

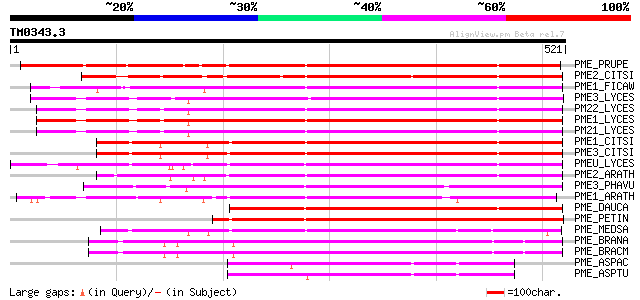
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0343.3
(521 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11) ... 552 e-157
PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 395 e-109
PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pect... 392 e-109
PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 386 e-107
PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 385 e-106
PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 383 e-106
PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 380 e-105
PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 374 e-103
PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 372 e-102
PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11) (P... 367 e-101
PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 354 3e-97
PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 351 2e-96
PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 337 3e-92
PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 317 5e-86
PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 293 7e-79
PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 285 2e-76
PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.1... 238 2e-62
PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 238 2e-62
PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 135 2e-31
PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 120 9e-27
>PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 522
Score = 552 bits (1422), Expect = e-157
Identities = 291/509 (57%), Positives = 362/509 (70%), Gaps = 12/509 (2%)
Query: 11 LILLLIMSLQPCTTPSSASTITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPFTK 70
L++LL P ++ +IT+ CL+VS F S I+ +Q V+ LS F
Sbjct: 20 LLVLLCAFFSSSFIPFASCSITDDLQTQCLKVSATEFAGSLKDTIDAVQQVASILSQFA- 78
Query: 71 VNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVL 130
N D RL+NAISDCL+L D + + L WS+SA+QN +G KNN TG LSSDLRTW+SA L
Sbjct: 79 -NAFGDFRLANAISDCLDLLDFSADELNWSLSASQNQKG-KNNSTGKLSSDLRTWLSAAL 136
Query: 131 SYQETCIDGFEGTNSNVKDHVSGGLDQVISLVKNELLLQVVPDSDPSNISNSGEFPSWVK 190
Q+TC +GFEGTNS V+ +S GL QV SLV+ ELL QV P+S+ +G+ PSWVK
Sbjct: 137 VNQDTCSNGFEGTNSIVQGLISAGLGQVTSLVQ-ELLTQVHPNSNQQG--PNGQIPSWVK 193
Query: 191 AEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVE 250
+D KLLQ D D +VA DG+GN+T V DAV AP+Y+M RYVI++K+G YKE VE
Sbjct: 194 TKDRKLLQADGVS--VDAIVAQDGTGNFTNVTDAVLAAPDYSMRRYVIYIKRGTYKENVE 251
Query: 251 IKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPE 310
IKKKKWN+M++GDGMD T ISGNRS+ D TTFR+ATFAVS GFIARDITFENTAGPE
Sbjct: 252 IKKKKWNLMMIGDGMDATIISGNRSFV-DGWTTFRSATFAVSGRGFIARDITFENTAGPE 310
Query: 311 NKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNC 370
QAVALRSDSDLSVFYRC I+GYQD+LY H+ RQFY++C+I GTVDFI G+A VFQNC
Sbjct: 311 KHQAVALRSDSDLSVFYRCNIRGYQDTLYTHTMRQFYRDCKISGTVDFIFGDATVVFQNC 370
Query: 371 LIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSN-GSIQTFLGRPWKN 429
I KK L +QKN+I AQ R PN+ +G S C I D D + ++ S T+LGRPWK
Sbjct: 371 QILAKKGLPNQKNSITAQGRKDPNEPTGISIQFCNITADSDLEAASVNSTPTYLGRPWKL 430
Query: 430 FSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLN 489
+SRTV M+SF+S +I PEGWLEW + ++LF+GEY N GPG+G+ +RVKW GY V N
Sbjct: 431 YSRTVIMQSFLSNVIRPEGWLEWNGDFAL-NSLFYGEYMNYGPGAGLGSRVKWPGYQVFN 489
