

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.16
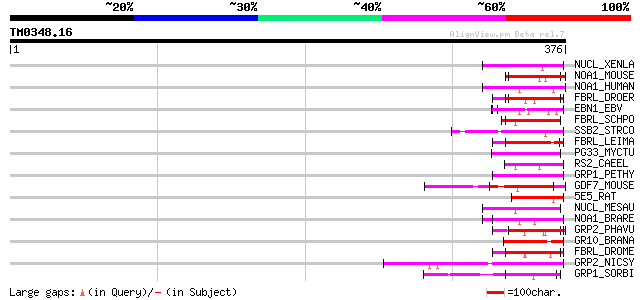
(376 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NUCL_XENLA (P20397) Nucleolin (Protein C23) 52 2e-06
NOA1_MOUSE (Q9CY66) Nucleolar protein family A member 1 (snoRNP ... 51 5e-06
NOA1_HUMAN (Q9NY12) Nucleolar protein family A member 1 (snoRNP ... 51 5e-06
FBRL_DROER (Q8I1F4) Fibrillarin 50 7e-06
EBN1_EBV (P03211) EBNA-1 nuclear protein 50 9e-06
FBRL_SCHPO (P35551) Fibrillarin 50 1e-05
SSB2_STRCO (Q9X8U3) Single-strand binding protein 2 (SSB 2) (Hel... 49 2e-05
FBRL_LEIMA (P35549) Fibrillarin 49 2e-05
PG33_MYCTU (Q50615) Hypothetical PE-PGRS family protein PE_PGRS33 48 4e-05
RS2_CAEEL (P51403) 40S ribosomal protein S2 48 5e-05
GRP1_PETHY (P09789) Glycine-rich cell wall structural protein 1 ... 47 6e-05
GDF7_MOUSE (P43029) Growth/differentiation factor 7 precursor (G... 47 6e-05
5E5_RAT (Q63003) 5E5 antigen 47 6e-05
NUCL_MESAU (P08199) Nucleolin (Protein C23) 47 1e-04
NOA1_BRARE (Q7ZVE0) Nucleolar protein family A member 1 (snoRNP ... 47 1e-04
GRP2_PHAVU (P10496) Glycine-rich cell wall structural protein 1.... 47 1e-04
GR10_BRANA (Q05966) Glycine-rich RNA-binding protein 10 47 1e-04
FBRL_DROME (Q9W1V3) Fibrillarin 47 1e-04
GRP2_NICSY (P27484) Glycine-rich protein 2 46 2e-04
GRP1_SORBI (Q99069) Glycine-rich RNA-binding protein 1 (Fragment) 46 2e-04
>NUCL_XENLA (P20397) Nucleolin (Protein C23)
Length = 650
Score = 52.0 bits (123), Expect = 2e-06
Identities = 28/58 (48%), Positives = 32/58 (54%), Gaps = 3/58 (5%)
Query: 321 RILQDLAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGA---GAMGGRRGRGGRRGRG 375
++ D A +G + GRGG G GG G GGR GGG G GG RGRGG GRG
Sbjct: 569 KVTLDFAKPKGDSQRGGRGGFGRGGGFRGGRGGRGGGGGRGFGGRGGGRGRGGFGGRG 626
Score = 43.1 bits (100), Expect = 0.001
Identities = 23/46 (50%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query: 330 QGIAYAAGRGGLGAGGRAG--GRSGGREGGGAGAMGGRRGRGGRRG 373
+G + GRGG G GG G GR GGR GG G GG RGG+ G
Sbjct: 591 RGGGFRGGRGGRGGGGGRGFGGRGGGRGRGGFGGRGGGGFRGGQGG 636
Score = 36.6 bits (83), Expect = 0.11
Identities = 22/41 (53%), Positives = 24/41 (57%), Gaps = 3/41 (7%)
Query: 337 GRGGLGAGGRAGGRSGGREGG--GAGAMGGRRGRGGRRGRG 375
G GG G GGR GGR G GG G G GG +G G R G+G
Sbjct: 604 GGGGRGFGGRGGGRGRGGFGGRGGGGFRGG-QGGGFRGGQG 643
Score = 33.1 bits (74), Expect = 1.2
Identities = 23/50 (46%), Positives = 25/50 (50%), Gaps = 3/50 (6%)
Query: 328 GTQGIAYAAGRGGLG-AGGRAGGRSGGREGGG--AGAMGGRRGRGGRRGR 374
G G GRG G GGR G GGR GGG G GG RG G++ R
Sbjct: 598 GRGGRGGGGGRGFGGRGGGRGRGGFGGRGGGGFRGGQGGGFRGGQGKKMR 647
>NOA1_MOUSE (Q9CY66) Nucleolar protein family A member 1 (snoRNP
protein GAR1) (H/ACA ribonucleoprotein GAR1)
Length = 231
Score = 50.8 bits (120), Expect = 5e-06
Identities = 25/40 (62%), Positives = 25/40 (62%), Gaps = 2/40 (5%)
Query: 339 GGLGAGGRAGGRSGGREGG--GAGAMGGRRGRGGRRGRGH 376
GG G GGR GGR GG GG G G G RG GG RGRGH
Sbjct: 192 GGRGGGGRGGGRGGGFRGGRGGGGGFRGGRGGGGFRGRGH 231
Score = 46.6 bits (109), Expect = 1e-04
Identities = 24/39 (61%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGA--MGGRRGRGGRRG 373
G GG G GGR GGR GG GGG G GGR G GG RG
Sbjct: 181 GGGGGGRGGRGGGRGGGGRGGGRGGGFRGGRGGGGGFRG 219
Score = 40.4 bits (93), Expect = 0.007
Identities = 25/38 (65%), Positives = 25/38 (65%), Gaps = 5/38 (13%)
Query: 338 RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
RGG G GGR GGR GGR GGG G G RG G R GRG
Sbjct: 180 RGG-GGGGR-GGRGGGRGGGGRG---GGRGGGFRGGRG 212
Score = 35.0 bits (79), Expect = 0.31
Identities = 21/43 (48%), Positives = 21/43 (48%), Gaps = 6/43 (13%)
Query: 337 GRGGLGAGGRAGG--RSGGR----EGGGAGAMGGRRGRGGRRG 373
GRGG GG GG R GG GGG G G RG GG G
Sbjct: 7 GRGGFNRGGGGGGFNRGGGSNNHFRGGGGGGGGSFRGGGGGGG 49
Score = 34.7 bits (78), Expect = 0.41
Identities = 20/38 (52%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Query: 338 RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
RGG G GG R GG GGG+ GGR G G GRG
Sbjct: 31 RGG-GGGGGGSFRGGGGGGGGSFRGGGRGGFGRGGGRG 67
Score = 33.9 bits (76), Expect = 0.69
Identities = 21/52 (40%), Positives = 21/52 (40%), Gaps = 6/52 (11%)
Query: 328 GTQGIAYAAGRGGLGAGG------RAGGRSGGREGGGAGAMGGRRGRGGRRG 373
G G G GG GG R GG GG G G GG RGG RG
Sbjct: 7 GRGGFNRGGGGGGFNRGGGSNNHFRGGGGGGGGSFRGGGGGGGGSFRGGGRG 58
Score = 32.7 bits (73), Expect = 1.5
Identities = 18/32 (56%), Positives = 18/32 (56%), Gaps = 2/32 (6%)
Query: 344 GGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G GG GGR G G G GG GRGG RG G
Sbjct: 177 GPPRGGGGGGRGGRGGGRGGG--GRGGGRGGG 206
Score = 31.6 bits (70), Expect = 3.4
Identities = 16/42 (38%), Positives = 20/42 (47%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRG 369
G G ++ G GG G R GGR G GGG G + +G
Sbjct: 35 GGGGGSFRGGGGGGGGSFRGGGRGGFGRGGGRGGFNKFQDQG 76
Score = 31.2 bits (69), Expect = 4.5
Identities = 15/30 (50%), Positives = 15/30 (50%)
Query: 346 RAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
R G G GGG G GGR G G GRG
Sbjct: 171 RPPGEKGPPRGGGGGGRGGRGGGRGGGGRG 200
Score = 30.8 bits (68), Expect = 5.9
Identities = 16/30 (53%), Positives = 17/30 (56%), Gaps = 2/30 (6%)
Query: 344 GGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
G + R GG GGG G GG RG GGR G
Sbjct: 174 GEKGPPRGGG--GGGRGGRGGGRGGGGRGG 201
>NOA1_HUMAN (Q9NY12) Nucleolar protein family A member 1 (snoRNP
protein GAR1) (H/ACA ribonucleoprotein GAR1)
Length = 217
Score = 50.8 bits (120), Expect = 5e-06
Identities = 31/62 (50%), Positives = 32/62 (51%), Gaps = 6/62 (9%)
Query: 321 RILQDLAGTQGIAYAAGRGGLGAG----GRAGGRSGGREGGGAGAMGGRRG--RGGRRGR 374
R L G +G GRGG G G GR GGR GG GG G GG RG GG RGR
Sbjct: 156 RFLPRPPGEKGPPRGGGRGGRGGGRGGGGRGGGRGGGFRGGRGGGGGGFRGGRGGGFRGR 215
Query: 375 GH 376
GH
Sbjct: 216 GH 217
Score = 35.8 bits (81), Expect = 0.18
Identities = 19/38 (50%), Positives = 19/38 (50%)
Query: 338 RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
RGG G G GG S GGG G GG GGR G G
Sbjct: 13 RGGGGGGFNRGGSSNHFRGGGGGGGGGNFRGGGRGGFG 50
Score = 34.7 bits (78), Expect = 0.41
Identities = 21/50 (42%), Positives = 22/50 (44%), Gaps = 2/50 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAM--GGRRGRGGRRGRG 375
G G G GG GG + GG GGG G GGR G G GRG
Sbjct: 7 GRGGFNRGGGGGGFNRGGSSNHFRGGGGGGGGGNFRGGGRGGFGRGGGRG 56
Score = 33.9 bits (76), Expect = 0.69
Identities = 22/51 (43%), Positives = 22/51 (43%), Gaps = 14/51 (27%)
Query: 337 GRGGLGAGGRAGG----------RSGGREGGGAGAMGGRRGR----GGRRG 373
GRGG GG GG R GG GGG GG RG GGR G
Sbjct: 7 GRGGFNRGGGGGGFNRGGSSNHFRGGGGGGGGGNFRGGGRGGFGRGGGRGG 57
Score = 30.4 bits (67), Expect = 7.7
Identities = 18/42 (42%), Positives = 18/42 (42%), Gaps = 7/42 (16%)
Query: 339 GGLGAGGRAGGRSGGREG-------GGAGAMGGRRGRGGRRG 373
GG G R GG G G GG G GG RGG RG
Sbjct: 6 GGRGGFNRGGGGGGFNRGGSSNHFRGGGGGGGGGNFRGGGRG 47
>FBRL_DROER (Q8I1F4) Fibrillarin
Length = 345
Score = 50.4 bits (119), Expect = 7e-06
Identities = 27/48 (56%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G GRGG G GGR GG GG GGG G GGR G GGR G G
Sbjct: 42 GRGGGGDRGGRGGFG-GGRGGGGRGGGGGGGRGGFGGRGGGGGRGGGG 88
Score = 48.9 bits (115), Expect = 2e-05
Identities = 24/39 (61%), Positives = 24/39 (61%)
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
GRGG G GG GG GGR GG G GGR G GG RG G
Sbjct: 24 GRGGGGGGGGGGGFGGGRGRGGGGDRGGRGGFGGGRGGG 62
Score = 47.4 bits (111), Expect = 6e-05
Identities = 23/39 (58%), Positives = 23/39 (58%)
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G GG G GG GR GG GGG G GG RGRGG RG
Sbjct: 12 GGGGGGGGGGFRGRGGGGGGGGGGGFGGGRGRGGGGDRG 50
Score = 46.2 bits (108), Expect = 1e-04
Identities = 24/42 (57%), Positives = 24/42 (57%), Gaps = 5/42 (11%)
Query: 339 GGLGAGGRAGGRSGGR-----EGGGAGAMGGRRGRGGRRGRG 375
GG G GGR GG GGR GGG G GG RG GGR G G
Sbjct: 57 GGRGGGGRGGGGGGGRGGFGGRGGGGGRGGGGRGGGGRGGGG 98
Score = 45.4 bits (106), Expect = 2e-04
