

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
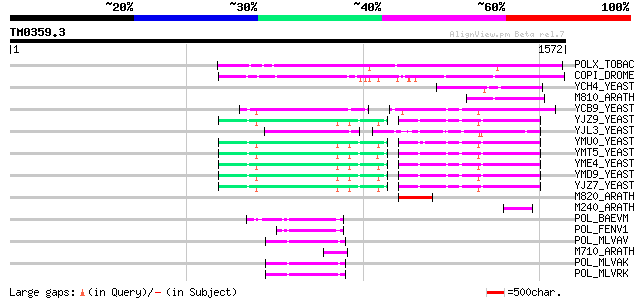
Query= TM0359.3
(1572 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 537 e-152
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 476 e-133
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 164 2e-39
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 156 4e-37
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 122 1e-26
YJZ9_YEAST (P47100) Transposon Ty1 protein B 108 2e-22
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 107 3e-22
YMU0_YEAST (Q04670) Transposon Ty1 protein B 105 7e-22
YMT5_YEAST (Q04214) Transposon Ty1 protein B 105 1e-21
YME4_YEAST (Q04711) Transposon Ty1 protein B 105 1e-21
YMD9_YEAST (Q03434) Transposon Ty1 protein B 104 2e-21
YJZ7_YEAST (P47098) Transposon Ty1 protein B 102 8e-21
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 100 3e-20
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 68 2e-10
POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.2... 66 9e-10
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 62 2e-08
POL_MLVAV (P03356) Pol polyprotein [Contains: Protease (EC 3.4.2... 59 1e-07
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 58 2e-07
POL_MLVAK (P03357) Pol polyprotein [Contains: Reverse transcript... 55 2e-06
POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC 3.4.2... 54 3e-06
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 537 bits (1384), Expect = e-152
Identities = 334/1010 (33%), Positives = 539/1010 (53%), Gaps = 49/1010 (4%)
Query: 590 WYLDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIVGTGTICVDSSP----CIDNV 645
W +D+ S H T + +F G V G KI G G IC+ ++ + +V
Sbjct: 294 WVVDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVLKDV 353
Query: 646 LLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGS-VLFNSKRKNNIYKIR--LSE 702
V L NL+S L GY+ F + R GS V+ + +Y+ + +
Sbjct: 354 RHVPDLRMNLISGIALDRDGYESYFANQKWRLTK---GSLVIAKGVARGTLYRTNAEICQ 410
Query: 703 LEAQNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRGLPNLKFASDALCEACQKG 762
E + +SV+ +WH+R+GH S + + L+K +L+ K + C+ C G
Sbjct: 411 GELNAAQDEISVD----LWHKRMGHMSEKGLQILAKKSLIS---YAKGTTVKPCDYCLFG 463
Query: 763 KFTKVPFNAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRK 822
K +V F + + L+L++ D+ GP++ ES+GG +Y + +DD SR WV L K
Sbjct: 464 KQHRVSFQTSSERKLNI-LDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTK 522
Query: 823 DESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGISHDFSCPRTPQQN 882
D+ VF F A V+ E ++ R+RSD+GGE+ + +FE S+GI H+ + P TPQ N
Sbjct: 523 DQVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHN 582
Query: 883 GVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRISVRPILNKTPYELWKNIK 942
GV ER NRT+ E R+ML+ + K FW EAV TACY+ NR P+ + P +W N +
Sbjct: 583 GVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNKE 642
Query: 943 PNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHV 1002
+ S+ FGC + K++ K D KS C+ +GY D G+R ++ K + S V
Sbjct: 643 VSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRDV 702
Query: 1003 RFDD---KLDSDQSKLVEK--------FADLSINVSDKGKAPEEVEPEEDEPEE--EAGP 1049
F + + +D S+ V+ S N + +EV + ++P E E G
Sbjct: 703 VFRESEVRTAADMSEKVKNGIIPNFVTIPSTSNNPTSAESTTDEVSEQGEQPGEVIEQGE 762
Query: 1050 SNSQTLKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEAL 1109
+ +++ ++ + E R S PS E +L + EP+S+ E L
Sbjct: 763 QLDEGVEEVEHPTQGEEQHQPLRRSERPRVESRRYPSTEYVL----ISDDREPESLKEVL 818
Query: 1110 ---QDKEWILAMEEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGEVVRNKARL 1166
+ + + AM+EE+ KN + LV+ P+ + KWVF+ K + ++VR KARL
Sbjct: 819 SHPEKNQLMKAMQEEMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARL 878
Query: 1167 VAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYV 1226
V +G+ Q++GID+ E F+PV ++ +IR ++S + + ++ + Q+DVK+AFL+G + EE+Y+
Sbjct: 879 VVKGFEQKKGIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYM 938
Query: 1227 HQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTY- 1285
QP GFE K V KL KSLYGLKQAPR WY + SF+ +++ D ++ K +
Sbjct: 939 EQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFS 998
Query: 1286 KDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQV--DQTPEGT 1343
+++ +I+ +YVDD++ ++ L + + F+M +G + LG+++ ++T
Sbjct: 999 ENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKL 1058
Query: 1344 YIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKE------DKSGKVCQKLYRGMIGSL 1397
++ Q KY + +L++FNM + TP+ L K+ ++ G + + Y +GSL
Sbjct: 1059 WLSQEKYIERVLERFNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGSL 1118
Query: 1398 LY-LTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSG 1456
+Y + +RPDI +V + +RF +P + H AVK ILRYL+GTT L + S+ L G
Sbjct: 1119 MYAMVCTRPDIAHAVGVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCF-GGSDPILKG 1177
Query: 1457 YCDADYAGDRTERKSTFGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWM 1516
Y DAD AGD RKS+ G +SW SK Q +ALST EAEYI+A +M+W+
Sbjct: 1178 YTDADMAGDIDNRKSSTGYLFTFSGGAISWQSKLQKCVALSTTEAEYIAATETGKEMIWL 1237
Query: 1517 KHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYV 1566
K L++ + + +YCD+ +AI LSKN + H+R KHI+V+YH+IR+ V
Sbjct: 1238 KRFLQELGLHQKEYVVYCDSQSAIDLSKNSMYHARTKHIDVRYHWIREMV 1287
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 476 bits (1224), Expect = e-133
Identities = 341/1095 (31%), Positives = 549/1095 (49%), Gaps = 133/1095 (12%)
Query: 592 LDSGCSRHMTGEKRMFRE-LKLKPGGEVGFGGNEKGKIV-----GTGTICVDSSPCIDNV 645
LDSG S H+ ++ ++ + +++ P ++ ++G+ + G + D +++V
Sbjct: 291 LDSGASDHLINDESLYTDSVEVVPPLKIAVA--KQGEFIYATKRGIVRLRNDHEITLEDV 348
Query: 646 LLVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEA 705
L NL+S+ +L + G + F+ KS +S+ V+ NS NN+ I +A
