ++ G ++ V
Sbjct: 120 CGWNSTLEGISAGVPIVACPLFAEQFYNEKLVVEVLGIGVSVGVEA-AVTWGLEDKCGAV 178
Query: 410 ISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHL 464
+ +++++KA+ +M G+E E+ RRR E+G +A ++ GGSSY ++ LI ++
Sbjct: 179 MKKEQVKKAIEIVMDKGKEGEERRRRAREIGEMAKRTIEEGGSSYLDMEMLIQYV 233
>ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40)
(Zeatin O-beta-D-xylosyltransferase)
Length = 454
Score = 175 bits (443), Expect = 3e-43
Identities = 144/477 (30%), Positives = 230/477 (48%), Gaps = 62/477 (12%)
Query: 9 PLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLHTV---- 64
P++ + P+L H+I +IA+ + H+ + T N+H H
Sbjct: 17 PVQGHLNPFLQLSHLIAAQNIAVHYVGTVTHIRQAKLRYHNAT-----SNIHFHAFEVPP 71
Query: 65 ---PFPSRQVGLPDG-IESFSSSADLQTTF-KVHQGIKLLREPIQLFMEHHPADCV---- 115
P P+ + P I SF +SA L+ K+ Q + + + L + A
Sbjct: 72 YVSPPPNPEDDFPSHLIPSFEASAHLREPVGKLLQSLSSQAKRVVLINDSLMASVAQDAA 131
Query: 116 ----VADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSSFLISGLPHGDIALN 171
V YCF L T P PL + F +P ++
Sbjct: 132 NFSNVERYCFQVFSALNTAGDFWEQMGKP-PL------------ADFHFPDIPSLQGCIS 178
Query: 172 ASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVR 231
A + LTA E RK ++G + N+ ++G YV+ ER GG + W LGP + +
Sbjct: 179 AQFTDFLTAQNE--FRKF--NNGDIYNTSRVIEG-PYVELLERFNGGKEVWALGPFTPL- 232
Query: 232 GTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASS 291
A EK + S+ + + WL ++PSSV+YV FG+ + +EQ+ E+A G+E S
Sbjct: 233 --AVEKKD-----SIGFSHPCMEWLDKQEPSSVIYVSFGTTTALRDEQIQELATGLEQSK 285
Query: 292 VEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQLLILGHPSVGAFL 347
+F+WV+ +G + E K LPEGFEE+ G+++R WAPQ+ IL H S G F+
Sbjct: 286 QKFIWVLRDADKGDIFDGSEAKRYELPEGFEERVEGMGLVVRDWAPQMEILSHSSTGGFM 345
Query: 348 THCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERER 407
+HCG NS LE+++ GVP+ TW + DQ L+T+V VG+ V ++W E+ +
Sbjct: 346 SHCGWNSCLESLTRGVPMATWAMHSDQPRNAVLVTDVLKVGLIV--KDW-------EQRK 396
Query: 408 VVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHL 464
++S IE AVRRLM +E ++IR+R +L + + ++ GG S + + IAH+
Sbjct: 397 SLVSASVIENAVRRLM-ETKEGDEIRKRAVKLKDEIHRSMDEGGVSRMEMASFIAHI 452
>ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203)
(Trans-zeatin O-beta-D-glucosyltransferase)
Length = 459
Score = 168 bits (426), Expect = 2e-41
Identities = 134/471 (28%), Positives = 222/471 (46%), Gaps = 53/471 (11%)
Query: 15 IPYLAAGHMIPLCDIAILFASRGQHVTIITTPSN----TTTIDSSLPNLHLHTVPFPSRQ 70
IP+ A GH+ ++ L ++ V + T ++ T ++ N+H H P
Sbjct: 19 IPFPAQGHLNQFLHLSRLIVAQNIPVHYVGTVTHIRQATLRYNNPTSNIHFHAFQVPPFV 78
Query: 71 VGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADC---VVADYCFPWIDDL 127
P+ + F S H LREP+ ++ + VV + D L
Sbjct: 79 SPPPNPEDDFPSHLIPSFEASAH-----LREPVGKLLQSLSSQAKRVVVIN------DSL 127
Query: 128 TTELHIPRLSFNPSPLFALCAMRAKRGSSSFL--ISGLPHGDIALNASPPEALTACVEPL 185
+ + + + + A S F + P GD P +L C+
Sbjct: 128 MASVAQDAANISNVENYTFHSFSAFNTSGDFWEEMGKPPVGDFHFPEFP--SLEGCIAAQ 185
Query: 186 LRK------ELK--SHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEK 237
+ E + ++G + N+ ++G YV+ E GG K W LGP + + A EK
Sbjct: 186 FKGFRTAQYEFRKFNNGDIYNTSRVIEG-PYVELLELFNGGKKVWALGPFNPL---AVEK 241
Query: 238 AERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWV 297
+ S+ + WL ++PSSV+Y+ FG+ + +EQ+ ++A G+E S +F+WV
Sbjct: 242 KD-----SIGFRHPCMEWLDKQEPSSVIYISFGTTTALRDEQIQQIATGLEQSKQKFIWV 296