Identities = 24/39 (61%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query: 337 GRGGLGAGGRAG--GRSGGREGGGAGAMGGRRGRGGRRG 373
GRGG G GGR G GR GG GG G GG RG GGR G
Sbjct: 63 GRGGGGGGGRGGFGGRGGGGGRGGGGRGGGGRGGGGRGG 101
Score = 44.3 bits (103), Expect = 5e-04
Identities = 25/51 (49%), Positives = 26/51 (50%), Gaps = 3/51 (5%)
Query: 328 GTQGIAYAAGRGGLGAGGRAG---GRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG G GR GG + GG G GG RG GGR G G
Sbjct: 18 GGGGFRGRGGGGGGGGGGGFGGGRGRGGGGDRGGRGGFGGGRGGGGRGGGG 68
Score = 43.1 bits (100), Expect = 0.001
Identities = 21/43 (48%), Positives = 22/43 (50%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
G +G GRGG G G GGR GG GGG GGR G G
Sbjct: 62 GGRGGGGGGGRGGFGGRGGGGGRGGGGRGGGGRGGGGRGGGAG 104
Score = 42.0 bits (97), Expect = 0.003
Identities = 26/50 (52%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query: 328 GTQGIAYAAGRGGLGAG-GRAGGRS-GGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G G GR GG GGR G G G GG RG GG GRG
Sbjct: 24 GRGGGGGGGGGGGFGGGRGRGGGGDRGGRGGFGGGRGGGGRGGGGGGGRG 73
Score = 40.4 bits (93), Expect = 0.007
Identities = 24/52 (46%), Positives = 25/52 (47%), Gaps = 4/52 (7%)
Query: 328 GTQGIAYAAGRGGLGAGGRA----GGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G + G GG G GG GG GG GGG G GR G G R GRG
Sbjct: 2 GKPGFSPRGGGGGGGGGGGGFRGRGGGGGGGGGGGFGGGRGRGGGGDRGGRG 53
Score = 40.4 bits (93), Expect = 0.007
Identities = 26/49 (53%), Positives = 26/49 (53%), Gaps = 3/49 (6%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAM-GGRRGRGGRRGRG 375
G G GRGG G G GG GGR GGG G GG GRGG GRG
Sbjct: 33 GGGGFGGGRGRGGGGDRGGRGGFGGGRGGGGRG--GGGGGGRGGFGGRG 79
>EBN1_EBV (P03211) EBNA-1 nuclear protein
Length = 641
Score = 50.1 bits (118), Expect = 9e-06
Identities = 28/49 (57%), Positives = 28/49 (57%), Gaps = 2/49 (4%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A G G GAGG G GG GGAGA GG RGRGG GRG
Sbjct: 292 AGGAGAGGAGGAGAGGAGG--AGAGGGAGAGGAGAGGGGRGRGGSGGRG 338
Score = 49.7 bits (117), Expect = 1e-05
Identities = 30/56 (53%), Positives = 31/56 (54%), Gaps = 7/56 (12%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGR--SGGREGGGAGAMG-----GRRGRGGRRGRG 375
AG G AG GG GAGG GR SGGR GG+G G GRRGRG R RG
Sbjct: 308 AGGAGAGGGAGAGGAGAGGGGRGRGGSGGRGRGGSGGRGRGGSGGRRGRGRERARG 363
Score = 48.1 bits (113), Expect = 4e-05
Identities = 25/48 (52%), Positives = 25/48 (52%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G A GG GAGG AGG G GGGAGA GG G G G G
Sbjct: 117 GAGGAGGAGAGGGAGAGGGAGGAGGAGAGGGAGAGGGAGGAGAGGGAG 164
Score = 48.1 bits (113), Expect = 4e-05
Identities = 24/45 (53%), Positives = 24/45 (53%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G GG GAGG AGG G GGGAGA GG G G G G
Sbjct: 147 GAGAGGGAGGAGAGGGAGGAGGAGAGGGAGAGGGAGGAGAGGGAG 191
Score = 47.8 bits (112), Expect = 5e-05
Identities = 28/50 (56%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMG-GRRGRGGRRGRG 375
AG G A A G GG GAGG AGG G GGAGA G G G GG G G
Sbjct: 255 AGGAGGAGAGGAGGAGAGGGAGGAGAGGGAGGAGAGGAGGAGAGGAGGAG 304
Score = 47.4 bits (111), Expect = 6e-05
Identities = 31/61 (50%), Positives = 31/61 (50%), Gaps = 12/61 (19%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMG---------GRRGRG---GRRGR 374
AG G A A G GG GAGG AG G GGG G G G RGRG GRRGR
Sbjct: 297 AGGAGGAGAGGAGGAGAGGGAGAGGAGAGGGGRGRGGSGGRGRGGSGGRGRGGSGGRRGR 356
Query: 375 G 375
G
Sbjct: 357 G 357
Score = 46.2 bits (108), Expect = 1e-04
Identities = 27/49 (55%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A AG GG GAGG AGG +GG GGAGA G G G G G
Sbjct: 204 AGGAGGAGGAGAGGAGAGGGAGG-AGGAGAGGAGAGGAGAGGAGAGGAG 251
Score = 45.4 bits (106), Expect = 2e-04
Identities = 26/50 (52%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAG-GRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A A G GG GAGG G G GG G GAG G G GG G G
Sbjct: 247 AGGAGGAGAGGAGGAGAGGAGGAGAGGGAGGAGAGGGAGGAGAGGAGGAG 296
Score = 45.1 bits (105), Expect = 3e-04
Identities = 25/49 (51%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A AG GG GAGG G +GG G GAG GG GG G G
Sbjct: 232 AGGAG-AGGAGAGGAGAGGAGGAGAGGAGGAGAGGAGGAGAGGGAGGAG 279
Score = 44.7 bits (104), Expect = 4e-04
Identities = 24/45 (53%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G GG GAGG AGG G GGGAGA GG G GG G
Sbjct: 174 GAGAGGGAGGAGAGGGAGGAGGAGAGGGAGA-GGAGGAGGAGAGG 217
Score = 43.9 bits (102), Expect = 7e-04
Identities = 23/45 (51%), Positives = 23/45 (51%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G A G GG GAGG AG G GGAGA GG GG G G
Sbjct: 114 GAGGAGGAGGAGAGGGAGAGGGAGGAGGAGAGGGAGAGGGAGGAG 158
Score = 43.9 bits (102), Expect = 7e-04
Identities = 26/52 (50%), Positives = 26/52 (50%), Gaps = 3/52 (5%)
Query: 327 AGTQGIAYAAGRGGLGAG---GRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A GG GAG G AGG G GGGAGA GG G GG G
Sbjct: 95 AGGAGAGGAGAGGGAGAGGGAGGAGGAGGAGAGGGAGAGGGAGGAGGAGAGG 146
Score = 43.5 bits (101), Expect = 9e-04
Identities = 23/49 (46%), Positives = 24/49 (48%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G AG GG G AGG +GG G GAG G G GG G G
Sbjct: 166 AGGAGAGGGAGAGGGAGGAGAGGGAGGAGGAGAGGGAGAGGAGGAGGAG 214
Score = 43.1 bits (100), Expect = 0.001
Identities = 22/43 (51%), Positives = 23/43 (53%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
G A A G GG GAGG G +GG G GAG G G GG G
Sbjct: 243 GGAGAGGAGGAGAGGAGGAGAGGAGGAGAGGGAGGAGAGGGAG 285
Score = 42.7 bits (99), Expect = 0.001
Identities = 25/49 (51%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMG-GRRGRGGRRGRG 375
G G A AG GG GAGG G +G GGAGA G G G GG G G
Sbjct: 222 GGAGGAGGAGAGGAGAGGAGAGGAGAGGAGGAGAGGAGGAGAGGAGGAG 270
Score = 42.4 bits (98), Expect = 0.002
Identities = 24/50 (48%), Positives = 24/50 (48%), Gaps = 2/50 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMG--GRRGRGGRRGRG 375
G G A GG GAGG AGG G GGAG G G G GG G G
Sbjct: 162 GAGGAGGAGAGGGAGAGGGAGGAGAGGGAGGAGGAGAGGGAGAGGAGGAG 211
Score = 42.4 bits (98), Expect = 0.002
Identities = 26/50 (52%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGR-EGGGAGAMGGRRGRGGRRGRG 375
AG G A AG G G G AGG +GG GGGAG GG G GG G G
Sbjct: 130 AGAGGGAGGAGGAGAGGGAGAGGGAGGAGAGGGAGGAGG-AGAGGGAGAG 178
Score = 42.4 bits (98), Expect = 0.002
Identities = 23/45 (51%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G AG GG GAGG AG G GGGAG GG G G G G
Sbjct: 88 GTGAGAGAGGAGAGG-AGAGGGAGAGGGAGGAGGAGGAGAGGGAG 131
Score = 42.4 bits (98), Expect = 0.002
Identities = 26/50 (52%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGR-EGGGAGAMGGRRGRGGRRGRG 375
AG G A AG G G G AGG +GG GGGAG GG G GG G G
Sbjct: 157 AGAGGGAGGAGGAGAGGGAGAGGGAGGAGAGGGAGGAGG-AGAGGGAGAG 205
Score = 42.0 bits (97), Expect = 0.003
Identities = 21/48 (43%), Positives = 21/48 (43%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G G GG G GGAGA GG G GG G
Sbjct: 153 GAGGAGAGGGAGGAGGAGAGGGAGAGGGAGGAGAGGGAGGAGGAGAGG 200
Score = 42.0 bits (97), Expect = 0.003
Identities = 22/48 (45%), Positives = 22/48 (45%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G A GG GAGG AGG G GGAG G G G G G
Sbjct: 135 GAGGAGGAGAGGGAGAGGGAGGAGAGGGAGGAGGAGAGGGAGAGGGAG 182
Score = 41.6 bits (96), Expect = 0.003
Identities = 22/49 (44%), Positives = 23/49 (46%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A A G GG G G G +GG GG GA G G GG G
Sbjct: 196 AGAGGGAGAGGAGGAGGAGAGGAGAGGGAGGAGGAGAGGAGAGGAGAGG 244
Score = 40.8 bits (94), Expect = 0.006
Identities = 24/50 (48%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Query: 327 AGTQGIAYAAGRGGLGAGGR-AGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A G G GAGG AGG G GGGAG G G GG G
Sbjct: 242 AGGAGAGGAGGAGAGGAGGAGAGGAGGAGAGGGAGGAGAGGGAGGAGAGG 291
Score = 40.4 bits (93), Expect = 0.007
Identities = 23/48 (47%), Positives = 24/48 (49%), Gaps = 2/48 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G A GG GAGG G +GG GGAGA GG G GG G
Sbjct: 189 GAGGAGGAGAGGGAGAGGAGG--AGGAGAGGAGAGGGAGGAGGAGAGG 234