Sbjct: 349 LFCKEAAGNLMSVKRLQEAGMSIEFD-KSGVTISKNGLMVVKNSGMLNNVPVINF---QA 404
Query: 706 QNVKCLLSVNEEQWVWHRRLGHASMRKISQLSKLNLVRG---LPNLKFASDALCEACQKG 762
++ N +WH R GH S K+ ++ + N+ L NL+ + + +CE C G
Sbjct: 405 YSINAKHKNNFR--LWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCE-ICEPCLNG 461
Query: 763 KFTKVPFNA-KNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTR 821
K ++PF K+ RPL ++H D+ GP+ ++ K Y ++ VD ++ + +
Sbjct: 462 KQARLPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKY 521
Query: 822 KDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGISHDFSCPRTPQQ 881
K + ++F F+A+ + ++V + D+G E+ +++ GIS+ + P TPQ
Sbjct: 522 KSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPHTPQL 581
Query: 882 NGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRISVRPIL--NKTPYELWK 939
NGV ER RT+ E ARTM+ + K FW EAV TA Y+ NRI R ++ +KTPYE+W
Sbjct: 582 NGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRIPSRALVDSSKTPYEMWH 641
Query: 940 NIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYN--------- 990
N KP + + FG YV + K++ KFD KS K + +GY GF+ ++
Sbjct: 642 NKKPYLKHLRVFGATVYV-HIKNKQGKFDDKSFKSIFVGY--EPNGFKLWDAVNEKFIVA 698
Query: 991 ----TDAKTIEESIHVRFD------------DKLDSDQSKLVE-------KFADLSINVS 1027
D + S V+F+ +D K+++ K D +
Sbjct: 699 RDVVVDETNMVNSRAVKFETVFLKDSKESENKNFPNDSRKIIQTEFPNESKECDNIQFLK 758
Query: 1028 DKGKAPEEVEPEEDE-------PEEEAGPSNSQTLKKSRITAAH-PKELILGNKDEPV-R 1078
D ++ + P + P E N Q LK S+ + + E +D+ +
Sbjct: 759 DSKESENKNFPNDSRKIIQTEFPNESKECDNIQFLKDSKESNKYFLNESKKRKRDDHLNE 818
Query: 1079 TRSAFRPSE----ETLLSLKGL----------VSLIEPKSIDEALQDKEWILAMEEELNQ 1124
++ + P+E ET LK + + +I +S E L+ K I + EE N
Sbjct: 819 SKGSGNPNESRESETAEHLKEIGIDNPTKNDGIEIINRRS--ERLKTKPQI-SYNEEDNS 875
Query: 1125 FSK---------NDV---------------WSLVKKPE-NVHVIGTKW------------ 1147
+K NDV W E N H I W
Sbjct: 876 LNKVVLNAHTIFNDVPNSFDEIQYRDDKSSWEEAINTELNAHKINNTWTITKRPENKNIV 935
Query: 1148 ----VFRNKLNEKGEVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHN 1203
VF K NE G +R KARLVA+G++Q+ IDY ETFAPVAR+ + R ++S + +N
Sbjct: 936 DSRWVFSVKYNELGNPIRYKARLVARGFTQKYQIDYEETFAPVARISSFRFILSLVIQYN 995
Query: 1204 IVLHQMDVKSAFLNGYISEEVYVHQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERLS 1263
+ +HQMDVK+AFLNG + EE+Y+ P G D+V KL K++YGLKQA R W+E
Sbjct: 996 LKVHQMDVKTAFLNGTLKEEIYMRLPQGI--SCNSDNVCKLNKAIYGLKQAARCWFEVFE 1053
Query: 1264 SFLLENEFVRGKVDTTLFC--KTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFE 1321
L E EFV VD ++ K ++ + V +YVDD++ + + + F + +F
Sbjct: 1054 QALKECEFVNSSVDRCIYILDKGNINENIYVLLYVDDVVIATGDMTRMNNFKRYLMEKFR 1113
Query: 1322 MSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMLESTVAKTPMHPTCILEKEDK 1381
M+ + E+K+F+GI+++ + Y+ QS Y K++L KFNM TP+ P+ I +
Sbjct: 1114 MTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFNMENCNAVSTPL-PSKINYELLN 1172
Query: 1382 SGKVCQKLYRGMIGSLLY-LTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTT 1440
S + C R +IG L+Y + +RPD+ +V++ +R+ S +KR+LRYLKGT
Sbjct: 1173 SDEDCNTPCRSLIGCLMYIMLCTRPDLTTAVNILSRYSSKNNSELWQNLKRVLRYLKGTI 1232
Query: 1441 NLGLMYKK--TSEYKLSGYCDADYAGDRTERKSTFGNC-QFLGSNLVSWASKRQSTIALS 1497
++ L++KK E K+ GY D+D+AG +RKST G + NL+ W +KRQ+++A S
Sbjct: 1233 DMKLIFKKNLAFENKIIGYVDSDWAGSEIDRKSTTGYLFKMFDFNLICWNTKRQNSVAAS 1292
Query: 1498 TAEAEYISAAICSTQMLWMKHQLEDYQI-LESNIPIYCDNTAAISLSKNPILHSRAKHIE 1556
+ EAEY++ + LW+K L I LE+ I IY DN IS++ NP H RAKHI+
Sbjct: 1293 STEAEYMALFEAVREALWLKFLLTSINIKLENPIKIYEDNQGCISIANNPSCHKRAKHID 1352
Query: 1557 VKYHFIRDYVQKGVL 1571
+KYHF R+ VQ V+
Sbjct: 1353 IKYHFAREQVQNNVI 1367
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 164 bits (415), Expect = 2e-39
Identities = 98/309 (31%), Positives = 167/309 (53%), Gaps = 17/309 (5%)
Query: 1209 MDVKSAFLNGYISEEVYVHQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLE 1268
MDV +AFLN + E +YV QPPGF +ER PD+V++L +YGLKQAP W E +++ L +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 1269 NEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGEL 1328
F R + + L+ ++ D + + +YVDD++ + + + + + + M +G++
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 1329 KYFLGIQVDQTPEG-------TYIHQSKYTKELLKKFNMLESTVAKT-PMHPTCILEKED 1380
FLG+ + Q+ G YI ++ E + F + ++ + + P+ T +D
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESE-INTFKLTQTPLCNSKPLFETTSPHLKD 179
Query: 1381 KSGKVCQKLYRGMIGSLLY-LTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGT 1439
+ Y+ ++G LL+ RPDI + V L +RF +PR HL + +R+LRYL T
Sbjct: 180 ITP------YQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTT 233
Query: 1440 TNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTFGNCQFLGSNLVSWASKR-QSTIALST 1498
++ L Y+ S+ L+ YCDA + ST G L V+W+SK+ + I + +
Sbjct: 234 RSMCLKYRSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPS 293
Query: 1499 AEAEYISAA 1507
EAEYI+A+
Sbjct: 294 TEAEYITAS 302
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 156 bits (395), Expect = 4e-37
Identities = 76/222 (34%), Positives = 129/222 (57%), Gaps = 1/222 (0%)
Query: 1294 IYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKE 1353
+YVDDI+ ++ +L + + F M +G + YFLGIQ+ P G ++ Q+KY ++
Sbjct: 5 LYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYAEQ 64
Query: 1354 LLKKFNMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLTASRPDILFSVHL 1413
+L ML+ TP+ P + + +R ++G+L YLT +RPDI ++V++
Sbjct: 65 ILNNAGMLDCKPMSTPL-PLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYAVNI 123
Query: 1414 CARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDADYAGDRTERKSTF 1473
+ +P +KR+LRY+KGT GL K S+ + +CD+D+AG + R+ST
Sbjct: 124 VCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRRSTT 183
Query: 1474 GNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLW 1515
G C FLG N++SW++KRQ T++ S+ E EY + A+ + ++ W