Query: 298 VPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQLLILGHPSVGAFLTHCGSN 353
+ +G E K LP+GFEE+ G+++R WAPQL IL H S G F++HCG N
Sbjct: 297 LREADKGDIFAGSEAKRYELPKGFEERVEGMGLVVRDWAPQLEILSHSSTGGFMSHCGWN 356
Query: 354 SALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRD 413
S LE+++ GVPI TWP+ DQ L+TEV VG+ V ++W+ + ++S
Sbjct: 357 SCLESITMGVPIATWPMHSDQPRNAVLVTEVLKVGLVV--KDWA-------QRNSLVSAS 407
Query: 414 KIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHL 464
+E VRRLM +E +++R+R L N + ++ GG S+ + + IAH+
Sbjct: 408 VVENGVRRLM-ETKEGDEMRQRAVRLKNAIHRSMDEGGVSHMEMGSFIAHI 457
>UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
5)
Length = 487
Score = 153 bits (386), Expect = 1e-36
Identities = 88/224 (39%), Positives = 129/224 (57%), Gaps = 22/224 (9%)
Query: 248 STDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVV--PAEKRGK 305
S ELL WL + SV+YV FGS + S EQ+ E+A G+E S F+WVV P K G
Sbjct: 257 SNCELLDWLDQQPKESVVYVSFGSGGTLSLEQMIELAWGLERSQQRFIWVVRQPTVKTGD 316
Query: 306 -----EEEEEEEKEKWLPEGF----EEKGMILRGWAPQLLILGHPSVGAFLTHCGSNSAL 356
+ + ++ + PEGF + G+++ W+PQ+ I+ HPSVG FL+HCG NS L
Sbjct: 317 AAFFTQGDGADDMSGYFPEGFLTRIQNVGLVVPQWSPQIHIMSHPSVGVFLSHCGWNSVL 376
Query: 357 EAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIE 416
E+++AGVPII WP+ +Q L+TE GV V + + V+ R++IE
Sbjct: 377 ESITAGVPIIAWPIYAEQRMNATLLTEELGVAVR----------PKNLPAKEVVKREEIE 426
Query: 417 KAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTAL 460
+ +RR+M EE +IR+RV EL + A+ GGSS+ ++AL
Sbjct: 427 RMIRRIM-VDEEGSEIRKRVRELKDSGEKALNEGGSSFNYMSAL 469
>UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
2) (Fragment)
Length = 346
Score = 152 bits (383), Expect = 2e-36
Identities = 90/280 (32%), Positives = 158/280 (56%), Gaps = 34/280 (12%)
Query: 185 LLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEK 244
+ +K ++ G+++N+F+EL+ ++ ++ H +GP V+ +
Sbjct: 84 IAKKFRQAKGIIVNTFLELESRA-IESFKVPPLYH----VGPILDVKSDGRN-------- 130
Query: 245 SVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVV----PA 300
+ E++ WL + SV+++CFGSM SFS +QL E+A +E S F+W + P
Sbjct: 131 ---THPEIMQWLDDQPEGSVVFLCFGSMGSFSEDQLKEIAYALENSGHRFLWSIRRPPPP 187
Query: 301 EKRGKEEEEEEEKEKWLPEGFEEKGMILR---GWAPQLLILGHPSVGAFLTHCGSNSALE 357
+K + E+ ++ LPEGF E+ + + GWAPQ+ +L HP++G F++HCG NS LE
Sbjct: 188 DKIASPTDYEDPRDV-LPEGFLERTVAVGKVIGWAPQVAVLAHPAIGGFVSHCGWNSVLE 246
Query: 358 AVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEK 417
++ GVPI TWP+ +Q + M G+GVE+ G R+ ++++ DKIE+
Sbjct: 247 SLWFGVPIATWPMYAEQQFNAFEMVVELGLGVEIDM-------GYRKESGIIVNSDKIER 299
Query: 418 AVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNL 457
A+R+LM E +++ R++V E+ + A+ GGSS+ +L
Sbjct: 300 AIRKLM---ENSDEKRKKVKEMREKSKMALIDGGSSFISL 336
>UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
1)
Length = 449
Score = 144 bits (364), Expect = 4e-34
Identities = 131/472 (27%), Positives = 212/472 (44%), Gaps = 60/472 (12%)
Query: 21 GHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLHTVPFPS---RQVGLP--- 74
GH++ + A L SR ++I N + + S + N + S R + LP
Sbjct: 2 GHLVSAVETAKLLLSRCHSLSITVLIFNNSVVTSKVHNYVDSQIASSSNRLRFIYLPRDE 61
Query: 75 DGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFPWIDDLTTELHIP 134
GI SFSS + Q V + + + E +V +C ID + E +P
Sbjct: 62 TGISSFSSLIEKQKPH-VKESVMKITEFGSSVESPRLVGFIVDMFCTAMID-VANEFGVP 119
Query: 135 RLSFNPSPLFALCAM---RAKRGSSSFLISGLPHGDIAL------NASPPEALTACV--- 182