Query: 490 NS-QAMDFTVNQFIKGDLWLPSTGVSYNA 517
S QA ++TV QFI+G+LWLPSTGV Y A
Sbjct: 490 ESTQAKNYTVAQFIEGNLWLPSTGVKYTA 518
>PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 510
Score = 395 bits (1016), Expect = e-109
Identities = 218/454 (48%), Positives = 284/454 (62%), Gaps = 25/454 (5%)
Query: 68 FTKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWIS 127
+T ++ + R A DC EL+++ L + N+ G D +TW+S
Sbjct: 80 YTLGSKCRNEREKAAWEDCRELYELTVLKLNQT----------SNSSPGCTKVDKQTWLS 129
Query: 128 AVLSYQETCIDGFEGTNSNVKDHVSGGL-DQVISLVKNELLLQVVPDSDPSNISNSGEFP 186
L+ ETC E + V ++V L + V L+ N L L VP ++PS FP
Sbjct: 130 TALTNLETCRASLE--DLGVPEYVLPLLSNNVTKLISNTLSLNKVPYNEPSYKDG---FP 184
Query: 187 SWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYK 246
+WVK D KLLQ P A++VVA DGSGN T+ +AV A +RYVI++K G Y
Sbjct: 185 TWVKPGDRKLLQTT---PRANIVVAQDGSGNVKTIQEAVAAASRAGGSRYVIYIKAGTYN 241
Query: 247 EYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENT 306
E +E+K K NIM VGDG+ T I+G++S G TTF++AT AV FIARDIT NT
Sbjct: 242 ENIEVKLK--NIMFVGDGIGKTIITGSKSVG-GGATTFKSATVAVVGDNFIARDITIRNT 298
Query: 307 AGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAV 366
AGP N QAVALRS SDLSVFYRC +GYQD+LY HSQRQFY+EC I GTVDFI GNAA V
Sbjct: 299 AGPNNHQAVALRSGSDLSVFYRCSFEGYQDTLYVHSQRQFYRECDIYGTVDFIFGNAAVV 358
Query: 367 FQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRP 426
QNC I +KP ++ NT+ AQ RT PNQ++G HNC + D +P S++TFLGRP
Sbjct: 359 LQNCNIFARKP-PNRTNTLTAQGRTDPNQSTGIIIHNCRVTAASDLKPVQSSVKTFLGRP 417
Query: 427 WKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYH 486
WK +SRTV++++F+ +I+P GW+EW + +TL++ EY N GPGS ANRVKWRGYH
Sbjct: 418 WKQYSRTVYIKTFLDSLINPAGWMEWSGDFAL-NTLYYAEYMNTGPGSSTANRVKWRGYH 476
Query: 487 VLNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
VL + SQ FTV FI G+ WLP+T V + +GL
Sbjct: 477 VLTSPSQVSQFTVGNFIAGNSWLPATNVPFTSGL 510
>PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 545
Score = 392 bits (1008), Expect = e-109
Identities = 222/509 (43%), Positives = 305/509 (59%), Gaps = 24/509 (4%)
Query: 20 QPCTTPSSASTITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPFTKVNQDNDNRL 79
Q S + I+E +GL+ D N + + L+ + + + D R+
Sbjct: 50 QSVNKESCLAMISEVTGLN--------MADHRNLLKSFLEKTTPRIQKAFETANDASRRI 101
Query: 80 SN-----AISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQE 134
+N A+ DC EL D++ E + SIS HQ N T DL W+S VL+
Sbjct: 102 NNPQERTALLDCAELMDLSKERVVDSISILF-HQ----NLTTRSHEDLHVWLSGVLTNHV 156
Query: 135 TCIDGFE-GTNSNVKDHVSGGLDQVISLVKNELLLQVVPDSDPSNISN--SGEFPSWVKA 191
TC+DG E G+ +K + L+++I + L + V SN+ +G FP+WV A
Sbjct: 157 TCLDGLEEGSTDYIKTLMESHLNELILRARTSLAIFVTLFPAKSNVIEPVTGNFPTWVTA 216
Query: 192 EDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEI 251
D +LLQ D+VVA DGSG+Y T+ +AV P+ + R ++ V+ GIY+E V+
Sbjct: 217 GDRRLLQTLGKDIEPDIVVAKDGSGDYETLNEAVAAIPDNSKKRVIVLVRTGIYEENVDF 276
Query: 252 KKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPEN 311
+K N+M+VG+GMD T I+G+R+ D TTF +AT A GFIA+DI F+NTAGPE
Sbjct: 277 GYQKKNVMLVGEGMDYTIITGSRNVV-DGSTTFDSATVAAVGDGFIAQDICFQNTAGPEK 335
Query: 312 KQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCL 371
QAVALR +D +V RC I YQD+LY H+ RQFY++ I GTVDFI GNAA VFQNC
Sbjct: 336 YQAVALRIGADETVINRCRIDAYQDTLYPHNYRQFYRDRNITGTVDFIFGNAAVVFQNCN 395
Query: 372 IQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFS 431
