

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
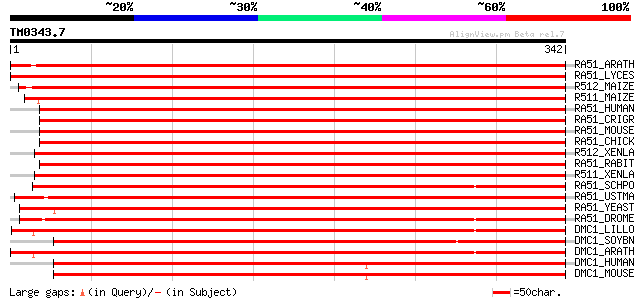
Query= TM0343.7
(342 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RA51_ARATH (P94102) DNA repair protein RAD51 homolog 1 (Rad51-li... 600 e-171
RA51_LYCES (Q40134) DNA repair protein RAD51 homolog 596 e-170
R512_MAIZE (Q9XED7) DNA repair protein RAD51 homolog B (Rad51-li... 593 e-169
R511_MAIZE (Q67EU8) DNA repair protein RAD51 homolog A (Rad51-li... 586 e-167
RA51_HUMAN (Q06609) DNA repair protein RAD51 homolog 1 (hRAD51) ... 478 e-134
RA51_CRIGR (P70099) DNA repair protein RAD51 homolog 1 476 e-134
RA51_MOUSE (Q08297) DNA repair protein RAD51 homolog 1 476 e-134
RA51_CHICK (P37383) DNA repair protein RAD51 homolog 474 e-133
R512_XENLA (Q91917) DNA repair protein RAD51 homolog 2 (XRAD51.2) 472 e-133
RA51_RABIT (O77507) DNA repair protein RAD51 homolog 1 472 e-133
R511_XENLA (Q91918) DNA repair protein RAD51 homolog 1 (XRAD51.1) 472 e-133
RA51_SCHPO (P36601) DNA repair protein rhp51 (RAD51 homolog) 458 e-129
RA51_USTMA (Q99133) DNA repair protein RAD51 451 e-126
RA51_YEAST (P25454) DNA repair protein RAD51 435 e-122
RA51_DROME (Q27297) DNA repair protein Rad51 homolog (RecA prote... 383 e-106
DMC1_LILLO (P37384) Meiotic recombination protein DMC1 homolog 343 3e-94
DMC1_SOYBN (Q96449) Meiotic recombination protein DMC1 homolog 342 6e-94
DMC1_ARATH (Q39009) Meiotic recombination protein DMC1 homolog 338 9e-93
DMC1_HUMAN (Q14565) Meiotic recombination protein DMC1/LIM15 hom... 318 1e-86
DMC1_MOUSE (Q61880) Meiotic recombination protein DMC1/LIM15 hom... 317 2e-86
>RA51_ARATH (P94102) DNA repair protein RAD51 homolog 1 (Rad51-like
protein 1) (AtRAD51)
Length = 342
Score = 600 bits (1546), Expect = e-171
Identities = 301/342 (88%), Positives = 324/342 (94%), Gaps = 3/342 (0%)
Query: 1 MEQQRHQKTAQQQPQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPR 60
MEQ+R+Q QQQ + EE QHGP P+EQLQA+GIAS+DVKKL+DAG+CTVE VAYTPR
Sbjct: 4 MEQRRNQNAVQQQ---DDEETQHGPFPVEQLQAAGIASVDVKKLRDAGLCTVEGVAYTPR 60
Query: 61 KDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGI 120
KDLLQIKGIS+AKVDKIVEAASKLVP+GFTSAS+LH QR+ IIQIT+GSRELDK+L+GGI
Sbjct: 61 KDLLQIKGISDAKVDKIVEAASKLVPLGFTSASQLHAQRQEIIQITSGSRELDKVLEGGI 120
Query: 121 ETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADR 180
ETGSITELYGEFRSGKTQLCHTLCVTCQLP+DQGGGEGKAMYIDAEGTFRPQRLLQIADR
Sbjct: 121 ETGSITELYGEFRSGKTQLCHTLCVTCQLPMDQGGGEGKAMYIDAEGTFRPQRLLQIADR 180
Query: 181 FGLNGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGE 240
FGLNGADVLENVAYARAYNTDHQSRLLLEAASMM+ETRFA++IVDSATALYRTDFSGRGE
Sbjct: 181 FGLNGADVLENVAYARAYNTDHQSRLLLEAASMMIETRFALLIVDSATALYRTDFSGRGE 240
Query: 241 LSARQMHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATT 300
LSARQMHLAKFLRSLQKLADEFGVA+VITNQVV+QVDGSA+FAGPQ KPIGGNIMAHATT
Sbjct: 241 LSARQMHLAKFLRSLQKLADEFGVAVVITNQVVAQVDGSALFAGPQFKPIGGNIMAHATT 300
Query: 301 TRLALRKGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
TRLALRKGR EERICKVISSPCL EAEARFQI EGV+D KD
Sbjct: 301 TRLALRKGRAEERICKVISSPCLPEAEARFQISTEGVTDCKD 342
>RA51_LYCES (Q40134) DNA repair protein RAD51 homolog
Length = 342
Score = 596 bits (1537), Expect = e-170
Identities = 297/342 (86%), Positives = 322/342 (93%)
Query: 1 MEQQRHQKTAQQQPQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPR 60
MEQQ + + Q +E E++QHGP P+EQLQASGIA++DVKKLKDAG+CTVESV Y PR
Sbjct: 1 MEQQHRNQKSMQDQNDEIEDVQHGPFPVEQLQASGIAALDVKKLKDAGLCTVESVVYAPR 60
Query: 61 KDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGI 120
K+LLQIKGISEAKVDKI+EAASKLVP+GFTSAS+LH QR IIQIT+GS+ELDKIL+GGI
Sbjct: 61 KELLQIKGISEAKVDKIIEAASKLVPLGFTSASQLHAQRLEIIQITSGSKELDKILEGGI 120
Query: 121 ETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADR 180