Sbjct: 184 GFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 122 bits (305), Expect = 1e-26
Identities = 114/388 (29%), Positives = 180/388 (46%), Gaps = 36/388 (9%)
Query: 651 LTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKI--------RLSE 702
+ ++LLS+S+LA++ F + + + + DG+VL + + Y + +S+
Sbjct: 518 IAYDLLSLSELANQNITACFTRNT---LERSDGTVLAPIVKHGDFYWLSKKYLIPSHISK 574
Query: 703 LEAQNVKCLLSVNEEQW-VWHRRLGHASMRKISQLSKLNLVRGLPNLKF----ASDALCE 757
L NV SVN+ + + HR LGHA+ R I + K N V L AS C
Sbjct: 575 LTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQCP 634
Query: 758 ACQKGKFTK---VPFNAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWT 814
C GK TK V + + P + LH D+FGPV Y + D+ +R+
Sbjct: 635 DCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQ 694
Query: 815 WVKFL-TRKDESHA-VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGISHD 872
WV L R++ES VF++ +A ++N+ R++ ++ D G E+ N F + GI+
Sbjct: 695 WVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITAC 754
Query: 873 FSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRISVRPILNK 932
++ + +GV ER NRTL RT+L +G+ H W AV + I N + V P +K
Sbjct: 755 YTTTADSRAHGVAERLNRTLLNDCRTLLHCSGLPNHLWFSAVEFSTIIRNSL-VSPKNDK 813
Query: 933 TPYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGY----SDRSKGFRF 988
+ + +I+ PFG V N H D+K + GY S S G+
Sbjct: 814 SARQHAGLAGLDITTILPFGQPVIVNN-----HNPDSKIHPRGIPGYALHPSRNSYGYII 868
Query: 989 YNTD-AKTIEESIHVRFDDKLDSDQSKL 1015
Y KT++ + +V DK QSKL
Sbjct: 869 YLPSLKKTVDTTNYVILQDK----QSKL 892
Score = 101 bits (252), Expect = 1e-20
Identities = 112/507 (22%), Positives = 231/507 (45%), Gaps = 52/507 (10%)
Query: 1075 EPVRTRSAFRPSEETLLSLKGLVSLIEPKSI---DEAL-------QDKEWILAMEEELNQ 1124
EP R++ + ++KG+ S+ ++ DEA+ + ++ A +E++Q
Sbjct: 1220 EPPRSKKRIN----LIAAIKGVKSIKPVRTTLRYDEAITYNKDNKEKDRYVEAYHKEISQ 1275
Query: 1125 FSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGEVVRNKARLVAQGYSQQEGIDYTETFA 1184
K + W K + + K + + K +KAR VA+G Q ++ +
Sbjct: 1276 LLKMNTWDTNKYYDRNDIDPKKVINSMFIFNKKRDGTHKARFVARGDIQHPDTYDSDMQS 1335
Query: 1185 PVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPP--GFEDERKPDHVF 1242
A+ +S +++++ + Q+D+ SA+L I EE+Y+ PP G D+ +
Sbjct: 1336 NTVHHYALMTSLSIALDNDYYITQLDISSAYLYADIKEELYIRPPPHLGLNDK-----LL 1390
Query: 1243 KLKKSLYGLKQAPRAWYERLSSFLL---ENEFVRGKVDTTLFCKTYKDDILIVQIYVDDI 1299
+L+KSLYGLKQ+ WYE + S+L+ + + VRG + +K+ + + ++VDD+
Sbjct: 1391 RLRKSLYGLKQSGANWYETIKSYLINCCDMQEVRG------WSCVFKNSQVTICLFVDDM 1444
Query: 1300 IFGSANQSLCKEFSEMMQAEFEMSMM------GELKY-FLGIQVD-QTPEGTYIHQSKYT 1351
I S + + K+ ++ +++ ++ E++Y LG+++ Q + + K
Sbjct: 1445 ILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRSKYMKLGMEKSL 1504
Query: 1352 KELLKKFNMLESTVAK---TPMHPTCILEKED---KSGKVCQKLY--RGMIGSLLYLTAS 1403
E L K N+ + K P P +++++ + +K++ + +IG Y+
Sbjct: 1505 TEKLPKLNVPLNPKGKKLRAPGQPGHYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYK 1564
Query: 1404 -RPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTS----EYKLSGYC 1458
R D+L+ ++ A+ P L +++++ T + L++ K + KL
Sbjct: 1565 FRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTKPDNKLVAIS 1624
Query: 1459 DADYAGDRTERKSTFGNCQFLGSNLVSWASKRQSTIALSTAEAEYISAAICSTQMLWMKH 1518
DA Y G++ KS GN L ++ S + S ST EAE + + + + H
Sbjct: 1625 DASY-GNQPYYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAVSEAIPLLNNLSH 1683
Query: 1519 QLEDYQILESNIPIYCDNTAAISLSKN 1545
+++ + D+ + IS+ K+
Sbjct: 1684 LVQELNKKPIIKGLLTDSRSTISIIKS 1710
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 108 bits (269), Expect = 2e-22
Identities = 109/433 (25%), Positives = 199/433 (45%), Gaps = 47/433 (10%)
Query: 1101 EPKSIDEALQDKE-WILAMEEELNQFSKNDVWSLV-----KKPENVHVIGTKWVFRNKLN 1154
E + ++ +++KE +I A +E+NQ K W K+ + VI + ++F K +
Sbjct: 1236 EAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKKRD 1295
Query: 1155 EKGEVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSA 1214
+KAR VA+G Q + + A+ +S ++++N + Q+D+ SA
Sbjct: 1296 GT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSA 1350
Query: 1215 FLNGYISEEVYVHQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLEN---EF 1271
+L I EE+Y+ PP D + +LKKSLYGLKQ+ WYE + S+L++ E
Sbjct: 1351 YLYADIKEELYIRPPPHLGMN---DKLIRLKKSLYGLKQSGANWYETIKSYLIQQCGMEE 1407
Query: 1272 VRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMG----- 1326
VRG + +K+ + + ++VDD++ S N + K E ++ +++ ++
Sbjct: 1408 VRG------WSCVFKNSQVTICLFVDDMVLFSKNLNSNKRIIEKLKMQYDTKIINLGESD 1461
Query: 1327 -ELKY-FLGIQVDQTPEGTY--IHQSKYTKELLKKFNM---LESTVAKTPMHPTCI---- 1375
E++Y LG+++ + G Y + E + K N+ + P P
Sbjct: 1462 EEIQYDILGLEI-KYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQ 1520
Query: 1376 -LEKEDKSGKVCQKLYRGMIGSLLYLTAS-RPDILFSVHLCARFQSDPRETHLTAVKRIL 1433
LE E+ K+ + +IG Y+ R D+L+ ++ A+ P + L ++
Sbjct: 1521 ELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELI 1580
Query: 1434 RYLKGTTNLGLMYKKTSEY----KLSGYCDADYAGDRTERKSTFGNCQFLGSNLVSWASK 1489
+++ T + L++ K+ KL DA Y G++ KS GN L ++ S
Sbjct: 1581 QFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKST 1639
Query: 1490 RQSTIALSTAEAE 1502
+ S ST EAE
Sbjct: 1640 KASLTCTSTTEAE 1652
Score = 100 bits (248), Expect = 4e-20
Identities = 123/538 (22%), Positives = 210/538 (38%), Gaps = 70/538 (13%)
Query: 592 LDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV---DSSPCIDNVLLV 648
LDSG SR + P V I G + D++ VL
Sbjct: 460 LDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHT 519
Query: 649 DGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKI--------RL 700
+ ++LLS+++LA F + + + DG+VL + + Y + +
Sbjct: 520 PNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLPSNI 576
Query: 701 SELEAQNVKCLLSVNEEQWVW-HRRLGHASMRKISQLSKLNLVRGLP----NLKFASDAL 755
S NV S + + + HR L HA+ + I K N + + A D
