

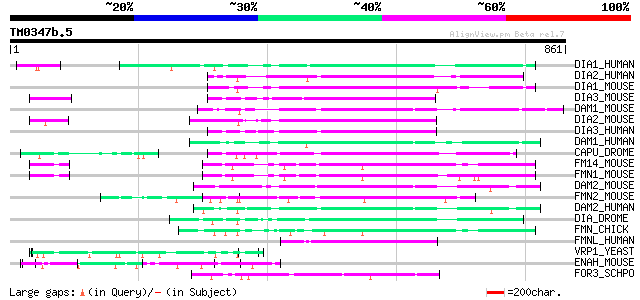
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0347b.5
(861 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DIA1_HUMAN (O60610) Diaphanous protein homolog 1 (Diaphanous-rel... 155 4e-37
DIA2_HUMAN (O60879) Diaphanous protein homolog 2 (Diaphanous-rel... 152 4e-36
DIA1_MOUSE (O08808) Diaphanous protein homolog 1 (Diaphanous-rel... 152 4e-36
DIA3_MOUSE (Q9Z207) Diaphanous protein homolog 3 (Diaphanous-rel... 147 2e-34
DAM1_MOUSE (Q8BPM0) Disheveled associated activator of morphogen... 140 2e-32
DIA2_MOUSE (O70566) Diaphanous protein homolog 2 (Diaphanous-rel... 139 3e-32
DIA3_HUMAN (Q9NSV4) Diaphanous protein homolog 3 (Diaphanous-rel... 137 2e-31
DAM1_HUMAN (Q9Y4D1) Disheveled associated activator of morphogen... 135 4e-31
CAPU_DROME (Q24120) Cappuccino protein 132 4e-30
FM14_MOUSE (Q05859) Formin 1 isoform IV (Limb deformity protein) 129 3e-29
FMN1_MOUSE (Q05860) Formin 1 isoforms I/II/III (Limb deformity p... 128 6e-29
DAM2_MOUSE (Q80U19) Disheveled associated activator of morphogen... 124 1e-27
FMN2_MOUSE (Q9JL04) Formin 2 119 3e-26
DAM2_HUMAN (Q86T65) Disheveled associated activator of morphogen... 116 2e-25
DIA_DROME (P48608) Diaphanous protein 114 1e-24
FMN_CHICK (Q05858) Formin (Limb deformity protein) 112 5e-24
FMNL_HUMAN (O95466) Formin-like protein (Protein C17orf1) 94 2e-18
VRP1_YEAST (P37370) Verprolin 87 2e-16
ENAH_MOUSE (Q03173) Enabled protein homolog (NPC derived proline... 74 2e-12
FOR3_SCHPO (O94532) Formin 3 70 2e-11
>DIA1_HUMAN (O60610) Diaphanous protein homolog 1 (Diaphanous-related
formin 1) (DRF1)
Length = 1248
Score = 155 bits (392), Expect = 4e-37
Identities = 164/668 (24%), Positives = 258/668 (38%), Gaps = 99/668 (14%)
Query: 171 GTVEPSRTSVSDAQNLNSAKLASNYRYRPS-PELQPLPPLSKPPDE---IHSPPAASSSS 226
G V + DA+ ++ A+ PS P P+PP P + I PP A S
Sbjct: 537 GEVAKLTKELEDAKKEMASLSAAAITVPPSVPSRAPVPPAPPLPGDSGTIIPPPPAPGDS 596
Query: 227 PSSSEDAESRETAFHSPHGSSVS-----HDDNNYYTPVSRHSSVANGSPAA---ATAVPF 278
+ P G+++S D P V SP++ TA+P
Sbjct: 597 TTPPPPPPPPPPPPPLPGGTAISPPPPLSGDATIPPPPPLPEGVGIPSPSSLPGGTAIPP 656
Query: 279 SKRTSPKSRLSASSPDIRHAMIPSIKHNPLPPPPAPPA----PPSTQLDGGPSRRPKFST 334
+R+ P + P P PPPP P PP L GGP P
Sbjct: 657 PPPLPGSARIPPPPPPL-----PGSAGIPPPPPPLPGEAGMPPPPPPLPGGPGIPPP--- 708
Query: 335 HPLAPNMASLNTPPPPPP---PPPPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASS 391
P PPPPP PPPPP V + +
Sbjct: 709 ---PPFPGGPGIPPPPPGMGMPPPPPFGFGVPAAPVLPF--------------------- 744
Query: 392 NGVSSVSVSKPSEEGESMMEGGKPKLKALHWDKVRATS-NRATVWNQLKSSSFQLNEDMM 450
G++ + KP + L+ +W K+ A ++ W ++K F+ NE
Sbjct: 745 -GLTPKKLYKPEVQ-----------LRRPNWSKLVAEDLSQDCFWTKVKEDRFENNELFA 792
Query: 451 ETLFGFNSTTNSAPKETTTTSVVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEV 510
+ F++ T + K+ +K V +E +VLD K +QN++I L + + E+
Sbjct: 793 KLTLTFSAQTKT--KKDQEGGEEKKSVQKKKVKELKVLDSKTAQNLSIFLGSFRMPYQEI 850
Query: 511 SEALLDGNPEGLGAELLETLVKMAPTKEEEIKLKNYDGDLSKLGSAERFLKAVLDIPLAF 570
+L+ N L +++ L+K P E+ L + L +E+F + +P
Sbjct: 851 KNVILEVNEAVLTESMIQNLIKQMPEPEQLKMLSELKDEYDDLAESEQFGVVMGTVPRLR 910
Query: 571 KRVEAMLYRANFETEVNYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGTNRG 630
R+ A+L++ F +V ++ ++ AA EEL+ S F LLE L GN MN G+
Sbjct: 911 PRLNAILFKLQFSEQVENIKPEIVSVTAACEELRKSESFSNLLEITLLVGNYMNAGSRNA 970
Query: 631 DAKSFKLDTLLKLVDIKGTDGKTTLLHFVVQEIIRSEGTGGESANESVQNQTNSQFNEDE 690
A F + L KL D K TD K TLLHF+ E
Sbjct: 971 GAFGFNISFLCKLRDTKSTDQKMTLLHFLA-----------------------------E 1001
Query: 691 FRKKGLQVVAGLSRDLGNVKKAAGMDSDVLSSYVSKLEMGLDKVRLVLQY--QKPDMQGS 748
+ V +L +V+KA+ + ++ L + +++ + V +Q D +
Sbjct: 1002 LCENDYPDVLKFPDELAHVEKASRVSAENLQKNLDQMKKQISDVERDVQNFPAATDEKDK 1061
Query: 749 FFNATEIFLKDAEEKIIVIKDDERRALFLVKEVTEYFHGDTAKEEVHPFRIFMIVRDFLN 808
F F+KDA+E+ ++ L KE+ EYF D K V F FM + +F N
Sbjct: 1062 FVEKMTSFVKDAQEQYNKLRMMHSNMETLYKELGEYFLFDPKKLSVEEF--FMDLHNFRN 1119
Query: 809 ILDQVCKE 816
+ Q KE
Sbjct: 1120 MFLQAVKE 1127
Score = 49.3 bits (116), Expect = 4e-05
Identities = 30/80 (37%), Positives = 36/80 (44%), Gaps = 12/80 (15%)
Query: 11 LSLSLFTPLNSTPNARRILHQPLFPAGSA----PPPS-----TPPPPPPDTPSSPDIPFF 61
LS + T S P+ + P P S PPP+ TPPPPPP P P +P
Sbjct: 556 LSAAAITVPPSVPSRAPVPPAPPLPGDSGTIIPPPPAPGDSTTPPPPPPPPPPPPPLPGG 615
Query: 62 NEYSPPPP---DQALTQPPP 78
SPPPP D + PPP
Sbjct: 616 TAISPPPPLSGDATIPPPPP 635
Score = 41.6 bits (96), Expect = 0.009
Identities = 21/54 (38%), Positives = 23/54 (41%), Gaps = 13/54 (24%)
Query: 32 PLFPAGSAPPPSTPP-------PPPPDTPSSPDIPFFNEYSPPPPDQALTQPPP 78
P P + PP PP PPPP P P IP PPPP + PPP
Sbjct: 684 PPLPGEAGMPPPPPPLPGGPGIPPPPPFPGGPGIP------PPPPGMGMPPPPP 731
Score = 40.4 bits (93), Expect = 0.020
Identities = 29/79 (36%), Positives = 31/79 (38%), Gaps = 14/79 (17%)
Query: 23 PNARRILHQPLFPAGSA----PPPSTP-----PPPPPDTPSSPDIPFFNEYS-----PPP 68
P + RI P GSA PPP P PPPPP P P IP + PPP
Sbjct: 661 PGSARIPPPPPPLPGSAGIPPPPPPLPGEAGMPPPPPPLPGGPGIPPPPPFPGGPGIPPP 720
Query: 69 PDQALTQPPPSASIAYPTA 87
P PPP P A
Sbjct: 721 PPGMGMPPPPPFGFGVPAA 739
Score = 35.4 bits (80), Expect = 0.65
Identities = 29/80 (36%), Positives = 33/80 (41%), Gaps = 19/80 (23%)
Query: 32 PLFPAGSA---PPP----STPPPPPP-----DTPSSPDIPFFNEYSPPP--PDQALTQPP 77
P P G+A PPP +T PPPPP PS +P PPP P A PP
Sbjct: 610 PPLPGGTAISPPPPLSGDATIPPPPPLPEGVGIPSPSSLPGGTAIPPPPPLPGSARIPPP 669
Query: 78 P-----SASIAYPTATKPAK 92
P SA I P P +
Sbjct: 670 PPPLPGSAGIPPPPPPLPGE 689
Score = 32.3 bits (72), Expect = 5.5
Identities = 22/61 (36%), Positives = 26/61 (42%), Gaps = 4/61 (6%)
Query: 35 PAGSAPPPSTPPPPPPDTPSSPDIPFFNEYS--PPPPDQALTQPPPSASIAYPTATKPAK 92
P PPP PPP P T SP P + + PPPP P +S+ TA P
Sbjct: 601 PPPPPPPP--PPPLPGGTAISPPPPLSGDATIPPPPPLPEGVGIPSPSSLPGGTAIPPPP 658
Query: 93 P 93
P
Sbjct: 659 P 659
>DIA2_HUMAN (O60879) Diaphanous protein homolog 2
(Diaphanous-related formin 2) (DRF2)
Length = 1101
Score = 152 bits (384), Expect = 4e-36
Identities = 130/508 (25%), Positives = 212/508 (41%), Gaps = 89/508 (17%)
Query: 307 PLP---PPPAPPAPPSTQLDGGPSRRPKFSTHPLAPNMASLNTPPPPP-----PPPPPPA 358
PLP PPP PPAPP L GG P P P M + PPPPP PPPPPP
Sbjct: 556 PLPGVGPPPPPPAPP---LPGGAPLPPP---PPPLPGMMGIPPPPPPPLLFGGPPPPPPL 609
Query: 359 ARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVSKPSEEGESMMEGGKPKLK 418
P I S++ P + M + +K
Sbjct: 610 GGVPPPPGI------------------------------SLNLPYGMKQKKMYKPEVSMK 639
Query: 419 ALHWDKVRATS-NRATVWNQLKSSSFQLNEDMMETLFGFNSTT------NSAPKETTTTS 471
++W K+ T + W ++K F+ N D+ L N T N+ E T
Sbjct: 640 RINWSKIEPTELSENCFWLRVKEDKFE-NPDLFAKL-ALNFATQIKVQKNAEALEEKKTG 697
Query: 472 VVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEVSEALLDGNPEGLGAELLETLV 531
+K V +E R+LDPK +QN++I L + + +++ +L+ N + L L++ LV
Sbjct: 698 PTKKKV-----KELRILDPKTAQNLSIFLGSYRMPYEDIRNVILEVNEDMLSEALIQNLV 752
Query: 532 KMAPTKEEEIKLKNYDGDLSKLGSAERFLKAVLDIPLAFKRVEAMLYRANFETEVNYLRK 591
K P ++ +L + L E+F + + + R+ ++L++ FE +N ++
Sbjct: 753 KHLPEQKILNELAELKNEYDDLCEPEQFGVVMSSVKMLQPRLSSILFKLTFEEHINNIKP 812
Query: 592 SFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGTNRGDAKSFKLDTLLKLVDIKGTDG 651
S + A EELK S F +LLE VL GN MN G+ + FK++ L K+ D K D
Sbjct: 813 SIIAVTLACEELKKSESFNRLLELVLLVGNYMNSGSRNAQSLGFKINFLCKIRDTKSADQ 872
Query: 652 KTTLLHFVVQEIIRSEGTGGESANESVQNQTNSQFNEDEFRKKGLQVVAGLSRDLGNVKK 711
KTTLLHF+ + E+++R + +L +V+
Sbjct: 873 KTTLLHFI------------------------ADICEEKYRD-----ILKFPEELEHVES 903
Query: 712 AAGMDSDVLSSYVSKLEMGLDKVRLVLQY--QKPDMQGSFFNATEIFLKDAEEKIIVIKD 769
A+ + + +L S ++ +E + + ++ Q + F F K A E+ +
Sbjct: 904 ASKVSAQILKSNLASMEQQIVHLERDIKKFPQAENQHDKFVEKMTSFTKTAREQYEKLST 963
Query: 770 DERRALFLVKEVTEYFHGDTAKEEVHPF 797
+ L + + EYF D+ + F
Sbjct: 964 MHNNMMKLYENLGEYFIFDSKTVSIEEF 991
Score = 41.6 bits (96), Expect = 0.009
Identities = 27/78 (34%), Positives = 28/78 (35%), Gaps = 14/78 (17%)
Query: 32 PLFPAGSAPPPSTPP--------PPPPDTPSS------PDIPFFNEYSPPPPDQALTQPP 77
PL G PPP PP PPPP P P P PPPP PP
Sbjct: 556 PLPGVGPPPPPPAPPLPGGAPLPPPPPPLPGMMGIPPPPPPPLLFGGPPPPPPLGGVPPP 615
Query: 78 PSASIAYPTATKPAKPAK 95
P S+ P K K K
Sbjct: 616 PGISLNLPYGMKQKKMYK 633
Score = 34.3 bits (77), Expect = 1.4
Identities = 18/50 (36%), Positives = 23/50 (46%), Gaps = 7/50 (14%)
Query: 36 AGSAPPPSTPPPP---PPDTPSSPDIPFFNEYSPPPPD----QALTQPPP 78
+G PP+ PP P PP P +P +P PPPP + PPP
Sbjct: 546 SGIPGPPAAPPLPGVGPPPPPPAPPLPGGAPLPPPPPPLPGMMGIPPPPP 595
>DIA1_MOUSE (O08808) Diaphanous protein homolog 1 (Diaphanous-related
formin 1) (DRF1) (mDIA1) (p140mDIA)
Length = 1255
Score = 152 bits (384), Expect = 4e-36