+ P+K +K Q+NTI AQ RT PNQ +G S NC I D +P + +++LGRPWK +S
Sbjct: 396 LIPRKQMKGQENTITAQGRTDPNQNTGTSIQNCEIFASADLEPVEDTFKSYLGRPWKEYS 455
Query: 432 RTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNS 491
RTV MES++S++I P GWLEW+ + TLF+GEY+N GPGSG + RVKW GYHV+ +
Sbjct: 456 RTVVMESYISDVIDPAGWLEWDRDFAL-KTLFYGEYRNGGPGSGTSERVKWPGYHVITSP 514
Query: 492 Q-AMDFTVNQFIKGDLWLPSTGVSYNAGL 519
+ A FTV + I+G WL STGV Y AGL
Sbjct: 515 EVAEQFTVAELIQGGSWLGSTGVDYTAGL 543
>PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 544
Score = 386 bits (991), Expect = e-107
Identities = 215/504 (42%), Positives = 299/504 (58%), Gaps = 24/504 (4%)
Query: 20 QPCTTPSSASTITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPFTKVNQDNDNRL 79
Q C + S TE G++ L+ +++ N I +++ + NQ ND R
Sbjct: 61 QLCLSYVSEIVTTESDGVTVLKKFLVKYVHQMNNAIPVVRKIK---------NQINDIRQ 111
Query: 80 SNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDG 139
A++DCLEL D + + + SI+A +++ ++ ++W+S VL+ TC+D
Sbjct: 112 QGALTDCLELLDQSVDLVSDSIAAIDKRSRSEH-------ANAQSWLSGVLTNHVTCLDE 164
Query: 140 FEGTNSNVKDHVSGGLDQVISLVKNEL--LLQVVPDSDPSNISNSGEFPSWVKAEDEKLL 197
+ + K+ LD++I+ K L L V +D G+ P WV + D KL+
Sbjct: 165 LTSFSLSTKNGTV--LDELITRAKVALAMLASVTTPNDEVLRQGLGKMPYWVSSRDRKLM 222
Query: 198 QQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWN 257
+ A+ VVA DG+G+Y T+ +AV AP+ N TRYVI+VK GIYKE V + KKK N
Sbjct: 223 ESSGKDIIANRVVAQDGTGDYQTLAEAVAAAPDKNKTRYVIYVKMGIYKENVVVTKKKMN 282
Query: 258 IMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVAL 317
+MIVGDGM+ T I+G+ + + TF + T A GFI +DI +NTAGPE QAVAL
Sbjct: 283 LMIVGDGMNATIITGSLNVVDGS--TFPSNTLAAVGQGFILQDICIQNTAGPEKDQAVAL 340
Query: 318 RSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKP 377
R +D+SV RC I YQD+LYAHSQRQFY++ + GTVDFI GNAA VFQ C I +KP
Sbjct: 341 RVGADMSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTVDFIFGNAAVVFQKCQIVARKP 400
Query: 378 LKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFME 437
K QKN + AQ RT PNQ +G S C II PD +P +T+LGRPWK SRTV M+
Sbjct: 401 NKRQKNMVTAQGRTDPNQATGTSIQFCDIIASPDLEPVMNEYKTYLGRPWKKHSRTVVMQ 460
Query: 438 SFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNN-SQAMDF 496
S++ I P GW EW + TL++GE+ N GPG+G + RVKW GYHV+ + ++AM F
Sbjct: 461 SYLDGHIDPSGWFEWRGDFAL-KTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPNEAMPF 519
Query: 497 TVNQFIKGDLWLPSTGVSYNAGLL 520
TV + I+G WL ST V+Y GL+
Sbjct: 520 TVAELIQGGSWLNSTSVAYVEGLV 543
>PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 385 bits (988), Expect = e-106
Identities = 215/498 (43%), Positives = 296/498 (59%), Gaps = 25/498 (5%)
Query: 26 SSASTITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPFTKVNQDNDNRLSNAISD 85
S+ T+ GLS L ++ N I ++ + NQ ND R A++D
Sbjct: 72 SNEIVTTDSDGLSILMKFLVNYVHQMNNAIPVVSKMK---------NQINDIRQEGALTD 122
Query: 86 CLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFEGTNS 145
CLEL D + + + SI+A +++ ++ ++W+S VL+ TC+D + S
Sbjct: 123 CLELLDQSVDLVSDSIAAIDKRTHSEH-------ANAQSWLSGVLTNHVTCLDELD---S 172
Query: 146 NVKDHVSG-GLDQVISLVKNEL--LLQVVPDSDPSNISNSGEFPSWVKAEDEKLLQQDAP 202
K ++G LD++IS K L L V +D G+ PSWV + D KL++
Sbjct: 173 FTKAMINGTNLDELISRAKVALAMLASVTTPNDDVLRPGLGKMPSWVSSRDRKLMESSGK 232
Query: 203 PPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVG 262
A+ VVA DG+G Y T+ +AV AP+ + TRYVI+VK+GIYKE VE+ +K +MIVG