Score = 40.4 bits (93), Expect = 0.007
Identities = 22/39 (56%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query: 339 GGLGAGGRAGGRSGGREG--GGAGAMGGRRGRGGRRGRG 375
GG GAG AGG G G GGAGA GG G GG G G
Sbjct: 87 GGTGAGAGAGGAGAGGAGAGGGAGAGGGAGGAGGAGGAG 125
Score = 40.4 bits (93), Expect = 0.007
Identities = 20/45 (44%), Positives = 20/45 (44%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G GG G G GG G GGAGA GG G GG G
Sbjct: 129 GAGAGGGAGGAGGAGAGGGAGAGGGAGGAGAGGGAGGAGGAGAGG 173
Score = 39.7 bits (91), Expect = 0.013
Identities = 21/45 (46%), Positives = 21/45 (46%), Gaps = 1/45 (2%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G GG G G AG G GGGAG GG G GG G G
Sbjct: 108 GAGAGGGAGGAGGAGGAGAGGGAGAGGGAGGAGG-AGAGGGAGAG 151
Score = 39.3 bits (90), Expect = 0.016
Identities = 21/49 (42%), Positives = 21/49 (42%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G AG GG G G AG G GG GA G G G G G
Sbjct: 193 AGGAGAGGGAGAGGAGGAGGAGAGGAGAGGGAGGAGGAGAGGAGAGGAG 241
Score = 39.3 bits (90), Expect = 0.016
Identities = 21/45 (46%), Positives = 22/45 (48%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G A A G G GAGG G +GG GGAG G G GG G
Sbjct: 238 GGAGAGGAGAGGAGGAGAGGAGGAGAGGAGGAGAGGGAGGAGAGG 282
Score = 39.3 bits (90), Expect = 0.016
Identities = 19/46 (41%), Positives = 19/46 (41%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
G G G GG G G GG G GG GA G G GG G
Sbjct: 180 GAGGAGAGGGAGGAGGAGAGGGAGAGGAGGAGGAGAGGAGAGGGAG 225
Score = 39.3 bits (90), Expect = 0.016
Identities = 25/50 (50%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREG-GGAGAMGGRRGRGGRRGRG 375
AG G A AG GG GAGG G +GG G GGAG GG G G G
Sbjct: 175 AGAGGGAGGAGAGG-GAGGAGGAGAGGGAGAGGAGGAGGAGAGGAGAGGG 223
Score = 38.5 bits (88), Expect = 0.028
Identities = 27/53 (50%), Positives = 28/53 (51%), Gaps = 7/53 (13%)
Query: 327 AGTQGIAYAAGRGGLGAGGR----AGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A AG GG GAGG AGG +G GGGAG G G GG G G
Sbjct: 148 AGAGGGAGGAGAGG-GAGGAGGAGAGGGAGA--GGGAGGAGAGGGAGGAGGAG 197
Score = 38.1 bits (87), Expect = 0.037
Identities = 23/49 (46%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A A G G G AGG +G GGGAG G G GG G G
Sbjct: 124 AGAGGGAGAGGGAGGAGGAGAGGGAGA--GGGAGGAGAGGGAGGAGGAG 170
Score = 37.7 bits (86), Expect = 0.048
Identities = 24/56 (42%), Positives = 28/56 (49%), Gaps = 9/56 (16%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAG---------AMGGRRGRGGRRGR 374
G +G + GRG G+GGR G SGGR G G A G RGRG +R R
Sbjct: 327 GGRGRGGSGGRGRGGSGGRGRGGSGGRRGRGRERARGGSRERARGRGRGRGEKRPR 382
Score = 37.4 bits (85), Expect = 0.063
Identities = 23/50 (46%), Positives = 23/50 (46%), Gaps = 1/50 (2%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMG-GRRGRGGRRGRG 375
AG G A A G G G G AGG G G G GA G G G GG G
Sbjct: 190 AGGAGGAGAGGGAGAGGAGGAGGAGAGGAGAGGGAGGAGGAGAGGAGAGG 239
Score = 36.6 bits (83), Expect = 0.11
Identities = 22/48 (45%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG G R GRE G+ RGRG RGRG
Sbjct: 332 GGSGGRGRGGSGGRGRGGSGGRRGRGRERARGGSRERARGRG--RGRG 377
Score = 31.6 bits (70), Expect = 3.4
Identities = 17/39 (43%), Positives = 17/39 (43%)
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G GG G R G G G G G GGR G G G G
Sbjct: 25 GSGGSGPQRRGGDNHGRGRGRGRGRGGGRPGAPGGSGSG 63
>FBRL_SCHPO (P35551) Fibrillarin
Length = 305
Score = 49.7 bits (117), Expect = 1e-05
Identities = 25/39 (64%), Positives = 25/39 (64%), Gaps = 2/39 (5%)
Query: 337 GRGGL--GAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
GRGG G GG GGR G R GG GA GGR GRGG RG
Sbjct: 17 GRGGFNGGRGGFGGGRGGARGGGRGGARGGRGGRGGARG 55
Score = 43.9 bits (102), Expect = 7e-04
Identities = 25/41 (60%), Positives = 26/41 (62%), Gaps = 3/41 (7%)
Query: 334 YAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRG-RGGRRG 373
+ GRGG G GGR G R GGR GG G GGR G RGGR G
Sbjct: 21 FNGGRGGFG-GGRGGARGGGR-GGARGGRGGRGGARGGRGG 59
Score = 42.4 bits (98), Expect = 0.002
Identities = 21/40 (52%), Positives = 22/40 (54%), Gaps = 2/40 (5%)
Query: 334 YAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
+ GRGG GGR G R G GG GA GGR G G RG
Sbjct: 28 FGGGRGGARGGGRGGARGG--RGGRGGARGGRGGSSGGRG 65
Score = 41.6 bits (96), Expect = 0.003
Identities = 27/43 (62%), Positives = 27/43 (62%), Gaps = 5/43 (11%)
Query: 337 GRGGL--GAGGRAGGRSGGREGGGAGAMGGRRG--RGGRRGRG 375
GRGG G GG GGR GG GG GA GG RG RGGR GRG
Sbjct: 10 GRGGSRGGRGGFNGGR-GGFGGGRGGARGGGRGGARGGRGGRG 51
Score = 35.8 bits (81), Expect = 0.18
Identities = 20/42 (47%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query: 328 GTQGIAYAAGRGGL--GAGGRAGGRSGGREGGGAGAMGGRRG 367
G +G A GRGG G GGR G R G GG +G GG +G
Sbjct: 30 GGRGGARGGGRGGARGGRGGRGGARGG--RGGSSGGRGGAKG 69
>SSB2_STRCO (Q9X8U3) Single-strand binding protein 2 (SSB 2)
(Helix-destabilizing protein 2)
Length = 199
Score = 49.3 bits (116), Expect = 2e-05
Identities = 32/86 (37%), Positives = 42/86 (48%), Gaps = 14/86 (16%)
Query: 300 LEVDDALLPGTQARALTKRALRILQDLAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGA 359
L+VD+ G R+ T + + G QG Y G GG G GG GG GG++GGGA
Sbjct: 101 LDVDEV---GASLRSATAKVTKTSGQGRGGQG-GYGGGGGGQGGGGWGGGPGGGQQGGGA 156
Query: 360 GA----------MGGRRGRGGRRGRG 375
A GG++G GG+ G G
Sbjct: 157 PADDPWATGGAPAGGQQGGGGQGGGG 182
>FBRL_LEIMA (P35549) Fibrillarin
Length = 297
Score = 48.9 bits (115), Expect = 2e-05
Identities = 26/40 (65%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query: 337 GRGGLGAG-GRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
GRGG G G GR GGR GGR GG G GG RG GGR G G
Sbjct: 18 GRGGFGRGGGRGGGRGGGRGGGRGGGRGGGRG-GGRGGAG 56
Score = 48.9 bits (115), Expect = 2e-05
Identities = 25/48 (52%), Positives = 25/48 (52%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G RGG G GR GGR GGR GG G GG RG G GRG
Sbjct: 6 GRGGGGRGGSRGGRGGFGRGGGRGGGRGGGRGGGRGGGRGGGRGGGRG 53
Score = 47.4 bits (111), Expect = 6e-05
Identities = 23/45 (51%), Positives = 26/45 (57%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRR 372
G++G GRGG GGR GGR GGR GG G GG RG G +
Sbjct: 14 GSRGGRGGFGRGGGRGGGRGGGRGGGRGGGRGGGRGGGRGGAGAK 58
Score = 40.8 bits (94), Expect = 0.006
Identities = 25/49 (51%), Positives = 25/49 (51%), Gaps = 10/49 (20%)
Query: 337 GRGGLGAGGRAG--------GRSGGREGGGAGAMGGRR--GRGGRRGRG 375
G G G GGR G GR GGR GG G GG R GRGG RG G
Sbjct: 3 GGFGRGGGGRGGSRGGRGGFGRGGGRGGGRGGGRGGGRGGGRGGGRGGG 51
Score = 33.1 bits (74), Expect = 1.2
Identities = 21/38 (55%), Positives = 21/38 (55%), Gaps = 8/38 (21%)
Query: 338 RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
RGG G GG GGR G R GG G G RGG RG G
Sbjct: 2 RGGFGRGG--GGRGGSR--GGRGGFG----RGGGRGGG 31
>PG33_MYCTU (Q50615) Hypothetical PE-PGRS family protein PE_PGRS33
Length = 498
Score = 48.1 bits (113), Expect = 4e-05
Identities = 23/47 (48%), Positives = 26/47 (54%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
+GT G A G GGL G G +GGR GGG G +GG G GG G
Sbjct: 177 SGTAGFGGAGGAGGLLYGAGGAGGAGGRAGGGVGGIGGAGGAGGNGG 223
Score = 38.5 bits (88), Expect = 0.028
Identities = 24/48 (50%), Positives = 25/48 (52%), Gaps = 6/48 (12%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAG----GRSGGREGGGAGAMGGRRGRGG 370
AG G A G GGL GG+AG G GG G G GA GG G GG
Sbjct: 412 AGGFGFGGAGGAGGL--GGKAGLIGDGGDGGAGGNGTGAKGGDGGAGG 457
Score = 37.4 bits (85), Expect = 0.063
Identities = 23/49 (46%), Positives = 25/49 (50%), Gaps = 2/49 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAM-GGRRGRGGRRGRG 375
G G+ G G GAGG AG G GGAGA+ GG G GG G G
Sbjct: 353 GAGGVGSTTGGAG-GAGGNAGLLVGAGGAGGAGALGGGATGVGGAGGNG 400
Score = 37.4 bits (85), Expect = 0.063
Identities = 23/53 (43%), Positives = 26/53 (48%), Gaps = 7/53 (13%)
Query: 328 GTQGIAYAAGRGG-----LGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G+ A G GG GAGG G GG GGAG +GG+ G G G G
Sbjct: 389 GATGVGGAGGNGGTAGLLFGAGGAGGFGFGG--AGGAGGLGGKAGLIGDGGDG 439
Score = 36.6 bits (83), Expect = 0.11