Identities = 137/526 (26%), Positives = 212/526 (40%), Gaps = 91/526 (17%)
Query: 307 PLPPPPAPPA---PPSTQLDGGPSRRPKFSTHPLAPNMASLNTPPPPPP----PPPPPAA 359
P PPPP P + PP L GGP P P AP + PPPPP PPPPP
Sbjct: 684 PPPPPPLPGSVGVPPPPPLPGGPGLPPPPPPFPGAPGI------PPPPPGMGVPPPPPFG 737
Query: 360 RKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVSKPSEEGESMMEGGKPKLKA 419
V + + G++ V KP + L+
Sbjct: 738 FGVPAAPVLPF----------------------GLTPKKVYKPEVQ-----------LRR 764
Query: 420 LHWDKVRATS-NRATVWNQLKSSSFQLNEDMMETLFGFNSTTN-SAPKETTTTSVVRKPV 477
+W K A ++ W ++K F+ NE + F++ T S K+ +K V
Sbjct: 765 PNWSKFVAEDLSQDCFWTKVKEDRFENNELFAKLTLAFSAQTKTSKAKKDQEGGEEKKSV 824
Query: 478 FPPVEQENRVLDPKKSQNIAILLRALNVTRDEVSEALLDGNPEGLGAELLETLVKMAPTK 537
+E +VLD K +QN++I L + + E+ +L+ N L +++ L+K P
Sbjct: 825 QKKKVKELKVLDSKTAQNLSIFLGSFRMPYQEIKNVILEVNEAVLTESMIQNLIKQMPEP 884
Query: 538 EEEIKLKNYDGDLSKLGSAERFLKAVLDIPLAFKRVEAMLYRANFETEVNYLRKSFQTLE 597
E+ L + L +E+F + +P R+ A+L++ F +V ++ ++
Sbjct: 885 EQLKMLSELKEEYDDLAESEQFGVVMGTVPRLRPRLNAILFKLQFSEQVENIKPEIVSVT 944
Query: 598 AASEELKNSRLFFKLLEAVLRTGNRMNVGTNRGDAKSFKLDTLLKLVDIKGTDGKTTLLH 657
AA EEL+ S F LLE L GN MN G+ A F + L KL D K D K TLLH
Sbjct: 945 AACEELRKSENFSSLLELTLLVGNYMNAGSRNAGAFGFNISFLCKLRDTKSADQKMTLLH 1004
Query: 658 FVVQ-------EIIRSEGTGGESANESVQNQTNSQFNEDEFRKKGLQVVAGLSRDLGNVK 710
F+ + E+++ S + N Q + D+ +K+ +A + RD+ N
Sbjct: 1005 FLAELCENDHPEVLKFPDELAHVEKASRVSAENLQKSLDQMKKQ----IADVERDVQNFP 1060
Query: 711 KAAGMDSDVLSSYVSKLEMGLDKVRLVLQYQKPDMQGSFFNATEIFLKDAEEKIIVIKDD 770
A D + F F+KDA+E+ ++
Sbjct: 1061 AAT------------------------------DEKDKFVEKMTSFVKDAQEQYNKLRMM 1090
Query: 771 ERRALFLVKEVTEYFHGDTAKEEVHPFRIFMIVRDFLNILDQVCKE 816
L KE+ +YF D K V F FM + +F N+ Q KE
Sbjct: 1091 HSNMETLYKELGDYFVFDPKKLSVEEF--FMDLHNFRNMFLQAVKE 1134
Score = 43.1 bits (100), Expect = 0.003
Identities = 21/47 (44%), Positives = 23/47 (48%), Gaps = 11/47 (23%)
Query: 37 GSAPPPSTP-----PPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPP 78
G PPP P PPPPP P +P IP PPPP + PPP
Sbjct: 695 GVPPPPPLPGGPGLPPPPPPFPGAPGIP------PPPPGMGVPPPPP 735
Score = 43.1 bits (100), Expect = 0.003
Identities = 30/115 (26%), Positives = 42/115 (36%), Gaps = 14/115 (12%)
Query: 259 VSRHSSVANGSPAAATAVPFSKRTSPKSRLSASSPDIRHAMIPSIKHNPLPPPPAPPAPP 318
+S+ A A+ +AV + S + + + P +P +PPPP PP P
Sbjct: 542 LSKELEDAKNEMASLSAVVVAPSVSSSAAVPPAPP------LPGDSGTVIPPPPPPPPLP 595
Query: 319 STQLDGGPSRRPKFSTHPLAPNMASLNTPPPP--------PPPPPPPAARKVWSP 365
+ P P P P PPPP PPPPP P + P
Sbjct: 596 GGVVPPSPPLPPGTCIPPPPPLPGGACIPPPPQLPGSAAIPPPPPLPGVASIPPP 650
Score = 39.7 bits (91), Expect = 0.035
Identities = 25/74 (33%), Positives = 28/74 (37%), Gaps = 12/74 (16%)
Query: 32 PLFPAGSAPPPSTP------PPPPPDTPSS------PDIPFFNEYSPPPPDQALTQPPPS 79
P P G+ PP P PPPP P S P +P PPPP T PP
Sbjct: 603 PPLPPGTCIPPPPPLPGGACIPPPPQLPGSAAIPPPPPLPGVASIPPPPPLPGATAIPPP 662
Query: 80 ASIAYPTATKPAKP 93
+ TA P P
Sbjct: 663 PPLPGATAIPPPPP 676
Score = 38.9 bits (89), Expect = 0.059
Identities = 22/71 (30%), Positives = 25/71 (34%)
Query: 23 PNARRILHQPLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASI 82
P I P P +A PP P P P P +P PPPP T PP +
Sbjct: 618 PGGACIPPPPQLPGSAAIPPPPPLPGVASIPPPPPLPGATAIPPPPPLPGATAIPPPPPL 677
Query: 83 AYPTATKPAKP 93
T P P
Sbjct: 678 PGGTGIPPPPP 688
Score = 38.1 bits (87), Expect = 0.10
Identities = 28/93 (30%), Positives = 34/93 (36%), Gaps = 4/93 (4%)
Query: 10 LLSLSLFTPLNSTPNARRILHQPLFPAGSA----PPPSTPPPPPPDTPSSPDIPFFNEYS 65
+ SLS S ++ + P P S PPP PP P P SP +P
Sbjct: 553 MASLSAVVVAPSVSSSAAVPPAPPLPGDSGTVIPPPPPPPPLPGGVVPPSPPLPPGTCIP 612
Query: 66 PPPPDQALTQPPPSASIAYPTATKPAKPAKKVA 98
PPPP PP + A P P VA
Sbjct: 613 PPPPLPGGACIPPPPQLPGSAAIPPPPPLPGVA 645
Score = 37.0 bits (84), Expect = 0.22
Identities = 23/69 (33%), Positives = 26/69 (37%), Gaps = 12/69 (17%)
Query: 37 GSAPPPSTP-------PPPPP-----DTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAY 84
G PPS P PPPPP P P +P PPPP + PP +
Sbjct: 596 GGVVPPSPPLPPGTCIPPPPPLPGGACIPPPPQLPGSAAIPPPPPLPGVASIPPPPPLPG 655
Query: 85 PTATKPAKP 93
TA P P
Sbjct: 656 ATAIPPPPP 664
>DIA3_MOUSE (Q9Z207) Diaphanous protein homolog 3
(Diaphanous-related formin 3) (DRF3) (mDIA2) (p134mDIA2)
Length = 1171
Score = 147 bits (370), Expect = 2e-34
Identities = 104/356 (29%), Positives = 166/356 (46%), Gaps = 25/356 (7%)
Query: 308 LPPPPAPPAPPSTQLDGGPSRRPKFSTHPLAPNMASLNTPPPPPPPPPPPAARKVWSPAI 367
LPP P PS + + GPS P P P ++ PPPPPPPPPPP P +
Sbjct: 533 LPPGTKIPLQPSVEGEAGPSALP-----PAPPALSGGVPPPPPPPPPPPPPL-----PGM 582
Query: 368 TTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVSKPSEEGESMMEGGKPKL--KALHWDKV 425
V G SS+ ++ P G + KP++ + L+W K+
Sbjct: 583 PMPFGGPVPPPPPLGFL-------GGQSSIPLNLPF--GLKPKKEFKPEISMRRLNWLKI 633
Query: 426 RATS-NRATVWNQLKSSSFQLNEDMMETLFGFNSTTNSAPKETTTTSVVRKPVFPPVEQE 484
+ W ++ + ++ N D++ L N+ ++ T K V +E
Sbjct: 634 GPNEMSENCFWIKVNENKYE-NRDLLCKLE--NTFCCQEKEKRNTNDFDEKKVIKKRMKE 690
Query: 485 NRVLDPKKSQNIAILLRALNVTRDEVSEALLDGNPEGLGAELLETLVKMAPTKEEEIKLK 544
+ LDPK +QN++I L + V +++ +L+ + L +++ L+K P +E+ L
Sbjct: 691 LKFLDPKIAQNLSIFLSSFRVPYEKIRTMILEVDETQLSESMIQNLIKHLPDEEQLKSLS 750
Query: 545 NYDGDLSKLGSAERFLKAVLDIPLAFKRVEAMLYRANFETEVNYLRKSFQTLEAASEELK 604
+ D + L E+F + ++ R+ A+L++ FE +VN ++ + A EE+K
Sbjct: 751 QFRSDYNSLCEPEQFAVVMSNVKRLRPRLSAILFKLQFEEQVNNIKPDIMAVSTACEEIK 810
Query: 605 NSRLFFKLLEAVLRTGNRMNVGTNRGDAKSFKLDTLLKLVDIKGTDGKTTLLHFVV 660
S+ F KLLE VL GN MN G+ F L +L KL D K D KTTLLHF+V
Sbjct: 811 KSKGFSKLLELVLLMGNYMNAGSRNAQTFGFDLSSLCKLKDTKSADQKTTLLHFLV 866
Score = 45.4 bits (106), Expect = 6e-04
Identities = 25/64 (39%), Positives = 27/64 (42%), Gaps = 2/64 (3%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKPA 91
P G PPP PPPPPP P P +PF PPPP S + P KP
Sbjct: 559 PALSGGVPPPPPPPPPPPPPLPGMP-MPFGGPV-PPPPPLGFLGGQSSIPLNLPFGLKPK 616
Query: 92 KPAK 95
K K
Sbjct: 617 KEFK 620
>DAM1_MOUSE (Q8BPM0) Disheveled associated activator of morphogenesis
1
Length = 1071
Score = 140 bits (352), Expect = 2e-32
Identities = 148/583 (25%), Positives = 237/583 (40%), Gaps = 107/583 (18%)
Query: 292 SPDIRHAMIPSIKHNPLPPPPAPPAPPSTQLDGGPSRRPKFSTHPLAPNMASLNTPPPPP 351
SP PS L PPP PP L GG + P P + + PPPP
Sbjct: 532 SPGAPGGPFPSSGLGSLLPPPPPPL-----LSGG-------ALPPPPPPLPAGGPPPPPG 579
Query: 352 PPP-----PPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVSKPSEEG 406
PPP PPP A P T+ ++ + +
Sbjct: 580 PPPLGGVLPPPGA-----PVSLTLKKKNIPQPTNA------------------------- 609
Query: 407 ESMMEGGKPKLKALHWDKVRATSNRATVWNQLKSSS-FQLN--EDMMETLFGFNS---TT 460
LK+ +W K+ TVW ++ + F++ ED+ T + TT
Sbjct: 610 ----------LKSFNWSKLPENKLDGTVWTEIDDTKVFKILDLEDLERTFSAYQRQQVTT 659
Query: 461 NSAPKETTTTSVVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEVSEALLD-GNP 519
A T S K +E V+D +++QN ILL L ++ DE+ A+L
Sbjct: 660 KEADAIDDTLSSKLKV------KELSVIDGRRAQNCNILLSRLKLSNDEIKRAILTMDEQ 713
Query: 520 EGLGAELLETLVKMAPTKEEEIKLKNYDGDLSKLGSAERFLKAVLDIPLAFKRVEAMLYR 579
E L ++LE L+K P K + L+ + +L ++ A+RFL + I +R++++ ++
Sbjct: 714 EDLPKDMLEQLLKFVPEKSDIDLLEEHKHELDRMAKADRFLFEMSRINHYQQRLQSLYFK 773
Query: 580 ANFETEVNYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGTNRGDAKSFKLDT 639
F V ++ + + + SEE+ SR +LLE VL GN MN G RG+A FK+ +
Sbjct: 774 KKFAERVAEVKPKVEAIRSGSEEVFRSRALKQLLEVVLAFGNYMNKG-QRGNAYGFKISS 832
Query: 640 LLKLVDIKGT-DGKTTLLHFVVQEIIRSEGTGGESANESVQNQTNSQFNEDEFRKKGLQV 698
L K+ D K + D TLLH+++ + E+++ K
Sbjct: 833 LNKIADTKSSIDKNITLLHYLITIV------------------------ENKYPK----- 863
Query: 699 VAGLSRDLGNVKKAAGMDSDVLSSYVSKLEMGLDKVRLVLQYQK---PDMQGSFFNATEI 755
V LS +L ++ +AA ++ L +S L GL V L+YQK P F +
Sbjct: 864 VLNLSEELRDIPQAAKVNMTELDKEISTLRSGLKAVETELEYQKSQPPQPGDKFVSVVSQ 923
Query: 756 FLKDAEEKIIVIKDDERRALFLVKEVTEYFHGDTAKEEVHPFRIFMIVRDFLNILDQVCK 815
F+ A ++D A L + ++F + K + P F I FL + + +
Sbjct: 924 FITLASFSFSDVEDLLAEAKELFTKAVKHFGEEAGK--IQPDEFFGIFDQFLQAVAEAKQ 981
Query: 816 EVGRMQDRTVIGSARSFRIAANASLPVLNRYHARQDRSSDEES 858
E M+ R R+ R+ A R+ + S EES
Sbjct: 982 ENENMRKRKEEEERRA-RLEAQLKEQRERERKVRKAKESSEES 1023
Score = 33.5 bits (75), Expect = 2.5
Identities = 19/52 (36%), Positives = 22/52 (41%), Gaps = 11/52 (21%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIA 83
PL G+ PPP PPP P P P P PP PPP A ++
Sbjct: 555 PLLSGGALPPP--PPPLPAGGPPPP---------PGPPPLGGVLPPPGAPVS 595
>DIA2_MOUSE (O70566) Diaphanous protein homolog 2
(Diaphanous-related formin 2) (DRF2) (mDia3)
Length = 1098
Score = 139 bits (350), Expect = 3e-32
Identities = 109/391 (27%), Positives = 164/391 (41%), Gaps = 39/391 (9%)