Sbjct: 577 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 636
Query: 756 CEACQKGKFTK---VPFNAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSR 812
C C GK TK + + ++ P + LH D+FGPV Y + D+ ++
Sbjct: 637 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 696
Query: 813 WTWVKFL--TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIS 870
+ WV L R+D VF+T +A ++N+ ++ ++ D G E+ N + GI+
Sbjct: 697 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 756
Query: 871 HDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRIS----- 925
++ + +GV ER NRTL + RT LQ +G+ H W A+ + + N ++
Sbjct: 757 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSK 816
Query: 926 -------------VRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTK----------D 962
+ +L + + PN S HP G Y L+
Sbjct: 817 KSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALHPSRNSYGYIIYLP 875
Query: 963 RLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADL 1022
L K ++ +L G R F N DA T +E ++ S +++ DL
Sbjct: 876 SLKKTVDTTNYVILQGKESRLDQF---NYDALTFDEDLNRLTASYQSFIASNEIQQSDDL 932
Query: 1023 SINVSDKGKAPEEVEPEED--------EPEEEAGPS----NSQTLKKSRITAAHPKEL 1068
+I ++ E+ PE+ P + PS +S+ + K+ I A P+E+
Sbjct: 933 NIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRA--PREV 988
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 107 bits (266), Expect = 3e-22
Identities = 120/507 (23%), Positives = 224/507 (43%), Gaps = 82/507 (16%)
Query: 1028 DKGKAPEEVEPEEDEPEEEAG-----PSNSQTLKKSRITAAHPKELILGNKDEPVRTRSA 1082
DK + E E D+ + P N +T+ +I A + E I N D ++ +
Sbjct: 1231 DKNNSLTSYELERDKKRSKKNRVKLIPDNMETVSAPKIRAIYYNEAISKNPD--LKEKHE 1288
Query: 1083 FRPSEETLLSLKGLVSLIEPKSIDEALQDKEWILAMEEELNQFSKNDVWSLVKKPENVHV 1142
++ + K L +L + K D ++ +S++++ P+N+ +
Sbjct: 1289 YKQAYH-----KELQNLKDMKVFDVDVK--------------YSRSEI------PDNL-I 1322
Query: 1143 IGTKWVFRNKLNEKGEVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNH 1202
+ T +F K N KAR+V +G +Q Y+ I++ + + N
Sbjct: 1323 VPTNTIFTKKRNGI-----YKARIVCRGDTQSPDT-YSVITTESLNHNHIKIFLMIANNR 1376
Query: 1203 NIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERL 1262
N+ + +D+ AFL + EE+Y+ P D R V KL K+LYGLKQ+P+ W + L
Sbjct: 1377 NMFMKTLDINHAFLYAKLEEEIYIPHP---HDRRC---VVKLNKALYGLKQSPKEWNDHL 1430
Query: 1263 SSFL-----LENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQ 1317
+L +N + G T +D L++ +YVDD + ++N+ EF ++
Sbjct: 1431 RQYLNGIGLKDNSYTPGLYQT-------EDKNLMIAVYVDDCVIAASNEQRLDEFINKLK 1483
Query: 1318 AEFEMSMMGEL------KYFLGIQ---------VDQTPEGTYIHQ--SKYTKEL--LKKF 1358
+ FE+ + G L LG+ +D T + ++I++ KY +EL ++K
Sbjct: 1484 SNFELKITGTLIDDVLDTDILGMDLVYNKRLGTIDLTLK-SFINRMDKKYNEELKKIRKS 1542
Query: 1359 NMLESTVAKTPMHPTCILEKEDKSGKVCQKLYRGMIGSLLYLT-ASRPDILFSVHLCARF 1417
++ + K + E++ + KL + ++G L Y+ R DI F+V AR
Sbjct: 1543 SIPHMSTYKIDPKKDVLQMSEEEFRQGVLKLQQ-LLGELNYVRHKCRYDIEFAVKKVARL 1601
Query: 1418 QSDPRETHLTAVKRILRYLKGTTNLGLMYKK--TSEYKLSGYCDADYAGDRTERKSTFGN 1475
+ P E + +I++YL ++G+ Y + + K+ DA G + +S G
Sbjct: 1602 VNYPHERVFYMIYKIIQYLVRYKDIGIHYDRDCNKDKKVIAITDAS-VGSEYDAQSRIGV 1660
Query: 1476 CQFLGSNLVSWASKRQSTIALSTAEAE 1502
+ G N+ + S + + +S+ EAE
Sbjct: 1661 ILWYGMNIFNVYSNKSTNRCVSSTEAE 1687
Score = 89.4 bits (220), Expect = 7e-17
Identities = 73/276 (26%), Positives = 123/276 (44%), Gaps = 11/276 (3%)
Query: 722 HRRLGHASMRKISQLSKLN-LVRGLPNLKFASDALCEACQKGKFTK---VPFNAKNVVST 777
H+R+GH +++I K N L +K ++ C+ C+ K TK + N +
Sbjct: 562 HKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTD 621
Query: 778 SRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRW--TWVKFLTRKDESHAVFSTFIAQ 835
P +D+FGPV + + KRY +++VD+ +R+ T F + A I
Sbjct: 622 HEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQY 681
Query: 836 VQNEKACRIVRVRSDHGGEFENDKFESLFDSYGISHDFSCPRTPQQNGVVERKNRTLQEM 895
V+ + ++ + SD G EF ND+ E F S GI H + + NG ER RT+
Sbjct: 682 VETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITD 741
Query: 896 ARTMLQETGMAKHFWAEAVNTACYIPNRISVRPILNKTPYELWKN--IKPNISYFHPFGC 953
A T+L+++ + FW AV +A I N + + K P + + + F PFG
Sbjct: 742 ATTLLRQSNLRVKFWEYAVTSATNIRNYLEHKS-TGKLPLKAISRQPVTVRLMSFLPFGE 800
Query: 954 VCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFY 989
+ N + K ++L S G++F+
Sbjct: 801 KGIIWNHNHK--KLKPSGLPSIILCKDPNSYGYKFF 834
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 105 bits (263), Expect = 7e-22
Identities = 110/438 (25%), Positives = 203/438 (46%), Gaps = 57/438 (13%)
Query: 1101 EPKSIDEALQDKE-WILAMEEELNQFSKNDVWSLV-----KKPENVHVIGTKWVFRNKLN 1154
E + ++ +++KE +I A +E+NQ K W K+ + VI + ++F K +
Sbjct: 809 EAITYNKDIKEKEKYIQAYHKEVNQLLKMKTWDTDRYYDRKEIDPKRVINSMFIFNRKRD 868
Query: 1155 EKGEVVRNKARLVAQGYSQQEGIDYTETFAPVARLE-----AIRLLISFSVNHNIVLHQM 1209
+KAR VA+G I + +T+ P + A+ +S ++++N + Q+
Sbjct: 869 GT-----HKARFVARG-----DIQHPDTYDPGMQSNTVHHYALMTSLSLALDNNYYITQL 918
Query: 1210 DVKSAFLNGYISEEVYVHQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLEN 1269
D+ SA+L I EE+Y+ PP D + +LKKSLYGLKQ+ WYE + S+L++
Sbjct: 919 DISSAYLYADIKEELYIRPPPHLGMN---DKLIRLKKSLYGLKQSGANWYETIKSYLIKQ 975
Query: 1270 ---EFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMM- 1325
E VRG + +K+ + + ++VDD+I S + + K+ ++ +++ ++
Sbjct: 976 CGMEEVRG------WSCVFKNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIIN 1029
Query: 1326 -----GELKY-FLGIQVDQTPEGTY--IHQSKYTKELLKKFNM---LESTVAKTPMHPTC 1374
E++Y LG+++ + G Y + E + K N+ + P P
Sbjct: 1030 LGESDNEIQYDILGLEI-KYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGL 1088
Query: 1375 I-----LEKEDKSGKVCQKLYRGMIGSLLYLTAS-RPDILFSVHLCARFQSDPRETHLTA 1428
LE E+ K+ + +IG Y+ R D+L+ ++ A+ P + L
Sbjct: 1089 YIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDM 1148
Query: 1429 VKRILRYLKGTTNLGLMYKKTSEY----KLSGYCDADYAGDRTERKSTFGNCQFLGSNLV 1484
+++++ T + L++ K+ KL DA Y G++ KS GN L ++