ETGSITE+YGEFR GKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADR
Sbjct: 121 ETGSITEIYGEFRCGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADR 180
Query: 181 FGLNGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGE 240
+GLNG DVLENVAYARAYNTDHQSRLLLEAASMMVETRFA+MIVDSATALYRTDFSGRGE
Sbjct: 181 YGLNGPDVLENVAYARAYNTDHQSRLLLEAASMMVETRFALMIVDSATALYRTDFSGRGE 240
Query: 241 LSARQMHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATT 300
LSARQMHLAKFLRSLQKLADEFGVA+VITNQVV+QVDGSAVFAGPQ+KPIGGNIMAHA+T
Sbjct: 241 LSARQMHLAKFLRSLQKLADEFGVAVVITNQVVAQVDGSAVFAGPQIKPIGGNIMAHAST 300
Query: 301 TRLALRKGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
TRLALRKGR EERICKV+SSPCLAEAEARFQI EGV+DVKD
Sbjct: 301 TRLALRKGRAEERICKVVSSPCLAEAEARFQISVEGVTDVKD 342
>R512_MAIZE (Q9XED7) DNA repair protein RAD51 homolog B (Rad51-like
protein B) (RAD51B) (ZmRAD51b)
Length = 340
Score = 593 bits (1528), Expect = e-169
Identities = 298/337 (88%), Positives = 322/337 (95%), Gaps = 3/337 (0%)
Query: 6 HQKTAQQQPQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQ 65
HQK + P E E +HGP PIEQLQASGIA++DVKKLKDAG+CTVESVAY+PRKDLLQ
Sbjct: 7 HQKAS---PPIEEEATEHGPFPIEQLQASGIAALDVKKLKDAGLCTVESVAYSPRKDLLQ 63
Query: 66 IKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSI 125
IKGISEAKVDKI+EAASKLVP+GFTSAS+LH QR IIQ+TTGSRELD+ILDGGIETGSI
Sbjct: 64 IKGISEAKVDKIIEAASKLVPLGFTSASQLHAQRLEIIQLTTGSRELDQILDGGIETGSI 123
Query: 126 TELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNG 185
TE+YGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKA+YIDAEGTFRPQR+LQIADRFGLNG
Sbjct: 124 TEMYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKALYIDAEGTFRPQRILQIADRFGLNG 183
Query: 186 ADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQ 245
ADVLENVAYARAYNTDHQSRLLLEAASMMVETRFA+M+VDSATALYRTDFSGRGELSARQ
Sbjct: 184 ADVLENVAYARAYNTDHQSRLLLEAASMMVETRFALMVVDSATALYRTDFSGRGELSARQ 243
Query: 246 MHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLAL 305
MHLAKFLRSLQKLADEFGVA+VITNQVV+QVDG+A+FAGPQ+KPIGGNIMAHA+TTRL L
Sbjct: 244 MHLAKFLRSLQKLADEFGVAVVITNQVVAQVDGAAMFAGPQIKPIGGNIMAHASTTRLFL 303
Query: 306 RKGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
RKGRGEERICKVISSPCLAEAEARFQI +EGV+DVKD
Sbjct: 304 RKGRGEERICKVISSPCLAEAEARFQISSEGVTDVKD 340
>R511_MAIZE (Q67EU8) DNA repair protein RAD51 homolog A (Rad51-like
protein A) (RAD51A) (ZmRAD51a)
Length = 340
Score = 586 bits (1510), Expect = e-167
Identities = 293/336 (87%), Positives = 321/336 (95%), Gaps = 3/336 (0%)
Query: 10 AQQQPQE---EGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQI 66
AQQQ + E EE++HGP PIEQLQASGIA++DVKKLKD+G+ TVE+VAYTPRKDLLQI
Sbjct: 5 AQQQQKAAAAEQEEVEHGPFPIEQLQASGIAALDVKKLKDSGLHTVEAVAYTPRKDLLQI 64
Query: 67 KGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSIT 126
KGISEAK DKI+EAASK+VP+GFTSAS+LH QR IIQ+TTGSRELDKIL+GGIETGSIT
Sbjct: 65 KGISEAKADKIIEAASKIVPLGFTSASQLHAQRLEIIQVTTGSRELDKILEGGIETGSIT 124
Query: 127 ELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGA 186
E+YGEFRSGKTQLCHT CVTCQLPLDQGGGEGKA+YIDAEGTFRPQRLLQIADRFGLNGA
Sbjct: 125 EIYGEFRSGKTQLCHTPCVTCQLPLDQGGGEGKALYIDAEGTFRPQRLLQIADRFGLNGA 184
Query: 187 DVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQM 246
DVLENVAYARAYNTDHQSRLLLEAASMM+ETRFA+M+VDSATALYRTDFSGRGELSARQM
Sbjct: 185 DVLENVAYARAYNTDHQSRLLLEAASMMIETRFALMVVDSATALYRTDFSGRGELSARQM 244
Query: 247 HLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALR 306
H+AKFLRSLQKLADEFGVA+VITNQVV+QVDGSA+FAGPQ KPIGGNIMAHA+TTRLALR
Sbjct: 245 HMAKFLRSLQKLADEFGVAVVITNQVVAQVDGSAMFAGPQFKPIGGNIMAHASTTRLALR 304
Query: 307 KGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
KGRGEERICKVISSPCLAEAEARFQ+ +EG++DVKD
Sbjct: 305 KGRGEERICKVISSPCLAEAEARFQLASEGIADVKD 340
>RA51_HUMAN (Q06609) DNA repair protein RAD51 homolog 1 (hRAD51)