Query: 280 KRTSPKSRLSASSPDIRHAMIPSIKHNPLPPPPAP---PAPPSTQLDGGPSRRPKFSTHP 336
KR L +R +PS P PPPP P P PP P P P
Sbjct: 519 KRDEKIKELETEIQQLRGQGVPSAIPGPPPPPPLPGAGPCPPPPPPPPPPPPLPGVVPPP 578
Query: 337 LAPNMASLNTPPPPPPP-----PPPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASS 391
P PPPPPPP PPPP V+ P + S IE
Sbjct: 579 PPPLPGMPGIPPPPPPPLSGVPPPPPPPGGVF-PLL--------------SGPIE---LP 620
Query: 392 NGVSSVSVSKPSEEGESMMEGGKPKLKALHWDKVRATS-NRATVWNQLKSSSFQLNEDMM 450
G+ + KP +K ++W K+ + VW +LK ++ N D+
Sbjct: 621 YGMKQKKLYKPDIP-----------MKRINWSKIEPKELSENCVWLKLKEEKYE-NADLF 668
Query: 451 ETLFGFNSTTNSAPKETTTTSVVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEV 510
L + + T R +E R+LD K +QN++I L + + +E+
Sbjct: 669 AKLALTFPSQMKGQRNTEAAEENRSGPPKKKVKELRILDTKTAQNLSIFLGSYRMPYEEI 728
Query: 511 SEALLDGNPEGLGAELLETLVKMAPTKEEEIKLKNYDGDLSKLGSAERFLKAVLDIPLAF 570
+L+ N E L L++ LVK P + +L + L E+F + + +
Sbjct: 729 KNIILEVNEEMLSEALIQNLVKYLPDQNALRELAQLKSEYDDLCEPEQFGVVMSTVKMLR 788
Query: 571 KRVEAMLYRANFETEVNYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGTNRG 630
R+ ++L++ FE VN ++ S + A EELK S F +LLE +L GN MN G+
Sbjct: 789 PRLTSILFKLTFEEHVNNIKPSIIAVTLACEELKKSESFKRLLELILLVGNYMNSGSRNA 848
Query: 631 DAKSFKLDTLLKLVDIKGTDGKTTLLHFVVQ 661
+ FK++ L K+ D K D K+TLLHF+ +
Sbjct: 849 QSLGFKINFLCKIKDTKSADQKSTLLHFLAE 879
Score = 47.4 bits (111), Expect = 2e-04
Identities = 25/66 (37%), Positives = 29/66 (43%), Gaps = 7/66 (10%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPS-----SPDIPFFNEYSPPPPD--QALTQPPPSASIAY 84
PL AG PPP PPPPPP P P +P PPPP + PPP +
Sbjct: 551 PLPGAGPCPPPPPPPPPPPPLPGVVPPPPPPLPGMPGIPPPPPPPLSGVPPPPPPPGGVF 610
Query: 85 PTATKP 90
P + P
Sbjct: 611 PLLSGP 616
Score = 37.4 bits (85), Expect = 0.17
Identities = 22/63 (34%), Positives = 24/63 (37%), Gaps = 11/63 (17%)
Query: 37 GSAPPPSTP-----PPPPPDTPSSPDIPFFNEYSPPPPD----QALTQPPPSASIAYPTA 87
G PPP P PPPPP P P +P PPPP + PPP P
Sbjct: 545 GPPPPPPLPGAGPCPPPPPPPPPPPPLP--GVVPPPPPPLPGMPGIPPPPPPPLSGVPPP 602
Query: 88 TKP 90
P
Sbjct: 603 PPP 605
Score = 35.0 bits (79), Expect = 0.85
Identities = 18/49 (36%), Positives = 20/49 (40%)
Query: 45 PPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKPAKP 93
PPPPPP + P P PPPP + PPP P P P
Sbjct: 546 PPPPPPLPGAGPCPPPPPPPPPPPPLPGVVPPPPPPLPGMPGIPPPPPP 594
>DIA3_HUMAN (Q9NSV4) Diaphanous protein homolog 3
(Diaphanous-related formin 3) (DRF3) (Fragment)
Length = 853
Score = 137 bits (344), Expect = 2e-31
Identities = 101/359 (28%), Positives = 164/359 (45%), Gaps = 29/359 (8%)
Query: 308 LPPPPAPPAPPSTQLDGGPSRRPKFSTHPLAPNMASLNTPPPPPPPPPPPAARKVWSPAI 367
LP P PPS + G S P P P + PPPPPPPPPPP P +
Sbjct: 297 LPADCNIPLPPSKEGGTGHSALP-----PPPPLPSGGGVPPPPPPPPPPPL------PGM 345
Query: 368 TTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVS-KPSEEGESMMEGGKPKLKALHWDKVR 426
S V G +S + + KP +E + + ++ L+W K+R
Sbjct: 346 RMPFSGPVPPPPPLG--FLGGQNSPPLPILPFGLKPKKEFKPEIS-----MRRLNWLKIR 398
Query: 427 ATS-NRATVWNQLKSSSFQLNEDMM---ETLFGFNSTTNSAPKETTTTSVVRKPVFPPVE 482
W ++ + ++ N D++ E F ++ ++K +
Sbjct: 399 PHEMTENCFWIKVNENKYE-NVDLLCKLENTFCCQQKERREEEDIEEKKSIKKKI----- 452
Query: 483 QENRVLDPKKSQNIAILLRALNVTRDEVSEALLDGNPEGLGAELLETLVKMAPTKEEEIK 542
+E + LD K +QN++I L + V +E+ +L+ + L +++ L+K P +E+
Sbjct: 453 KELKFLDSKIAQNLSIFLSSFRVPYEEIRMMILEVDETRLAESMIQNLIKHLPDQEQLNS 512
Query: 543 LKNYDGDLSKLGSAERFLKAVLDIPLAFKRVEAMLYRANFETEVNYLRKSFQTLEAASEE 602
L + + S L E+F+ + ++ R+ A+L++ FE +VN ++ + A EE
Sbjct: 513 LSQFKSEYSNLCEPEQFVVVMSNVKRLRPRLSAILFKLQFEEQVNNIKPDIMAVSTACEE 572
Query: 603 LKNSRLFFKLLEAVLRTGNRMNVGTNRGDAKSFKLDTLLKLVDIKGTDGKTTLLHFVVQ 661
+K S+ F KLLE VL GN MN G+ F L +L KL D K D KTTLLHF+V+
Sbjct: 573 IKKSKSFSKLLELVLLMGNYMNAGSRNAQTFGFNLSSLCKLKDTKSADQKTTLLHFLVE 631
Score = 42.4 bits (98), Expect = 0.005
Identities = 24/64 (37%), Positives = 26/64 (40%), Gaps = 1/64 (1%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKPA 91
P P+G PP PPPPPP P +PF PPPP L P KP
Sbjct: 322 PPLPSGGGVPPPPPPPPPPPLPGM-RMPFSGPVPPPPPLGFLGGQNSPPLPILPFGLKPK 380
Query: 92 KPAK 95
K K
Sbjct: 381 KEFK 384
>DAM1_HUMAN (Q9Y4D1) Disheveled associated activator of
morphogenesis 1
Length = 1078
Score = 135 bits (341), Expect = 4e-31
Identities = 138/557 (24%), Positives = 224/557 (39%), Gaps = 91/557 (16%)
Query: 279 SKRTSPKSRLSASSPDIRHAMIPSIKHNPLPPPPAPPAPPSTQLDGGPSRRPKFSTHPLA 338
S+R S SP PS L PPP PP P L P PL
Sbjct: 519 SRRAVCASIPGGPSPGAPGGPFPSSVPGSLLPPPPPPPLPGGMLPPPPP--------PLP 570
Query: 339 PNMASLNTPPPPPPPPPPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSVS 398
P PPPPP PPP + P G + S
Sbjct: 571 PG-------GPPPPPGPPPLGAIMPPP---------------------GAPMGLALKKKS 602
Query: 399 VSKPSEEGESMMEGGKPKLKALHWDKVRATSNRATVWNQLKSSS-FQL--NEDMMETLFG 455
+ +P+ LK+ +W K+ TVW ++ + F++ ED+ T
Sbjct: 603 IPQPTN-----------ALKSFNWSKLPENKLEGTVWTEIDDTKVFKILDLEDLERTFSA 651
Query: 456 FNST----TNSAPKETTTTSVVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEVS 511
+ NS K+ ++ +E V+D +++QN ILL L ++ DE+
Sbjct: 652 YQRQQDFFVNSNSKQKEADAIDDTLSSKLKVKELSVIDGRRAQNCNILLSRLKLSNDEIK 711
Query: 512 EALLD-GNPEGLGAELLETLVKMAPTKEEEIKLKNYDGDLSKLGSAERFLKAVLDIPLAF 570
A+L E L ++LE L+K P K + L+ + +L ++ A+RFL + I
Sbjct: 712 RAILTMDEQEDLPKDMLEQLLKFVPEKSDIDLLEEHKHELDRMAKADRFLFEMSRINHYQ 771
Query: 571 KRVEAMLYRANFETEVNYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGTNRG 630
+R++++ ++ F V ++ + + + SEE+ S +LLE VL GN MN G RG
Sbjct: 772 QRLQSLYFKKKFAERVAEVKPKVEAIRSGSEEVFRSGALKQLLEVVLAFGNYMNKG-QRG 830
Query: 631 DAKSFKLDTLLKLVDIKGT-DGKTTLLHFVVQEIIRSEGTGGESANESVQNQTNSQFNED 689
+A FK+ +L K+ D K + D TLLH+++ V+N+ S N
Sbjct: 831 NAYGFKISSLNKIADTKSSIDKNITLLHYLI---------------TIVENKYPSVLN-- 873
Query: 690 EFRKKGLQVVAGLSRDLGNVKKAAGMDSDVLSSYVSKLEMGLDKVRLVLQYQK---PDMQ 746
L+ +L ++ +AA ++ L +S L GL V L+YQK P
Sbjct: 874 ------------LNEELRDIPQAAKVNMTELDKEISTLRSGLKAVETELEYQKSQPPQPG 921
Query: 747 GSFFNATEIFLKDAEEKIIVIKDDERRALFLVKEVTEYFHGDTAKEEVHPFRIFMIVRDF 806
F + F+ A ++D A L + ++F + K + P F I F
Sbjct: 922 DKFVSVVSQFITVASFSFSDVEDLLAEAKDLFTKAVKHFGEEAGK--IQPDEFFGIFDQF 979
Query: 807 LNILDQVCKEVGRMQDR 823
L + + +E M+ +
Sbjct: 980 LQAVSEAKQENENMRKK 996
Score = 36.2 bits (82), Expect = 0.38
Identities = 25/69 (36%), Positives = 28/69 (40%), Gaps = 8/69 (11%)
Query: 34 FPAGSAPPPSTPPPPPPDTPSS---PDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKP 90
FP+ S P PPPPPP P P P PPPP PPP +I P
Sbjct: 540 FPS-SVPGSLLPPPPPPPLPGGMLPPPPPPLPPGGPPPP----PGPPPLGAIMPPPGAPM 594
Query: 91 AKPAKKVAI 99
KK +I
Sbjct: 595 GLALKKKSI 603
Score = 32.7 bits (73), Expect = 4.2
Identities = 16/37 (43%), Positives = 16/37 (43%), Gaps = 1/37 (2%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPP 68
P P G PPP PP PP P P P PPP
Sbjct: 555 PPLPGGMLPPPP-PPLPPGGPPPPPGPPPLGAIMPPP 590
>CAPU_DROME (Q24120) Cappuccino protein
Length = 1059
Score = 132 bits (332), Expect = 4e-30
Identities = 132/522 (25%), Positives = 215/522 (40%), Gaps = 87/522 (16%)
Query: 307 PLPPPPAPPAPPSTQLDGGPSRRPKFSTHPLAPNMASLNTPPPP---------PPPPPPP 357
P PPPP PP PP G P P P P S PPPP PPPPPP
Sbjct: 501 PPPPPPPPPPPPPLANYGAPPPPP-----PPPPGSGSAPPPPPPAPIEGGGGIPPPPPPM 555
Query: 358 AARKV----------------WSPAITTMHSSSVSKQQQS-----SSSIEGDASSNGVSS 396
+A W TM S+V+ + + + + S
Sbjct: 556 SASPSKTTISPAPLPDPAEGNWFHRTNTMRKSAVNPPKPMRPLYWTRIVTSAPPAPRPPS 615
Query: 397 VSVSKPSEEGESMMEGGKPKLKALHWDKVRATSNRAT--VWNQLKSSSFQLNEDMMETLF 454
V+ S S E P A + T+ AT +W +++ + ++ E
Sbjct: 616 VANSTDSTENSGSSPDEPP---AANGADAPPTAPPATKEIWTEIEETPLDNIDEFTELFS 672
Query: 455 GFNSTTNSAPKETTTTSVVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEVSEAL 514
S PKE + +VLDP++S+N+ I+ R+L+V E+ A+
Sbjct: 673 RQAIAPVSKPKELKVKRA----------KSIKVLDPERSRNVGIIWRSLHVPSSEIEHAI 722
Query: 515 LDGNPEGLGAELLETLVKMAPTKEEEIKLKNYDGDLSKLGSAERFLKAVLDIPLAFKRVE 574
+ + E L+ + + T++E ++K G L E+FL + I +A +R+
Sbjct: 723 YHIDTSVVSLEALQHMSNIQATEDELQRIKEAAGGDIPLDHPEQFLLDISLISMASERIS 782
Query: 575 AMLYRANFETEVNYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGT-NRGDAK 633
++++A FE V L + +T+ S++L S + +L GN MN G RG A
Sbjct: 783 CIVFQAEFEESVTLLFRKLETVSQLSQQLIESEDLKLVFSIILTLGNYMNGGNRQRGQAD 842
Query: 634 SFKLDTLLKLVDIKGTDGKTTLLHFVVQEIIRSEGTGGESANESVQNQTNSQFNEDEFRK 693
F LD L KL D+K + TTLLHF+V+ I RK
Sbjct: 843 GFNLDILGKLKDVKSKESHTTLLHFIVRTYIAQR------------------------RK 878
Query: 694 KG---LQVVAGLSRDLGNVKKAAGMDSDVLSSYV---SKLEMGLDKVRL-VLQYQKPDMQ 746
+G L++ + + +V++AA MD + + + +K +G + VL +P++
Sbjct: 879 EGVHPLEIRLPIP-EPADVERAAQMDFEEVQQQIFDLNKKFLGCKRTTAKVLAASRPEIM 937
Query: 747 GSFFNATEIFLKDAEEKIIVIKD--DERRALFLVKEVTEYFH 786