Sbjct: 233 DIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKRGIYKENVEVSSRKMKLMIVG 292
Query: 263 DGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSD 322
DGM T I+GN + D TTF +AT A GFI +DI +NTAGP QAVALR +D
Sbjct: 293 DGMHATIITGNLNVV-DGSTTFHSATLAAVGKGFILQDICIQNTAGPAKHQAVALRVGAD 351
Query: 323 LSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQK 382
SV RC I YQD+LYAHSQRQFY++ + GT+DFI GNAA VFQ C + +KP K Q+
Sbjct: 352 KSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGNAAVVFQKCKLVARKPGKYQQ 411
Query: 383 NTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSE 442
N + AQ RT PNQ +G S C II D +P T+LGRPWK +SRTV MES++
Sbjct: 412 NMVTAQGRTDPNQATGTSIQFCNIIASSDLEPVLKEFPTYLGRPWKKYSRTVVMESYLGG 471
Query: 443 MIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNN-SQAMDFTVNQF 501
+I+P GW EW+ + TL++GE+ N GPG+G + RVKW GYH + + ++AM FTV +
Sbjct: 472 LINPAGWAEWDGDFAL-KTLYYGEFMNNGPGAGTSKRVKWPGYHCITDPAEAMPFTVAKL 530
Query: 502 IKGDLWLPSTGVSYNAGL 519
I+G WL STGV+Y GL
Sbjct: 531 IQGGSWLRSTGVAYVDGL 548
>PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 546
Score = 383 bits (984), Expect = e-106
Identities = 215/498 (43%), Positives = 300/498 (60%), Gaps = 25/498 (5%)
Query: 26 SSASTITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPFTKVNQDNDNRLSNAISD 85
S+ TE G S L ++ N I +++ + NQ ND R A++D
Sbjct: 68 SNEIVTTESDGHSILMKFLVNYVHQMNNAIPVVRKMK---------NQINDIRQHGALTD 118
Query: 86 CLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFEGTNS 145
CLEL D + + SI+A +++ ++ ++W+S VL+ TC+D + S
Sbjct: 119 CLELLDQSVDFASDSIAAIDKRSRSEH-------ANAQSWLSGVLTNHVTCLDELD---S 168
Query: 146 NVKDHVSG-GLDQVISLVKNEL--LLQVVPDSDPSNISNSGEFPSWVKAEDEKLLQQDAP 202
K ++G L+++IS K L L + + ++ G+ PSWV + D KL++
Sbjct: 169 FTKAMINGTNLEELISRAKVALAMLASLTTQDEDVFMTVLGKMPSWVSSMDRKLMESSGK 228
Query: 203 PPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVG 262
A+ VVA DG+G+Y T+ +AV AP+ + TRYVI+VK+G YKE VE+ K N+MIVG
Sbjct: 229 DIIANAVVAQDGTGDYQTLAEAVAAAPDKSKTRYVIYVKRGTYKENVEVASNKMNLMIVG 288
Query: 263 DGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSD 322
DGM TTI+G+ + D TTFR+AT A GFI +DI +NTAGP QAVALR +D
Sbjct: 289 DGMYATTITGSLNVV-DGSTTFRSATLAAVGQGFILQDICIQNTAGPAKDQAVALRVGAD 347
Query: 323 LSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQK 382
+SV RC I YQD+LYAHSQRQFY++ + GTVDFI GNAA VFQ C + +KP K Q+
Sbjct: 348 MSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTVDFIFGNAAVVFQKCQLVARKPGKYQQ 407
Query: 383 NTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSE 442
N + AQ RT PNQ +G S C II D +P T+LGRPWK +SRTV MES++
Sbjct: 408 NMVTAQGRTDPNQATGTSIQFCNIIASSDLEPVLKEFPTYLGRPWKEYSRTVVMESYLGG 467
Query: 443 MIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNN-SQAMDFTVNQF 501
+I+P GW EW+ + TL++GE+ N GPG+G + RVKW GYHV+ + ++AM FTV +
Sbjct: 468 LINPAGWAEWDGDFAL-KTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPAKAMPFTVAKL 526
Query: 502 IKGDLWLPSTGVSYNAGL 519
I+G WL STGV+Y GL
Sbjct: 527 IQGGSWLRSTGVAYVDGL 544
>PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 380 bits (977), Expect = e-105
Identities = 213/498 (42%), Positives = 297/498 (58%), Gaps = 25/498 (5%)
Query: 26 SSASTITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPFTKVNQDNDNRLSNAISD 85
S+ ++ GLS L+ + N I +++ + NQ ND R A++D
Sbjct: 72 SNEIVTSDSDGLSILKKFLVYSVHQMNNAIPVVRKIK---------NQINDIREQGALTD 122