Identities = 22/53 (41%), Positives = 24/53 (44%), Gaps = 5/53 (9%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMG-----GRRGRGGRRGRG 375
G G+ AG G G G AGG G +GG GA G G G GG G G
Sbjct: 422 GAGGLGGKAGLIGDGGDGGAGGNGTGAKGGDGGAGGGAILVGNGGNGGNAGSG 474
Score = 36.6 bits (83), Expect = 0.11
Identities = 25/59 (42%), Positives = 26/59 (43%), Gaps = 12/59 (20%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGG-----------AGAMGGRRGRGGRRGRG 375
G G+ Y AG G GAGGRAGG GG G G AG GG G G G
Sbjct: 187 GAGGLLYGAGGAG-GAGGRAGGGVGGIGGAGGAGGNGGLLFGAGGAGGVGGLAADAGDG 244
Score = 36.2 bits (82), Expect = 0.14
Identities = 24/56 (42%), Positives = 26/56 (45%), Gaps = 6/56 (10%)
Query: 326 LAGTQGIAYAAGRGGLGAGGRAG-GRSGGREG-----GGAGAMGGRRGRGGRRGRG 375
L G G A GR G G GG G G +GG G GGAG +GG G G G
Sbjct: 192 LYGAGGAGGAGGRAGGGVGGIGGAGGAGGNGGLLFGAGGAGGVGGLAADAGDGGAG 247
Score = 35.8 bits (81), Expect = 0.18
Identities = 22/49 (44%), Positives = 22/49 (44%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A G GG A G AG G GG GG G GGR G G
Sbjct: 160 AGLFGNGGAGGAGGNVASGTAGFGGAGGAGGLLYGAGGAGGAGGRAGGG 208
Score = 35.4 bits (80), Expect = 0.24
Identities = 24/60 (40%), Positives = 26/60 (43%), Gaps = 11/60 (18%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAG-----------GRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G A A G G G GG G G +GG GGAG GG G+ G G G
Sbjct: 377 AGGAGGAGALGGGATGVGGAGGNGGTAGLLFGAGGAGGFGFGGAGGAGGLGGKAGLIGDG 436
Score = 35.0 bits (79), Expect = 0.31
Identities = 22/47 (46%), Positives = 23/47 (48%), Gaps = 5/47 (10%)
Query: 329 TQGIAYAAGRGGL--GAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
T G A G GL GAGG G G GGGA +GG G GG G
Sbjct: 361 TGGAGGAGGNAGLLVGAGGAGGA---GALGGGATGVGGAGGNGGTAG 404
Score = 34.3 bits (77), Expect = 0.53
Identities = 20/50 (40%), Positives = 23/50 (46%), Gaps = 2/50 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGR--EGGGAGAMGGRRGRGGRRGRG 375
G+ G A G GG G G G +GG GG AG + G G GG G
Sbjct: 338 GSGGSALLWGDGGAGGAGGVGSTTGGAGGAGGNAGLLVGAGGAGGAGALG 387
Score = 33.9 bits (76), Expect = 0.69
Identities = 21/49 (42%), Positives = 24/49 (48%), Gaps = 3/49 (6%)
Query: 328 GTQGIAY-AAGRGGLGAGGRAGGRSGGREGGGAGAMG--GRRGRGGRRG 373
G G+A+ GG G G AGG G GGAG +G G G GG G
Sbjct: 293 GGDGVAFLGTAPGGPGGAGGAGGLFGVGGAGGAGGIGLVGNGGAGGSGG 341
Score = 33.9 bits (76), Expect = 0.69
Identities = 20/49 (40%), Positives = 22/49 (44%), Gaps = 3/49 (6%)
Query: 328 GTQGIAYAAGRGGLGAGGR---AGGRSGGREGGGAGAMGGRRGRGGRRG 373
G G+A AG GG G G G +GG G G GG G GG G
Sbjct: 233 GVGGLAADAGDGGAGGDGGLFFGVGGAGGAGGTGTNVTGGAGGAGGNGG 281
Score = 33.5 bits (75), Expect = 0.90
Identities = 19/48 (39%), Positives = 21/48 (43%), Gaps = 1/48 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G+ + AG G G GG A G GG G G G GG G G
Sbjct: 220 GNGGLLFGAGGAG-GVGGLAADAGDGGAGGDGGLFFGVGGAGGAGGTG 266
Score = 32.7 bits (73), Expect = 1.5
Identities = 21/54 (38%), Positives = 24/54 (43%), Gaps = 8/54 (14%)
Query: 328 GTQGIAYAAGRGGLGAGGRAG-GRSGGRE-------GGGAGAMGGRRGRGGRRG 373
G G+ A G GG+G G G G SGG GGAG +G G G G
Sbjct: 315 GLFGVGGAGGAGGIGLVGNGGAGGSGGSALLWGDGGAGGAGGVGSTTGGAGGAG 368
Score = 32.7 bits (73), Expect = 1.5
Identities = 17/47 (36%), Positives = 18/47 (38%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
AG G G GG G G G G G+G G G GG G
Sbjct: 441 AGGNGTGAKGGDGGAGGGAILVGNGGNGGNAGSGTPNGSAGTGGAGG 487
Score = 32.7 bits (73), Expect = 1.5
Identities = 21/49 (42%), Positives = 23/49 (46%), Gaps = 2/49 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSG-GREGGGAGAMGGRRGRGGRRGRG 375
G G+ AG G GAG GG +G G GG G G G GG G G
Sbjct: 369 GNAGLLVGAGGAG-GAGALGGGATGVGGAGGNGGTAGLLFGAGGAGGFG 416
Score = 32.3 bits (72), Expect = 2.0
Identities = 20/50 (40%), Positives = 22/50 (44%), Gaps = 3/50 (6%)
Query: 328 GTQGIAYAAGRGGL--GAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG+ G GG G + G GG GA G G GG G G
Sbjct: 124 GAPGTGANGGDGGILIGNGGAGGSGAAGMPGGNGGA-AGLFGNGGAGGAG 172
Score = 32.0 bits (71), Expect = 2.6
Identities = 23/52 (44%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query: 328 GTQGIAYAAGR-GGLGAGGRA--GGRSGGREG-GGAGAMGGRRGRGGRRGRG 375
G G+ + AG GG+G G A G GG G GGAG + G G GG G G
Sbjct: 278 GNGGLLFGAGGVGGVGGDGVAFLGTAPGGPGGAGGAGGLFGVGGAGGAGGIG 329
Score = 32.0 bits (71), Expect = 2.6
Identities = 16/43 (37%), Positives = 20/43 (46%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
G G A G GG G +G +G GGAG + G+ G G
Sbjct: 454 GAGGGAILVGNGGNGGNAGSGTPNGSAGTGGAGGLLGKNGMNG 496
Score = 31.6 bits (70), Expect = 3.4
Identities = 22/50 (44%), Positives = 23/50 (46%), Gaps = 11/50 (22%)
Query: 337 GRGGLGAGGRAG--GRSGGREG----GGAGAMGGR-----RGRGGRRGRG 375
G GG G G AG G +GG G GGAG GG G GG G G
Sbjct: 140 GNGGAGGSGAAGMPGGNGGAAGLFGNGGAGGAGGNVASGTAGFGGAGGAG 189
Score = 31.6 bits (70), Expect = 3.4
Identities = 20/53 (37%), Positives = 22/53 (40%), Gaps = 4/53 (7%)
Query: 327 AGTQGIAYAAGRGGLG----AGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG G G GG+G AGG G G GG G + G GG G G
Sbjct: 198 AGGAGGRAGGGVGGIGGAGGAGGNGGLLFGAGGAGGVGGLAADAGDGGAGGDG 250
Score = 30.8 bits (68), Expect = 5.9
Identities = 17/42 (40%), Positives = 19/42 (44%), Gaps = 3/42 (7%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGG---GAGAMGGRRGRG 369
G+ A G GG G G G GG GAG +GG G G
Sbjct: 255 GVGGAGGAGGTGTNVTGGAGGAGGNGGLLFGAGGVGGVGGDG 296
Score = 30.4 bits (67), Expect = 7.7
Identities = 20/49 (40%), Positives = 22/49 (44%), Gaps = 8/49 (16%)
Query: 335 AAGRGGL----GAGGRAG----GRSGGREGGGAGAMGGRRGRGGRRGRG 375
A G GGL GAGG G G G GG+ + G G GG G G
Sbjct: 310 AGGAGGLFGVGGAGGAGGIGLVGNGGAGGSGGSALLWGDGGAGGAGGVG 358
Score = 30.4 bits (67), Expect = 7.7
Identities = 18/45 (40%), Positives = 19/45 (42%)
Query: 326 LAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
LA G A G GGL G G +GG G GG G GG
Sbjct: 237 LAADAGDGGAGGDGGLFFGVGGAGGAGGTGTNVTGGAGGAGGNGG 281
>RS2_CAEEL (P51403) 40S ribosomal protein S2
Length = 272
Score = 47.8 bits (112), Expect = 5e-05
Identities = 27/49 (55%), Positives = 27/49 (55%), Gaps = 9/49 (18%)
Query: 336 AGRGGL--GAGGRAGGRSGGREGG-------GAGAMGGRRGRGGRRGRG 375
A RGG G GGR GGR G R G G G GGR GRGGR GRG
Sbjct: 2 ADRGGFQSGFGGRGGGRGGARPAGDRPAGRGGRGGRGGRGGRGGRAGRG 50
>GRP1_PETHY (P09789) Glycine-rich cell wall structural protein 1
precursor
Length = 384
Score = 47.4 bits (111), Expect = 6e-05
Identities = 27/49 (55%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGA-GAMGGRRGRGGRRGRG 375
G G A G GGLG GG AGG GG GGGA G +GG G GG G G
Sbjct: 88 GLGGGGGAGGGGGLGGGGGAGGGFGGGAGGGAGGGLGGGGGLGGGGGGG 136
Score = 43.1 bits (100), Expect = 0.001
Identities = 25/49 (51%), Positives = 28/49 (57%), Gaps = 2/49 (4%)
Query: 328 GTQGIAYAAGRGG-LGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G+ +G GG GAGG GG +GG GGG GA GG G GG G G
Sbjct: 301 GGGGVGGGSGHGGGFGAGGGVGGGAGGGLGGGGGA-GGGGGIGGGHGGG 348
Score = 42.4 bits (98), Expect = 0.002
Identities = 24/48 (50%), Positives = 25/48 (52%), Gaps = 1/48 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G A GGLG GG GG GG GGG G +GG G GG G G
Sbjct: 110 GFGGGAGGGAGGGLGGGGGLGGGGGGGAGGGGG-VGGGAGSGGGFGAG 156
Score = 42.0 bits (97), Expect = 0.003
Identities = 22/50 (44%), Positives = 24/50 (48%)
Query: 326 LAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
+ G G G GG GG GG +GG GGG G GG G GG G G
Sbjct: 225 IGGGSGHGGGFGAGGGVGGGVGGGAAGGGGGGGGGGGGGGGGLGGGSGHG 274
Score = 42.0 bits (97), Expect = 0.003
Identities = 23/48 (47%), Positives = 27/48 (55%), Gaps = 3/48 (6%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G +G ++ GRG AGG GG +GG GGG G GG G GG G G
Sbjct: 55 GGRGPSFGRGRG---AGGGFGGGAGGGAGGGLGGGGGLGGGGGAGGGG 99
Score = 42.0 bits (97), Expect = 0.003
Identities = 26/51 (50%), Positives = 28/51 (53%), Gaps = 3/51 (5%)
Query: 327 AGTQGIAYAAGR--GGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
AG+ G A G GG GAGG GG GG GGG G +GG G GG G G