F + E F++ A++ + + DE R LFL E ++H
Sbjct: 938 EPFKSKMEEFVEGADKSMAKLHQSLDECRDLFL--ETMRFYH 977
Score = 52.8 bits (125), Expect = 4e-06
Identities = 58/240 (24%), Positives = 80/240 (33%), Gaps = 97/240 (40%)
Query: 17 TPLNSTPNARRILHQPLFPAGSAPPPSTP------PPPPPDTPSSPDIPFFNEYSPPPPD 70
T ++ +A+ +P A PPP P PPPPP P P P N +PPPP
Sbjct: 465 TKVSDNESAKEDGEKPHAVAPPPPPPPPPLHAFVAPPPPPPPPPPPPPPLANYGAPPPP- 523
Query: 71 QALTQPPPSASIAYPTATKPAKPAKKVAIAASAAVVTVAVLSALAFFLYKHRANSNSNQT 130
PPP S + P PA
Sbjct: 524 ----PPPPPGSGSAPPPPPPAP-------------------------------------- 541
Query: 131 SSENHKLVVDGGGGRGNSRRVLDDSPRAPPPPPPSSFLYIGTVEPSRTSVSDAQNLNSAK 190
++GGGG PPPPPP S PS+T++S A + A+
Sbjct: 542 --------IEGGGG-------------IPPPPPPMS------ASPSKTTISPAPLPDPAE 574
Query: 191 LASNYRYR----------PSPELQPL---------PPLSKPPDEIHSPPAASSSSPSSSE 231
N+ +R P ++PL PP +PP +S + +S S E
Sbjct: 575 --GNWFHRTNTMRKSAVNPPKPMRPLYWTRIVTSAPPAPRPPSVANSTDSTENSGSSPDE 632
Score = 33.9 bits (76), Expect = 1.9
Identities = 24/96 (25%), Positives = 37/96 (38%), Gaps = 16/96 (16%)
Query: 72 ALTQPPPSASIAYPTATKPAKPAKKVAIAASAAVVTVAVLSALAFFLYKHRANSNSNQTS 131
A+TQ PP + + P + PA + S+ + ++F + S +N +
Sbjct: 63 AVTQTPPGVTSSTPNESGVTGPAGPLGATTSSPSLETQSTVIISFKSSQTPVQSQTNSAA 122
Query: 132 SENHKLVVDGGGGRGNSRRVLDDSPRAPPPPPPSSF 167
SEN V DD+ P PPPP F
Sbjct: 123 SEN----------------VEDDTAPLPLPPPPPGF 142
Score = 32.7 bits (73), Expect = 4.2
Identities = 54/239 (22%), Positives = 95/239 (39%), Gaps = 37/239 (15%)
Query: 218 SPPAASSSSPSSSEDAESRETAFHSPHGSSVSHDDNNYYTPVSRHSSVANGSPAAATAVP 277
+PP +SS+P+ ES T P G++ S + S+V ++ T P
Sbjct: 67 TPPGVTSSTPN-----ESGVTGPAGPLGATTSSPS------LETQSTVIISFKSSQT--P 113
Query: 278 FSKRTSPKSRLSASSPDIRHAMIPSIKHNPLPPPPAPPAPPSTQLDGGPSRRPKFSTHPL 337
+T+ SA+S ++ P PLPPPP P+T L + K ++ +
Sbjct: 114 VQSQTN-----SAASENVEDDTAPL----PLPPPPPGFGTPTTPLLSSNVLK-KVASFTV 163
Query: 338 APNMASLNTPPPPPPPPPPPAARKVWSPAITTMHSSSVSKQ---QQSSSSIEGDASSNGV 394
+ A N+ PP P + +P +++ +++ + ++ + G A S
Sbjct: 164 EKSSAGNNSSNPPNLCPTSDETTLLATPCSSSLTVATLPPEIAVGAAAGGVAGGAGSRRG 223
Query: 395 SSVSVSKPSEEGESMMEGGKPKLKALHWDKVRATSNRATVWNQLKSSSFQLNEDMMETL 453
SS K S EG + W +T+ + KSSS N+++ TL
Sbjct: 224 SSYVPEKLSFAAYEKFEG----QMLIKW-------LISTMQSNPKSSSGDANQELFNTL 271
>FM14_MOUSE (Q05859) Formin 1 isoform IV (Limb deformity protein)
Length = 1206
Score = 129 bits (324), Expect = 3e-29
Identities = 129/535 (24%), Positives = 225/535 (41%), Gaps = 76/535 (14%)
Query: 300 IPSIKHNPLPPPPAPPAPPSTQLDGGPSRRPKFSTHPLAPNMASL----NTPPPPPPPPP 355
IP + PPPP PP P GP P P PN+ +L PPPPPPPPP
Sbjct: 677 IPPVCPVSPPPPPPPPPPTPVPPSDGPPPPP--PPPPPLPNVLALPNSGGPPPPPPPPPP 734
Query: 356 PPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVSKPSEEGESMMEGGKP 415
PP P ++ SSS S+ + KP+ E M
Sbjct: 735 PPGLAPPPPPGLSFGLSSSSSQYPR--------------------KPAIEPSCPM----- 769
Query: 416 KLKALHWDKV----RATSNRATVWNQLKSSSFQLNEDMMETLFGFNSTTNSAPKETTTTS 471
K L+W ++ ++ T+W+ L+ + D E + F+ T K+ + +
Sbjct: 770 --KPLYWTRIQINDKSQDAAPTLWDSLEEPHIR---DTSEFEYLFSKDTTQQKKKPLSEA 824
Query: 472 VVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEVSEALLDGNPEGLGAELLETLV 531
+K V++ ++LD K+SQ + IL+ +L++ ++ +A+ + + E L L
Sbjct: 825 YEKK---NKVKKIIKLLDGKRSQTVGILISSLHLEMKDIQQAIFTVDDSVVDLETLAALY 881
Query: 532 KMAPTKEEEIKLKNY-----DGDLSKLGSAERFLKAVLDIPLAFKRVEAMLYRANFETEV 586
+ ++E K++ Y + DL L E+FL + IP +R + +++RA F +
Sbjct: 882 ENRAQEDELTKIRKYYETSKEEDLKLLDKPEQFLHELAQIPNFAERAQCIIFRAVFSEGI 941
Query: 587 NYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGT-NRGDAKSFKLDTLLKLVD 645
L + + + AS+ L + + +L +L GN MN G RG A + L+ L KL D
Sbjct: 942 TSLHRKVEIVTRASKGLLHMKSVKDILALILAFGNYMNGGNRTRGQADGYSLEILPKLKD 1001
Query: 646 IKGTDGKTTLLHFVVQEIIR---SEGTGGESANESVQNQTNSQFNEDEFRKKGLQVVAGL 702
+K D L+ +VV+ +R E +S + Q ++ +F L
Sbjct: 1002 VKSRDNGMNLVDYVVKYYLRYYDQEAGTDKSVFPLPEPQDFFLASQVKFE--------DL 1053
Query: 703 SRDLGNVKKAAGMDSDVLSSYVSKLEMGLDKVRLVLQYQKPDMQGSFFNATEIFLKDAEE 762
+DL +K+ +LE +++LV + + F + E F K A++
Sbjct: 1054 LKDLRKLKR--------------QLEASEQQMKLVCKESPREYLQPFKDKLEEFFKKAKK 1099
Query: 763 KIIVIKDDERRALFLVKEVTEYF--HGDTAKEEVHPFRIFMIVRDFLNILDQVCK 815
+ + + A + YF T ++EV P +FM+ +F + + K
Sbjct: 1100 EHKMEESHLENAQKSFETTVGYFGMKPKTGEKEVTPSYVFMVWFEFCSDFKTIWK 1154
Score = 45.4 bits (106), Expect = 6e-04
Identities = 27/65 (41%), Positives = 32/65 (48%), Gaps = 9/65 (13%)
Query: 32 PLFPAGSAPP--PSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASI-AYPTAT 88
PL PA PP P +PPPPPP P +P P PPPP PPP ++ A P +
Sbjct: 670 PLPPAPPIPPVCPVSPPPPPPPPPPTPVPPSDGPPPPPPP------PPPLPNVLALPNSG 723
Query: 89 KPAKP 93
P P
Sbjct: 724 GPPPP 728
Score = 44.3 bits (103), Expect = 0.001
Identities = 22/71 (30%), Positives = 35/71 (48%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKPA 91
P+ P+ PPP PPPP P+ + P+ PPPP PPP +++ ++ +
Sbjct: 696 PVPPSDGPPPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGLAPPPPPGLSFGLSSSSS 755
Query: 92 KPAKKVAIAAS 102
+ +K AI S
Sbjct: 756 QYPRKPAIEPS 766
Score = 43.5 bits (101), Expect = 0.002
Identities = 24/70 (34%), Positives = 30/70 (42%), Gaps = 10/70 (14%)
Query: 35 PAGSAPPPSTPPPPPPDTPSSPDIPFFNEYS-------PPPPDQALTQPPP---SASIAY 84
P PP PPPPPP P P++ PPPP L PPP S ++
Sbjct: 693 PPTPVPPSDGPPPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGLAPPPPPGLSFGLSS 752
Query: 85 PTATKPAKPA 94
++ P KPA
Sbjct: 753 SSSQYPRKPA 762
Score = 41.2 bits (95), Expect = 0.012
Identities = 26/68 (38%), Positives = 28/68 (40%), Gaps = 15/68 (22%)
Query: 35 PAGSAPPPSTPP---PPPP------DTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYP 85
PA PPP PP PPPP P +P IP SPPPP PPP P
Sbjct: 645 PAPPTPPPLPPPLIPPPPPLPPGLGPLPPAPPIPPVCPVSPPPP------PPPPPPTPVP 698
Query: 86 TATKPAKP 93
+ P P
Sbjct: 699 PSDGPPPP 706
>FMN1_MOUSE (Q05860) Formin 1 isoforms I/II/III (Limb deformity
protein)
Length = 1468
Score = 128 bits (322), Expect = 6e-29
Identities = 136/561 (24%), Positives = 229/561 (40%), Gaps = 92/561 (16%)
Query: 300 IPSIKHNPLPPPPAPPAPPSTQLDGGPSRRPKFSTHPLAPNMASL----NTPPPPPPPPP 355
IP + PPPP PP P GP P P PN+ +L PPPPPPPPP
Sbjct: 903 IPPVCPVSPPPPPPPPPPTPVPPSDGPPPPP--PPPPPLPNVLALPNSGGPPPPPPPPPP 960
Query: 356 PPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVSKPSEEGESMMEGGKP 415
PP P ++ SSS S+ + KP+ E M
Sbjct: 961 PPGLAPPPPPGLSFGLSSSSSQYPR--------------------KPAIEPSCPM----- 995
Query: 416 KLKALHWDKV----RATSNRATVWNQLKSSSFQLNEDMMETLFGFNSTTNSAPKETTTTS 471
K L+W ++ ++ T+W+ L+ + D E + F+ T K+ + +
Sbjct: 996 --KPLYWTRIQINDKSQDAAPTLWDSLEEPHIR---DTSEFEYLFSKDTTQQKKKPLSEA 1050
Query: 472 VVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEVSEALLDGNPEGLGAELLETLV 531
+K V++ ++LD K+SQ + IL+ +L++ ++ +A+ + + E L L
Sbjct: 1051 YEKK---NKVKKIIKLLDGKRSQTVGILISSLHLEMKDIQQAIFTVDDSVVDLETLAALY 1107
Query: 532 KMAPTKEEEIKLKNY-----DGDLSKLGSAERFLKAVLDIPLAFKRVEAMLYRANFETEV 586
+ ++E K++ Y + DL L E+FL + IP +R + +++RA F +
Sbjct: 1108 ENRAQEDELTKIRKYYETSKEEDLKLLDKPEQFLHELAQIPNFAERAQCIIFRAVFSEGI 1167
Query: 587 NYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGT-NRGDAKSFKLDTLLKLVD 645
L + + + AS+ L + + +L +L GN MN G RG A + L+ L KL D
Sbjct: 1168 TSLHRKVEIVTRASKGLLHMKSVKDILALILAFGNYMNGGNRTRGQADGYSLEILPKLKD 1227
Query: 646 IKGTDGKTTLLHFVVQEIIRSEGTGGESANESVQNQTNSQFNEDEFRKKGL-----QVVA 700
+K D L+ +VV+ +R +Q E R K L +
Sbjct: 1228 VKSRDNGMNLVDYVVKYYLR------------YYDQCKHHDQEASCRGKDLFSLYFHIAV 1275
Query: 701 GLSRDLG-NVKKAAGMDSDV----------LSSYVS-------------KLEMGLDKVRL 736
R G +K+ AG D V L+S V +LE +++L
Sbjct: 1276 HPQRKSGLELKQEAGTDKSVFPLPEPQDFFLASQVKFEDLLKDLRKLKRQLEASEQQMKL 1335
Query: 737 VLQYQKPDMQGSFFNATEIFLKDAEEKIIVIKDDERRALFLVKEVTEYF--HGDTAKEEV 794
V + + F + E F K A+++ + + A + YF T ++EV
Sbjct: 1336 VCKESPREYLQPFKDKLEEFFKKAKKEHKMEESHLENAQKSFETTVGYFGMKPKTGEKEV 1395
Query: 795 HPFRIFMIVRDFLNILDQVCK 815
P +FM+ +F + + K
Sbjct: 1396 TPSYVFMVWFEFCSDFKTIWK 1416
Score = 45.4 bits (106), Expect = 6e-04
Identities = 27/65 (41%), Positives = 32/65 (48%), Gaps = 9/65 (13%)
Query: 32 PLFPAGSAPP--PSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASI-AYPTAT 88
PL PA PP P +PPPPPP P +P P PPPP PPP ++ A P +
Sbjct: 896 PLPPAPPIPPVCPVSPPPPPPPPPPTPVPPSDGPPPPPPP------PPPLPNVLALPNSG 949
Query: 89 KPAKP 93
P P
Sbjct: 950 GPPPP 954
Score = 44.3 bits (103), Expect = 0.001
Identities = 22/71 (30%), Positives = 35/71 (48%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKPA 91
P+ P+ PPP PPPP P+ + P+ PPPP PPP +++ ++ +
Sbjct: 922 PVPPSDGPPPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGLAPPPPPGLSFGLSSSSS 981
Query: 92 KPAKKVAIAAS 102
+ +K AI S