F S L M + +F + D L N+ P +A+ +
Sbjct: 120 SYIFYTSGAAFLNFMLHVQKIHDEENFNPTEFNASDGELQVPGLVNSFPSKAMPTAILSK 179
Query: 183 ---EPLL---RKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKE 236
PLL R+ ++ GV+IN+F EL+ H + + +GP VR +
Sbjct: 180 QWFPPLLENTRRYGEAKGVIINTFFELESHAIESFKDPPI-----YPVGPILDVRSNGRN 234
Query: 237 KAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVW 296
+ E++ WL + PSSV+++CFGS SFS +Q+ E+AC +E S F+W
Sbjct: 235 -----------TNQEIMQWLDDQPPSSVVFLCFGSNGSFSKDQVKEIACALEDSGHRFLW 283
Query: 297 VVPAEKRG----KEEEEEEEKEKWLPEGFEEKGMILR---GWAPQLLILGHPSVGAFLTH 349
+ A+ R + + E+ ++ LPEGF E+ + GWAPQ+ +L HP+ G ++H
Sbjct: 284 SL-ADHRAPGFLESPSDYEDLQEVLPEGFLERTSGIEKVIGWAPQVAVLAHPATGGLVSH 342
Query: 350 CGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVV 409
G NS LE++ GVP+ TWP+ +Q + M G+ VE+ + R +
Sbjct: 343 SGWNSILESIWFGVPVATWPMYAEQQFNAFQMVIELGLAVEIKMD-------YRNDSGEI 395
Query: 410 ISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALI 461
+ D+IE+ +R LM + R++V E+ + A+ GGSSY L LI
Sbjct: 396 VKCDQIERGIRCLM---KHDSDRRKKVKEMSEKSRGALMEGGSSYCWLDNLI 444
>IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (EC
2.4.1.121) (IAA-Glu synthetase) ((Uridine
5'-diphosphate-glucose:indol-3-ylacetyl)-beta-D-glucosyl
transferase)
Length = 471
Score = 144 bits (363), Expect = 5e-34
Identities = 139/489 (28%), Positives = 196/489 (39%), Gaps = 69/489 (14%)
Query: 15 IPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLHTVPFPSRQVGLP 74
+P+ GHM P+ A AS+G T++TT T D + H + G
Sbjct: 8 VPFPGQGHMNPMVQFAKRLASKGVATTLVTTRFIQRTAD-----VDAHPAMVEAISDGHD 62
Query: 75 DGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPAD---CVVADYCFPWIDDLTTEL 131
+G F+S+A + + + AD CVV D W+ + +
Sbjct: 63 EG--GFASAAGVAEYLEKQAAAASASLASLVEARASSADAFTCVVYDSYEDWVLPVARRM 120
Query: 132 HIPRLSFNPSPLFALCAMRAKRGSSSFLISGLPHGDIALNASPPEALTACVEPLL----- 186
+P + F+ CA+ A S +P G A + A E L
Sbjct: 121 GLPAVPFSTQS----CAVSAVYYHFSQGRLAVPPGAAADGSDGGAGAAALSEAFLGLPEM 176
Query: 187 -RKELKS----HG---------------------VLINSFVELDGHEYVKHYERTTGGHK 220
R EL S HG VL NSF EL+ + T K
Sbjct: 177 ERSELPSFVFDHGPYPTIAMQAIKQFAHAGKDDWVLFNSFEELE----TEVLAGLTKYLK 232
Query: 221 AWLLGPASLVRGTAKEKAERGH----EKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFS 276
A +GP + + G V D WL TK SV YV FGS+ S
Sbjct: 233 ARAIGPCVPLPTAGRTAGANGRITYGANLVKPEDACTKWLDTKPDRSVAYVSFGSLASLG 292
Query: 277 NEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEKGMILRGWAPQLL 336
N Q E+A G+ A+ F+WVV A +E + L E ++ W PQL
Sbjct: 293 NAQKEELARGLLAAGKPFLWVVRA------SDEHQVPRYLLAEATATGAAMVVPWCPQLD 346
Query: 337 ILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEW 396
+L HP+VG F+THCG NS LEA+S GVP++ + DQ + + G GV
Sbjct: 347 VLAHPAVGCFVTHCGWNSTLEALSFGVPMVAMALWTDQPTNARNVELAWGAGVR------ 400
Query: 397 SMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQN 456
RR+ V R ++E+ VR +M GGE A R+ E + A AAV PGGSS +N
Sbjct: 401 ----ARRDAGAGVFLRGEVERCVRAVMDGGEAASAARKAAGEWRDRARAAVAPGGSSDRN 456
Query: 457 LTALIAHLK 465
L + ++
Sbjct: 457 LDEFVQFVR 465
>UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
6) (Fragment)
Length = 394
Score = 142 bits (359), Expect = 1e-33
Identities = 87/284 (30%), Positives = 154/284 (53%), Gaps = 26/284 (9%)
Query: 185 LLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEK 244