Sbjct: 147 AGSGGGFGAGGGVGGGAGAGGGVGG-GGGFGGGGGGGVGGGSGHGGGFGAG 196
Score = 41.6 bits (96), Expect = 0.003
Identities = 22/50 (44%), Positives = 23/50 (46%)
Query: 326 LAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
+ G G G GG GG GG GG GGG G GG G GG G G
Sbjct: 183 VGGGSGHGGGFGAGGGVGGGAGGGLGGGVGGGGGGGSGGGGGIGGGSGHG 232
Score = 41.6 bits (96), Expect = 0.003
Identities = 25/50 (50%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGR--SGGREGGGAGAMGGRRGRGGRRGRG 375
G G A G GG+G G +GG +GG GGGAGA GG G GG G G
Sbjct: 130 GGGGGGGAGGGGGVGGGAGSGGGFGAGGGVGGGAGAGGGVGGGGGFGGGG 179
Score = 41.2 bits (95), Expect = 0.004
Identities = 24/49 (48%), Positives = 27/49 (54%), Gaps = 2/49 (4%)
Query: 328 GTQGIAYAAGRGG-LGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G+ +G GG GAGG GG +GG GGG G GG G GG G G
Sbjct: 179 GGGGVGGGSGHGGGFGAGGGVGGGAGGGLGGGVGG-GGGGGSGGGGGIG 226
Score = 41.2 bits (95), Expect = 0.004
Identities = 22/50 (44%), Positives = 23/50 (46%)
Query: 326 LAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
L G G+ G GG G G GG GG GG G GG G GG G G
Sbjct: 83 LGGGGGLGGGGGAGGGGGLGGGGGAGGGFGGGAGGGAGGGLGGGGGLGGG 132
Score = 40.8 bits (94), Expect = 0.006
Identities = 22/39 (56%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G GGLG GG G GG GGGAG+ GG G GG G G
Sbjct: 125 GGGGLGGGGGGGAGGGGGVGGGAGS-GGGFGAGGGVGGG 162
Score = 40.8 bits (94), Expect = 0.006
Identities = 22/45 (48%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G GG AGG GG GG GGG G +GG G GG G G
Sbjct: 238 GGGVGGGVGGGAAGGGGGGGGGG--GGGGGGLGGGSGHGGGFGAG 280
Score = 40.8 bits (94), Expect = 0.006
Identities = 24/48 (50%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G + G GG GAGG GG GG GGG G GG G GG G G
Sbjct: 105 GGAGGGFGGGAGG-GAGGGLGG-GGGLGGGGGGGAGGGGGVGGGAGSG 150
Score = 40.4 bits (93), Expect = 0.007
Identities = 24/52 (46%), Positives = 25/52 (47%), Gaps = 2/52 (3%)
Query: 326 LAGTQGIAYAAGRGGLGAGGRAGGRSGGRE--GGGAGAMGGRRGRGGRRGRG 375
L G G G GG GG AGG GG GGG G +GG G GG G G
Sbjct: 267 LGGGSGHGGGFGAGGGVGGGAAGGVGGGGGFGGGGGGGVGGGSGHGGGFGAG 318
Score = 40.0 bits (92), Expect = 0.010
Identities = 24/51 (47%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Query: 328 GTQGIAYAAGRGG-LGAGGRAGGRSGGREG-GGAGAMGGRRGRGGRRGRGH 376
G G+ AG GG GAGG GG +G G GG G GG G G G GH
Sbjct: 139 GGGGVGGGAGSGGGFGAGGGVGGGAGAGGGVGGGGGFGGGGGGGVGGGSGH 189
Score = 39.3 bits (90), Expect = 0.016
Identities = 24/48 (50%), Positives = 26/48 (54%), Gaps = 3/48 (6%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G+ G GG G GG AGG +GG GGG G GG G GG G G
Sbjct: 97 GGGGLGGGGGAGG-GFGGGAGGGAGGGLGGGGGLGGG--GGGGAGGGG 141
Score = 38.9 bits (89), Expect = 0.022
Identities = 24/49 (48%), Positives = 27/49 (54%), Gaps = 4/49 (8%)
Query: 331 GIAYAAGRG-GLGAGGRAGGRSGGREGGGA---GAMGGRRGRGGRRGRG 375
G + AG G G GAGG GG GG GGG+ G +GG G GG G G
Sbjct: 190 GGGFGAGGGVGGGAGGGLGGGVGGGGGGGSGGGGGIGGGSGHGGGFGAG 238
Score = 38.5 bits (88), Expect = 0.028
Identities = 23/46 (50%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query: 331 GIAYAAGRGG-LGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G+ AG GG +G GG GG GG GGG+G GG G GG G G
Sbjct: 158 GVGGGAGAGGGVGGGGGFGGGGGGGVGGGSG-HGGGFGAGGGVGGG 202
Score = 38.5 bits (88), Expect = 0.028
Identities = 26/53 (49%), Positives = 27/53 (50%), Gaps = 4/53 (7%)
Query: 328 GTQGIAYAAGRGG-LGAGGRAGGRSGGRE--GGGAGAMGGRRGRGG-RRGRGH 376
G GI +G GG GAGG GG GG GGG G GG G GG G GH
Sbjct: 221 GGGGIGGGSGHGGGFGAGGGVGGGVGGGAAGGGGGGGGGGGGGGGGLGGGSGH 273
Score = 38.1 bits (87), Expect = 0.037
Identities = 22/48 (45%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G GG+G GG GG GG GGG+G GG G GG G G
Sbjct: 278 GAGGGVGGGAAGGVGGGGGFGGGGGGGVGGGSG-HGGGFGAGGGVGGG 324
Score = 37.7 bits (86), Expect = 0.048
Identities = 22/48 (45%), Positives = 23/48 (47%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G+ G G G G G AGG GG GGGA GG G GG G G
Sbjct: 218 GSGGGGGIGGGSGHGGGFGAGGGVGGGVGGGAAGGGGGGGGGGGGGGG 265
Score = 37.4 bits (85), Expect = 0.063
Identities = 22/48 (45%), Positives = 22/48 (45%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GGLG G GG G G G GA GG G GG G G
Sbjct: 254 GGGGGGGGGGGGGLGGGSGHGGGFGAGGGVGGGAAGGVGGGGGFGGGG 301
Score = 37.4 bits (85), Expect = 0.063
Identities = 23/49 (46%), Positives = 23/49 (46%), Gaps = 1/49 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G G G G G G AGG GG GGG G GG G GG G GH
Sbjct: 298 GGGGGGGVGGGSGHGGGFGAGGGVGGGAGGGLGG-GGGAGGGGGIGGGH 345
Score = 37.0 bits (84), Expect = 0.082
Identities = 23/55 (41%), Positives = 25/55 (44%), Gaps = 7/55 (12%)
Query: 328 GTQGIAYAAGRGGLGAG-------GRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G + G GG+G G G GG GG GG AG GG G GG G G
Sbjct: 212 GGGGGGGSGGGGGIGGGSGHGGGFGAGGGVGGGVGGGAAGGGGGGGGGGGGGGGG 266
Score = 36.6 bits (83), Expect = 0.11
Identities = 22/48 (45%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G GG G GG GG GG GGG+G GG G GG G G
Sbjct: 240 GVGGGVGGGAAGGGGGGGGGGGGGGGGLGGGSG-HGGGFGAGGGVGGG 286
Score = 36.2 bits (82), Expect = 0.14
Identities = 23/51 (45%), Positives = 24/51 (46%), Gaps = 3/51 (5%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGA---MGGRRGRGGRRGRG 375
G G G GG GAGG G G GGG GA +GG G GG G G
Sbjct: 122 GLGGGGGLGGGGGGGAGGGGGVGGGAGSGGGFGAGGGVGGGAGAGGGVGGG 172
Score = 36.2 bits (82), Expect = 0.14
Identities = 24/61 (39%), Positives = 25/61 (40%), Gaps = 13/61 (21%)
Query: 328 GTQGIAYAAGRGGLGAGGRAG-------------GRSGGREGGGAGAMGGRRGRGGRRGR 374
G G A G GG G GG G G GG GG AG +GG G GG G
Sbjct: 244 GVGGGAAGGGGGGGGGGGGGGGGLGGGSGHGGGFGAGGGVGGGAAGGVGGGGGFGGGGGG 303
Query: 375 G 375
G
Sbjct: 304 G 304
Score = 36.2 bits (82), Expect = 0.14
Identities = 20/45 (44%), Positives = 20/45 (44%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G A G GG G GG GG GG GG GG G G G G
Sbjct: 248 GAAGGGGGGGGGGGGGGGGLGGGSGHGGGFGAGGGVGGGAAGGVG 292
Score = 36.2 bits (82), Expect = 0.14
Identities = 23/48 (47%), Positives = 24/48 (49%), Gaps = 3/48 (6%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G A GGLG GG GG GG GG G +GG G GG G G
Sbjct: 70 GFGGGAGGGAGGGLGGGGGLGG-GGGA--GGGGGLGGGGGAGGGFGGG 114
Score = 36.2 bits (82), Expect = 0.14
Identities = 22/53 (41%), Positives = 24/53 (44%), Gaps = 5/53 (9%)
Query: 328 GTQGIAYAAGRGGLGAGGRAG-----GRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG +G G GG GG G +GG G GG G G
Sbjct: 168 GVGGGGGFGGGGGGGVGGGSGHGGGFGAGGGVGGGAGGGLGGGVGGGGGGGSG 220
Score = 35.8 bits (81), Expect = 0.18
Identities = 22/48 (45%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G A GG+G GG G GG GGG+G GG G GG G G
Sbjct: 198 GVGGGAGGGLGGGVGGGGGGGSGGGGGIGGGSG-HGGGFGAGGGVGGG 244
Score = 35.0 bits (79), Expect = 0.31
Identities = 24/50 (48%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRG--RGGRRGRG 375
G G G GG G GG GG SG GGG GA GG G GG G G
Sbjct: 206 GLGGGVGGGGGGGSGGGGGIGGGSG--HGGGFGAGGGVGGGVGGGAAGGG 253
Score = 35.0 bits (79), Expect = 0.31
Identities = 19/45 (42%), Positives = 20/45 (44%), Gaps = 2/45 (4%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G+ G GG G G GG GG GG G GG G G G G
Sbjct: 82 GLGGGGGLGGGGGAGGGGGLGGG--GGAGGGFGGGAGGGAGGGLG 124
Score = 35.0 bits (79), Expect = 0.31
Identities = 20/40 (50%), Positives = 20/40 (50%), Gaps = 1/40 (2%)
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGG-RRGRGGRRGRG 375
G GG G GG G G GGG GA GG G GG G G
Sbjct: 293 GGGGFGGGGGGGVGGGSGHGGGFGAGGGVGGGAGGGLGGG 332
Score = 34.7 bits (78), Expect = 0.41
Identities = 21/48 (43%), Positives = 21/48 (43%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G G G G G AGG GG GGG G G G GG G G
Sbjct: 176 GGGGGGGVGGGSGHGGGFGAGGGVGGGAGGGLGGGVGGGGGGGSGGGG 223
Score = 34.3 bits (77), Expect = 0.53
Identities = 22/50 (44%), Positives = 23/50 (46%), Gaps = 6/50 (12%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSG-----GREGGGAGAMGGRRGRGGRRGRG 375