(HsRAD51)
Length = 339
Score = 478 bits (1229), Expect = e-134
Identities = 233/324 (71%), Positives = 283/324 (86%)
Query: 19 EEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIV 78
EE GP PI +L+ GI + DVKKL++AG TVE+VAY P+K+L+ IKGISEAK DKI+
Sbjct: 16 EEESFGPQPISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKIL 75
Query: 79 EAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQ 138
A+KLVPMGFT+A+E H +R IIQITTGS+ELDK+L GGIETGSITE++GEFR+GKTQ
Sbjct: 76 AEAAKLVPMGFTTATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITEMFGEFRTGKTQ 135
Query: 139 LCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAY 198
+CHTL VTCQLP+D+GGGEGKAMYID EGTFRP+RLL +A+R+GL+G+DVL+NVAYARA+
Sbjct: 136 ICHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAF 195
Query: 199 NTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKL 258
NTDHQ++LL +A++MMVE+R+A++IVDSATALYRTD+SGRGELSARQMHLA+FLR L +L
Sbjct: 196 NTDHQTQLLYQASAMMVESRYALLIVDSATALYRTDYSGRGELSARQMHLARFLRMLLRL 255
Query: 259 ADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVI 318
ADEFGVA+VITNQVV+QVDG+A+FA KPIGGNI+AHA+TTRL LRKGRGE RICK+
Sbjct: 256 ADEFGVAVVITNQVVAQVDGAAMFAADPKKPIGGNIIAHASTTRLYLRKGRGETRICKIY 315
Query: 319 SSPCLAEAEARFQILAEGVSDVKD 342
SPCL EAEA F I A+GV D KD
Sbjct: 316 DSPCLPEAEAMFAINADGVGDAKD 339
>RA51_CRIGR (P70099) DNA repair protein RAD51 homolog 1
Length = 339
Score = 476 bits (1225), Expect = e-134
Identities = 232/324 (71%), Positives = 282/324 (86%)
Query: 19 EEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIV 78
EE GP PI +L+ GI++ DVKKL++AG TVE+VAY P+K+L+ IKGISEAK DKI+
Sbjct: 16 EEESFGPQPISRLEQCGISANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKIL 75
Query: 79 EAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQ 138
A+KLVPMGFT+A+E H +R IIQITTGS+ELDK+L GGIETGSITE++GEFR+GKTQ
Sbjct: 76 AEAAKLVPMGFTTATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITEMFGEFRTGKTQ 135
Query: 139 LCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAY 198
+CHTL VTCQLP+D+GGGEGKAMYID EGTFRP+RLL +A+R+GL+G+DVL+NVAYAR +
Sbjct: 136 ICHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARGF 195
Query: 199 NTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKL 258
NTDHQ++LL +A++MMVE+R+A++IVDSATALYRTD+SGRGELSARQMHLA+FLR L +L
Sbjct: 196 NTDHQTQLLYQASAMMVESRYALLIVDSATALYRTDYSGRGELSARQMHLARFLRMLLRL 255
Query: 259 ADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVI 318
ADEFGVA+VITNQVV+QVDG+A+F KPIGGNI+AHA+TTRL LRKGRGE RICKV
Sbjct: 256 ADEFGVAVVITNQVVAQVDGAAMFTADPKKPIGGNIIAHASTTRLYLRKGRGETRICKVY 315
Query: 319 SSPCLAEAEARFQILAEGVSDVKD 342
SPCL EAEA F I A+GV D KD
Sbjct: 316 DSPCLPEAEAMFAINADGVGDAKD 339
>RA51_MOUSE (Q08297) DNA repair protein RAD51 homolog 1
Length = 339
Score = 476 bits (1224), Expect = e-134
Identities = 232/324 (71%), Positives = 282/324 (86%)
Query: 19 EEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIV 78
EE GP PI +L+ GI + DVKKL++AG TVE+VAY P+K+L+ IKGISEAK DKI+
Sbjct: 16 EEESFGPQPISRLEQCGINANDVKKLEEAGYHTVEAVAYAPKKELINIKGISEAKADKIL 75
Query: 79 EAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQ 138
A+KLVPMGFT+A+E H +R IIQITTGS+ELDK+L GGIETGSITE++GEFR+GKTQ
Sbjct: 76 TEAAKLVPMGFTTATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITEMFGEFRTGKTQ 135
Query: 139 LCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAY 198
+CHTL VTCQLP+D+GGGEGKAMYID EGTFRP+RLL +A+R+GL+G+DVL+NVAYAR +
Sbjct: 136 ICHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARGF 195
Query: 199 NTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKL 258
NTDHQ++LL +A++MMVE+R+A++IVDSATALYRTD+SGRGELSARQMHLA+FLR L +L
Sbjct: 196 NTDHQTQLLYQASAMMVESRYALLIVDSATALYRTDYSGRGELSARQMHLARFLRMLLRL 255
Query: 259 ADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVI 318