Sbjct: 982 QYPRKPAIEPS 992
Score = 43.5 bits (101), Expect = 0.002
Identities = 24/70 (34%), Positives = 30/70 (42%), Gaps = 10/70 (14%)
Query: 35 PAGSAPPPSTPPPPPPDTPSSPDIPFFNEYS-------PPPPDQALTQPPP---SASIAY 84
P PP PPPPPP P P++ PPPP L PPP S ++
Sbjct: 919 PPTPVPPSDGPPPPPPPPPPLPNVLALPNSGGPPPPPPPPPPPPGLAPPPPPGLSFGLSS 978
Query: 85 PTATKPAKPA 94
++ P KPA
Sbjct: 979 SSSQYPRKPA 988
Score = 41.2 bits (95), Expect = 0.012
Identities = 26/68 (38%), Positives = 28/68 (40%), Gaps = 15/68 (22%)
Query: 35 PAGSAPPPSTPP---PPPP------DTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYP 85
PA PPP PP PPPP P +P IP SPPPP PPP P
Sbjct: 871 PAPPTPPPLPPPLIPPPPPLPPGLGPLPPAPPIPPVCPVSPPPP------PPPPPPTPVP 924
Query: 86 TATKPAKP 93
+ P P
Sbjct: 925 PSDGPPPP 932
>DAM2_MOUSE (Q80U19) Disheveled associated activator of
morphogenesis 2
Length = 1068
Score = 124 bits (310), Expect = 1e-27
Identities = 129/558 (23%), Positives = 227/558 (40%), Gaps = 81/558 (14%)
Query: 285 KSRLSASSPDIRHA--MIPSI--KHNPLPPPPAPPAPPSTQLDGGPSRRPKFSTH---PL 337
K +L+ S ++R A + + +HN P PP GGP T P
Sbjct: 487 KDKLARESQELRQARGQVAELVARHNESSTGPVSSPPPP----GGPLTLSSSRTTNDLPP 542
Query: 338 APNMASLNTPPPPPPPPPPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSV 397
P ++ PPPP PP PP + P SS SS E
Sbjct: 543 PPPPLPFDSCPPPPAPPLPPGGPPI-PPGAPPCFSSGPPPSHDPFSSNEAPLRKK----- 596
Query: 398 SVSKPSEEGESMMEGGKPKLKALHWDKVRATSNRATVWNQLKSSSFQLNEDMMETLFGFN 457
+ +PS LK+ +W K+ TVWN++ S D+ + F+
Sbjct: 597 RIPQPSHP-----------LKSFNWVKLNEERVSGTVWNEIDDSQVFRILDLEDFEKMFS 645
Query: 458 STTNSAPKETTTTSVVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEVSEALLD- 516
+ KE +T + + +E V+D +++QN ILL L ++ DE+ +A+L
Sbjct: 646 AYQRHQQKELGSTEDIY--ITSRKVKELSVIDGRRAQNCIILLSKLKLSNDEIRQAILRM 703
Query: 517 GNPEGLGAELLETLVKMAPTKEEEIKLKNYDGDLSKLGSAERFLKAVLDIPLAFKRVEAM 576
E L ++LE L+K P K + L+ + ++ ++ A+RFL + I +R++A+
Sbjct: 704 DEQEDLAKDMLEQLLKFIPEKSDIDLLEEHKHEIERMARADRFLYEMSRIDHYQQRLQAL 763
Query: 577 LYRANFETEVNYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGTNRGDAKSFK 636
++ F+ + + + + AS EL S+ ++LE VL GN MN G RG A F+
Sbjct: 764 FFKKKFQERLAEAKPKVEAILLASRELTLSQRLKQMLEVVLAIGNFMNKG-QRGGAYGFR 822
Query: 637 LDTLLKLVDIKGT-DGKTTLLHFVVQEIIRSEGTGGESANESVQNQTNSQFNEDEFRKKG 695
+ +L K+ D K + D +LLH+++ + +K
Sbjct: 823 VASLNKIADTKSSIDRNISLLHYLIMIL-----------------------------EKH 853
Query: 696 LQVVAGLSRDLGNVKKAAGMDSDVLSSYVSKLEMGLDKVRLVLQYQKPD----------M 745
+ + +L ++ +AA ++ L VS L GL V + L+YQ+ +
Sbjct: 854 FPDILNMPSELKHLSEAAKVNLAELEKEVSILRRGLRAVEVELEYQRHQARDPNDKFVPV 913
Query: 746 QGSFFNATEIFLKDAEEKIIVIKDDERRALFLVKEVTEYFHGDTAKEEVHPFRIFMIVRD 805
F + + E+++ +D +AL H + ++ P F I
Sbjct: 914 MSDFITVSSFSFSELEDQLNEARDKFAKAL---------THFGEQESKMQPDEFFGIFDT 964
Query: 806 FLNILDQVCKEVGRMQDR 823
FL + +++ M+ R
Sbjct: 965 FLQAFLEARQDLEAMRRR 982
Score = 37.0 bits (84), Expect = 0.22
Identities = 21/62 (33%), Positives = 24/62 (37%), Gaps = 1/62 (1%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPSSPDI-PFFNEYSPPPPDQALTQPPPSASIAYPTATKP 90
P P S PPP PP PP P P P F+ PP D + P P + P
Sbjct: 545 PPLPFDSCPPPPAPPLPPGGPPIPPGAPPCFSSGPPPSHDPFSSNEAPLRKKRIPQPSHP 604
Query: 91 AK 92
K
Sbjct: 605 LK 606
Score = 32.7 bits (73), Expect = 4.2
Identities = 28/90 (31%), Positives = 34/90 (37%), Gaps = 8/90 (8%)
Query: 161 PPPPSSFLYIGTVEPSRTSVSDAQNLNSAKLASNYRYRPSPELQPLPPLSKPPDEIHSPP 220
PPPP L T+ SRT+ +L + P P PLPP PP +PP
Sbjct: 522 PPPPGGPL---TLSSSRTT----NDLPPPPPPLPFDSCPPPPAPPLPP-GGPPIPPGAPP 573
Query: 221 AASSSSPSSSEDAESRETAFHSPHGSSVSH 250
SS P S + S E SH
Sbjct: 574 CFSSGPPPSHDPFSSNEAPLRKKRIPQPSH 603
>FMN2_MOUSE (Q9JL04) Formin 2
Length = 1567
Score = 119 bits (298), Expect = 3e-26
Identities = 112/449 (24%), Positives = 197/449 (42%), Gaps = 41/449 (9%)
Query: 300 IPSIKHNPLPPPPAPPAPPSTQLDGG----PSRRPKFSTHPLAPNMASLNTPPPPPPPP- 354
+P + P PP P PP L G P P + P P + + PPP PPPP
Sbjct: 1015 LPGVGIPPPPPLPGMGIPPPPPLPGSGIPPPPALPGVAIPP-PPPLPGMGVPPPAPPPPG 1073
Query: 355 ----PPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVSKPSEEGESMM 410
PPP P + + SS++ Q + + G+ + +++ + +
Sbjct: 1074 AGIPPPPLLPGSGPPHSSQVGSSTLPAAPQGCGFLFPPLPT-GLFGLGMNQDRVARKQPI 1132
Query: 411 EGGKPKLKALHWDKVRATSNR----ATVWNQLKSSSFQLNEDMMETLFGFNSTTNSAPKE 466
E +P +K L+W +++ S R + +W +++ S +E E LF +
Sbjct: 1133 EPCRP-MKPLYWTRIQLHSKRDSSPSLIWEKIEEPSIDCHE--FEELFSKTAVKERKKPI 1189
Query: 467 TTTTSVVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEVSEALLDGNPEGLGAEL 526
+ T S + +Q ++L K+SQ + IL+ +L++ ++ A+++ + + E
Sbjct: 1190 SDTISKTK------AKQVVKLLSNKRSQAVGILMSSLHLDMKDIQHAVVNLDNSVVDLET 1243
Query: 527 LETLVKMAPTKEEEIKLKNYDG------DLSKLGSAERFLKAVLDIPLAFKRVEAMLYRA 580
L+ L + +E K++ + + L E+FL + IP +RV +L+++
Sbjct: 1244 LQALYENRAQSDELEKIEKHSRSSKDKENAKSLDKPEQFLYELSLIPNFSERVFCILFQS 1303
Query: 581 NFETEVNYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGT-NRGDAKSFKLDT 639
F + +R+ + L+ E LKN ++L VL GN MN G RG A F LD
Sbjct: 1304 TFSESICSIRRKLELLQKLCETLKNGPGVMQVLGLVLAFGNYMNAGNKTRGQADGFGLDI 1363
Query: 640 LLKLVDIKGTDGKTTLLHFVVQEIIRS--EGTGGESA----NESVQNQTNSQFNEDEFRK 693
L KL D+K +D +LL ++V +R+ E G E E + SQ ++F+K
Sbjct: 1364 LPKLKDVKSSDNSRSLLSYIVSYYLRNFDEDAGKEQCVFPLAEPQELFQASQMKFEDFQK 1423
Query: 694 KGLQVVAGLSRDLGNVKKAAGMDSDVLSS 722
+ L +DL + AG V S+
Sbjct: 1424 D----LRKLKKDLKACEAEAGKVYQVSSA 1448
Score = 49.7 bits (117), Expect = 3e-05
Identities = 63/240 (26%), Positives = 82/240 (33%), Gaps = 50/240 (20%)
Query: 141 GGGGRGNSRRVLDDSPRAPPPPPPSSFLYIGTVEPSRTSVS-DAQNLNSAKLASNYRYRP 199
G + L SP P S I V P R SV DAQ + SA
Sbjct: 746 GSPAAASGESALLTSPSGPQTKFCSEISLI--VSPRRISVQLDAQQIQSAS--------- 794
Query: 200 SPELQPLPPLSKPPDEIHSPPAASSSSPSSSEDAESRETAFHSPHGSSVSHDDNNYYT-- 257
LPP PP + S S PS ++E+ H SVS N
Sbjct: 795 -----QLPP---PPPLLGSDSQGQPSQPSLHTESETS-------HEHSVSSSFGNNCNVP 839
Query: 258 -----PVSRHSSVANGSPAAATAVPFSKRTSPKSRLSASSPDIRHAMIPSIKHNPLPP-- 310
P + SS G A P + + S ++P ++ + + + P PP
Sbjct: 840 PAPPLPCTESSSFMPGLGMAIPPPPCLSDITVPALPSPTAPALQFSNLQGPEMLPAPPQP 899
Query: 311 --------PPAPPAPPSTQLDGGPSRRPKFSTHPLAPNMASLNTPPPPP------PPPPP 356
PP PPAPP + P P P + + PPPPP PPPPP
Sbjct: 900 PPLPGLGVPPPPPAPPLPGMGIPPPPPLPGMGIPPPPPLPGMGIPPPPPLPGVGIPPPPP 959
Score = 40.4 bits (93), Expect = 0.020
Identities = 48/185 (25%), Positives = 72/185 (37%), Gaps = 35/185 (18%)
Query: 206 LPPLSKPPDEIHSPPAASS------SSPSSSEDAESRE-TAFHSPHGSSVSHDDNNYYT- 257
LPP PP+ + PAA+S +SPS + E + SP SV D +
Sbjct: 734 LPPPKAPPEGLPGSPAAASGESALLTSPSGPQTKFCSEISLIVSPRRISVQLDAQQIQSA 793
Query: 258 -----PVSRHSSVANGSPAAATAVPFSKRTSPKSRLSAS---------SPDI----RHAM 299
P S + G P + ++ TS + +S+S +P + +
Sbjct: 794 SQLPPPPPLLGSDSQGQP-SQPSLHTESETSHEHSVSSSFGNNCNVPPAPPLPCTESSSF 852
Query: 300 IPSIKHNPLPPP-------PAPPAPPSTQLDGGPSRRPK-FSTHPLAPNMASLNTPPPPP 351
+P + PPP PA P+P + L + P+ P P + L PPPPP
Sbjct: 853 MPGLGMAIPPPPCLSDITVPALPSPTAPALQFSNLQGPEMLPAPPQPPPLPGLGVPPPPP 912
Query: 352 PPPPP 356
PP P
Sbjct: 913 APPLP 917
Score = 40.0 bits (92), Expect = 0.026
Identities = 24/69 (34%), Positives = 27/69 (38%), Gaps = 5/69 (7%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPP-PDQALTQPPPSASIAYPTATKP 90
PL G PPP PP P P P +P PPP P + PPP + P P
Sbjct: 901 PLPGLGVPPPPPAPPLPGMGIPPPPPLPGMGIPPPPPLPGMGIPPPPPLPGVGIP----P 956
Query: 91 AKPAKKVAI 99
P V I
Sbjct: 957 PPPLPGVGI 965
Score = 37.4 bits (85), Expect = 0.17
Identities = 77/324 (23%), Positives = 107/324 (32%), Gaps = 59/324 (18%)
Query: 49 PPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKPAKPAKKVAIAASAAVVTV 108
PP T + I + SP + LT PPP A P P PA A + +A++T
Sbjct: 712 PPRTLEAKSI----QTSPTEEGRILTLPPPKA----PPEGLPGSPA---AASGESALLTS 760
Query: 109 AVLSALAFFLYKHRANSNSNQTSSENHKLVVDGGGGRGNSRRVLDDSPRAPPPPPPSSFL 168
F + S S + +D ++++ S PPPP S
Sbjct: 761 PSGPQTKF------CSEISLIVSPRRISVQLD-------AQQIQSASQLPPPPPLLGSDS 807
Query: 169 YIGTVEPSRTSVSDAQNLNS--AKLASNYRYRPSPELQPLPPLSKPPD---EIHSPPAAS 223
+PS + S+ + +S + +N P+P L S P I PP S
Sbjct: 808 QGQPSQPSLHTESETSHEHSVSSSFGNNCNVPPAPPLPCTESSSFMPGLGMAIPPPPCLS 867
Query: 224 SSSPSSSEDAESRETAFHSPHGSSVSHDDNNYYTPVSRHSSVANGSPAAATAVPFSKRTS 283
+ + + F + G + P G P A P
Sbjct: 868 DITVPALPSPTAPALQFSNLQGPEMLP------APPQPPPLPGLGVPPPPPAPPLPGMGI 921
Query: 284 PKSRLSASSPDIRHAMIPSIKHNPLPPPPAP----PAPPSTQLDGGPSRRPKFSTH-PLA 338
P P + IP PPPP P P PP G P P P
Sbjct: 922 PPP------PPLPGMGIP-------PPPPLPGMGIPPPPPLPGVGIPPPPPLPGVGIPPP 968
Query: 339 PNMASLNTPPPPP------PPPPP 356
P + + PPPPP PPPPP
Sbjct: 969 PPLPGVGIPPPPPLPGVGIPPPPP 992
Score = 36.6 bits (83), Expect = 0.29
Identities = 25/70 (35%), Positives = 28/70 (39%), Gaps = 13/70 (18%)