G+ G GG G GG GG SG G GG G GG G GG G G
Sbjct: 290 GVGGGGGFGG-GGGGGVGGGSGHGGGFGAGGGVGGGAGGGLGGGGGAGGG 338
Score = 33.9 bits (76), Expect = 0.69
Identities = 20/52 (38%), Positives = 21/52 (39%), Gaps = 13/52 (25%)
Query: 337 GRGGLGAG-------------GRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
GR G G G G GG GG GG G +GG G GG G G
Sbjct: 45 GRRGCGGGRFGGRGPSFGRGRGAGGGFGGGAGGGAGGGLGGGGGLGGGGGAG 96
Score = 33.5 bits (75), Expect = 0.90
Identities = 23/52 (44%), Positives = 24/52 (45%), Gaps = 2/52 (3%)
Query: 326 LAGTQGIAYAAGRGGLGAGGRAGGRSGGR-EGGGAGAMGG-RRGRGGRRGRG 375
+ G G G GG GG GG GG GGG GA GG G GG G G
Sbjct: 159 VGGGAGAGGGVGGGGGFGGGGGGGVGGGSGHGGGFGAGGGVGGGAGGGLGGG 210
Score = 32.3 bits (72), Expect = 2.0
Identities = 22/49 (44%), Positives = 24/49 (48%), Gaps = 4/49 (8%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSG---GREGGGAGAMGGRRGRGGRRGRGH 376
G+ AG GGLG GG AGG G G GG +G G G G GH
Sbjct: 320 GVGGGAG-GGLGGGGGAGGGGGIGGGHGGGFGVGVGIGIGVGVGAGAGH 367
Score = 30.4 bits (67), Expect = 7.7
Identities = 17/44 (38%), Positives = 22/44 (49%), Gaps = 1/44 (2%)
Query: 326 LAGTQGIAYAAGRG-GLGAGGRAGGRSGGREGGGAGAMGGRRGR 368
+ G G + G G G+G G AG G G G+G+ GG GR
Sbjct: 341 IGGGHGGGFGVGVGIGIGVGVGAGAGHGVGVGSGSGSGGGGNGR 384
>GDF7_MOUSE (P43029) Growth/differentiation factor 7 precursor
(GDF-7)
Length = 461
Score = 47.4 bits (111), Expect = 6e-05
Identities = 26/43 (60%), Positives = 27/43 (62%), Gaps = 3/43 (6%)
Query: 326 LAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGR 368
LAGT+G A G GG G GG GG GG GGGAG GRRGR
Sbjct: 318 LAGTRG---AQGSGGGGGGGGGGGGGGGGGGGGAGRGHGRRGR 357
Score = 44.7 bits (104), Expect = 4e-04
Identities = 36/104 (34%), Positives = 45/104 (42%), Gaps = 12/104 (11%)
Query: 282 DLSAVAALRRALEVLELSLEVDDALLPGTQARALTKRALRILQDLAGTQGIAYAAGRGGL 341
D AA RAL V+ + ++L +A+A RALR + G + + L
Sbjct: 252 DGGGTAAEERALLVISSRTQRKESLFREIRAQA---RALRAAAEPPPDPGPGAGSRKANL 308
Query: 342 G---------AGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G AG R SGG GGG G GG G GG GRGH
Sbjct: 309 GGRRRRRTALAGTRGAQGSGGGGGGGGGGGGGGGGGGGGAGRGH 352
>5E5_RAT (Q63003) 5E5 antigen
Length = 825
Score = 47.4 bits (111), Expect = 6e-05
Identities = 25/40 (62%), Positives = 25/40 (62%), Gaps = 5/40 (12%)
Query: 341 LGAGGRAGGRSGGREGGGAGAMGGRRG-----RGGRRGRG 375
LG GR G GGR GGGAGA GG RG RGGRRG G
Sbjct: 562 LGGRGRRGSWRGGRRGGGAGASGGGRGGRGRGRGGRRGSG 601
Score = 37.0 bits (84), Expect = 0.082
Identities = 23/47 (48%), Positives = 24/47 (50%), Gaps = 8/47 (17%)
Query: 336 AGRGGLGAGGRAGGRSGGREGGG-------AGAMGGRRGRGGRRGRG 375
AG G G GGR GR GGR G G AG+ RRG RRG G
Sbjct: 580 AGASGGGRGGRGRGR-GGRRGSGLSGTREDAGSPSARRGEQRRRGHG 625
>NUCL_MESAU (P08199) Nucleolin (Protein C23)
Length = 713
Score = 46.6 bits (109), Expect = 1e-04
Identities = 27/55 (49%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Query: 321 RILQDLAGTQGIAYAAGRGGL--GAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
++ D A +G GRGG G GGR GGR GGR G G GG GRGG RG
Sbjct: 638 KVTLDWAKPKGEGGFGGRGGGRGGFGGRGGGRGGGRGGFGGRGRGGFGGRGGFRG 692
>NOA1_BRARE (Q7ZVE0) Nucleolar protein family A member 1 (snoRNP
protein GAR1) (H/ACA ribonucleoprotein GAR1)
Length = 226
Score = 46.6 bits (109), Expect = 1e-04
Identities = 26/52 (50%), Positives = 28/52 (53%), Gaps = 4/52 (7%)
Query: 328 GTQGIAYAAGRGGLGAG----GRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G +G + GRG G G G GG GGR GGG G GGR G GGR RG
Sbjct: 173 GGRGGGFRGGRGANGGGRGGFGGRGGGFGGRGGGGGGFRGGRGGGGGRGFRG 224
Score = 44.7 bits (104), Expect = 4e-04
Identities = 27/57 (47%), Positives = 28/57 (48%), Gaps = 2/57 (3%)
Query: 321 RILQDLAGTQGIAYAAGRGGLGAGGRAGGRSGGR--EGGGAGAMGGRRGRGGRRGRG 375
R L G +G GG G GGR GG GGR GGG G GGR G G RG G
Sbjct: 150 RFLPRPPGEKGPPRGGRGGGGGRGGRGGGFRGGRGANGGGRGGFGGRGGGFGGRGGG 206
Score = 42.0 bits (97), Expect = 0.003
Identities = 24/50 (48%), Positives = 27/50 (54%), Gaps = 6/50 (12%)
Query: 332 IAYAAGRGGLGAG---GRAGGRSGGREGGGAGAMGGRRG---RGGRRGRG 375
+++ G GG G G G GGR GG GG G GG RG GGR GRG
Sbjct: 1 MSFRGGGGGRGGGFNRGGGGGRGGGFGGGRGGGFGGGRGGGFGGGRGGRG 50
Score = 42.0 bits (97), Expect = 0.003
Identities = 23/46 (50%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
G +G + G GG GG GGR GG GG G GG GRGGR G
Sbjct: 8 GGRGGGFNRGGGGGRGGGFGGGRGGGFGGGRGGGFGG--GRGGRGG 51
Score = 40.4 bits (93), Expect = 0.007
Identities = 24/42 (57%), Positives = 24/42 (57%), Gaps = 3/42 (7%)
Query: 333 AYAAGRGGLGA-GGRAGGRSGGREG--GGAGAMGGRRGRGGR 371
A GRGG G GG GGR GG G GG G GGR RGGR
Sbjct: 185 ANGGGRGGFGGRGGGFGGRGGGGGGFRGGRGGGGGRGFRGGR 226
>GRP2_PHAVU (P10496) Glycine-rich cell wall structural protein 1.8
precursor (GRP 1.8)
Length = 465
Score = 46.6 bits (109), Expect = 1e-04
Identities = 24/46 (52%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRG 373
G GI Y G GG GAGG G +GG +GGG G GG G GG G
Sbjct: 135 GEHGIGYGGG-GGSGAGGGGGYNAGGAQGGGYGTGGGAGGGGGGGG 179
Score = 44.7 bits (104), Expect = 4e-04
Identities = 22/40 (55%), Positives = 25/40 (62%), Gaps = 3/40 (7%)
Query: 339 GGLGAGGRAGGRSGGREGGGAG---AMGGRRGRGGRRGRG 375
GG GAGG GG +GG +GGGAG GG G GG G+G
Sbjct: 285 GGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGGGGGQG 324
Score = 44.7 bits (104), Expect = 4e-04
Identities = 26/51 (50%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGA---MGGRRGRGGRRGRG 375
G QG Y G GG G GG GG GG GGG GA GG G GG G G
Sbjct: 159 GAQGGGYGTG-GGAGGGGGGGGDHGGGYGGGQGAGGGAGGGYGGGGEHGGG 208
Score = 44.3 bits (103), Expect = 5e-04
Identities = 24/52 (46%), Positives = 25/52 (47%), Gaps = 3/52 (5%)
Query: 328 GTQGIAYAAGRGGLGAGGRA---GGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G QG Y G G G GG A GG GG GG G GG G GG G G+
Sbjct: 90 GDQGAGYGGGGGSGGGGGVAYGGGGERGGYGGGQGGGAGGGYGAGGEHGIGY 141
Score = 43.5 bits (101), Expect = 9e-04
Identities = 25/48 (52%), Positives = 26/48 (54%), Gaps = 4/48 (8%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G QG AG GG G GG GG GG +GGGAG G G GG G G
Sbjct: 187 GGQGAGGGAG-GGYGGGGEHGGGGGGGQGGGAG---GGYGAGGEHGGG 230
Score = 42.4 bits (98), Expect = 0.002
Identities = 21/40 (52%), Positives = 24/40 (59%), Gaps = 3/40 (7%)
Query: 339 GGLGAGGRAGGRSGGREGGGAG---AMGGRRGRGGRRGRG 375
GG GAGG GG +GG +GGGAG GG G G G+G
Sbjct: 241 GGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQG 280
Score = 42.4 bits (98), Expect = 0.002
Identities = 21/40 (52%), Positives = 24/40 (59%), Gaps = 3/40 (7%)
Query: 339 GGLGAGGRAGGRSGGREGGGAG---AMGGRRGRGGRRGRG 375
GG GAGG GG +GG +GGGAG GG G G G+G
Sbjct: 263 GGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQG 302
Score = 42.4 bits (98), Expect = 0.002
Identities = 21/40 (52%), Positives = 24/40 (59%), Gaps = 3/40 (7%)
Query: 339 GGLGAGGRAGGRSGGREGGGAG---AMGGRRGRGGRRGRG 375
GG GAGG GG +GG +GGGAG GG G G G+G
Sbjct: 219 GGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQG 258
Score = 42.0 bits (97), Expect = 0.003
Identities = 24/54 (44%), Positives = 27/54 (49%), Gaps = 8/54 (14%)
Query: 328 GTQGIAYAAGRGG-----LGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G G Y G+GG G GG GG GG +GGGA GG G GG G G+
Sbjct: 335 GEHGGGYGGGQGGGDGGGYGTGGEHGGGYGGGQGGGA---GGGYGTGGEHGGGY 385
Score = 40.0 bits (92), Expect = 0.010
Identities = 25/53 (47%), Positives = 28/53 (52%), Gaps = 5/53 (9%)
Query: 328 GTQGIAYAAGRGG---LGAGGRAGGRS-GGREGGGAGAMGGRRGRGGRRGRGH 376
G G Y G+GG GAGG G GG EGGG G+ GG G GG G G+
Sbjct: 379 GEHGGGYGGGQGGGGGYGAGGDHGAAGYGGGEGGGGGS-GGGYGDGGAHGGGY 430
Score = 39.7 bits (91), Expect = 0.013
Identities = 22/51 (43%), Positives = 25/51 (48%), Gaps = 2/51 (3%)
Query: 328 GTQGIAYAAG--RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G G Y AG GG GG+ GG GG GG GG G+GG G G+
Sbjct: 281 GGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGGGGGQGGGAGGGY 331
Score = 39.7 bits (91), Expect = 0.013
Identities = 22/49 (44%), Positives = 22/49 (44%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G G G GG GG GG GG G GA GG G GG G GH