++R ++ G+++N+F+EL+ H + + + +GP + + ++ + G E
Sbjct: 122 IIRGLREAKGIMVNTFMELESHALNSLKDDQSKIPPIYPVGP---ILKLSNQENDVGPEG 178
Query: 245 SVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVV---PAE 301
S E++ WL + PSSV+++CFGSM F +Q E+AC +E S F+W + P +
Sbjct: 179 S-----EIIEWLDDQPPSSVVFLCFGSMGGFDMDQAKEIACALEQSRHRFLWSLRRPPPK 233
Query: 302 KRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQLLILGHPSVGAFLTHCGSNSALE 357
+ + + E ++ LP GF E+ G ++ GWAPQ+ IL HP++G F++HCG NS LE
Sbjct: 234 GKIETSTDYENLQEILPVGFSERTAGMGKVV-GWAPQVAILEHPAIGGFVSHCGWNSILE 292
Query: 358 AVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEK 417
++ VPI TWP+ +Q + M G+ VE+ + ++ +++S D IE+
Sbjct: 293 SIWFSVPIATWPLYAEQQFNAFTMVTELGLAVEIKMD-------YKKESEIILSADDIER 345
Query: 418 AVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALI 461
++ +M E +IR+RV E+ + + A+ SS L LI
Sbjct: 346 GIKCVM---EHHSEIRKRVKEMSDKSRKALMDDESSSFWLDRLI 386
>HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (EC
2.4.1.218) (Arbutin synthase)
Length = 480
Score = 133 bits (334), Expect = 1e-30
Identities = 138/517 (26%), Positives = 224/517 (42%), Gaps = 90/517 (17%)
Query: 6 EQRPLKLYFIPYLAAGHMIPLCDIAI-LFASRGQHVTIITT----PSNTT-TIDSSLPNL 59
E + + IP GH+IPL + A L G VT + PS T+ SLP+
Sbjct: 3 ESKTPHVAIIPSPGMGHLIPLVEFAKRLVHLHGLTVTFVIAGEGPPSKAQRTVLDSLPS- 61
Query: 60 HLHTVPFPSRQVGLPDGIESFSSSADLQT--TFKVHQGIKLLREPIQLFMEH-HPADCVV 116
+ +V P P + SSS +++ + V + LR+ F+E +V
Sbjct: 62 SISSVFLP------PVDLTDLSSSTRIESRISLTVTRSNPELRKVFDSFVEGGRLPTALV 115
Query: 117 ADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSSFLISGLPHGDIALNA---- 172
D D+ E H+P F P+ L SF + LP D ++
Sbjct: 116 VDLFGTDAFDVAVEFHVPPYIFYPTTANVL----------SFFLH-LPKLDETVSCEFRE 164
Query: 173 -SPPEALTACVEPLLRKEL-----------------------KSHGVLINSFVELDGHEY 208
+ P L CV P+ K+ ++ G+L+N+F EL+ +
Sbjct: 165 LTEPLMLPGCV-PVAGKDFLDPAQDRKDDAYKWLLHNTKRYKEAEGILVNTFFELEPNAI 223
Query: 209 VKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVC 268
E + +GP + K++A++ E E L WL + SVLYV
Sbjct: 224 KALQEPGLDKPPVYPVGPLVNI---GKQEAKQTEES------ECLKWLDNQPLGSVLYVS 274
Query: 269 FGSMCSFSNEQLHEMACGIEASSVEFVWVVP-----AEKRGKEEEEEEEKEKWLPEGFEE 323
FGS + + EQL+E+A G+ S F+WV+ A + + + +LP GF E
Sbjct: 275 FGSGGTLTCEQLNELALGLADSEQRFLWVIRSPSGIANSSYFDSHSQTDPLTFLPPGFLE 334
Query: 324 K----GMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEK 379
+ G ++ WAPQ +L HPS G FLTHCG NS LE+V +G+P+I WP+ +Q
Sbjct: 335 RTKKRGFVIPFWAPQAQVLAHPSTGGFLTHCGWNSTLESVVSGIPLIAWPLYAEQKMNAV 394
Query: 380 LMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHEL 439
L++E + R + ++ R+++ + V+ LM GEE + +R ++ EL
Sbjct: 395 LLSEDIRAALR-----------PRAGDDGLVRREEVARVVKGLM-EGEEGKGVRNKMKEL 442
Query: 440 GNLANAAVQPGGSSYQNLTALI----AHLKTLRDSNS 472
A ++ G+S + L+ + AH K L + +
Sbjct: 443 KEAACRVLKDDGTSTKALSLVALKWKAHKKELEQNGN 479
>LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.210)
(Limonoid glucosyltransferase) (Limonoid GTase) (LGTase)
Length = 511
Score = 130 bits (327), Expect = 8e-30
Identities = 129/497 (25%), Positives = 207/497 (40%), Gaps = 69/497 (13%)
Query: 21 GHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLHTVPFPS---RQVGLPDG- 76
GH+ PL + L AS+G +T+ TTP + N P R DG
Sbjct: 18 GHVNPLLRLGRLLASKGFFLTL-TTPESFGKQMRKAGNFTYEPTPVGDGFIRFEFFEDGW 76
Query: 77 ---------IESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFPWIDDL 127