Query: 32 PLFPAGSAPPPSTP----PPPPP----DTPSSPDIPFFNEYSPPP---PDQALTQPPPSA 80
PL G PPP P PPPPP P P +P PPP P A+ PPP
Sbjct: 1003 PLPGVGIPPPPPLPGVGIPPPPPLPGMGIPPPPPLP--GSGIPPPPALPGVAIPPPPPLP 1060
Query: 81 SIAYPTATKP 90
+ P P
Sbjct: 1061 GMGVPPPAPP 1070
Score = 36.2 bits (82), Expect = 0.38
Identities = 25/71 (35%), Positives = 31/71 (43%), Gaps = 13/71 (18%)
Query: 32 PLFPAGSAPPPSTPP---PPPPDTPSSPDIPFFNEYSPPPPDQALTQPP------PSASI 82
PL +G PPP+ P PPPP P P +PPPP + PP P S
Sbjct: 1036 PLPGSGIPPPPALPGVAIPPPPPLPGMGVPP----PAPPPPGAGIPPPPLLPGSGPPHSS 1091
Query: 83 AYPTATKPAKP 93
++T PA P
Sbjct: 1092 QVGSSTLPAAP 1102
Score = 36.2 bits (82), Expect = 0.38
Identities = 23/59 (38%), Positives = 25/59 (41%), Gaps = 11/59 (18%)
Query: 309 PPPPAP----PAPPSTQLDGGPSRRPKFSTH-PLAPNMASLNTPPPPP------PPPPP 356
PPPP P P PP G P P P P + + PPPPP PPPPP
Sbjct: 967 PPPPLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPP 1025
Score = 35.8 bits (81), Expect = 0.50
Identities = 24/72 (33%), Positives = 29/72 (39%), Gaps = 14/72 (19%)
Query: 33 LFPAGSAPPP----STPPPPPPDTPSSPDIPFFNEYSPPP-PDQALTQPPPSASIAYPTA 87
+ PA PPP PPPPP +P +P PPP P + PPP + P
Sbjct: 892 MLPAPPQPPPLPGLGVPPPPP-----APPLPGMGIPPPPPLPGMGIPPPPPLPGMGIP-- 944
Query: 88 TKPAKPAKKVAI 99
P P V I
Sbjct: 945 --PPPPLPGVGI 954
Score = 34.7 bits (78), Expect = 1.1
Identities = 28/88 (31%), Positives = 33/88 (36%), Gaps = 13/88 (14%)
Query: 32 PLFPAGSAPPPSTP------PPPPPDT--PSSPDIPFFNEYSPPP-PDQALTQPPPSASI 82
PL G PPP P PPP P P P +P PPP P + PPP +
Sbjct: 970 PLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGM 1029
Query: 83 AYPTATKPAKPAKKVAIAASAAVVTVAV 110
P P P I A+ VA+
Sbjct: 1030 GIP----PPPPLPGSGIPPPPALPGVAI 1053
Score = 34.7 bits (78), Expect = 1.1
Identities = 26/77 (33%), Positives = 29/77 (36%), Gaps = 13/77 (16%)
Query: 32 PLFPAGSAPPPSTP------PPPPPDT--PSSPDIPFFNEYSPPP-PDQALTQPPPSASI 82
PL G PPP P PPP P P P +P PPP P + PPP +
Sbjct: 948 PLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGV 1007
Query: 83 AYPTATKPAKPAKKVAI 99
P P P V I
Sbjct: 1008 GIP----PPPPLPGVGI 1020
Score = 34.3 bits (77), Expect = 1.4
Identities = 26/77 (33%), Positives = 29/77 (36%), Gaps = 13/77 (16%)
Query: 32 PLFPAGSAPPPSTP------PPPPPDT--PSSPDIPFFNEYSPPP-PDQALTQPPPSASI 82
PL G PPP P PPP P P P +P PPP P + PPP +
Sbjct: 926 PLPGMGIPPPPPLPGMGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGVGIPPPPPLPGV 985
Query: 83 AYPTATKPAKPAKKVAI 99
P P P V I
Sbjct: 986 GIP----PPPPLPGVGI 998
>DAM2_HUMAN (Q86T65) Disheveled associated activator of
morphogenesis 2
Length = 1068
Score = 116 bits (291), Expect = 2e-25
Identities = 124/565 (21%), Positives = 226/565 (39%), Gaps = 96/565 (16%)
Query: 285 KSRLSASSPDIRHA------MIPSIKHNPLPPPPAPPAPPSTQLDGGPSRRPKFSTHPLA 338
K +L+ S ++R A ++ + P +PP P GGP L+
Sbjct: 487 KDKLARESQELRQARGQVAELVAQLSELSTGPVSSPPPP------GGPLT--------LS 532
Query: 339 PNMASLNTPPPPPP------PPPPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSN 392
+M + + PPPPPP PPPPP P T + +S
Sbjct: 533 SSMTTNDLPPPPPPLPFACCPPPPPPPLPPGGPP-TPPGAPPCLGMGLPLPQDPYPSSDV 591
Query: 393 GVSSVSVSKPSEEGESMMEGGKPKLKALHWDKVRATSNRATVWNQLKSSSFQ--LNEDMM 450
+ V +PS LK+ +W K+ TVWN++ L+ +
Sbjct: 592 PLRKKRVPQPSHP-----------LKSFNWVKLNEERVPGTVWNEIDDMQVFRILDLEDF 640
Query: 451 ETLFGFNSTTNSAPKETTTTSVVRKPVFPPVEQENRVLDPKKSQNIAILLRALNVTRDEV 510
E +F T + + V +E V+D +++QN ILL L ++ +E+
Sbjct: 641 EKMFSAYQRHQKELGSTEDIYLASRKV-----KELSVIDGRRAQNCIILLSKLKLSNEEI 695
Query: 511 SEALLD-GNPEGLGAELLETLVKMAPTKEEEIKLKNYDGDLSKLGSAERFLKAVLDIPLA 569
+A+L E L ++LE L+K P K + L+ + ++ ++ A+RFL + I
Sbjct: 696 RQAILKMDEQEDLAKDMLEQLLKFIPEKSDIDLLEEHKHEIERMARADRFLYEMSRIDHY 755
Query: 570 FKRVEAMLYRANFETEVNYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGTNR 629
+R++A+ ++ F+ + + + + AS EL S+ ++LE +L GN MN G R
Sbjct: 756 QQRLQALFFKKKFQERLAEAKPKVEAILLASRELVRSKRLRQMLEVILAIGNFMNKG-QR 814
Query: 630 GDAKSFKLDTLLKLVDIKGT-DGKTTLLHFVVQEIIRSEGTGGESANESVQNQTNSQFNE 688
G A F++ +L K+ D K + D +LLH+++ +
Sbjct: 815 GGAYGFRVASLNKIADTKSSIDRNISLLHYLIMIL------------------------- 849
Query: 689 DEFRKKGLQVVAGLSRDLGNVKKAAGMDSDVLSSYVSKLEMGLDKVRLVLQYQKPDMQ-- 746
+K + + +L ++ +AA ++ L V L GL V + L+YQ+ ++
Sbjct: 850 ----EKHFPDILNMPSELQHLPEAAKVNLAELEKEVGNLRRGLRAVEVELEYQRRQVREP 905
Query: 747 --------GSFFNATEIFLKDAEEKIIVIKDDERRALFLVKEVTEYFHGDTAKEEVHPFR 798
F + + E+++ +D +AL H ++ P
Sbjct: 906 SDKFVPVMSDFITVSSFSFSELEDQLNEARDKFAKAL---------MHFGEHDSKMQPDE 956
Query: 799 IFMIVRDFLNILDQVCKEVGRMQDR 823
F I FL + +++ M+ R
Sbjct: 957 FFGIFDTFLQAFSEARQDLEAMRRR 981
>DIA_DROME (P48608) Diaphanous protein
Length = 1091
Score = 114 bits (285), Expect = 1e-24
Identities = 126/567 (22%), Positives = 223/567 (39%), Gaps = 79/567 (13%)
Query: 249 SHDDNNYYTPVSRHSSVANGSPAAATAVPFSKRTSPKSRLSASSPDIRHAMIPSIKHNPL 308
S Y + + S + A A + + + ++A SP+ +P + + P+
Sbjct: 459 SKRSEEYEKKIEQLESAKQEAEAKAAHLEEKVKLMEANGVAAPSPN----KLPKV-NIPM 513
Query: 309 PPPP-----APPAPPST---QLDGGPSRRPKFSTHPLAPNMASLNTPPPPPP----PPPP 356
PPPP APP PP + GGP P P P A PPPPPP PPPP
Sbjct: 514 PPPPPGGGGAPPPPPPPMPGRAGGGPPPPPP----PPMPGRAGGPPPPPPPPGMGGPPPP 569
Query: 357 PAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVSKPSEEGESMMEGGKPK 416
P P + + +G+ KP ++ + K
Sbjct: 570 PM------PGMMRPGGGPPPPPMMMGPMVP--VLPHGL------KPKKKWDV-----KNP 610
Query: 417 LKALHWDK-VRATSNRATVWNQLKSSSFQLNEDMMETLFGFNSTTNSAPKETTTTSVVRK 475
+K +W V A + W + + ++ + E F+S P + V K
Sbjct: 611 MKRANWKAIVPAKMSDKAFWVKCQEDKLAQDDFLAELAVKFSSK----PVKKEQKDAVDK 666
Query: 476 PV-FPPVEQENRVLDPKKSQNIAILLRAL--NVTRDEVSEALLDGNPEGLGAELLETLVK 532
P + RVLD K +QN+AI+L +++ +++ LL + + L + +L+ L++
Sbjct: 667 PTTLTKKNVDLRVLDSKTAQNLAIMLGGSLKHLSYEQIKICLLRCDTDILSSNILQQLIQ 726
Query: 533 MAPTKEEEIKLKNYDGDLSKLGSAERFLKAVLDIPLAFKRVEAMLYRANFETEVNYLRKS 592
P E +L+ L E+F + +I R+ + ++ + V ++
Sbjct: 727 YLPPPEHLKRLQEIKAKGEPLPPIEQFAATIGEIKRLSPRLHNLNFKLTYADMVQDIKPD 786
Query: 593 FQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGTNRGDAKSFKLDTLLKLVDIKGTDGK 652
AA EE++NS+ F K+LE +L GN MN G+ A F++ L KL + K D K
Sbjct: 787 IVAGTAACEEIRNSKKFSKILELILLLGNYMNSGSKNEAAFGFEISYLTKLSNTKDADNK 846
Query: 653 TTLLHFVVQEIIRSEGTGGESANESVQNQTNSQFNEDEFRKKGLQVVAGLSRDLGNVKKA 712
TLLH++ + +K DL +V KA
Sbjct: 847 QTLLHYLADLV-----------------------------EKKFPDALNFYDDLSHVNKA 877
Query: 713 AGMDSDVLSSYVSKLEMGLDKVRLVLQYQKPDM--QGSFFNATEIFLKDAEEKIIVIKDD 770
+ ++ D + + ++ + + LQ K F F ++ +++ V+
Sbjct: 878 SRVNMDAIQKAMRQMNSAVKNLETDLQNNKVPQCDDDKFSEVMGKFAEECRQQVDVLGKM 937
Query: 771 ERRALFLVKEVTEYFHGDTAKEEVHPF 797
+ + L K+++EY+ D +K + F
Sbjct: 938 QLQMEKLYKDLSEYYAFDPSKYTMEEF 964
Score = 42.7 bits (99), Expect = 0.004
Identities = 26/82 (31%), Positives = 28/82 (33%), Gaps = 8/82 (9%)
Query: 20 NSTPNARRILHQPLFPAGSAPPPSTPP--------PPPPDTPSSPDIPFFNEYSPPPPDQ 71
N P + P G APPP PP PPPP P P PPPP
Sbjct: 504 NKLPKVNIPMPPPPPGGGGAPPPPPPPMPGRAGGGPPPPPPPPMPGRAGGPPPPPPPPGM 563
Query: 72 ALTQPPPSASIAYPTATKPAKP 93
PPP + P P P
Sbjct: 564 GGPPPPPMPGMMRPGGGPPPPP 585
Score = 32.7 bits (73), Expect = 4.2
Identities = 21/66 (31%), Positives = 23/66 (34%), Gaps = 7/66 (10%)
Query: 32 PLFPAGSAPPPSTPPPPPPDTPSSPDIP-FFNEYSPPPPDQALTQPPPSASIAYPTATKP 90
P P + PP PPPP P P +P PPP PP P
Sbjct: 545 PPMPGRAGGPPPPPPPPGMGGPPPPPMPGMMRPGGGPPP------PPMMMGPMVPVLPHG 598
Query: 91 AKPAKK 96
KP KK
Sbjct: 599 LKPKKK 604
>FMN_CHICK (Q05858) Formin (Limb deformity protein)
Length = 1213
Score = 112 bits (279), Expect = 5e-24
Identities = 145/613 (23%), Positives = 237/613 (38%), Gaps = 132/613 (21%)
Query: 262 HSSVANGSPAAATAVPFSKRT---SPKSRLSASSPDIRHAMIPSIKHNPLPPPPAPP--- 315
HS G + VP S+ PK L S P P P PPPP PP
Sbjct: 622 HSISTQGENKDSYDVPSSESVLSCQPKQMLPPSPP-------PPPPPPPPPPPPPPPFSD 674
Query: 316 ------APPSTQLDGGP-SRRPKFS-------------------THPLAPNMASLNTPPP 349
PP L GP S P F+ P P + L P P
Sbjct: 675 SSLPGLVPPPPPLPTGPTSVTPHFAFGPPLPPQLSEGCRDFQAPAPPAPPPLPGLGPPVP 734
Query: 350 PP------PPPPPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNGVSSVSVSKPS 403
PP PPPPPP P +S++S SS KP+
Sbjct: 735 PPLPGSGLPPPPPP-------PGPGLFFNSTLS------------------SSQGPRKPA 769
Query: 404 EEGESMMEGGKPKLKALHWDKVRATSNRAT----VWNQLKSSSFQLNEDMMETLFGFNST 459
+E +P +K L+W +++ +R T +W L+ L+ E LF +
Sbjct: 770 ------IEPSRP-MKPLYWTRIQLQGSRKTAIPTLWESLEEPDI-LDTTEFEYLFSKD-- 819
Query: 460 TNSAPKETTTTSVVRKPVFPPVEQENR------VLDPKKSQNIAILLRALNVTRDEVSEA 513
TT RKP+ E++ + +LD K+SQ + IL+ +L++ ++ +A
Sbjct: 820 ---------TTQEKRKPLSETYEKKTKAKKIIKLLDGKRSQTVGILISSLHLEMKDIQQA 870
Query: 514 LLDGNPEGLGAELLETLVKMAPTKEEEIKLKNY-----DGDLSKLGSAERFLKAVLDIPL 568