Sbjct: 412 GGGGSGGGYGDGGAHGGGYGGGAGGGGGYGAGGAHGGGYGGGGGIGGGH 460
Score = 39.3 bits (90), Expect = 0.016
Identities = 22/55 (40%), Positives = 25/55 (45%), Gaps = 2/55 (3%)
Query: 324 QDLAGTQGIAYAAG--RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
Q G G Y G GG G GG+ GG GG GG G G+GG G G+
Sbjct: 189 QGAGGGAGGGYGGGGEHGGGGGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGY 243
Score = 38.9 bits (89), Expect = 0.022
Identities = 25/59 (42%), Positives = 27/59 (45%), Gaps = 2/59 (3%)
Query: 319 ALRILQDLAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMG--GRRGRGGRRGRG 375
A R L L G+ + G G GA G GG GG GGG G G G G GG G G
Sbjct: 23 ARRALLTLDAGYGLGHGTGGGYGGAAGSYGGGGGGGSGGGGGYAGEHGVVGYGGGSGGG 81
Score = 38.1 bits (87), Expect = 0.037
Identities = 24/48 (50%), Positives = 25/48 (52%), Gaps = 4/48 (8%)
Query: 328 GTQGIAYAAGRGGLGAGGR--AGGRSGGREGGGAGAMGGRRGRGGRRG 373
G G Y G+GG GAGG GG GG GGG G GG G GG G
Sbjct: 357 GEHGGGYGGGQGG-GAGGGYGTGGEHGGGYGGGQGG-GGGYGAGGDHG 402
Score = 38.1 bits (87), Expect = 0.037
Identities = 21/48 (43%), Positives = 22/48 (45%), Gaps = 2/48 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G Y G+G GG GG GG E GG G G G GG G G
Sbjct: 179 GDHGGGYGGGQGA--GGGAGGGYGGGGEHGGGGGGGQGGGAGGGYGAG 224
Score = 37.7 bits (86), Expect = 0.048
Identities = 25/52 (48%), Positives = 26/52 (49%), Gaps = 3/52 (5%)
Query: 327 AGTQGIAYAAGRGGLGAGG--RAGGRSGGREGGG-AGAMGGRRGRGGRRGRG 375
AG + A G G GAGG AGG GG GGG G GG G GG G G
Sbjct: 245 AGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGG 296
Score = 37.7 bits (86), Expect = 0.048
Identities = 25/52 (48%), Positives = 26/52 (49%), Gaps = 3/52 (5%)
Query: 327 AGTQGIAYAAGRGGLGAGG--RAGGRSGGREGGG-AGAMGGRRGRGGRRGRG 375
AG + A G G GAGG AGG GG GGG G GG G GG G G
Sbjct: 223 AGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGG 274
Score = 37.4 bits (85), Expect = 0.063
Identities = 21/51 (41%), Positives = 24/51 (46%), Gaps = 2/51 (3%)
Query: 328 GTQGIAYAAG--RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G G Y AG GG GG+ GG GG GG G G+GG G G+
Sbjct: 237 GGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGY 287
Score = 37.4 bits (85), Expect = 0.063
Identities = 24/61 (39%), Positives = 26/61 (42%), Gaps = 12/61 (19%)
Query: 328 GTQGIAYAAG--RGGLGAGGRAGG----------RSGGREGGGAGAMGGRRGRGGRRGRG 375
G G Y AG GG G GG+ GG GG GG G GG G GG G G
Sbjct: 303 GGAGGGYGAGGEHGGGGGGGQGGGAGGGYAAVGEHGGGYGGGQGGGDGGGYGTGGEHGGG 362
Query: 376 H 376
+
Sbjct: 363 Y 363
Score = 37.4 bits (85), Expect = 0.063
Identities = 21/51 (41%), Positives = 24/51 (46%), Gaps = 2/51 (3%)
Query: 328 GTQGIAYAAG--RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G G Y AG GG GG+ GG GG GG G G+GG G G+
Sbjct: 259 GGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGY 309
Score = 37.4 bits (85), Expect = 0.063
Identities = 20/40 (50%), Positives = 20/40 (50%), Gaps = 1/40 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRG 367
G G Y G GG G G AGG GG GGG G GG G
Sbjct: 424 GAHGGGYGGGAGG-GGGYGAGGAHGGGYGGGGGIGGGHGG 462
Score = 37.4 bits (85), Expect = 0.063
Identities = 21/44 (47%), Positives = 23/44 (51%), Gaps = 1/44 (2%)
Query: 333 AYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
A A GG G GG AGG GG G G GG +G GG G G+
Sbjct: 157 AGGAQGGGYGTGGGAGG-GGGGGGDHGGGYGGGQGAGGGAGGGY 199
Score = 37.4 bits (85), Expect = 0.063
Identities = 21/51 (41%), Positives = 24/51 (46%), Gaps = 2/51 (3%)
Query: 328 GTQGIAYAAG--RGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G G Y AG GG GG+ GG GG GG G G+GG G G+
Sbjct: 215 GGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGY 265
Score = 36.6 bits (83), Expect = 0.11
Identities = 22/54 (40%), Positives = 25/54 (45%), Gaps = 8/54 (14%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSG--------GREGGGAGAMGGRRGRGGRRGRGH 376
G AAG G G GG +GG G G GG G GG G GG +G G+
Sbjct: 43 GYGGAAGSYGGGGGGGSGGGGGYAGEHGVVGYGGGSGGGQGGGVGYGGDQGAGY 96
Score = 36.2 bits (82), Expect = 0.14
Identities = 22/51 (43%), Positives = 24/51 (46%), Gaps = 6/51 (11%)
Query: 331 GIAYAAGRGGLGAGGRAGG-RSGGREGGGA-----GAMGGRRGRGGRRGRG 375
G + G GG GG GG +GG GGGA G GG G GG G G
Sbjct: 268 GGEHGGGAGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGG 318
Score = 36.2 bits (82), Expect = 0.14
Identities = 25/56 (44%), Positives = 26/56 (45%), Gaps = 8/56 (14%)
Query: 328 GTQGIAYAAGRGGLGAGGRA-------GGRSGGREGGGAGAMGGRRGRGGRRGRGH 376
G G AAG GG GG GG GG GGGAG GG G GG G G+
Sbjct: 396 GAGGDHGAAGYGGGEGGGGGSGGGYGDGGAHGGGYGGGAGG-GGGYGAGGAHGGGY 450
Score = 35.8 bits (81), Expect = 0.18
Identities = 22/51 (43%), Positives = 24/51 (46%), Gaps = 6/51 (11%)
Query: 331 GIAYAAGRGGLGAGGRAGG-RSGGREGGGA-----GAMGGRRGRGGRRGRG 375
G + G GG GG GG +GG GGGA G GG G GG G G
Sbjct: 202 GGEHGGGGGGGQGGGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAGGEHGGG 252
Score = 35.4 bits (80), Expect = 0.24
Identities = 26/62 (41%), Positives = 30/62 (47%), Gaps = 13/62 (20%)
Query: 328 GTQGIAYAAG--RGGLGAG---GRAGGRSGGRE-------GGGAGAMGGRR-GRGGRRGR 374
G G+AY G RGG G G G GG G E GGG+GA GG GG +G
Sbjct: 104 GGGGVAYGGGGERGGYGGGQGGGAGGGYGAGGEHGIGYGGGGGSGAGGGGGYNAGGAQGG 163
Query: 375 GH 376
G+
Sbjct: 164 GY 165
Score = 35.0 bits (79), Expect = 0.31
Identities = 22/52 (42%), Positives = 24/52 (45%), Gaps = 4/52 (7%)
Query: 328 GTQGIAYAAG--RGGLGAGGRAGGRSGGREGGG--AGAMGGRRGRGGRRGRG 375
G G Y G GG GG+ GG GG GG G GG +G GG G G
Sbjct: 347 GGDGGGYGTGGEHGGGYGGGQGGGAGGGYGTGGEHGGGYGGGQGGGGGYGAG 398
Score = 35.0 bits (79), Expect = 0.31
Identities = 20/40 (50%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
G A+ G GG GAGG G +GG GGG G GG G G
Sbjct: 423 GGAHGGGYGG-GAGGGGGYGAGGAHGGGYGGGGGIGGGHG 461
Score = 34.3 bits (77), Expect = 0.53
Identities = 19/45 (42%), Positives = 21/45 (46%)
Query: 331 GIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G YA G +G GG +GG GG G G G G GG G G
Sbjct: 62 GGGYAGEHGVVGYGGGSGGGQGGGVGYGGDQGAGYGGGGGSGGGG 106
Score = 34.3 bits (77), Expect = 0.53
Identities = 24/61 (39%), Positives = 26/61 (42%), Gaps = 12/61 (19%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAG---------GRSGGREGGGAGAMGG---RRGRGGRRGR 374
AG G+ G G G GG G G GG GGG A GG R G GG +G
Sbjct: 66 AGEHGVVGYGGGSGGGQGGGVGYGGDQGAGYGGGGGSGGGGGVAYGGGGERGGYGGGQGG 125
Query: 375 G 375
G
Sbjct: 126 G 126
Score = 33.9 bits (76), Expect = 0.69
Identities = 19/48 (39%), Positives = 21/48 (43%), Gaps = 1/48 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
GT G Y G G GG G GG G G +G G GG +G G
Sbjct: 39 GTGG-GYGGAAGSYGGGGGGGSGGGGGYAGEHGVVGYGGGSGGGQGGG 85
Score = 33.5 bits (75), Expect = 0.90
Identities = 21/51 (41%), Positives = 21/51 (41%), Gaps = 3/51 (5%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGG---AGAMGGRRGRGGRRGRG 375
G G G G G G GG SGG G G G GG G GG G G
Sbjct: 393 GGYGAGGDHGAAGYGGGEGGGGGSGGGYGDGGAHGGGYGGGAGGGGGYGAG 443
Score = 33.5 bits (75), Expect = 0.90
Identities = 21/49 (42%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Query: 328 GTQGIAYAAGRGGLGA-GGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G QG G +G GG GG GG +GGG G GG G G G+G
Sbjct: 321 GGQGGGAGGGYAAVGEHGGGYGGGQGGGDGGGYGT-GGEHGGGYGGGQG 368
Score = 32.7 bits (73), Expect = 1.5
Identities = 24/58 (41%), Positives = 27/58 (46%), Gaps = 9/58 (15%)
Query: 327 AGTQGIAYAAGRGG------LGAGGRAGGRSGGREGGGAG---AMGGRRGRGGRRGRG 375
AG + A G G GAGG GG GG +GGGAG A G G G G+G
Sbjct: 289 AGGEHGGGAGGGQGGGAGGGYGAGGEHGGGGGGGQGGGAGGGYAAVGEHGGGYGGGQG 346
Score = 32.7 bits (73), Expect = 1.5
Identities = 21/51 (41%), Positives = 22/51 (42%), Gaps = 2/51 (3%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGG--GAGAMGGRRGRGGRRGRGH 376
G G G G G GG GG G GG GA GG G GG G G+
Sbjct: 370 GAGGGYGTGGEHGGGYGGGQGGGGGYGAGGDHGAAGYGGGEGGGGGSGGGY 420
Score = 32.0 bits (71), Expect = 2.6
Identities = 23/51 (45%), Positives = 24/51 (46%), Gaps = 7/51 (13%)