ADEFGVA+VITNQVV+QVDG+A+FA KPIGGNI+AHA+TTRL LRKGRGE RICK+
Sbjct: 256 ADEFGVAVVITNQVVAQVDGAAMFAADPKKPIGGNIIAHASTTRLYLRKGRGETRICKIY 315
Query: 319 SSPCLAEAEARFQILAEGVSDVKD 342
SPCL EAEA F I A+GV D KD
Sbjct: 316 DSPCLPEAEAMFAINADGVGDAKD 339
>RA51_CHICK (P37383) DNA repair protein RAD51 homolog
Length = 339
Score = 474 bits (1219), Expect = e-133
Identities = 233/324 (71%), Positives = 281/324 (85%)
Query: 19 EEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIV 78
EE GP PI +L+ GI + DVKKL++AG TVESVA+ P+K+LL IKGISEAK DKI+
Sbjct: 16 EEESFGPEPISRLEQCGINANDVKKLEEAGYHTVESVAHAPKKELLNIKGISEAKADKIL 75
Query: 79 EAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQ 138
A+KLVPMGFT+A+E H +R IIQITTGS+ELDK+L GGIETGSITEL+GEFR+GKTQ
Sbjct: 76 AEAAKLVPMGFTTATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITELFGEFRTGKTQ 135
Query: 139 LCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAY 198
LCHTL VTCQLP+D+GGGEGKAMYID EGTFRP+RLL +A+R+GL+G+DVL+NVAYAR +
Sbjct: 136 LCHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARGF 195
Query: 199 NTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKL 258
NTDHQ++LL +A++MM E+R+A++IVDSATALYRTD+SGRGELSARQMHLA+FLR L +L
Sbjct: 196 NTDHQTQLLYQASAMMAESRYALLIVDSATALYRTDYSGRGELSARQMHLARFLRMLLRL 255
Query: 259 ADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVI 318
ADEFGVA+VITNQVV+QVDG+A+FA KPIGGNI+AHA+TTRL LRKGRGE RICK+
Sbjct: 256 ADEFGVAVVITNQVVAQVDGAAMFAADPKKPIGGNIIAHASTTRLYLRKGRGETRICKIY 315
Query: 319 SSPCLAEAEARFQILAEGVSDVKD 342
SPCL EAEA F I A+GV D K+
Sbjct: 316 DSPCLPEAEAMFAINADGVGDAKE 339
>R512_XENLA (Q91917) DNA repair protein RAD51 homolog 2 (XRAD51.2)
Length = 336
Score = 472 bits (1215), Expect = e-133
Identities = 233/327 (71%), Positives = 281/327 (85%)
Query: 16 EEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVD 75
E EE GP I +L+ GI + DVKKL+DAG TVE+VAY P+K+LL IKGISEAK +
Sbjct: 10 EATEEENFGPQAITRLEQCGINANDVKKLEDAGFHTVEAVAYAPKKELLNIKGISEAKAE 69
Query: 76 KIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSG 135
KI+ A+KLVPMGFT+A+E H +R IIQI TGS+ELDK+L GGIETGSITE++GEFR+G
Sbjct: 70 KILAEAAKLVPMGFTTATEFHQRRSEIIQIGTGSKELDKLLQGGIETGSITEMFGEFRTG 129
Query: 136 KTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYA 195
KTQLCHTL VTCQLP+D+GGGEGKAMYID EGTFRP+RLL +A+R+GL+G+DVL+NVAYA
Sbjct: 130 KTQLCHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYA 189
Query: 196 RAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSL 255
RA+NTDHQ++LL +A++MM E+R+A++IVDSATALYRTD+SGRGELSARQMHLA+FLR L
Sbjct: 190 RAFNTDHQTQLLYQASAMMAESRYALLIVDSATALYRTDYSGRGELSARQMHLARFLRML 249
Query: 256 QKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERIC 315
+LADEFGVA+VITNQVV+QVDG+A+FA KPIGGNI+AHA+TTRL LRKGRGE RIC
Sbjct: 250 LRLADEFGVAVVITNQVVAQVDGAAMFAADPKKPIGGNIIAHASTTRLYLRKGRGETRIC 309
Query: 316 KVISSPCLAEAEARFQILAEGVSDVKD 342
K+ SPCL EAEA F I A+GV D KD
Sbjct: 310 KIYDSPCLPEAEAMFAINADGVGDAKD 336
>RA51_RABIT (O77507) DNA repair protein RAD51 homolog 1
Length = 339
Score = 472 bits (1214), Expect = e-133
Identities = 229/324 (70%), Positives = 280/324 (85%)
Query: 19 EEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIV 78
EE GP P+ +L+ GI + DVKKL++AG T E+VAY P+K+L+ IKGISEAK DKI+
Sbjct: 16 EEESFGPQPVSRLEQCGINANDVKKLEEAGFHTEEAVAYAPKKELINIKGISEAKADKIL 75
Query: 79 EAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQ 138
A+KLVPMGFT+A+E H +R IIQITTGS+ELDK+L GGIETGSITE++GEFR+GKTQ
Sbjct: 76 TEAAKLVPMGFTTATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITEMFGEFRTGKTQ 135
Query: 139 LCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAY 198
+CHTL VTCQLP+D+GGGEGKAMYID EGTFRP+RLL +A+R+GL+G+DVL+NVAYAR +
Sbjct: 136 ICHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARGF 195
Query: 199 NTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKL 258