Query: 86 CLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFEGTNS 145
CLEL D++ + + SI+A +++ ++ ++W+S VL+ TC+D + S
Sbjct: 123 CLELLDLSVDLVCDSIAAIDKRSRSEH-------ANAQSWLSGVLTNHVTCLDELD---S 172
Query: 146 NVKDHVSG-GLDQVISLVKNEL--LLQVVPDSDPSNISNSGEFPSWVKAEDEKLLQQDAP 202
K ++G LD++IS K L L V +D G+ PSWV + D KL++
Sbjct: 173 FTKAMINGTNLDELISRAKVALAMLASVTTPNDEVLRPGLGKMPSWVSSRDRKLMESSGK 232
Query: 203 PPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVG 262
A+ VVA DG+G Y T+ +AV AP+ + TRYVI+VK+G YKE VE+ +K N+MI+G
Sbjct: 233 DIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKRGTYKENVEVSSRKMNLMIIG 292
Query: 263 DGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSD 322
DGM T I+G+ + D TTF +AT A GFI +DI +NTAGP QAVALR +D
Sbjct: 293 DGMYATIITGSLNVV-DGSTTFHSATLAAVGKGFILQDICIQNTAGPAKHQAVALRVGAD 351
Query: 323 LSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQK 382
SV RC I YQD+LYAHSQRQFY++ + GT+DFI GNAA VFQ C + +KP K Q+
Sbjct: 352 KSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGNAAVVFQKCQLVARKPGKYQQ 411
Query: 383 NTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSE 442
N + AQ RT PNQ +G S C II PD +P T+LGRPWK +SRTV MES++
Sbjct: 412 NMVTAQGRTDPNQATGTSIQFCDIIASPDLKPVVKEFPTYLGRPWKKYSRTVVMESYLGG 471
Query: 443 MIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNN-SQAMDFTVNQF 501
+I P GW EW + TL++GE+ N GPG+G + RVKW GYHV+ + ++AM FTV +
Sbjct: 472 LIDPSGWAEWHGDFAL-KTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPAEAMSFTVAKL 530
Query: 502 IKGDLWLPSTGVSYNAGL 519
I+G WL ST V+Y GL
Sbjct: 531 IQGGSWLRSTDVAYVDGL 548
>PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 374 bits (961), Expect = e-103
Identities = 203/454 (44%), Positives = 276/454 (60%), Gaps = 22/454 (4%)
Query: 82 AISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGF- 140
A+ DCLE D + L ++ + + K+ + DL+T +SA ++ Q TC+DGF
Sbjct: 137 ALHDCLETIDETLDELHKAVEDLEEYPNKKS--LSQHADDLKTLMSAAMTNQGTCLDGFS 194
Query: 141 -EGTNSNVKDHVSGGLDQVISLVKNEL-LLQVVPDSDPSNISNSGE------------FP 186
+ N +V+D +S G V + N L +++ + D+D + S +P
Sbjct: 195 HDDANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTDMMIMRTSNNRKLTEETSTVDGWP 254
Query: 187 SWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYK 246
+W+ D +LLQ + P A VVAADGSGN+ TV AV AP+ RY+I +K G+Y+
Sbjct: 255 AWLSPGDRRLLQSSSVTPNA--VVAADGSGNFKTVAAAVAAAPQGGTKRYIIRIKAGVYR 312
Query: 247 EYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENT 306
E VE+ KK NIM +GDG T I+G+R+ D TTF++AT AV GF+ARDITF+NT
Sbjct: 313 ENVEVTKKHKNIMFIGDGRTRTIITGSRNVV-DGSTTFKSATAAVVGEGFLARDITFQNT 371
Query: 307 AGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAV 366
AGP QAVALR +DLS FY C + YQD+LY HS RQF+ C I GTVDFI GNAAAV
Sbjct: 372 AGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIFGNAAAV 431
Query: 367 FQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRP 426
QNC I +KP QKN + AQ RT PNQ +G I D +P GS T+LGRP
Sbjct: 432 LQNCDIHARKPNSGQKNMVTAQGRTDPNQNTGIVIQKSRIGATSDLKPVQGSFPTYLGRP 491
Query: 427 WKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYH 486
WK +SRTV M+S ++++IHP GW EW+ N +TLF+GE+QN G G+G + RVKW+G+
Sbjct: 492 WKEYSRTVIMQSSITDLIHPAGWHEWDGNFAL-NTLFYGEHQNSGAGAGTSGRVKWKGFR 550
Query: 487 VLNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
V+ + ++A FT FI G WL STG ++ GL
Sbjct: 551 VITSATEAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 584