Sbjct: 1149 TYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVI 1207
Query: 1485 SWASKRQSTIALSTAEAE 1502
S + S ST EAE
Sbjct: 1208 GGKSTKASLTCTSTTEAE 1225
Score = 100 bits (248), Expect = 4e-20
Identities = 123/538 (22%), Positives = 210/538 (38%), Gaps = 70/538 (13%)
Query: 592 LDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV---DSSPCIDNVLLV 648
LDSG SR + P V I G + D++ VL
Sbjct: 33 LDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHT 92
Query: 649 DGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKI--------RL 700
+ ++LLS+++LA F + + + DG+VL + + Y + +
Sbjct: 93 PNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 701 SELEAQNVKCLLSVNEEQWVW-HRRLGHASMRKISQLSKLNLVRGLP----NLKFASDAL 755
S NV S + + + HR L HA+ + I K N + + A D
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 209
Query: 756 CEACQKGKFTK---VPFNAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSR 812
C C GK TK + + ++ P + LH D+FGPV Y + D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 813 WTWVKFL--TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIS 870
+ WV L R+D VF+T +A ++N+ ++ ++ D G E+ N + GI+
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 871 HDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRIS----- 925
++ + +GV ER NRTL + RT LQ +G+ H W A+ + + N ++
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSK 389
Query: 926 -------------VRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTK----------D 962
+ +L + + PN S HP G Y L+
Sbjct: 390 KSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALHPSRNSYGYIIYLP 448
Query: 963 RLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADL 1022
L K ++ +L G R F N DA T +E ++ S +++ DL
Sbjct: 449 SLKKTVDTTNYVILQGKESRLDQF---NYDALTFDEDLNRLTASYQSFIASNEIQQSDDL 505
Query: 1023 SINVSDKGKAPEEVEPEED--------EPEEEAGPS----NSQTLKKSRITAAHPKEL 1068
+I ++ E+ PE+ P + PS +S+ + K+ I A P+E+
Sbjct: 506 NIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRA--PREV 561
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 105 bits (261), Expect = 1e-21
Identities = 107/433 (24%), Positives = 199/433 (45%), Gaps = 47/433 (10%)
Query: 1101 EPKSIDEALQDKE-WILAMEEELNQFSKNDVWSLVK-----KPENVHVIGTKWVFRNKLN 1154
E + ++ +++KE +I A +E+NQ K W K + + VI + ++F K +
Sbjct: 809 EAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDKYYDRKEIDPKRVINSMFIFNRKRD 868
Query: 1155 EKGEVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSA 1214
+KAR VA+G Q + + A+ +S ++++N + Q+D+ SA
Sbjct: 869 GT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSA 923
Query: 1215 FLNGYISEEVYVHQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLEN---EF 1271
+L I EE+Y+ PP D + +LKKSLYGLKQ+ WYE + S+L++ E
Sbjct: 924 YLYADIKEELYIRPPPHLGMN---DKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEE 980
Query: 1272 VRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMMG----- 1326
VRG + +++ + + ++VDD++ S N + K + ++ +++ ++
Sbjct: 981 VRG------WSCVFENSQVTICLFVDDMVLFSKNLNSNKRIIDKLKMQYDTKIINLGESD 1034
Query: 1327 -ELKY-FLGIQVDQTPEGTY--IHQSKYTKELLKKFNM---LESTVAKTPMHPTCI---- 1375
E++Y LG+++ + G Y + E + K N+ + P P
Sbjct: 1035 EEIQYDILGLEI-KYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQ 1093
Query: 1376 -LEKEDKSGKVCQKLYRGMIGSLLYLTAS-RPDILFSVHLCARFQSDPRETHLTAVKRIL 1433
LE E+ K+ + +IG Y+ R D+L+ ++ A+ P + L ++
Sbjct: 1094 ELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELI 1153
Query: 1434 RYLKGTTNLGLMYKKTSEY----KLSGYCDADYAGDRTERKSTFGNCQFLGSNLVSWASK 1489
+++ T + L++ K+ KL DA Y G++ KS GN L ++ S
Sbjct: 1154 QFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKST 1212
Query: 1490 RQSTIALSTAEAE 1502
+ S ST EAE
Sbjct: 1213 KASLTCTSTTEAE 1225
Score = 99.0 bits (245), Expect = 9e-20
Identities = 123/538 (22%), Positives = 210/538 (38%), Gaps = 70/538 (13%)
Query: 592 LDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV---DSSPCIDNVLLV 648
LDSG SR + P V I G + D++ VL
Sbjct: 33 LDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHT 92
Query: 649 DGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKI--------RL 700
+ ++LLS+++LA F + + + DG+VL + + Y + +
Sbjct: 93 PNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 701 SELEAQNVKCLLSVNEEQWVW-HRRLGHASMRKISQLSKLNLVRGLP----NLKFASDAL 755
S NV S + + + HR L HA+ + I K N + + A D
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 209
Query: 756 CEACQKGKFTK---VPFNAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSR 812
C C GK TK + + ++ P + LH D+FGPV Y + D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 813 WTWVKFL--TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIS 870
+ WV L R+D VF+T +A ++N+ ++ ++ D G E+ N + GI+
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 871 HDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRIS----- 925
++ + +GV ER NRTL + RT LQ +G+ H W A+ + + N ++
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSK 389
Query: 926 -------------VRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTK----------D 962
+ +L + + PN S HP G Y L+
Sbjct: 390 KSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALHPSRNSYGYIIYLP 448
Query: 963 RLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADL 1022
L K ++ +L G R F N DA T +E ++ S +++ DL
Sbjct: 449 SLKKTVDTTNYVILQGKESRLDQF---NYDALTFDEDLNRLTASYHSFIASNEIQQSNDL 505
Query: 1023 SINVSDKGKAPEEVEPEE--------DEPEEEAGPS----NSQTLKKSRITAAHPKEL 1068
+I ++ E+ PE+ P + PS +S+ + K+ I A P+E+
Sbjct: 506 NIESDHDFQSDIELHPEQLRNVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRA--PREV 561
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 105 bits (261), Expect = 1e-21
Identities = 105/433 (24%), Positives = 203/433 (46%), Gaps = 47/433 (10%)
Query: 1101 EPKSIDEALQDKE-WILAMEEELNQFSKNDVWSLVK-----KPENVHVIGTKWVFRNKLN 1154