+L + + E LE L + K+E K++ Y + +L L E+FL + IP
Sbjct: 871 ILCVDDSVVDLETLEALYENRAQKDELEKIEQYYQTSKEEELKLLDKPEQFLYELSQIPN 930
Query: 569 AFKRVEAMLYRANFETEVNYLRKSFQTLEAASEELKNSRLFFKLLEAVLRTGNRMNVGT- 627
+R + +++++ F + + + + S+ L N ++L +L GN MN G
Sbjct: 931 FTERAQCIIFQSVFSEGITSVHRKVDIITRVSKALLNMTSVKEILGLILAFGNYMNGGNR 990
Query: 628 NRGDAKSFKLDTLLKLVDIKGTDGKTTLLHFVVQEIIR---SEGTGGESANESVQNQTNS 684
RG A F L+ L KL D+K D + L+ +VV +R E +S + Q
Sbjct: 991 TRGQADGFGLEILPKLKDVKSRDNRINLVDYVVIYYLRHCDKEAGTDKSIFPLPEPQDFF 1050
Query: 685 QFNEDEFRKKGLQVVAGLSRDLGNVKKAAGMDSDVLSSYVSKLEMGLDKVRLVLQYQKPD 744
Q ++ +F L +DL +K+ LE +++LV + +
Sbjct: 1051 QASQVKFE--------DLIKDLRKLKR--------------DLEASEKQMKLVCRESSEE 1088
Query: 745 MQGSFFNATEIFLKDAEEKIIVIKDDERRALFLVKEVTEYF--HGDTAKEEVHPFRIFMI 802
F E F + A+E+ + A +E YF ++E+ P +F +
Sbjct: 1089 HLQPFKEKLEEFFQKAKEERKKEESSLENAQKCFEETVGYFGIKPKPGEKEITPNYVFTV 1148
Query: 803 VRDFLNILDQVCK 815
+F + + K
Sbjct: 1149 WYEFCSDFKTIWK 1161
Score = 39.7 bits (91), Expect = 0.035
Identities = 20/50 (40%), Positives = 24/50 (48%), Gaps = 3/50 (6%)
Query: 23 PNARRILH-QP--LFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPP 69
P++ +L QP + P PPP PPPPPP P D PPPP
Sbjct: 637 PSSESVLSCQPKQMLPPSPPPPPPPPPPPPPPPPPFSDSSLPGLVPPPPP 686
Score = 32.0 bits (71), Expect = 7.2
Identities = 19/62 (30%), Positives = 27/62 (42%), Gaps = 8/62 (12%)
Query: 41 PPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKPAKPAKKVAIA 100
PP+ PP P P P +P PPPP PPP + + + ++ +K AI
Sbjct: 720 PPAPPPLPGLGPPVPPPLP--GSGLPPPP------PPPGPGLFFNSTLSSSQGPRKPAIE 771
Query: 101 AS 102
S
Sbjct: 772 PS 773
>FMNL_HUMAN (O95466) Formin-like protein (Protein C17orf1)
Length = 463
Score = 94.0 bits (232), Expect = 2e-18
Identities = 65/247 (26%), Positives = 121/247 (48%), Gaps = 7/247 (2%)
Query: 420 LHWDKVRATSNRATVWNQLKSSSFQLNEDMMETLFGFNSTTNSAPKETTTTSVVRKPVFP 479
L+W ++ + TV+ +L DM + F T + P + S ++
Sbjct: 4 LNWVALKPSQITGTVFTELNDEKVLQELDMSDFEEQFK-TKSQGP--SLDLSALKSKAAQ 60
Query: 480 PVEQENRVLDPKKSQNIAILLRALNVTRDEVSEALLDGNPEGLGAELLETLVKMAPTKEE 539
+ +++ +++N+AI LR N+ + + +A+ + + LG + LE L++ PT+ E
Sbjct: 61 KAPSKATLIEANRAKNLAITLRKGNLGAERICQAIEAYDLQALGLDFLELLMRFLPTEYE 120
Query: 540 EIKLKNYDGD---LSKLGSAERFLKAVLDIPLAFKRVEAMLYRANFETEVNYLRKSFQTL 596
+ ++ + + +L +RF+ IP +R+ + + NF L +
Sbjct: 121 RSLITRFEREQRPMEELSEEDRFMLCFSRIPRLPERMTTLTFLGNFPDTAQLLMPQLNAI 180
Query: 597 EAASEELKNSRLFFKLLEAVLRTGNRMNVGTNRGDAKSFKLDTLLKLVDIKGTDGKTTLL 656
AAS +K+S ++LE VL GN MN + RG A F+L +L L+++K TD K TLL
Sbjct: 181 IAASMSIKSSDKLRQILEIVLAFGNYMN-SSKRGAAYGFRLQSLDALLEMKSTDRKQTLL 239
Query: 657 HFVVQEI 663
H++V+ I
Sbjct: 240 HYLVKVI 246
>VRP1_YEAST (P37370) Verprolin
Length = 817
Score = 87.0 bits (214), Expect = 2e-16
Identities = 104/406 (25%), Positives = 155/406 (37%), Gaps = 95/406 (23%)
Query: 35 PAGSAPP-----PSTPPPPPPDTPSSPD--IPFFNEYSPPPPDQALTQPPPSASIAYPTA 87
P+ SAPP PS PP P+ P SP +P S PP P +S A P
Sbjct: 116 PSASAPPIPGAVPSVAAPPIPNAPLSPAPAVPSIPSSSAPPI------PDIPSSAAPPIP 169
Query: 88 TKPAKPAKKVAIAASAAVVTVAVLSALAFFLYKHRANSNSNQTSSENHKLVVDGGGGRGN 147
P+ PA + ++ ++A + HR S+Q S N L +
Sbjct: 170 IVPSSPAPPLPLSGASAPKVPQNRPHMPSVRPAHR----SHQRKSSNISLPSVSAPPLPS 225
Query: 148 SRRV--LDDSPRAPPPPP--------------------PSSFL------YIGTVEPSRTS 179
+ + + P+APPPPP PSS + ++ + R+
Sbjct: 226 ASLPTHVSNPPQAPPPPPTPTIGLDSKNIKPTDNAVSPPSSEVPAGGLPFLAEINARRSE 285
Query: 180 VSDAQNLNSAKLASNYRYRPSPELQPLPPLSKPPDEIHSPPAASSSSPSSS----EDAES 235
+ ++S K+ + SP PLP + P H+PP ++ P S A
Sbjct: 286 RGAVEGVSSTKIQTENH--KSPSQPPLPSSAPPIPTSHAPPLPPTAPPPPSLPNVTSAPK 343
Query: 236 RETAFHSPHGSSVSHDDNNYYTPVSRHSSVANGSPAAATAVPFSKRTSPKSRLSASSPDI 295
+ T+ +P P +++++ S + A P +P + S P
Sbjct: 344 KATSAPAP-------------PPPPLPAAMSSASTNSVKATPVPPTLAPPLPNTTSVPPN 390
Query: 296 RHAMIPSIKHNPLPPPPAPPAPPSTQLDGGPSRRPKFSTHPLAPNMASLNTPPPPPP--- 352
+ + +P+ P PPPP PP ST S S+ PLAP PPPPPP
Sbjct: 391 KASSMPA----PPPPPPPPPGAFST------SSALSASSIPLAP------LPPPPPPSVA 434
Query: 353 -----PPPPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGDASSNG 393
PPPP P +TT S+ SKQ + SSS A + G
Sbjct: 435 TSVPSAPPPP-------PTLTTNKPSASSKQSKISSSSSSSAVTPG 473
Score = 79.3 bits (194), Expect = 4e-14
Identities = 86/354 (24%), Positives = 132/354 (36%), Gaps = 69/354 (19%)
Query: 36 AGSAPPPSTPPPPPP------DTPSSPDIPFFNEYSPPPPDQALT---QPPPSASIAYPT 86
A S P P PPPPPP S+ IP PPPP A + PPP ++ T
Sbjct: 392 ASSMPAPPPPPPPPPGAFSTSSALSASSIPLAPLPPPPPPSVATSVPSAPPPPPTL---T 448
Query: 87 ATKPAKPAKKVAIAASAAVVTVAVLSALAFF--LYKHR-----ANSNSNQTSSENHKLVV 139
KP+ +K+ I++S++ V L F + K R ++ T+ + + V+
Sbjct: 449 TNKPSASSKQSKISSSSSSSAVTPGGPLPFLAEIQKKRDDRFVVGGDTGYTTQDKQEDVI 508
Query: 140 DGGGGRGNSRRVLDDSPRAPPPPPPSSFLYIGTVEPSRTSVSDAQNLNSAKLASNYRYRP 199
G + ++ R SP PP S + + + + ++ + +SN P
Sbjct: 509 --GSSKDDNVRPSPISPSINPPKQSS--------QNGMSFLDEIESKLHKQTSSNAFNAP 558
Query: 200 SPELQ----PLPPLSKPPDEIHSPPAASSSSPSSSEDAESRETAFHSPHGSSVSHDDNNY 255
P PLPP S PP PP S +P++S D H ++
Sbjct: 559 PPHTDAMAPPLPP-SAPP-----PPITSLPTPTASGD----------------DHTNDKS 596
Query: 256 YTPVSRHSSVANGSPAAATAVPFSKRTSPKSRLSASSPDIRHAMIPSIKHNPLPPPPAPP 315
T + + A P P S S+ + + + H ++PS P PP
Sbjct: 597 ETVLGMKKAKAPALPGHVPPPPVPPVLSDDSKNNVPAASLLHDVLPSSNLEKPPSPPVAA 656
Query: 316 APP--------------STQLDGGPSRRPKFSTHPLAPNMASLNTPPPPPPPPP 355
APP ST + P P S +++ T PPPPP P
Sbjct: 657 APPLPTFSAPSLPQQSVSTSIPSPPPVAPTLSVRTETESISKNPTKSPPPPPSP 710
Score = 59.3 bits (142), Expect = 4e-08
Identities = 89/363 (24%), Positives = 129/363 (35%), Gaps = 79/363 (21%)
Query: 31 QPLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKP 90
QP P+ + P P++ PP P T +PPPP P + + P P
Sbjct: 307 QPPLPSSAPPIPTSHAPPLPPT------------APPPPSLPNVTSAPKKATSAPAPPPP 354
Query: 91 AKPAKKVAIAASAAVVTVAVLSALAFFLYKHRANSNSNQTSSENHKLVVDGGGGRGNSRR 150
PA ++ A++ +V V LA L N TS +K
Sbjct: 355 PLPAA-MSSASTNSVKATPVPPTLAPPL--------PNTTSVPPNKA------------- 392
Query: 151 VLDDSPRAPPPPPPSSFLYIGTVEPSRTSVSDAQNLNSAKLASNYRYRPSPELQPLPPLS 210
P PPPPPP P S S A + +S LA PLPP
Sbjct: 393 --SSMPAPPPPPPPP---------PGAFSTSSALSASSIPLA------------PLPP-- 427
Query: 211 KPPDEIHSPPAASSSSPSSSEDAESRETAFHSPHGSSVSHDDNNYYTPVSRHSSVANGSP 270
PP PP+ ++S PS+ + T + P SS + + S S+V G P
Sbjct: 428 -PP-----PPSVATSVPSAPPPPPTLTT--NKPSASS----KQSKISSSSSSSAVTPGGP 475
Query: 271 AAATAVPFSKRTSP---KSRLSASSPDIRHAMIPSIKHNPLPPPPAPPA--PPSTQLDGG 325
A KR ++ D + +I S K + + P P P+ PP G
Sbjct: 476 LPFLAEIQKKRDDRFVVGGDTGYTTQDKQEDVIGSSKDDNVRPSPISPSINPPKQSSQNG 535
Query: 326 PSRRPKFST--HPLAPNMASLNTPPPPPPPPPPPAARKVWSPAITTMHSSSVSKQQQSSS 383
S + + H + A N PPP PP P IT++ + + S ++
Sbjct: 536 MSFLDEIESKLHKQTSSNA-FNAPPPHTDAMAPPLPPSAPPPPITSLPTPTASGDDHTND 594
Query: 384 SIE 386
E
Sbjct: 595 KSE 597
Score = 44.7 bits (104), Expect = 0.001
Identities = 83/382 (21%), Positives = 117/382 (29%), Gaps = 85/382 (22%)
Query: 39 APPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKPAKPAKKVA 98
A P+ PPPPPP S P P +++ Q K K K
Sbjct: 2 AGAPAPPPPPPPPALGG---------SAPKPAKSVMQ---GRDALLGDIRKGMKLKKAET 49
Query: 99 IAASAAVVTVAVLSALAFFLYKHRANSNSNQTSSENHKLVVDGGGGRG------------ 146
SA +V V+S+ A+ +S SS+ + G G
Sbjct: 50 NDRSAPIVGGGVVSS---------ASGSSGTVSSKGPSMSAPPIPGMGAPQLGDILAGGI 100
Query: 147 ------NSRRVLDDSPRAPPPPPPSSFLYIGTVEPSRTSVSDAQNLNSAKLASNYRYRPS 200
N+ SP A PP P + + +S A + S +S P
Sbjct: 101 PKLKHINNNASTKPSPSASAPPIPGAVPSVAAPPIPNAPLSPAPAVPSIPSSS---APPI 157
Query: 201 PEL-------------QPLPPL--------SKPPDEIHSP---PAASSSSPSSSEDAESR 236
P++ P PPL P + H P PA S SS +
Sbjct: 158 PDIPSSAAPPIPIVPSSPAPPLPLSGASAPKVPQNRPHMPSVRPAHRSHQRKSSNISLPS 217
Query: 237 ETAFHSPHGSSVSHDDNNYYTP----------------VSRHSSVANGSPAAATAVPFSK 280
+A P S +H N P + ++ S A +PF
Sbjct: 218 VSAPPLPSASLPTHVSNPPQAPPPPPTPTIGLDSKNIKPTDNAVSPPSSEVPAGGLPFLA 277
Query: 281 RTSPKSRLSASSPDIRHAMIPSIKHNPLPPPPAPPAPPSTQLDGGPSRRPKFSTHPLAPN 340
+ + + + I + H PP P + P P P P PN
Sbjct: 278 EINARRSERGAVEGVSSTKIQTENHKSPSQPPLPSSAPPIPTSHAPPLPPTAPPPPSLPN 337
Query: 341 MASL---NTPPPPPPPPPPPAA 359
+ S T P PPPPP PAA
Sbjct: 338 VTSAPKKATSAPAPPPPPLPAA 359
Score = 40.0 bits (92), Expect = 0.026
Identities = 63/254 (24%), Positives = 87/254 (33%), Gaps = 47/254 (18%)
Query: 158 APPPPPPSSFLYIGTVEPSRTSVSDAQNLNSAKLASNYRYRPSPELQPLPPLSKPPDEIH 217
APPPPPP L +P+++ + L +++ L K
Sbjct: 6 APPPPPPPPALGGSAPKPAKSVMQGRDALLG-------------DIRKGMKLKKAETNDR 52
Query: 218 SPPAASSSSPSSSEDAESRETAFHSPHGSSVSHDDNNYYTPVSRHSSVANGSPAAATAVP 277
S P SS+ S + S G S+S +A G P
Sbjct: 53 SAPIVGGGVVSSA----SGSSGTVSSKGPSMSAPPIPGMGAPQLGDILAGGIPKLKH--- 105