Score = 372 bits (956), Expect = e-102
Identities = 201/454 (44%), Positives = 276/454 (60%), Gaps = 22/454 (4%)
Query: 82 AISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGF- 140
A+ DCLE D + L ++ + + K+ + DL+T +SA ++ Q TC+DGF
Sbjct: 137 ALHDCLETIDETLDELHKAVEDLEEYPNKKS--LSQHADDLKTLMSAAMTNQGTCLDGFS 194
Query: 141 -EGTNSNVKDHVSGGLDQVISLVKNEL-LLQVVPDSDPSNISNSGE------------FP 186
+ N +V+D +S G V + N L +++ + D+D + S +P
Sbjct: 195 HDDANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTDMMIMRTSNNRKLIEETSTVDGWP 254
Query: 187 SWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYK 246
+W+ D +LLQ + P +VVVAADGSGN+ TV +V AP+ RY+I +K G+Y+
Sbjct: 255 AWLSTGDRRLLQSSSVTP--NVVVAADGSGNFKTVAASVAAAPQGGTKRYIIRIKAGVYR 312
Query: 247 EYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENT 306
E VE+ KK NIM +GDG T I+G+R+ D TTF++AT AV GF+ARDITF+NT
Sbjct: 313 ENVEVTKKHKNIMFIGDGRTRTIITGSRNVV-DGSTTFKSATVAVVGEGFLARDITFQNT 371
Query: 307 AGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAV 366
AGP QAVALR +DLS FY C + YQD+LY HS RQF+ C I GTVDFI GNAAAV
Sbjct: 372 AGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIFGNAAAV 431
Query: 367 FQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRP 426
QNC I +KP QKN + AQ R PNQ +G I D +P GS T+LGRP
Sbjct: 432 LQNCDIHARKPNSGQKNMVTAQGRADPNQNTGIVIQKSRIGATSDLKPVQGSFPTYLGRP 491
Query: 427 WKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYH 486
WK +SRTV M+S ++++IHP GW EW+ N +TLF+GE+QN G G+G + RVKW+G+
Sbjct: 492 WKEYSRTVIMQSSITDVIHPAGWHEWDGNFAL-NTLFYGEHQNAGAGAGTSGRVKWKGFR 550
Query: 487 VLNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
V+ + ++A FT FI G WL STG ++ GL
Sbjct: 551 VITSATEAQAFTPGSFIAGSSWLGSTGFPFSLGL 584
>PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 583
Score = 367 bits (941), Expect = e-101
Identities = 218/545 (40%), Positives = 304/545 (55%), Gaps = 43/545 (7%)
Query: 1 MAISSTLTPTLILLLIMSLQPCTTPSSASTITEFSGLSCLRVSPNRFIDSANKVINLLQN 60
MAISS+ + +L P S+ +++FS + + S VI L N
Sbjct: 56 MAISSSAHAIVKSACSNTLHPELCYSAIVNVSDFS----------KKVTSQKDVIELSLN 105
Query: 61 VS--------YNLSPFTKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKN 112
++ Y + K + R A+ DCLE D + L ++ + + K+
Sbjct: 106 ITVKAVRRNYYAVKELIKTRKGLTPREKVALHDCLETMDETLDELHTAVEDLELYPNKKS 165
Query: 113 NGTGNLSSDLRTWISAVLSYQETCIDGFEGTNSNVK---------DHV----SGGLDQVI 159
DL+T IS+ ++ QETC+DGF ++ K HV S L +
Sbjct: 166 --LKEHVEDLKTLISSAITNQETCLDGFSHDEADKKVRKVLLKGQKHVEKMCSNALAMIC 223
Query: 160 SL----VKNELLLQVVPDSDPSNISNSGEFPSWVKAEDEKLLQQDAPPPPADVVVAADGS 215
++ + NE+ L P ++ + ++GE+P W+ A D +LLQ P DVVVAADGS
Sbjct: 224 NMTDTDIANEMKLSA-PANNRKLVEDNGEWPEWLSAGDRRLLQSSTVTP--DVVVAADGS 280
Query: 216 GNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRS 275
G+Y TV +AV+ APE + RYVI +K G+Y+E V++ KKK NIM +GDG T I+ +R+
Sbjct: 281 GDYKTVSEAVRKAPEKSSKRYVIRIKAGVYRENVDVPKKKTNIMFMGDGKSNTIITASRN 340
Query: 276 YGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQ 335
D TTF +AT A +ARDITF+NTAG QAVAL SDLS FYRC + YQ
Sbjct: 341 V-QDGSTTFHSATVVRVAGKVLARDITFQNTAGASKHQAVALCVGSDLSAFYRCDMLAYQ 399
Query: 336 DSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQ 395
D+LY HS RQF+ +C + GTVDFI GN AAVFQ+C I ++P QKN + AQ RT PNQ
Sbjct: 400 DTLYVHSNRQFFVQCLVAGTVDFIFGNGAAVFQDCDIHARRPGSGQKNMVTAQGRTDPNQ 459