E + ++ +++KE +I A +E+NQ K + W K + + VI + ++F K +
Sbjct: 809 EAITYNKDIKEKEKYIEAYHKEVNQLLKMNTWDTDKYYDRKEIDPKRVINSMFIFNRKRD 868
Query: 1155 EKGEVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSA 1214
+KAR VA+G Q + + A+ +S ++++N + Q+D+ SA
Sbjct: 869 GT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSA 923
Query: 1215 FLNGYISEEVYVHQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLEN---EF 1271
+L I EE+Y+ PP D + +LKKSLYGLKQ+ WYE + S+L++ E
Sbjct: 924 YLYADIKEELYIRPPPHLGMN---DKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEE 980
Query: 1272 VRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMM------ 1325
VRG + +K+ + + ++VDD+I S + + K+ ++ +++ ++
Sbjct: 981 VRG------WSCVFKNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESD 1034
Query: 1326 GELKY-FLGIQVDQTPEGTY--IHQSKYTKELLKKFNM---LESTVAKTPMHPTCILEKE 1379
E++Y LG+++ + G Y + E + K N+ + P P ++++
Sbjct: 1035 NEIQYDILGLEI-KYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQD 1093
Query: 1380 D---KSGKVCQKLY--RGMIGSLLYLTAS-RPDILFSVHLCARFQSDPRETHLTAVKRIL 1433
+ + +K++ + +IG Y+ R D+L+ ++ A+ P L ++
Sbjct: 1094 ELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELI 1153
Query: 1434 RYLKGTTNLGLMYKKTS----EYKLSGYCDADYAGDRTERKSTFGNCQFLGSNLVSWASK 1489
+++ T + L++ K + KL DA Y G++ KS GN L ++ S
Sbjct: 1154 QFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPYYKSQIGNIYLLNGKVIGGKST 1212
Query: 1490 RQSTIALSTAEAE 1502
+ S ST EAE
Sbjct: 1213 KASLTCTSTTEAE 1225
Score = 100 bits (248), Expect = 4e-20
Identities = 123/538 (22%), Positives = 210/538 (38%), Gaps = 70/538 (13%)
Query: 592 LDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV---DSSPCIDNVLLV 648
LDSG SR + P V I G + D++ VL
Sbjct: 33 LDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHT 92
Query: 649 DGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKI--------RL 700
+ ++LLS+++LA F + + + DG+VL + + Y + +
Sbjct: 93 PNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 701 SELEAQNVKCLLSVNEEQWVW-HRRLGHASMRKISQLSKLNLVRGLP----NLKFASDAL 755
S NV S + + + HR L HA+ + I K N + + A D
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 209
Query: 756 CEACQKGKFTK---VPFNAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSR 812
C C GK TK + + ++ P + LH D+FGPV Y + D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 813 WTWVKFL--TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIS 870
+ WV L R+D VF+T +A ++N+ ++ ++ D G E+ N + GI+
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 871 HDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRIS----- 925
++ + +GV ER NRTL + RT LQ +G+ H W A+ + + N ++
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSK 389
Query: 926 -------------VRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTK----------D 962
+ +L + + PN S HP G Y L+
Sbjct: 390 KSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALHPSRNSYGYIIYLP 448
Query: 963 RLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADL 1022
L K ++ +L G R F N DA T +E ++ S +++ DL
Sbjct: 449 SLKKTVDTTNYVILQGKESRLDQF---NYDALTFDEDLNRLTASYQSFIASNEIQQSDDL 505
Query: 1023 SINVSDKGKAPEEVEPEED--------EPEEEAGPS----NSQTLKKSRITAAHPKEL 1068
+I ++ E+ PE+ P + PS +S+ + K+ I A P+E+
Sbjct: 506 NIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRA--PREV 561
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 104 bits (259), Expect = 2e-21
Identities = 105/433 (24%), Positives = 202/433 (46%), Gaps = 47/433 (10%)
Query: 1101 EPKSIDEALQDKE-WILAMEEELNQFSKNDVWSLV-----KKPENVHVIGTKWVFRNKLN 1154
E + ++ +++KE +I A +E+NQ K W K+ + VI + ++F K +
Sbjct: 809 EAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKKRD 868
Query: 1155 EKGEVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSA 1214
+KAR VA+G Q + + A+ +S ++++N + Q+D+ SA
Sbjct: 869 GT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSA 923
Query: 1215 FLNGYISEEVYVHQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLEN---EF 1271
+L I EE+Y+ PP D + +LKKSLYGLKQ+ WYE + S+L++ E
Sbjct: 924 YLYADIKEELYIRPPPHLGMN---DKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEE 980
Query: 1272 VRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMM------ 1325
VRG + +K+ + + ++VDD+I S + + K+ ++ +++ ++
Sbjct: 981 VRG------WSCVFKNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESD 1034
Query: 1326 GELKY-FLGIQVDQTPEGTY--IHQSKYTKELLKKFNM---LESTVAKTPMHPTCILEKE 1379
E++Y LG+++ + G Y + E + K N+ + P P ++++
Sbjct: 1035 NEIQYDILGLEI-KYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQD 1093
Query: 1380 D---KSGKVCQKLY--RGMIGSLLYLTAS-RPDILFSVHLCARFQSDPRETHLTAVKRIL 1433
+ + +K++ + +IG Y+ R D+L+ ++ A+ P L ++
Sbjct: 1094 ELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELI 1153
Query: 1434 RYLKGTTNLGLMYKKTS----EYKLSGYCDADYAGDRTERKSTFGNCQFLGSNLVSWASK 1489
+++ T + L++ K + KL DA Y G++ KS GN L ++ S
Sbjct: 1154 QFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPYYKSQIGNIYLLNGKVIGGKST 1212
Query: 1490 RQSTIALSTAEAE 1502
+ S ST EAE
Sbjct: 1213 KASLTCTSTTEAE 1225
Score = 100 bits (248), Expect = 4e-20
Identities = 123/538 (22%), Positives = 210/538 (38%), Gaps = 70/538 (13%)
Query: 592 LDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV---DSSPCIDNVLLV 648
LDSG SR + P V I G + D++ VL
Sbjct: 33 LDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHT 92
Query: 649 DGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKI--------RL 700
+ ++LLS+++LA F + + + DG+VL + + Y + +
Sbjct: 93 PNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 701 SELEAQNVKCLLSVNEEQWVW-HRRLGHASMRKISQLSKLNLVRGLP----NLKFASDAL 755
S NV S + + + HR L HA+ + I K N + + A D
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDRSSAIDYQ 209
Query: 756 CEACQKGKFTK---VPFNAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSR 812