NTDHQ++LL +A++MMVE+R+A++IVDSATALYRTD+SGRGELSARQMHLA+FLR L +L
Sbjct: 196 NTDHQTQLLYQASAMMVESRYALLIVDSATALYRTDYSGRGELSARQMHLARFLRMLLRL 255
Query: 259 ADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVI 318
ADEFGV +VITNQVV+QVDG+A+FA KPIGGNI+AHA+TTRL LRKGRGE RICK+
Sbjct: 256 ADEFGVTVVITNQVVAQVDGAAMFAADPKKPIGGNIIAHASTTRLYLRKGRGETRICKIY 315
Query: 319 SSPCLAEAEARFQILAEGVSDVKD 342
SPCL EAEA F I A+GV D KD
Sbjct: 316 DSPCLPEAEAMFAINADGVGDAKD 339
>R511_XENLA (Q91918) DNA repair protein RAD51 homolog 1 (XRAD51.1)
Length = 336
Score = 472 bits (1214), Expect = e-133
Identities = 231/327 (70%), Positives = 282/327 (85%)
Query: 16 EEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVD 75
E EE GP I +L+ GI + DVKKL++AG TVE+VAY P+K+LL IKGISEAK +
Sbjct: 10 EATEEEHFGPQAISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELLNIKGISEAKAE 69
Query: 76 KIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSG 135
KI+ A+KLVPMGFT+A+E H +R IIQI+TGS+ELDK+L GG+ETGSITE++GEFR+G
Sbjct: 70 KILAEAAKLVPMGFTTATEFHQRRSEIIQISTGSKELDKLLQGGVETGSITEMFGEFRTG 129
Query: 136 KTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYA 195
KTQLCHTL VTCQLP+D+GGGEGKAMYID EGTFRP+RLL +A+R+GL+G+DVL+NVAYA
Sbjct: 130 KTQLCHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYA 189
Query: 196 RAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSL 255
RA+NTDHQ++LL +A++MM E+R+A++IVDSATALYRTD+SGRGELSARQMHLA+FLR L
Sbjct: 190 RAFNTDHQTQLLYQASAMMAESRYALLIVDSATALYRTDYSGRGELSARQMHLARFLRML 249
Query: 256 QKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERIC 315
+LADEFGVA+VITNQVV+QVDG+A+FA KPIGGNI+AHA+TTRL LRKGRGE RIC
Sbjct: 250 LRLADEFGVAVVITNQVVAQVDGAAMFAADPKKPIGGNIIAHASTTRLYLRKGRGETRIC 309
Query: 316 KVISSPCLAEAEARFQILAEGVSDVKD 342
K+ SPCL EAEA F I A+GV D KD
Sbjct: 310 KIYDSPCLPEAEAMFAINADGVGDAKD 336
>RA51_SCHPO (P36601) DNA repair protein rhp51 (RAD51 homolog)
Length = 365
Score = 458 bits (1179), Expect = e-129
Identities = 218/328 (66%), Positives = 279/328 (84%), Gaps = 1/328 (0%)
Query: 15 QEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKV 74
Q+E +E GP+P++ L+ +GI + D+KK+ +AG TVES+AYTP++ LL IKGISEAK
Sbjct: 34 QDEEDEAAAGPMPLQMLEGNGITASDIKKIHEAGYYTVESIAYTPKRQLLLIKGISEAKA 93
Query: 75 DKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRS 134
DK++ ASKLVPMGFT+A+E H++R +I ITTGS++LD +L GG+ETGSITEL+GEFR+
Sbjct: 94 DKLLGEASKLVPMGFTTATEYHIRRSELITITTGSKQLDTLLQGGVETGSITELFGEFRT 153
Query: 135 GKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAY 194
GK+Q+CHTL VTCQLP+D GGGEGK +YID EGTFRP RLL +ADR+GLNG +VL+NVAY
Sbjct: 154 GKSQICHTLAVTCQLPIDMGGGEGKCLYIDTEGTFRPVRLLAVADRYGLNGEEVLDNVAY 213
Query: 195 ARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRS 254
ARAYN DHQ LL +AA+MM E+RF++++VDS TALYRTDFSGRGELSARQMHLA+F+R+
Sbjct: 214 ARAYNADHQLELLQQAANMMSESRFSLLVVDSCTALYRTDFSGRGELSARQMHLARFMRT 273
Query: 255 LQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERI 314
LQ+LADEFG+A+VITNQVV+QVDG + P+ KPIGGNI+AH++TTRL+LRKGRGE+RI
Sbjct: 274 LQRLADEFGIAVVITNQVVAQVDGISFNPDPK-KPIGGNILAHSSTTRLSLRKGRGEQRI 332
Query: 315 CKVISSPCLAEAEARFQILAEGVSDVKD 342
CK+ SPCL E+EA F I ++GV D K+
Sbjct: 333 CKIYDSPCLPESEAIFAINSDGVGDPKE 360
>RA51_USTMA (Q99133) DNA repair protein RAD51
Length = 339
Score = 451 bits (1160), Expect = e-126
Identities = 222/339 (65%), Positives = 273/339 (80%), Gaps = 2/339 (0%)
Query: 4 QRHQKTAQQQPQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDL 63
Q Q AQ + GE GPLP+ +L+ GI+S D KKL ++G TVES+A+TP+K L
Sbjct: 3 QNAQDPAQIGEDDMGEAF--GPLPVSKLEEFGISSSDCKKLAESGYNTVESIAFTPKKQL 60
Query: 64 LQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETG 123
L +KG+SEAK DKI+ A++LVPMGFT+A+E H +R +I ITTGS+ LD IL GG+ETG
Sbjct: 61 LLVKGVSEAKADKILAEAARLVPMGFTTATEFHARRNELISITTGSKNLDAILGGGMETG 120