++ + + +L + + IK E E+ P C++ + PW+ D+
Sbjct: 77 DEDDPRREDLDQYMAQLELIGKQVIPKIIKKSAE------EYRPVSCLINNPFIPWVSDV 130
Query: 128 TTELHIPRLSFNPSPLFALCAM-RAKRGSSSFLISGLPHGDIALNASP-------PEALT 179
L +P A G F P D+ L P P L
Sbjct: 131 AESLGLPSAMLWVQSCACFAAYYHYFHGLVPFPSEKEPEIDVQLPCMPLLKHDEMPSFLH 190
Query: 180 ACVE-PLLRKEL--------KSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLV 230
P LR+ + K +L+++F EL+ E + + K + P +
Sbjct: 191 PSTPYPFLRRAILGQYENLGKPFCILLDTFYELE-KEIIDYMA------KICPIKPVGPL 243
Query: 231 RGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEAS 290
K + + DE + WL K PSSV+Y+ FG++ EQ+ E+ + S
Sbjct: 244 FKNPKAPTLTVRD-DCMKPDECIDWLDKKPPSSVVYISFGTVVYLKQEQVEEIGYALLNS 302
Query: 291 SVEFVWVV--PAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQLLILGHPSVG 344
+ F+WV+ P E G + + LP+GF EK G +++ W+PQ +L HPSV
Sbjct: 303 GISFLWVMKPPPEDSGVKIVD-------LPDGFLEKVGDKGKVVQ-WSPQEKVLAHPSVA 354
Query: 345 AFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRE 404
F+THCG NS +E++++GVP+IT+P GDQ + +V G+ + R E
Sbjct: 355 CFVTHCGWNSTMESLASGVPVITFPQWGDQVTDAMYLCDVFKTGLRL---------CRGE 405
Query: 405 RERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHL 464
E +ISRD++EK + G +A + + A AV GGSS +N+ A + +
Sbjct: 406 AENRIISRDEVEKCLLEAT-AGPKAVALEENALKWKKEAEEAVADGGSSDRNIQAFVDEV 464
Query: 465 KTLRDSNSVSSSMPPLH 481
+ SS +H
Sbjct: 465 RRTSVEIITSSKSKSIH 481
>HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.218)
(Arbutin synthase)
Length = 470
Score = 129 bits (324), Expect = 2e-29
Identities = 92/279 (32%), Positives = 140/279 (49%), Gaps = 33/279 (11%)
Query: 192 SHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDE 251
+ G+++N+F +L+ E G + +GP L+R + K V E
Sbjct: 204 AEGIMVNTFNDLEPGPLKALQEEDQGKPPVYPIGP--LIRADSSSK---------VDDCE 252
Query: 252 LLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVV--PAEKRGKEE-- 307
L WL + SVL++ FGS + S+ Q E+A G+E S F+WVV P +K
Sbjct: 253 CLKWLDDQPRGSVLFISFGSGGAVSHNQFIELALGLEMSEQRFLWVVRSPNDKIANATYF 312
Query: 308 --EEEEEKEKWLPEGFEEKG----MILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSA 361
+ + + +LPEGF E+ +++ WAPQ IL H S G FLTHCG NS LE+V
Sbjct: 313 SIQNQNDALAYLPEGFLERTKGRCLLVPSWAPQTEILSHGSTGGFLTHCGWNSILESVVN 372
Query: 362 GVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRR 421
GVP+I WP+ +Q ++TE G++V + E +I R +I AV+
Sbjct: 373 GVPLIAWPLYAEQKMNAVMLTE----GLKVALRP-------KAGENGLIGRVEIANAVKG 421
Query: 422 LMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTAL 460
LM GEE ++ R + +L + A+ A+ GSS + L L
Sbjct: 422 LM-EGEEGKKFRSTMKDLKDAASRALSDDGSSTKALAEL 459
>UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 453
Score = 115 bits (288), Expect = 2e-25
Identities = 121/484 (25%), Positives = 208/484 (42%), Gaps = 110/484 (22%)
Query: 28 DIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLHTVPFPSRQVGLPDGIESFSSSADLQ 87
DI F S +T I +P+N +I S++ + G P+G FS +
Sbjct: 34 DIIFSFFSTSSSITTIFSPTNLISIGSNIKPYAVWD--------GSPEGFV-FSGNP--- 81
Query: 88 TTFKVHQGIKLLREPIQLFMEHHPAD-----------------CVVADYCFPWIDDLTTE 130
REPI+ F+ P + C++ D + D + +
Sbjct: 82 ------------REPIEYFLNAAPDNFDKAMKKAVEDTGVNISCLLTDAFLWFAADFSEK 129
Query: 131 LHIPRLSFNPSPLFALC-----------------AMRAKR------GSSSFLISGLPHGD 167
+ +P + + +LC A +A++ G S+ S LP
Sbjct: 130 IGVPWIPVWTAASCSLCLHVYTDEIRSRFAEFDIAEKAEKTIDFIPGLSAISFSDLPEEL 189