Query: 278 FSKRTSPKSRLSASSPDIRHAM----IPSIKHNPLPPPPAPPAPPSTQLDGGPSRRPKFS 333
+ S K SAS+P I A+ P I + PL P PA P+ PS S
Sbjct: 106 INNNASTKPSPSASAPPIPGAVPSVAAPPIPNAPLSPAPAVPSIPS-------------S 152
Query: 334 THPLAPNMASLNTPPPP-----PPPPPPPAARKVWSPAITTMHSSSVSKQQQSSSSIEGD 388
+ P P++ S PP P P PP P + H SV +S
Sbjct: 153 SAPPIPDIPSSAAPPIPIVPSSPAPPLPLSGASAPKVPQNRPHMPSVRPAHRSH-----Q 207
Query: 389 ASSNGVSSVSVSKP 402
S+ +S SVS P
Sbjct: 208 RKSSNISLPSVSAP 221
Score = 31.6 bits (70), Expect = 9.4
Identities = 30/103 (29%), Positives = 39/103 (37%), Gaps = 28/103 (27%)
Query: 347 PPPPPPPPPPPAARKVWSPAITTMHSSSV-------------SKQQQSSSSIEG------ 387
P PPPPPPPP PA + M ++ S+ I G
Sbjct: 5 PAPPPPPPPPALGGSAPKPAKSVMQGRDALLGDIRKGMKLKKAETNDRSAPIVGGGVVSS 64
Query: 388 -DASSNGVSS--VSVSKPSEEG------ESMMEGGKPKLKALH 421
SS VSS S+S P G ++ GG PKLK ++
Sbjct: 65 ASGSSGTVSSKGPSMSAPPIPGMGAPQLGDILAGGIPKLKHIN 107
>ENAH_MOUSE (Q03173) Enabled protein homolog (NPC derived
proline-rich protein 1) (NDPP-1)
Length = 802
Score = 73.9 bits (180), Expect = 2e-12
Identities = 98/365 (26%), Positives = 131/365 (35%), Gaps = 58/365 (15%)
Query: 20 NSTPNARRILHQ-PLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPP 78
N+ P++ L PL S PPS PPP S+P S PD A+ P
Sbjct: 256 NAAPSSDSSLSSAPLPEYSSCQPPSAPPPSYAKVISAP-------VSDATPDYAVVTALP 308
Query: 79 SASIAYPTATKPAKPAKKVAIAASAAVVTVAVLSALAFFLYKHRANSNSNQTSSENHKLV 138
PT+T P P + A + ++ + A L + R + +++ SS +
Sbjct: 309 ------PTSTPPTPPLRHAATRFATSLGS-AFHPVLPHYATVPRPLNKNSRPSSPVNTPS 361
Query: 139 VDGGGGRGNSRRVLDDSPRAPPPP-----PPSSFLYIGTVEPSRTSVSDAQNLNSAKLAS 193
+ + + SP P PP PP V P S S + + A
Sbjct: 362 SQPPAAKSCAWPTSNFSPLPPSPPIMISSPPGKATGPRPVLPVCVS-SPVPQMPPSPTAP 420
Query: 194 N-------YRYRPSPELQPLPPLSKPPDEIHSPPAASSSSPSSSEDAESRETAFHSPHGS 246
N Y P P P P PP PP P P +
Sbjct: 421 NGSLDSVTYPVSPPPTSGPAAPPPPPPPPPPPPPPPLPPPPL--------------PPLA 466
Query: 247 SVSHDDNNYYTP-----VSRHSSVANGSPAAATAVPFSKRTSPKSRL---SASSPDIRHA 298
S+SH + P S SS + P+ + P S T L SAS P ++ A
Sbjct: 467 SLSHCGSQASPPPGTPLASTPSSKPSVLPSPSAGAPASAETPLNPELGDSSASEPGLQAA 526
Query: 299 MIPSIKHNP---LPPPPAPPAPPSTQLDGGP---SRRPKFSTHPLAPNMASLNTPPPPPP 352
P+ P + PPAPP PP L GP S P P P + S PPPPPP
Sbjct: 527 SQPAESPTPQGLVLGPPAPPPPPP--LPSGPAYASALPPPPGPPPPPPLPSTGPPPPPPP 584
Query: 353 PPPPP 357
PPP P
Sbjct: 585 PPPLP 589
Score = 60.8 bits (146), Expect = 1e-08
Identities = 65/232 (28%), Positives = 95/232 (40%), Gaps = 31/232 (13%)
Query: 206 LPPLSKPPDEIHSPPAASSSSPSSSEDAESRETAFHS--PHGSSVSHDDNNYYTPVSRHS 263
LPP S PP +PP +++ A S +AFH PH ++V N P S +
Sbjct: 307 LPPTSTPP----TPPLRHAAT----RFATSLGSAFHPVLPHYATVPRPLNKNSRPSSPVN 358
Query: 264 SVANGSPAAAT-AVPFSKRTS--PKSRLSASSPDIR----HAMIPSIKHNPLPPPPAPPA 316
+ ++ PAA + A P S + P + SSP + ++P +P+P P P
Sbjct: 359 TPSSQPPAAKSCAWPTSNFSPLPPSPPIMISSPPGKATGPRPVLPVCVSSPVPQMPPSPT 418
Query: 317 PPSTQLDGGPSRRPKFSTHPLAPNMASLNTPPPPPPPPPPPAARKVWSPAITTMHSSS-- 374
P+ LD S S P + A PPPPPPPPPPP P + H S
Sbjct: 419 APNGSLD---SVTYPVSPPPTSGPAAPPPPPPPPPPPPPPPLPPPPLPPLASLSHCGSQA 475
Query: 375 -------VSKQQQSSSSIEGDASSNGVSSVSVSKPSEEGESMMEGGKPKLKA 419
++ S S+ S+ +S E G+S +P L+A
Sbjct: 476 SPPPGTPLASTPSSKPSVLPSPSAGAPASAETPLNPELGDS--SASEPGLQA 525
Score = 56.2 bits (134), Expect = 4e-07
Identities = 73/307 (23%), Positives = 107/307 (34%), Gaps = 39/307 (12%)
Query: 40 PPPSTPPPPP-----------------PDTPSSPDIPF-FNEYSPP--PPDQALTQPPPS 79
PP STPP PP P P +P N+ S P P + +QPP +
Sbjct: 308 PPTSTPPTPPLRHAATRFATSLGSAFHPVLPHYATVPRPLNKNSRPSSPVNTPSSQPPAA 367
Query: 80 ASIAYPTAT-KPAKPAKKVAIAASAA-------VVTVAVLSALAFFLYKHRANSNSNQTS 131
S A+PT+ P P+ + I++ V+ V V S + + + + S
Sbjct: 368 KSCAWPTSNFSPLPPSPPIMISSPPGKATGPRPVLPVCVSSPVP----QMPPSPTAPNGS 423
Query: 132 SENHKLVVDGGGGRGNSRRVLDDSPRAPPPPPPSSFLYIGTVEPSRTSVSDAQNLNSAKL 191
++ V G + P PPPPPP + + S A L
Sbjct: 424 LDSVTYPVSPPPTSGPAAPPPPPPPPPPPPPPPLPPPPLPPLASLSHCGSQASPPPGTPL 483
Query: 192 ASNYRYRPSPELQPLPPLSKPPDEIHSPPAASSSSPSSSEDAESRETAFHSPHGSSVSHD 251
AS +PS P + +P SS+ A S+ +P G +
Sbjct: 484 ASTPSSKPSVLPSPSAGAPASAETPLNPELGDSSASEPGLQAASQPAESPTPQGLVLG-- 541
Query: 252 DNNYYTPVSRHSSVANGSPAAATAVPFSKRTSPKSRLSASSPDIRHAMIPSIKHNPLPPP 311
P PA A+A+P P L ++ P P + + PPP
Sbjct: 542 -----PPAPPPPPPLPSGPAYASALPPPPGPPPPPPLPSTGPPPPPPPPPPLPNQAPPPP 596
Query: 312 PAPPAPP 318
P PPAPP
Sbjct: 597 PPPPAPP 603
Score = 47.0 bits (110), Expect = 2e-04
Identities = 30/87 (34%), Positives = 40/87 (45%), Gaps = 5/87 (5%)
Query: 18 PLNSTPNARRILHQPLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPP 77
PL S P L P P P PST PPPPP P P +P +PPPP P
Sbjct: 550 PLPSGPAYASALPPPPGPPPPPPLPSTGPPPPP--PPPPPLP---NQAPPPPPPPPAPPL 604
Query: 78 PSASIAYPTATKPAKPAKKVAIAASAA 104
P++ I + ++ +P +A A + A
Sbjct: 605 PASGIFSGSTSEDNRPLTGLAAAIAGA 631
Score = 42.7 bits (99), Expect = 0.004
Identities = 53/224 (23%), Positives = 71/224 (31%), Gaps = 37/224 (16%)
Query: 14 SLFTPLNSTPNARRILHQPLFPAGSAPPPSTPPPPPPDTPSSPDIPFFNEYSPPPPDQAL 73
S+ P++ P + P P PPP PPPP P + + PPP L
Sbjct: 426 SVTYPVSPPPTSGPAAPPPPPPPPPPPPPPPLPPPP--LPPLASLSHCGSQASPPPGTPL 483
Query: 74 TQPPPSASIAYPTATKPAKPAKKVAIAASAAVVTVAVLSALAFFLYKHRANSNSNQTSSE 133
P S P+ + A + + + N +S+
Sbjct: 484 ASTPSSKPSVLPSPSAGAPASAETPL------------------------NPELGDSSAS 519
Query: 134 NHKLVVDGGGGRGNSRRVLDDSPRAPPPPP--PSSFLYIGTVEPSRTSVSDAQNLNSAKL 191
L + + L P APPPPP PS Y + P L
Sbjct: 520 EPGLQAASQPAESPTPQGLVLGPPAPPPPPPLPSGPAYASALPPPPGPPP------PPPL 573
Query: 192 ASNYRYRPSPELQPLP---PLSKPPDEIHSPPAASSSSPSSSED 232
S P P PLP P PP PA+ S S+SED
Sbjct: 574 PSTGPPPPPPPPPPLPNQAPPPPPPPPAPPLPASGIFSGSTSED 617
Score = 39.7 bits (91), Expect = 0.035
Identities = 24/69 (34%), Positives = 26/69 (36%), Gaps = 14/69 (20%)
Query: 33 LFPAGSAPPPSTPP--------PPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAY 84
L P PPP P PPPP P P +P PPPP PPP + A
Sbjct: 540 LGPPAPPPPPPLPSGPAYASALPPPPGPPPPPPLPSTGPPPPPPP------PPPLPNQAP 593
Query: 85 PTATKPAKP 93
P P P
Sbjct: 594 PPPPPPPAP 602
>FOR3_SCHPO (O94532) Formin 3
Length = 1461
Score = 70.5 bits (171), Expect = 2e-11
Identities = 98/424 (23%), Positives = 176/424 (41%), Gaps = 60/424 (14%)
Query: 282 TSPKSRLSASSPDIRHAMIPSIKHNPLPPP--------PAP-PAPPSTQLDGGPSRRPKF 332
+SP L S + +K P PPP PAP P PP + GGP P
Sbjct: 708 SSPSPLLPDVSDTVEEQQKLLLKSPPPPPPAVIVPTPAPAPIPVPPPAPIMGGPPPPPP- 766
Query: 333 STHPLAPNMASLNTPPPPPPPPPPPA----ARKVWSPAITT-----MHSSSVSKQQ--QS 381
P +A PPPPPPPPPA + ++PA + +S++++Q Q
Sbjct: 767 -----PPGVAGAG---PPPPPPPPPAVSAGGSRYYAPAPQAEPEPKIDETSLTEEQKIQL 818
Query: 382 SSSIEGDASSNGVSSVSVSKPSEEGESMMEGGKP--KLKALHWDKVRATSNRATVWNQLK 439
+ + +++ + ++ K + S+ + KP LK +HW +V
Sbjct: 819 EEARKQRKAADDAARAAIEKYTSI-PSLRDLHKPTRPLKRVHWQRVDPLPGPNVF----- 872
Query: 440 SSSFQLNEDMMETLFGFNSTTNSAPKETTTTSVVRKPVFPPVEQENRVLDPKKSQNIAIL 499
+ F LN D+ +F N + ++ T +Q + +L QN+AI+
Sbjct: 873 -TKFCLNFDITAKVFIDNGLLDFLDEKFDNTPREDFVAVEISDQRSSLLPDTVEQNMAII 931
Query: 500 LRALN--VTRDEVSEALLDGNPEGLGAELLE-TLVKMAPTKEEEIKLKNYDGDLSKLGSA 556
LR+++ D V + L++ P+ L A +L +A T Y D +K
Sbjct: 932 LRSVSNMPVEDLVQKFLVE--PDFLPASILYFDRASLASTNAYTDPFIPYSTDYTKKNPK 989
Query: 557 ER-------------FLKAVLDIPLAF-KRVEAMLYRANFETEVNYLRKSFQTLEAASEE 602
E F+ V+++ F +R++A+ +R+ ++ L + + S+
Sbjct: 990 EPTADVNSLSYFEKFFVLFVVNLRHYFQERMKALKFRSTLFGDLEILEVRMKEVIDTSDS 1049
Query: 603 LKNSRLFFKLLEAVLRTGNRMNVGTNRGDAKSFKLDTLLKLVDIKGTDGKTTLLHFVVQE 662
+ + F + + +L GN N +R A +F L + +L ++ + TL+H+ E
Sbjct: 1050 IMEDKNFAEFFQVLLIIGNYFNEPYDR--ASAFSLYMIYRLETLRDSSSALTLMHY-FDE 1106
Query: 663 IIRS 666
IIR+
Sbjct: 1107 IIRT 1110
Score = 33.9 bits (76), Expect = 1.9
Identities = 20/57 (35%), Positives = 22/57 (38%), Gaps = 9/57 (15%)
Query: 40 PPPST--PPPPPPDTPSSPDIPFFNEYSPPPPDQALTQPPPSASIAYPTATKPAKPA 94
PPP+ P P P P P P PPP PPP + A P P PA
Sbjct: 735 PPPAVIVPTPAPAPIPVPPPAPIMGGPPPPP-------PPPGVAGAGPPPPPPPPPA 784
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.129 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 108,716,326
Number of Sequences: 164201
Number of extensions: 5437415
Number of successful extensions: 81171
Number of sequences better than 10.0: 1822
Number of HSP's better than 10.0 without gapping: 514
Number of HSP's successfully gapped in prelim test: 1363
Number of HSP's that attempted gapping in prelim test: 35357
Number of HSP's gapped (non-prelim): 17539
length of query: 861
length of database: 59,974,054
effective HSP length: 119
effective length of query: 742
effective length of database: 40,434,135
effective search space: 30002128170
effective search space used: 30002128170
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0347b.5