Query: 124 SITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGL 183
SITELYGEFR+GK+QLCHTL VTCQLP+D GGGEGK +YID E TFRP RLL +A+RFGL
Sbjct: 121 SITELYGEFRTGKSQLCHTLAVTCQLPVDMGGGEGKCLYIDTENTFRPTRLLAVAERFGL 180
Query: 184 NGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSA 243
NG +VL+NVAYARAYN DHQ +LL++A++MM E+RF+++IVDS T+LYRTDFSGRGELSA
Sbjct: 181 NGEEVLDNVAYARAYNADHQLQLLMQASAMMAESRFSLLIVDSLTSLYRTDFSGRGELSA 240
Query: 244 RQMHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRL 303
RQMHLAKFLR L +LADEFGVA+VITNQVV+QVDG+ F KPIGGNI+AHA+TTRL
Sbjct: 241 RQMHLAKFLRGLMRLADEFGVAVVITNQVVAQVDGATAFTADAKKPIGGNIVAHASTTRL 300
Query: 304 ALRKGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
+LRKGRG +RIC++ SPCL EA+A F I EG+ D D
Sbjct: 301 SLRKGRGNQRICRIADSPCLPEADAVFAIGPEGIIDPVD 339
>RA51_YEAST (P25454) DNA repair protein RAD51
Length = 400
Score = 435 bits (1118), Expect = e-122
Identities = 214/340 (62%), Positives = 268/340 (77%), Gaps = 4/340 (1%)
Query: 7 QKTAQQQPQEEGEEIQHGPL----PIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKD 62
Q+ A+ Q + E E L PIE+LQ +GI DVKKL+++G+ T E+VAY PRKD
Sbjct: 58 QEQAEAQGEMEDEAYDEAALGSFVPIEKLQVNGITMADVKKLRESGLHTAEAVAYAPRKD 117
Query: 63 LLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIET 122
LL+IKGISEAK DK++ A++LVPMGF +A++ H++R +I +TTGS+ LD +L GG+ET
Sbjct: 118 LLEIKGISEAKADKLLNEAARLVPMGFVTAADFHMRRSELICLTTGSKNLDTLLGGGVET 177
Query: 123 GSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFG 182
GSITEL+GEFR+GK+QLCHTL VTCQ+PLD GGGEGK +YID EGTFRP RL+ IA RFG
Sbjct: 178 GSITELFGEFRTGKSQLCHTLAVTCQIPLDIGGGEGKCLYIDTEGTFRPVRLVSIAQRFG 237
Query: 183 LNGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELS 242
L+ D L NVAYARAYN DHQ RLL AA MM E+RF++++VDS ALYRTDFSGRGELS
Sbjct: 238 LDPDDALNNVAYARAYNADHQLRLLDAAAQMMSESRFSLIVVDSVMALYRTDFSGRGELS 297
Query: 243 ARQMHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTR 302
ARQMHLAKF+R+LQ+LAD+FGVA+V+TNQVV+QVDG F KPIGGNIMAH++TTR
Sbjct: 298 ARQMHLAKFMRALQRLADQFGVAVVVTNQVVAQVDGGMAFNPDPKKPIGGNIMAHSSTTR 357
Query: 303 LALRKGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
L +KG+G +R+CKV+ SPCL EAE F I +GV D ++
Sbjct: 358 LGFKKGKGCQRLCKVVDSPCLPEAECVFAIYEDGVGDPRE 397
>RA51_DROME (Q27297) DNA repair protein Rad51 homolog (RecA protein
homolog)
Length = 336
Score = 383 bits (984), Expect = e-106
Identities = 191/336 (56%), Positives = 253/336 (74%), Gaps = 2/336 (0%)
Query: 7 QKTAQQQPQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQI 66
+K Q Q+E EE + GPL + +L I + D+K L+ A + TVESVA +K L+ I
Sbjct: 2 EKLTNVQAQQEEEE-EEGPLSVTKLIGGSITAKDIKLLQQASLHTVESVANATKKQLMAI 60
Query: 67 KGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSIT 126
G+ KV++I+ A+KLVP+GF SA + R ++Q++TGS+ELDK+L GGIETGSIT
Sbjct: 61 PGLGGGKVEQIITEANKLVPLGFLSARTFYQMRADVVQLSTGSKELDKLLGGGIETGSIT 120
Query: 127 ELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGA 186
E++GEFR GKTQLCHTL VTCQLP+ Q GGEGK MYID E TFRP+RL IA R+ LN +
Sbjct: 121 EIFGEFRCGKTQLCHTLAVTCQLPISQKGGEGKCMYIDTENTFRPERLAAIAQRYKLNES 180
Query: 187 DVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQM 246
+VL+NVA+ RA+N+D Q++L+ AA M+ E+R+A++IVDSA ALYR+D+ GRGEL+ARQ
Sbjct: 181 EVLDNVAFTRAHNSDQQTKLIQMAAGMLFESRYALLIVDSAMALYRSDYIGRGELAARQN 240
Query: 247 HLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALR 306
HL FLR LQ+LADEFGVA+VITNQV + +DG+ + KPIGG+IMAH++TTRL LR
Sbjct: 241 HLGLFLRMLQRLADEFGVAVVITNQVTASLDGAPGMFDAK-KPIGGHIMAHSSTTRLYLR 299
Query: 307 KGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
KG+GE RICK+ SPCL E+EA F IL +G+ D ++
Sbjct: 300 KGKGETRICKIYDSPCLPESEAMFAILPDGIGDARE 335
>DMC1_LILLO (P37384) Meiotic recombination protein DMC1 homolog
Length = 349
Score = 343 bits (881), Expect = 3e-94