C C GK TK + + ++ P + LH D+FGPV Y + D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 813 WTWVKFL--TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIS 870
+ WV L R+D VF+T +A ++N+ ++ ++ D G E+ N + GI+
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 871 HDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRIS----- 925
++ + +GV ER NRTL + RT LQ +G+ H W A+ + + N ++
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSK 389
Query: 926 -------------VRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTK----------D 962
+ +L + + PN S HP G Y L+
Sbjct: 390 KSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALHPSRNSYGYIIYLP 448
Query: 963 RLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADL 1022
L K ++ +L G R F N DA T +E ++ S +++ DL
Sbjct: 449 SLKKTVDTTNYVILQGKESRLDQF---NYDALTFDEDLNRLTASYQSFIASNEIQQSDDL 505
Query: 1023 SINVSDKGKAPEEVEPEED--------EPEEEAGPS----NSQTLKKSRITAAHPKEL 1068
+I ++ E+ PE+ P + PS +S+ + K+ I A P+E+
Sbjct: 506 NIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRA--PREV 561
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 102 bits (254), Expect = 8e-21
Identities = 105/433 (24%), Positives = 201/433 (46%), Gaps = 47/433 (10%)
Query: 1101 EPKSIDEALQDKE-WILAMEEELNQFSKNDVWSLV-----KKPENVHVIGTKWVFRNKLN 1154
E + ++ +++KE +I A +E+NQ K W K+ + VI + ++F K +
Sbjct: 1236 EAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKKRD 1295
Query: 1155 EKGEVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLISFSVNHNIVLHQMDVKSA 1214
+KAR VA+G Q T + A+ +S ++++N + Q+D+ SA
Sbjct: 1296 GT-----HKARFVARGDIQHPDTYDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISSA 1350
Query: 1215 FLNGYISEEVYVHQPPGFEDERKPDHVFKLKKSLYGLKQAPRAWYERLSSFLLEN---EF 1271
+L I EE+Y+ PP D + +LKKS YGLKQ+ WYE + S+L++ E
Sbjct: 1351 YLYADIKEELYIRPPPHLGMN---DKLIRLKKSHYGLKQSGANWYETIKSYLIKQCGMEE 1407
Query: 1272 VRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQSLCKEFSEMMQAEFEMSMM------ 1325
VRG + +K+ + + ++VDD+I S + + K+ ++ +++ ++
Sbjct: 1408 VRG------WSCVFKNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESD 1461
Query: 1326 GELKY-FLGIQVDQTPEGTY--IHQSKYTKELLKKFNM---LESTVAKTPMHPTCILEKE 1379
E++Y LG+++ + G Y + E + K N+ + P P ++++
Sbjct: 1462 NEIQYDILGLEI-KYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQD 1520
Query: 1380 D---KSGKVCQKLY--RGMIGSLLYLTAS-RPDILFSVHLCARFQSDPRETHLTAVKRIL 1433
+ + +K++ + +IG Y+ R D+L+ ++ A+ P L ++
Sbjct: 1521 ELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELI 1580
Query: 1434 RYLKGTTNLGLMYKKTS----EYKLSGYCDADYAGDRTERKSTFGNCQFLGSNLVSWASK 1489
+++ T + L++ K + KL DA Y G++ KS GN L ++ S
Sbjct: 1581 QFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPYYKSQIGNIFLLNGKVIGGKST 1639
Query: 1490 RQSTIALSTAEAE 1502
+ S ST EAE
Sbjct: 1640 KASLTCTSTTEAE 1652
Score = 99.8 bits (247), Expect = 5e-20
Identities = 123/538 (22%), Positives = 210/538 (38%), Gaps = 70/538 (13%)
Query: 592 LDSGCSRHMTGEKRMFRELKLKPGGEVGFGGNEKGKIVGTGTICV---DSSPCIDNVLLV 648
LDSG SR + P V I G + D++ VL
Sbjct: 460 LDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHT 519
Query: 649 DGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKI--------RL 700
+ ++LLS+++LA F + + + DG+VL + + Y + +
Sbjct: 520 PNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVQYGDFYWVSKRYLLPSNI 576
Query: 701 SELEAQNVKCLLSVNEEQWVW-HRRLGHASMRKISQLSKLNLVRGLP----NLKFASDAL 755
S NV S + + + HR L HA+ + I K N + + A D
Sbjct: 577 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 636
Query: 756 CEACQKGKFTK---VPFNAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSR 812
C C GK TK + + ++ P + LH D+FGPV Y + D+ ++
Sbjct: 637 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 696
Query: 813 WTWVKFL--TRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIS 870
+ WV L R+D VF+T +A ++N+ ++ ++ D G E+ N + GI+
Sbjct: 697 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 756
Query: 871 HDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRIS----- 925
++ + +GV ER NRTL + RT LQ +G+ H W A+ + + N ++
Sbjct: 757 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSK 816
Query: 926 -------------VRPILNKTPYELWKNIKPNISYFHPFGCVCYVLNTK----------D 962
+ +L + + PN S HP G Y L+
Sbjct: 817 KSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALHPSRNSYGYIIYLP 875
Query: 963 RLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFADL 1022
L K ++ +L G R F N DA T +E ++ S +++ DL
Sbjct: 876 SLKKTVDTTNYVILQGKESRLDQF---NYDALTFDEDLNRLTASYQSFIASNEIQESNDL 932
Query: 1023 SINVSDKGKAPEEVEPEED--------EPEEEAGPS----NSQTLKKSRITAAHPKEL 1068
+I ++ E+ PE+ P + PS +S+ + K+ I A P+E+
Sbjct: 933 NIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRA--PREV 988
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 100 bits (249), Expect = 3e-20
Identities = 47/97 (48%), Positives = 67/97 (68%)
Query: 1101 EPKSIDEALQDKEWILAMEEELNQFSKNDVWSLVKKPENVHVIGTKWVFRNKLNEKGEVV 1160
EPKS+ AL+D W AM+EEL+ S+N W LV P N +++G KWVF+ KL+ G +
Sbjct: 27 EPKSVIFALKDPGWCQAMQEELDALSRNKTWILVPPPVNQNILGCKWVFKTKLHSDGTLD 86
Query: 1161 RNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLIS 1197
R KARLVA+G+ Q+EGI + ET++PV R IR +++
Sbjct: 87 RLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTILN 123
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 68.2 bits (165), Expect = 2e-10
Identities = 31/82 (37%), Positives = 49/82 (58%)
Query: 1398 LYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGY 1457
+YLT +RPD+ F+V+ ++F S R + AV ++L Y+KGT GL Y TS+ +L +
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 1458 CDADYAGDRTERKSTFGNCQFL 1479
D+D+A R+S G C +
Sbjct: 61 ADSDWASCPDTRRSVTGFCSLV 82
>POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1189
Score = 65.9 bits (159), Expect = 9e-10