Query: 168 IALNASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPA 227
I ++ ALT + + K K+ V +NSF E+D + ++ R+T +GP
Sbjct: 190 IMEDSQSIFALT--LHNMGLKLHKATAVAVNSFEEID--PIITNHLRSTNQLNILNIGPL 245
Query: 228 SLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGI 287
+ + + +E L WL T+K SSV+Y+ FG++ + ++ +A +
Sbjct: 246 QTLSSSIPPE-----------DNECLKWLQTQKESSVVYLSFGTVINPPPNEMAALASTL 294
Query: 288 EASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQLLILGHPSV 343
E+ + F+W + +E K LPE F ++ G I+ WAPQL +L +P++
Sbjct: 295 ESRKIPFLWSL-----------RDEARKHLPENFIDRTSTFGKIV-SWAPQLHVLENPAI 342
Query: 344 GAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRR 403
G F+THCG NS LE++ VP+I P GDQ +++ +V +GV V GG
Sbjct: 343 GVFVTHCGWNSTLESIFCRVPVIGRPFFGDQKVNARMVEDVWKIGVGV------KGG--- 393
Query: 404 ERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAH 463
V + D+ + V L+ ++ +++R+ V L A AV+ GSS +N +L+A
Sbjct: 394 -----VFTEDETTR-VLELVLFSDKGKEMRQNVGRLKEKAKDAVKANGSSTRNFESLLAA 447
Query: 464 LKTL 467
L
Sbjct: 448 FNKL 451
>UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
7) (Fragment)
Length = 287
Score = 109 bits (273), Expect = 1e-23
Identities = 91/311 (29%), Positives = 147/311 (47%), Gaps = 46/311 (14%)
Query: 154 GSSSFLISGLPHGDIALNASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYE 213
G S I LP G + N + + + R ++ VL+NSF ELD +
Sbjct: 7 GMSKIQIRDLPEGVLFGNLE--SLFSQMLHNMGRMLPRAAAVLMNSFEELDPTIVS---D 61
Query: 214 RTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMC 273
+ + +GP +LV V T ++WL +KP+SV Y+ FGS+
Sbjct: 62 LNSKFNNILCIGPFNLV----------SPPPPVPDTYGCMAWLDKQKPASVAYISFGSVA 111
Query: 274 SFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILR 329
+ +L +A +EAS V F+W + ++ + LP GF ++ G++L
Sbjct: 112 TPPPHELVALAEALEASKVPFLWSL-----------KDHSKVHLPNGFLDRTKSHGIVL- 159
Query: 330 GWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGV 389
WAPQ+ IL H ++G F+THCG NS LE++ GVP+I P GDQ +++ +V +G+
Sbjct: 160 SWAPQVEILEHAALGVFVTHCGWNSILESIVGGVPMICRPFFGDQRLNGRMVEDVWEIGL 219
Query: 390 EVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQP 449
M GG V+ I+ + L+ G + +++R + L LA A +P
Sbjct: 220 -------LMDGG------VLTKNGAIDGLNQILLQG--KGKKMRENIKRLKELAKGATEP 264
Query: 450 GGSSYQNLTAL 460
GSS ++ T L
Sbjct: 265 KGSSSKSFTEL 275
>UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 433
Score = 104 bits (259), Expect = 6e-22
Identities = 81/281 (28%), Positives = 131/281 (45%), Gaps = 50/281 (17%)
Query: 168 IALNASPPEALTACVEPLLRKEL--------KSHGVLINSFVELDGHEYVKHYERTTGGH 219
+++N PPE +E + L K+ V++NSF ELD + +
Sbjct: 169 LSINDIPPEVTAEDLEGPMSSMLYNMALNLHKADAVVLNSFQELDRDPLINK-DLQKNLQ 227
Query: 220 KAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQ 279
K + +GP LV ++++ E G + WL +K SV+Y+ FG++ + +
Sbjct: 228 KVFNIGP--LVLQSSRKLDESG----------CIQWLDKQKEKSVVYLSFGTVTTLPPNE 275
Query: 280 LHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQL 335
+ +A +E F+W + K LP+GF E+ G I+ WAPQL
Sbjct: 276 IGSIAEALETKKTPFIWSL-----------RNNGVKNLPKGFLERTKEFGKIV-SWAPQL 323
Query: 336 LILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEE 395
IL H SVG F+THCG NS LE +S GVP+I P GDQ +++ V +G+++
Sbjct: 324 EILAHKSVGVFVTHCGWNSILEGISFGVPMICRPFFGDQKLNSRMVESVWEIGLQIEGGI 383
Query: 396 WSMGG-------------GRRERERVVISRDKIEKAVRRLM 423
++ G G+ RE V ++K +AV ++M
Sbjct: 384 FTKSGIISALDTFFNEEKGKILRENVEGLKEKALEAVNQMM 424
>UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Anthocyanin rhamnosyl transferase)
Length = 473
Score = 103 bits (258), Expect = 8e-22
Identities = 108/453 (23%), Positives = 188/453 (40%), Gaps = 78/453 (17%)
Query: 10 LKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSL---PNLHLHTVPF 66
L + P+ A GH+ P +A +S G V+ T N + + S L P H+ +
Sbjct: 12 LHVVMFPFFAFGHISPFVQLANKLSSYGVKVSFFTASGNASRVKSMLNSAPTTHIVPLTL 71
Query: 67 PSRQVGLPDGIESFS----SSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFP 122
P + GLP G ES + +SA+L + + L++ I+ + H V+ D+
Sbjct: 72 PHVE-GLPPGAESTAELTPASAEL-----LKVALDLMQPQIKTLLSHLKPHFVLFDFAQE 125
Query: 123 WIDDLTTELHIPRLSFNP----SPLFALCAMRAKRGSSSFLISGLPHGDIALNASPPEAL 178
W+ + L I + ++ S F C R + + + P+
Sbjct: 126 WLPKMANGLGIKTVYYSVVVALSTAFLTCPARVLEPKKYPSLEDMKKPPLGF----PQTS 181
Query: 179 TACVE-----------------PLLRKELKS-----HGVLINSFVELDGHEYVKHYERTT 216
V P L ++S +L + +++G Y+K+ E
Sbjct: 182 VTSVRTFEARDFLYVFKSFHNGPTLYDRIQSGLRGCSAILAKTCSQMEG-PYIKYVEAQF 240
Query: 217 GGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFS 276
+L+GP +V K E ++ +WL+ + +V+Y FGS +
Sbjct: 241 N-KPVFLIGP--VVPDPPSGKLE----------EKWATWLNKFEGGTVIYCSFGSETFLT 287
Query: 277 NEQLHEMACGIEASSVEFVWVV--PAEKRGKEEEEEEEKEKWLPEGF----EEKGMILRG 330
++Q+ E+A G+E + + F V+ PA + E + LPEGF ++KG+I G
Sbjct: 288 DDQVKELALGLEQTGLPFFLVLNFPANV-----DVSAELNRALPEGFLERVKDKGIIHSG 342
Query: 331 WAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVE 390
W Q IL H SVG ++ H G +S +EA+ ++ P GDQ KL++ GVE
Sbjct: 343 WVQQQNILAHSSVGCYVCHAGFSSVIEALVNDCQVVMLPQKGDQILNAKLVSGDMEAGVE 402
Query: 391 VGAEEWSMGGGRRERERVVISRDKIEKAVRRLM 423
+ R E ++ I++AV ++M
Sbjct: 403 I----------NRRDEDGYFGKEDIKEAVEKVM 425
>UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-Mc2 allele)
Length = 471
Score = 103 bits (256), Expect = 1e-21
Identities = 70/214 (32%), Positives = 98/214 (45%), Gaps = 32/214 (14%)
Query: 253 LSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEE 312
L+WL + V YV FG++ ++L E+A G+EAS+ F+W + E+
Sbjct: 279 LAWLGRQPARGVAYVSFGTVACPRPDELRELAAGLEASAAPFLWSL-----------RED 327
Query: 313 KEKWLPEGFEEKGM-----ILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIIT 367
LP GF ++ ++ WAPQ+ +L HPSVGAF+TH G S LE VS+GVP+
Sbjct: 328 SWTLLPPGFLDRAAGTGSGLVVPWAPQVAVLRHPSVGAFVTHAGWASVLEGVSSGVPMAC 387
Query: 368 WPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGE 427
P GDQ + + V G G S G + AV L+ GE
Sbjct: 388 RPFFGDQRMNARSVAHVWGFGAAFEGAMTSAG---------------VAAAVEELL-RGE 431
Query: 428 EAEQIRRRVHELGNLANAAVQPGGSSYQNLTALI 461
E +R R EL L A PGG +N +
Sbjct: 432 EGAGMRARAKELQALVAEAFGPGGECRKNFDRFV 465
>UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-W22 allele)
Length = 471
Score = 101 bits (251), Expect = 5e-21
Identities = 69/214 (32%), Positives = 97/214 (45%), Gaps = 32/214 (14%)
Query: 253 LSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEE 312
L+WL + V YV FG++ ++L E+A G+EAS F+W + E+
Sbjct: 279 LAWLGRQPARGVAYVSFGTVACPRPDELRELAAGLEASGAPFLWSL-----------RED 327
Query: 313 KEKWLPEGFEEKGM-----ILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIIT 367
LP GF ++ ++ WAPQ+ +L HPSVGAF+TH G S LE VS+GVP+
Sbjct: 328 SWTLLPPGFLDRAAGTGSGLVVPWAPQVAVLRHPSVGAFVTHAGWASVLEGVSSGVPMAC 387
Query: 368 WPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGE 427
P GDQ + + V G G S G + AV L+ GE