Identities = 180/344 (52%), Positives = 243/344 (70%), Gaps = 4/344 (1%)
Query: 2 EQQRHQKTAQQQ---PQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYT 58
E++R + Q Q QE EE + I++L + GI + DVKKL+DAGI T +
Sbjct: 7 EERRFESPGQLQLLDRQEAEEEEEDCFESIDKLISQGINAGDVKKLQDAGIYTCNGLMMH 66
Query: 59 PRKDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDG 118
+K+L IKG+SEAKVDKI EAA KLV +G+ + S++ ++R+S+I+ITTGS+ LD++L G
Sbjct: 67 TKKNLTGIKGLSEAKVDKICEAAEKLVNVGYITGSDVLLKRKSVIRITTGSQALDELLGG 126
Query: 119 GIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIA 178
GIET ITE +GEFRSGKTQ+ HTLCV+ QLP+ GG GK YID EGTFRP R++ IA
Sbjct: 127 GIETLQITEAFGEFRSGKTQIAHTLCVSTQLPVSMHGGNGKVAYIDTEGTFRPDRIVPIA 186
Query: 179 DRFGLNGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGR 238
+RFG++ + VL+N+ YARAY +HQ LLL A+ M E F ++IVDS AL+R DFSGR
Sbjct: 187 ERFGMDASAVLDNIIYARAYTYEHQYNLLLALAAKMSEEPFRLLIVDSVIALFRVDFSGR 246
Query: 239 GELSARQMHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHA 298
GEL+ RQ LA+ L L K+A+EF VA+ +TNQV++ G + P+ KP GG+++AHA
Sbjct: 247 GELAERQQKLAQMLSRLTKIAEEFNVAVYMTNQVIADPGGGMFISDPK-KPAGGHVLAHA 305
Query: 299 TTTRLALRKGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
T RL LRKG+GE+R+CK+ +P L E+EA FQI GV+D KD
Sbjct: 306 ATVRLMLRKGKGEQRVCKIFDAPNLPESEAVFQITPGGVADAKD 349
>DMC1_SOYBN (Q96449) Meiotic recombination protein DMC1 homolog
Length = 345
Score = 342 bits (878), Expect = 6e-94
Identities = 177/315 (56%), Positives = 228/315 (72%), Gaps = 1/315 (0%)
Query: 28 IEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIVEAASKLVPM 87
I++L A GI + DVKKL+DAGI T + +K+L IKG+SEAKVDKI EAA KLV
Sbjct: 32 IDKLIAQGINAGDVKKLQDAGIYTCNGLMMHTKKNLTGIKGLSEAKVDKICEAAEKLVNF 91
Query: 88 GFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTC 147
G+ + S+ ++R+S+I+ITTGS+ LD++L GG+ET +ITE +GEFRSGKTQL HTLCV+
Sbjct: 92 GYITGSDALLKRKSVIRITTGSQALDELLGGGVETSAITEAFGEFRSGKTQLAHTLCVST 151
Query: 148 QLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAYNTDHQSRLL 207
QLP + GG GK YID EGTFRP R++ IA+RFG++ VL+N+ YARAY +HQ LL
Sbjct: 152 QLPTNMRGGNGKVAYIDTEGTFRPDRIVPIAERFGMDPGAVLDNIIYARAYTYEHQYNLL 211
Query: 208 LEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKLADEFGVAIV 267
L A+ M E F ++IVDS AL+R DFSGRGEL+ RQ LA+ L L K+A+EF VA+
Sbjct: 212 LGLAAKMSEEPFRLLIVDSVIALFRVDFSGRGELADRQQKLAQMLSRLIKIAEEFNVAVY 271
Query: 268 ITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVISSPCLAEAE 327
+TNQV+S G VF KP GG+++AHA T RL RKG+GE+RICKV +P L EAE
Sbjct: 272 MTNQVISD-PGGGVFVTDPKKPAGGHVLAHAATVRLMFRKGKGEQRICKVFDAPNLPEAE 330
Query: 328 ARFQILAEGVSDVKD 342
A FQI A G++D KD
Sbjct: 331 AVFQITAGGIADAKD 345
>DMC1_ARATH (Q39009) Meiotic recombination protein DMC1 homolog
Length = 344
Score = 338 bits (868), Expect = 9e-93
Identities = 177/344 (51%), Positives = 240/344 (69%), Gaps = 3/344 (0%)
Query: 1 MEQQRHQKTAQQQ--PQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYT 58
M + ++T+Q Q +EE +E + I++L A GI + DVKKL++AGI T +
Sbjct: 2 MASLKAEETSQMQLVEREENDEDEDLFEMIDKLIAQGINAGDVKKLQEAGIHTCNGLMMH 61
Query: 59 PRKDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDG 118
+K+L IKG+SEAKVDKI EAA K+V G+ + S+ ++R+S+++ITTG + LD +L G
Sbjct: 62 TKKNLTGIKGLSEAKVDKICEAAEKIVNFGYMTGSDALIKRKSVVKITTGCQALDDLLGG 121
Query: 119 GIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIA 178
GIET +ITE +GEFRSGKTQL HTLCVT QLP + GG GK YID EGTFRP R++ IA
Sbjct: 122 GIETSAITEAFGEFRSGKTQLAHTLCVTTQLPTNMKGGNGKVAYIDTEGTFRPDRIVPIA 181
Query: 179 DRFGLNGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGR 238
+RFG++ VL+N+ YARAY +HQ LLL A+ M E F ++IVDS AL+R DF+GR
Sbjct: 182 ERFGMDPGAVLDNIIYARAYTYEHQYNLLLGLAAKMSEEPFRILIVDSIIALFRVDFTGR 241
Query: 239 GELSARQMHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHA 298
GEL+ RQ LA+ L L K+A+EF VA+ +TNQV++ G + P+ KP GG+++AHA