Query: 396 TSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETN 455
+G C I D +P S T+LGRPWK +SRTV M+S ++++I P GW EW N
Sbjct: 460 NTGIVIQKCRIGATSDLRPVQKSFPTYLGRPWKEYSRTVIMQSSITDVIQPAGWHEWNGN 519
Query: 456 TTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNS-QAMDFTVNQFIKGDLWLPSTGVS 514
DTLF+GEY N G G+ + RVKW+G+ V+ +S +A +T +FI G WL STG
Sbjct: 520 FAL-DTLFYGEYANTGAGAPTSGRVKWKGHKVITSSTEAQAYTPGRFIAGGSWLSSTGFP 578
Query: 515 YNAGL 519
++ GL
Sbjct: 579 FSLGL 583
>PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 587
Score = 354 bits (909), Expect = 3e-97
Identities = 201/465 (43%), Positives = 269/465 (57%), Gaps = 33/465 (7%)
Query: 82 AISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFE 141
A+ DCLE D + L ++ HQ K + DL+T IS+ ++ Q TC+DGF
Sbjct: 129 ALHDCLETIDETLDELH--VAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFS 186
Query: 142 GTNSNVKD---------HVSGGLDQVISLVKNELLLQVV------PDSDPSNISN----- 181
+++ K HV ++++KN + S +N +N
Sbjct: 187 YDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKSSTFTNNNNRKLKE 246
Query: 182 ------SGEFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTR 235
S +P W+ D +LLQ AD VA DGSG++TTV AV APE + R
Sbjct: 247 VTGDLDSDGWPKWLSVGDRRLLQGSTIK--ADATVADDGSGDFTTVAAAVAAAPEKSNKR 304
Query: 236 YVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIG 295
+VIH+K G+Y+E VE+ KKK NIM +GDG T I+G+R+ D TTF +AT A
Sbjct: 305 FVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVV-DGSTTFHSATVAAVGER 363
Query: 296 FIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGT 355
F+ARDITF+NTAGP QAVALR SD S FY+C + YQD+LY HS RQF+ +C I GT
Sbjct: 364 FLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITGT 423
Query: 356 VDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPS 415
VDFI GNAAAV Q+C I ++P QKN + AQ R+ PNQ +G NC I D
Sbjct: 424 VDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLAV 483
Query: 416 NGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSG 475
G+ T+LGRPWK +SRTV M+S +S++I PEGW EW + DTL + EY N+G G+G
Sbjct: 484 KGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFAL-DTLTYREYLNRGGGAG 542
Query: 476 VANRVKWRGYHVL-NNSQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
ANRVKW+GY V+ ++++A FT QFI G WL STG ++ L
Sbjct: 543 TANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFSLSL 587
>PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 581
Score = 351 bits (901), Expect = 2e-96
Identities = 191/464 (41%), Positives = 269/464 (57%), Gaps = 24/464 (5%)
Query: 70 KVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAV 129
+ N + D RL AI C +F A + L SISA G + S++ TW+SA
Sbjct: 128 RANAEQDARLQKAIDVCSSVFGDALDRLNDSISALGTVAGRIASSAS--VSNVETWLSAA 185
Query: 130 LSYQETCIDGFEGTNSNVKDHVSGGLDQVISLVKN-------------ELLLQVVPDSDP 176
L+ Q+TC+D NS G L ++ + ++N ++L + P
Sbjct: 186 LTDQDTCLDAVGELNSTA---ARGALQEIETAMRNSTEFASNSLAIVTKILGLLSRFETP 242
Query: 177 SNISNSGEFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRY 236
+ FP W+ A + +LL++ D VVA DGSG + T+ +A+K + + R+
Sbjct: 243 IHHRRLLGFPEWLGAAERRLLEEKNNDSTPDAVVAKDGSGQFKTIGEALKLVKKKSEERF 302
Query: 237 VIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGF 296
++VK+G Y E +++ K WN+MI GDG D T + G+R++ D TF TATFAV GF
Sbjct: 303 SVYVKEGRYVENIDLDKNTWNVMIYGDGKDKTFVVGSRNF-MDGTPTFETATFAVKGKGF 361
Query: 297 IARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTV 356