Identities = 74/280 (26%), Positives = 125/280 (44%), Gaps = 27/280 (9%)
Query: 671 NQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQNVKCLLSVNEEQWVWHRRLGHASM 730
+Q+ RA+ + N +++ KI L + EA L++ ++ W LG+ +
Sbjct: 806 DQEEARAIGATENKDTRNWEKEG---KIVLPQKEA------LAMIQQMHAW-THLGNRKL 855
Query: 731 RKISQLSKLNLVRGLPNLKFASDALCEACQK--GKFTKVPFNAKNVVSTSRPLELLHIDL 788
+ + + + + R ++ + A C+ CQ+ T+VP + +RP ID
Sbjct: 856 KLLIEKTDFLIPRASTLIEQVTSA-CKVCQQVNAGATRVPAGKRT--RGNRPGVYWEID- 911
Query: 789 FGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVR 848
F VK G K Y +V VD +S W F TR++ +H V + ++ V +
Sbjct: 912 FTEVKPHYAGYK-YLLVFVDTFSGWVEA-FPTRQETAHIVAKKILEEIFPRFGLPKV-IG 968
Query: 849 SDHGGEFENDKFESLFDSYGISHDFSCPRTPQQNGVVERKNRTLQEMARTMLQETGMA-- 906
SD+G F + + L GI+ C PQ +G VER NRT++E + ETG+
Sbjct: 969 SDNGPAFVSQVSQGLARILGINWKLHCAYRPQSSGQVERMNRTIKETLTKLTLETGLKDW 1028
Query: 907 KHFWAEAVNTACYIPNRISVRPILNKTPYELWKNIKPNIS 946
+ + A+ A PNR + TPYE+ P +S
Sbjct: 1029 RRLLSLALLRARNTPNRFGL------TPYEILYGGPPPLS 1062
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 61.6 bits (148), Expect = 2e-08
Identities = 58/195 (29%), Positives = 89/195 (44%), Gaps = 16/195 (8%)
Query: 756 CEACQK--GKFTKVPFNAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRW 813
C+ CQ+ T+VP + +RP ID F VK G K Y +V VD +S W
Sbjct: 737 CKVCQQVNAGATRVPEGKRT--RGNRPGVYWEID-FTEVKPHYAGYK-YLLVFVDTFSGW 792
Query: 814 TWVKFLTRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGISHDF 873
+ TR++ +H V + ++ V + SD+G F + + L + GI+
Sbjct: 793 VEA-YPTRQETAHMVAKKILEEIFPRFGLPKV-IGSDNGPAFVSQVSQGLARTLGINWKL 850
Query: 874 SCPRTPQQNGVVERKNRTLQEMARTMLQETGMA--KHFWAEAVNTACYIPNRISVRPILN 931
C PQ +G VER NRT++E + ETG+ + + A+ A PNR +
Sbjct: 851 HCAYRPQSSGQVERMNRTIKETLTKLTLETGLKDWRRLLSLALLRARNTPNRFGL----- 905
Query: 932 KTPYELWKNIKPNIS 946
TPYE+ P +S
Sbjct: 906 -TPYEILYGGPPPLS 919
>POL_MLVAV (P03356) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1196
Score = 58.9 bits (141), Expect = 1e-07
Identities = 62/231 (26%), Positives = 95/231 (40%), Gaps = 14/231 (6%)
Query: 724 RLGHASMRKISQL-----SKLNLVRGLPNLKFASDALCEACQKGKFTKVPFNAKNVVSTS 778
RL H +K+ L S ++ L++ +D+ C C + +K A V
Sbjct: 849 RLTHLGYQKMKALLDRGESPYYMLNRDKTLQYVADS-CTVCAQVNASKAKIGAGVRVRGH 907
Query: 779 RPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQN 838
RP ID F VK + G +Y +V VD +S W F T+++ + V + ++
Sbjct: 908 RPGSHWEID-FTEVKP-GLYGYKYLLVFVDTFSGWVEA-FPTKRETARVVSKKLLEEIFP 964
Query: 839 EKACRIVRVRSDHGGEFENDKFESLFDSYGISHDFSCPRTPQQNGVVERKNRTLQEMART 898
V + SD+G F + +S+ D GI C PQ +G VER NRT++E
Sbjct: 965 RFGMPQV-LGSDNGPAFTSQVSQSVADLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTK 1023
Query: 899 MLQETGMAKHFWAEAVNTACYIPNRISVRPILNKTPYELWKNIKPNISYFH 949
+ G W + A Y L TPYE+ P + FH
Sbjct: 1024 LTLAAGTRD--WVLLLPLALYRARNTPGPHGL--TPYEILYGAPPPLVNFH 1070
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 57.8 bits (138), Expect = 2e-07
Identities = 30/69 (43%), Positives = 38/69 (54%)
Query: 889 NRTLQEMARTMLQETGMAKHFWAEAVNTACYIPNRISVRPILNKTPYELWKNIKPNISYF 948
NRT+ E R+ML E G+ K F A+A NTA +I N+ I P E+W P SY
Sbjct: 2 NRTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSYL 61
Query: 949 HPFGCVCYV 957
FGCV Y+
Sbjct: 62 RRFGCVAYI 70
>POL_MLVAK (P03357) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 843
Score = 55.1 bits (131), Expect = 2e-06
Identities = 62/231 (26%), Positives = 95/231 (40%), Gaps = 15/231 (6%)
Query: 724 RLGHASMRKISQL-----SKLNLVRGLPNLKFASDALCEACQKGKFTKVPFNAKNVVSTS 778
RL H +K+ L S ++ L++ +D+ C C + +K A V
Sbjct: 497 RLTHLGYQKMKALLDRGESPYYMLNRDKTLQYVADS-CTVCAQVNASKAKIGAGVRVRGH 555
Query: 779 RPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQN 838
RP ID F VK + G +Y +V VD +S W F T+++ + V + ++
Sbjct: 556 RPGSHWEID-FTEVKP-GLYGYKYLLVFVDTFSGWVEA-FPTKRETARVVSKKLLEEIFP 612
Query: 839 EKACRIVRVRSDHGGEFENDKFESLFDSYGISHDFSCPRTPQQNGVVERKNRTLQEMART 898
V + SD+G F + +S+ D GI C PQ +G VER NRT++E
Sbjct: 613 RFGMPQV-LGSDNGPAFTSQVSQSVADLLGIDK-LHCAYRPQSSGQVERMNRTIKETLTK 670
Query: 899 MLQETGMAKHFWAEAVNTACYIPNRISVRPILNKTPYELWKNIKPNISYFH 949
+ G W + A Y L TPYE+ P + FH
Sbjct: 671 LTLAAGTRD--WVLLLPLALYRARNTPGPHGL--TPYEILYGAPPPLVNFH 717
>POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)] (Fragment)
Length = 581
Score = 54.3 bits (129), Expect = 3e-06
Identities = 61/231 (26%), Positives = 94/231 (40%), Gaps = 14/231 (6%)
Query: 724 RLGHASMRKISQL-----SKLNLVRGLPNLKFASDALCEACQKGKFTKVPFNAKNVVSTS 778
RL H +K+ L S ++ L++ +D+ C C + +K A V
Sbjct: 234 RLTHLGYQKMKALLDRGESPYYMLNRDKTLQYVADS-CTVCAQVNASKAKIGAGVRVRGH 292
Query: 779 RPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLTRKDESHAVFSTFIAQVQN 838
RP ID F VK + G +Y +V VD +S W F T+ + + V + ++
Sbjct: 293 RPGTHWEID-FTEVKP-GLYGYKYLLVFVDTFSGWVEA-FPTKHETAKIVTKKLLEEIFP 349
Query: 839 EKACRIVRVRSDHGGEFENDKFESLFDSYGISHDFSCPRTPQQNGVVERKNRTLQEMART 898
V + +D+G F + +S+ GI C PQ +G VER NRT++E
Sbjct: 350 RFGMPQV-LGTDNGPAFVSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTK 408
Query: 899 MLQETGMAKHFWAEAVNTACYIPNRISVRPILNKTPYELWKNIKPNISYFH 949
+ TG W + A Y L TPYE+ P + FH
Sbjct: 409 LTLATGTRD--WVLLLPLALYRARNTPGPHGL--TPYEILYGAPPPLVNFH 455
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.332 0.142 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 165,264,400
Number of Sequences: 164201
Number of extensions: 6619788
Number of successful extensions: 22124
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 21854
Number of HSP's gapped (non-prelim): 206
length of query: 1572
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1448
effective length of database: 39,613,130
effective search space: 57359812240
effective search space used: 57359812240
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0359.3