Sbjct: 242 GELADRQQKLAQMLSRLIKIAEEFNVAVYMTNQVIADPGGGMFISDPK-KPAGGHVLAHA 300
Query: 299 TTTRLALRKGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
T RL RKG+G+ R+CKV +P LAEAEA FQI G++D KD
Sbjct: 301 ATIRLLFRKGKGDTRVCKVYDAPNLAEAEASFQITQGGIADAKD 344
>DMC1_HUMAN (Q14565) Meiotic recombination protein DMC1/LIM15
homolog
Length = 340
Score = 318 bits (815), Expect = 1e-86
Identities = 166/317 (52%), Positives = 218/317 (68%), Gaps = 2/317 (0%)
Query: 28 IEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIVEAASKLVPM 87
I+ LQ GI D+KKLK GICT++ + T R+ L +KG+SEAKVDKI EAA+KL+
Sbjct: 24 IDLLQKHGINVADIKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKVDKIKEAANKLIEP 83
Query: 88 GFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTC 147
GF +A E +R+ + ITTGS+E DK+L GGIE+ +ITE +GEFR+GKTQL HTLCVT
Sbjct: 84 GFLTAFEYSEKRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFRTGKTQLSHTLCVTA 143
Query: 148 QLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAYNTDHQSRLL 207
QLP G GK ++ID E TFRP RL IADRF ++ VL+NV YARAY ++HQ LL
Sbjct: 144 QLPGAGGYPGGKIIFIDTENTFRPDRLRDIADRFNVDHDAVLDNVLYARAYTSEHQMELL 203
Query: 208 LEAASMMVETR--FAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKLADEFGVA 265
A+ E F ++I+DS AL+R DFSGRGEL+ RQ LA+ L LQK+++E+ VA
Sbjct: 204 DYVAAKFHEEAGIFKLLIIDSIMALFRVDFSGRGELAERQQKLAQMLSRLQKISEEYNVA 263
Query: 266 IVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVISSPCLAE 325
+ +TNQ+ + + F KPIGG+I+AHA+TTR++LRKGRGE RI K+ SP + E
Sbjct: 264 VFVTNQMTADPGATMTFQADPKKPIGGHILAHASTTRISLRKGRGELRIAKIYDSPEMPE 323
Query: 326 AEARFQILAEGVSDVKD 342
EA F I A G+ D K+
Sbjct: 324 NEATFAITAGGIGDAKE 340
>DMC1_MOUSE (Q61880) Meiotic recombination protein DMC1/LIM15
homolog
Length = 340
Score = 317 bits (813), Expect = 2e-86
Identities = 164/317 (51%), Positives = 218/317 (68%), Gaps = 2/317 (0%)
Query: 28 IEQLQASGIASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIVEAASKLVPM 87
I+ LQ GI D+KKLK GICT++ + T R+ L +KG+SEAKV+KI EAA+KL+
Sbjct: 24 IDLLQKHGINMADIKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKVEKIKEAANKLIEP 83
Query: 88 GFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTC 147
GF +A + +R+ + ITTGS+E DK+L GGIE+ +ITE +GEFR+GKTQL HTLCVT
Sbjct: 84 GFLTAFQYSERRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFRTGKTQLSHTLCVTA 143
Query: 148 QLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAYNTDHQSRLL 207
QLP G GK ++ID E TFRP RL IADRF ++ VL+NV YARAY ++HQ LL
Sbjct: 144 QLPGTGGYSGGKIIFIDTENTFRPDRLRDIADRFNVDHEAVLDNVLYARAYTSEHQMELL 203
Query: 208 LEAASMMVETR--FAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKLADEFGVA 265
A+ E F ++I+DS AL+R DFSGRGEL+ RQ LA+ L LQK+++E+ VA
Sbjct: 204 DYVAAKFHEEAGIFKLLIIDSIMALFRVDFSGRGELAERQQKLAQMLSRLQKISEEYNVA 263
Query: 266 IVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVISSPCLAE 325
+ +TNQ+ + + F KPIGG+I+AHA+TTR++LRKGRGE RI K+ SP + E
Sbjct: 264 VFVTNQMTADPGATMTFQADPKKPIGGHILAHASTTRISLRKGRGELRIAKIYDSPEMPE 323
Query: 326 AEARFQILAEGVSDVKD 342
EA F I A G+ D K+
Sbjct: 324 NEATFAITAGGIGDAKE 340
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,379,142
Number of Sequences: 164201
Number of extensions: 1474544
Number of successful extensions: 6361
Number of sequences better than 10.0: 339
Number of HSP's better than 10.0 without gapping: 126
Number of HSP's successfully gapped in prelim test: 213
Number of HSP's that attempted gapping in prelim test: 6075
Number of HSP's gapped (non-prelim): 368
length of query: 342
length of database: 59,974,054
effective HSP length: 111
effective length of query: 231
effective length of database: 41,747,743
effective search space: 9643728633
effective search space used: 9643728633
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0343.7