Sbjct: 388 RPFFGDQRMNARSVAHVWGFGAAFEGAMTSAG---------------VAAAVEELL-RGE 431
Query: 428 EAEQIRRRVHELGNLANAAVQPGGSSYQNLTALI 461
E ++R R L L A PGG +N +
Sbjct: 432 EGARMRARAKVLQALVAEAFGPGGECRKNFDRFV 465
>UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-McC allele)
Length = 471
Score = 100 bits (249), Expect = 8e-21
Identities = 68/214 (31%), Positives = 97/214 (44%), Gaps = 32/214 (14%)
Query: 253 LSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEE 312
L+WL + V YV FG++ ++L E+A G+E S F+W + E+
Sbjct: 279 LAWLGRQPARGVAYVSFGTVACPRPDELRELAAGLEDSGAPFLWSL-----------RED 327
Query: 313 KEKWLPEGFEEKGM-----ILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIIT 367
LP GF ++ ++ WAPQ+ +L HPSVGAF+TH G S LE +S+GVP+
Sbjct: 328 SWPHLPPGFLDRAAGTGSGLVVPWAPQVAVLRHPSVGAFVTHAGWASVLEGLSSGVPMAC 387
Query: 368 WPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGE 427
P GDQ + + V G G S G + AV L+ GE
Sbjct: 388 RPFFGDQRMNARSVAHVWGFGAAFEGAMTSAG---------------VATAVEELL-RGE 431
Query: 428 EAEQIRRRVHELGNLANAAVQPGGSSYQNLTALI 461
E ++R R EL L A PGG +N +
Sbjct: 432 EGARMRARAKELQALVAEAFGPGGECRKNFDRFV 465
>UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1)
Length = 455
Score = 98.2 bits (243), Expect = 4e-20
Identities = 73/240 (30%), Positives = 113/240 (46%), Gaps = 34/240 (14%)
Query: 224 LGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEM 283
LGP L+ G A+ A+ ++ L+WL + SV YV FG+ + ++L E+
Sbjct: 242 LGPYHLLPG-AEPTADTN--EAPADPHGCLAWLDRRPARSVAYVSFGTNATARPDELQEL 298
Query: 284 ACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK--GMILRGWAPQLLILGHP 341
A G+EAS F+W + RG P GF E+ G+++ WAPQ+ +L H
Sbjct: 299 AAGLEASGAPFLWSL----RGVVAAA--------PRGFLERAPGLVVP-WAPQVGVLRHA 345
Query: 342 SVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGG 401
+VGAF+TH G S +E VS+GVP+ P GDQ + + V G G
Sbjct: 346 AVGAFVTHAGWASVMEGVSSGVPMACRPFFGDQTMNARSVASVWGFGTAFDGP------- 398
Query: 402 RRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALI 461
++R + AV L+ GE+ E++R + EL + A +P G +N +
Sbjct: 399 --------MTRGAVANAVATLL-RGEDGERMRAKAQELQAMVGKAFEPDGGCRKNFDEFV 449
>UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Fragment)
Length = 154
Score = 77.4 bits (189), Expect = 8e-14
Identities = 52/149 (34%), Positives = 77/149 (50%), Gaps = 20/149 (13%)
Query: 317 LPEGFEEK----GMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLG 372
LPEGF EK GM++ WAPQ +L GAF+THCG NS E+V+ GVP+I P G
Sbjct: 13 LPEGFLEKTRGYGMVVP-WAPQAEVLALRQFGAFVTHCGWNSLWESVAGGVPLICRPFFG 71
Query: 373 DQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQI 432
DQ +++ +V +GV + GG + ++ D+I E+ +++
Sbjct: 72 DQRLNGRMVEDVLEIGVRI-------EGGVFTKSGLMSCFDQILSQ--------EKGKKL 116
Query: 433 RRRVHELGNLANAAVQPGGSSYQNLTALI 461
R + L A+ AV P GSS +N L+
Sbjct: 117 RENLRALRETADRAVGPKGSSTENFKTLV 145
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 60,035,457
Number of Sequences: 164201
Number of extensions: 2707479
Number of successful extensions: 11324
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 11140
Number of HSP's gapped (non-prelim): 142
length of query: 481
length of database: 59,974,054
effective HSP length: 114
effective length of query: 367
effective length of database: 41,255,140
effective search space: 15140636380
effective search space used: 15140636380
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0367.1