Query: 331 GIAYAAG--RGGLGAGGRAGGRSGGREGGG---AGAMGGRRGRGGRRGRGH 376
G Y AG G G GG GG GG GGG GA GG G G G G+
Sbjct: 392 GGGYGAGGDHGAAGYGGGEGG--GGGSGGGYGDGGAHGGGYGGGAGGGGGY 440
>GR10_BRANA (Q05966) Glycine-rich RNA-binding protein 10
Length = 169
Score = 46.6 bits (109), Expect = 1e-04
Identities = 25/41 (60%), Positives = 25/41 (60%), Gaps = 2/41 (4%)
Query: 335 AAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
A RGG G GGR GG GGR GGG G GG G G RRG G
Sbjct: 82 AQSRGGGGGGGRGGGGYGGRGGGGYGGGGG--GYGDRRGGG 120
Score = 42.4 bits (98), Expect = 0.002
Identities = 25/52 (48%), Positives = 28/52 (53%), Gaps = 8/52 (15%)
Query: 327 AGTQGIAYAAGRGGLGAGGRAGGRSGG--------REGGGAGAMGGRRGRGG 370
A ++G GRGG G GGR GG GG R GGG G+ GG RG GG
Sbjct: 82 AQSRGGGGGGGRGGGGYGGRGGGGYGGGGGGYGDRRGGGGYGSGGGGRGGGG 133
Score = 41.6 bits (96), Expect = 0.003
Identities = 22/43 (51%), Positives = 23/43 (53%), Gaps = 2/43 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
G G Y +G GG G GG G R GG GGG G GG G GG
Sbjct: 128 GRGGGGYGSGGGGYGGGG--GRRDGGGYGGGDGGYGGGSGGGG 168
Score = 41.2 bits (95), Expect = 0.004
Identities = 24/51 (47%), Positives = 25/51 (48%), Gaps = 3/51 (5%)
Query: 328 GTQGIAYAAGRGGLG---AGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G Y G GG G GG G GGR GGG G+ GG G GG R G
Sbjct: 100 GRGGGGYGGGGGGYGDRRGGGGYGSGGGGRGGGGYGSGGGGYGGGGGRRDG 150
Score = 40.8 bits (94), Expect = 0.006
Identities = 24/50 (48%), Positives = 24/50 (48%), Gaps = 2/50 (4%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGR--GGRRGRG 375
G G Y RGG G G GGR GG G G G GG GR GG G G
Sbjct: 107 GGGGGGYGDRRGGGGYGSGGGGRGGGGYGSGGGGYGGGGGRRDGGGYGGG 156
Score = 35.8 bits (81), Expect = 0.18
Identities = 20/39 (51%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G GG G+GG G GGR GG G GG G GG G G
Sbjct: 130 GGGGYGSGGGGYGGGGGRRDGG-GYGGGDGGYGGGSGGG 167
>FBRL_DROME (Q9W1V3) Fibrillarin
Length = 344
Score = 46.6 bits (109), Expect = 1e-04
Identities = 29/58 (50%), Positives = 29/58 (50%), Gaps = 11/58 (18%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGR----------RGRGGRRGRG 375
G G GRGG G GGR GG GG GGG GA GGR RG GGR G G
Sbjct: 41 GRGGGGDRGGRGGFG-GGRGGGGRGGGGGGGRGAFGGRGGGGGRGGGGRGGGGRGGGG 97
Score = 45.8 bits (107), Expect = 2e-04
Identities = 24/44 (54%), Positives = 25/44 (56%), Gaps = 5/44 (11%)
Query: 337 GRGGLGAGGRAG-----GRSGGREGGGAGAMGGRRGRGGRRGRG 375
GRGG G GG G GR GG + GG G GG RG GGR G G
Sbjct: 24 GRGGGGGGGGGGFGGGRGRGGGGDRGGRGGFGGGRGGGGRGGGG 67
Score = 45.4 bits (106), Expect = 2e-04
Identities = 24/39 (61%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query: 337 GRGGLGAGGRA--GGRSGGREGGGAGAMGGRRGRGGRRG 373
GRGG G GGR GGR GG GG G GG RG GGR G
Sbjct: 62 GRGGGGGGGRGAFGGRGGGGGRGGGGRGGGGRGGGGRGG 100
Score = 44.3 bits (103), Expect = 5e-04
Identities = 23/48 (47%), Positives = 24/48 (49%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGRRGRG 375
G G + G GG G GG GG GGG G GG RGRGG RG
Sbjct: 2 GKPGFSPRGGGGGGGGGGGGFRGRGGGGGGGGGGFGGGRGRGGGGDRG 49
Score = 42.7 bits (99), Expect = 0.001
Identities = 26/55 (47%), Positives = 26/55 (47%), Gaps = 7/55 (12%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRS-------GGREGGGAGAMGGRRGRGGRRGRG 375
G G G GG G GG GGR GGR G G G GG RG GG GRG
Sbjct: 18 GGGGFRGRGGGGGGGGGGFGGGRGRGGGGDRGGRGGFGGGRGGGGRGGGGGGGRG 72
Score = 40.8 bits (94), Expect = 0.006
Identities = 20/43 (46%), Positives = 21/43 (48%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
G +G GRG G G GGR GG GGG GGR G G
Sbjct: 61 GGRGGGGGGGRGAFGGRGGGGGRGGGGRGGGGRGGGGRGGGAG 103
Score = 38.5 bits (88), Expect = 0.028
Identities = 26/55 (47%), Positives = 26/55 (47%), Gaps = 9/55 (16%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGR-------RGRGGRRGRG 375
G G GRGG G G GG GGR GGG G GG GRGG GRG
Sbjct: 32 GGGGFGGGRGRGGGGDRGGRGGFGGGRGGGGRG--GGGGGGRGAFGGRGGGGGRG 84
Score = 33.5 bits (75), Expect = 0.90
Identities = 19/44 (43%), Positives = 21/44 (47%), Gaps = 1/44 (2%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGGR 371
G G GRGG G G GGR GG GGG G +GG+
Sbjct: 67 GGGGRGAFGGRGG-GGGRGGGGRGGGGRGGGGRGGGAGGFKGGK 109
>GRP2_NICSY (P27484) Glycine-rich protein 2
Length = 214
Score = 45.8 bits (107), Expect = 2e-04
Identities = 43/129 (33%), Positives = 55/129 (42%), Gaps = 9/129 (6%)
Query: 254 EEGQAVGGYMDWYAIVSHC-FIIPDERRIDL----SAVAA--LRRALEVLELSLEVDDAL 306
E GQ G + W++ FI PD+ DL S + + R E + EV+
Sbjct: 4 ESGQRAKGTVKWFSDQKGFGFITPDDGGEDLFVHQSGIRSEGFRSLAEGETVEFEVESGG 63
Query: 307 LPGTQARALTKRALRILQDLAGTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRR 366
T+A +T +Q G G GRGG G GG +GG GG GG G GG
Sbjct: 64 DGRTKAVDVTGPDGAAVQ--GGRGGGGGGGGRGGGGYGGGSGGYGGGGRGGSRGYGGGDG 121
Query: 367 GRGGRRGRG 375
G GG G G
Sbjct: 122 GYGGGGGYG 130
Score = 41.6 bits (96), Expect = 0.003
Identities = 24/52 (46%), Positives = 24/52 (46%), Gaps = 4/52 (7%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGR----SGGREGGGAGAMGGRRGRGGRRGRG 375
G G Y G GG G GGR G R G GGG G GG R GG G G
Sbjct: 92 GRGGGGYGGGSGGYGGGGRGGSRGYGGGDGGYGGGGGYGGGSRYGGGGGGYG 143
Score = 40.0 bits (92), Expect = 0.010
Identities = 22/43 (51%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
G++G Y G GG G GG GG G R GGG G GG G GG
Sbjct: 112 GSRG--YGGGDGGYGGGGGYGG--GSRYGGGGGGYGGGGGYGG 150
Score = 35.0 bits (79), Expect = 0.31
Identities = 17/34 (50%), Positives = 17/34 (50%)
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
G GG G G R GG GG GGG GG G G
Sbjct: 125 GGGGYGGGSRYGGGGGGYGGGGGYGGGGSGGGSG 158
Score = 34.3 bits (77), Expect = 0.53
Identities = 24/62 (38%), Positives = 24/62 (38%), Gaps = 17/62 (27%)
Query: 331 GIAYAAGRGGLGAGGR-AGGRSGGREG----------------GGAGAMGGRRGRGGRRG 373
G Y G GG G GG GG SGG G G G GGR G GG G
Sbjct: 132 GSRYGGGGGGYGGGGGYGGGGSGGGSGCFKCGESGHFARDCSQSGGGGGGGRFGGGGGGG 191
Query: 374 RG 375
G
Sbjct: 192 GG 193
>GRP1_SORBI (Q99069) Glycine-rich RNA-binding protein 1 (Fragment)
Length = 142
Score = 45.8 bits (107), Expect = 2e-04
Identities = 24/42 (57%), Positives = 25/42 (59%), Gaps = 5/42 (11%)
Query: 337 GRGGLGAGGRAGGRSGG-----REGGGAGAMGGRRGRGGRRG 373
GRGG G GG GGR GG R+GGG G GG G GG RG
Sbjct: 68 GRGGGGGGGYGGGRGGGGGYGRRDGGGGGYGGGGGGYGGGRG 109
Score = 45.4 bits (106), Expect = 2e-04
Identities = 32/90 (35%), Positives = 41/90 (45%), Gaps = 9/90 (10%)
Query: 281 IDLSAVAALRRALEVLELSLEVDDALLPGTQARALTKRALRILQDLAGTQGIAYAAGRGG 340
+ S A+R A+E + E+D + +A++ R G G Y GRGG
Sbjct: 33 VTFSTEEAMRSAIEGMN-GKELDGRNITVNEAQSRGGRG--------GGGGGGYGGGRGG 83
Query: 341 LGAGGRAGGRSGGREGGGAGAMGGRRGRGG 370
G GR G GG GGG G GGR G GG
Sbjct: 84 GGGYGRRDGGGGGYGGGGGGYGGGRGGYGG 113
Score = 37.7 bits (86), Expect = 0.048
Identities = 20/35 (57%), Positives = 20/35 (57%), Gaps = 1/35 (2%)
Query: 337 GRGGLGAGGRAGGRSGGREGGGAGAM-GGRRGRGG 370
G GG G GG GG GG GGG G GG RG GG
Sbjct: 98 GGGGGGYGGGRGGYGGGGYGGGGGGYGGGSRGGGG 132
Score = 34.3 bits (77), Expect = 0.53
Identities = 19/41 (46%), Positives = 19/41 (46%), Gaps = 4/41 (9%)
Query: 328 GTQGIAYAAGRGGLGAGGRAGGRSG----GREGGGAGAMGG 364
G G Y GRGG G GG GG G R GGG G G
Sbjct: 98 GGGGGGYGGGRGGYGGGGYGGGGGGYGGGSRGGGGYGNSDG 138
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.140 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,892,602
Number of Sequences: 164201
Number of extensions: 2116044
Number of successful extensions: 34467
Number of sequences better than 10.0: 491
Number of HSP's better than 10.0 without gapping: 360
Number of HSP's successfully gapped in prelim test: 137
Number of HSP's that attempted gapping in prelim test: 12138
Number of HSP's gapped (non-prelim): 8915
length of query: 376
length of database: 59,974,054
effective HSP length: 112
effective length of query: 264
effective length of database: 41,583,542
effective search space: 10978055088
effective search space used: 10978055088
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0348.16