IA+DI F N AG QAVALRS SD SVF+RC G+QD+LYAHS RQFY++C I GT+
Sbjct: 362 IAKDIGFVNNAGASKHQAVALRSGSDRSVFFRCSFDGFQDTLYAHSNRQFYRDCDITGTI 421
Query: 357 DFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSN 416
DFI GNAA VFQ+C I P++PL +Q NTI AQ + PNQ +G TI P +N
Sbjct: 422 DFIFGNAAVVFQSCKIMPRQPLPNQFNTITAQGKKDPNQNTGIIIQKSTITP----FGNN 477
Query: 417 GSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGV 476
+ T+LGRPWK+FS TV M+S + +++P GW+ W N T+F+ EYQN GPG+ V
Sbjct: 478 LTAPTYLGRPWKDFSTTVIMQSDIGALLNPVGWMSWVPNVEPPTTIFYAEYQNSGPGADV 537
Query: 477 ANRVKWRGYH-VLNNSQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
+ RVKW GY + + A +FTV FI+G WLP+ V +++ L
Sbjct: 538 SQRVKWAGYKPTITDRNAEEFTVQSFIQGPEWLPNAAVQFDSTL 581
>PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 586
Score = 337 bits (865), Expect = 3e-92
Identities = 200/542 (36%), Positives = 294/542 (53%), Gaps = 62/542 (11%)
Query: 7 LTPTLILLLIMSL----QPCTT-----PSSASTITEFSGLSCLRVSPNRFIDSANKVINL 57
LTP+ L I S+ + C + PSS +T E + ++S ID + + +L
Sbjct: 67 LTPSTSLKAICSVTRFPESCISSISKLPSSNTTDPE----TLFKLSLKVIIDELDSISDL 122
Query: 58 LQNVSYNLSPFTKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGN 117
+ +S + D R+ +A+ C +L + A + L ++SA + + K +
Sbjct: 123 PEKLS---------KETEDERIKSALRVCGDLIEDALDRLNDTVSAIDDEEKKKTLSSSK 173
Query: 118 LSSDLRTWISAVLSYQETCIDGF--------EGTNSNVKDHVSGGLDQVISLVKNELLLQ 169
+ DL+TW+SA ++ ETC D E NS + ++ + + N L +
Sbjct: 174 IE-DLKTWLSATVTDHETCFDSLDELKQNKTEYANSTITQNLKSAMSRSTEFTSNSLAIV 232
Query: 170 VVPDSDPSNIS---------------NSGEFPSWVKAEDEKLLQQDAPPPPADVVVAADG 214
S S++ S +F W + +LLQ P DV VA DG
Sbjct: 233 SKILSALSDLGIPIHRRRRLMSHHHQQSVDFEKWAR---RRLLQTAGLKP--DVTVAGDG 287
Query: 215 SGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNR 274
+G+ TV +AV P+ ++ +VI+VK G Y E V + K KWN+MI GDG T ISG++
Sbjct: 288 TGDVLTVNEAVAKVPKKSLKMFVIYVKSGTYVENVVMDKSKWNVMIYGDGKGKTIISGSK 347
Query: 275 SYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGY 334
++ D T+ TATFA+ GFI +DI NTAG QAVA RS SD SV+Y+C G+
Sbjct: 348 NFV-DGTPTYETATFAIQGKGFIMKDIGIINTAGAAKHQAVAFRSGSDFSVYYQCSFDGF 406
Query: 335 QDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPN 394
QD+LY HS RQFY++C + GT+DFI G+AA VFQ C I P++PL +Q NTI AQ + PN
Sbjct: 407 QDTLYPHSNRQFYRDCDVTGTIDFIFGSAAVVFQGCKIMPRQPLSNQFNTITAQGKKDPN 466
Query: 395 QTSGFSFHNCTIIPDPDFQPSNGSI--QTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEW 452
Q+SG S CTI +NG++ T+LGRPWK FS TV ME+ + ++ P GW+ W
Sbjct: 467 QSSGMSIQRCTI-------SANGNVIAPTYLGRPWKEFSTTVIMETVIGAVVRPSGWMSW 519
Query: 453 ETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYH-VLNNSQAMDFTVNQFIKGDLWLPST 511
+ ++ +GEY+N GPGS V RVKW GY V+++++A FTV + G W+P+T
Sbjct: 520 VSGVDPPASIVYGEYKNTGPGSDVTQRVKWAGYKPVMSDAEAAKFTVATLLHGADWIPAT 579
Query: 512 GV 513
GV
Sbjct: 580 GV 581
>PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 319
Score = 317 bits (812), Expect = 5e-86
Identities = 162/314 (51%), Positives = 205/314 (64%), Gaps = 3/314 (0%)
Query: 207 DVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMD 266
+VVVAADGSG+Y TV +AV APE + TRYVI +K G+Y+E V++ KKK NIM +GDG
Sbjct: 8 NVVVAADGSGDYKTVSEAVAAAPEDSKTRYVIRIKAGVYRENVDVPKKKKNIMFLGDGRT 67
Query: 267 VTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVF 326
T I+ +++ D TTF +AT A GF+ARDITF+NTAG QAVALR SDLS F
Sbjct: 68 STIITASKNV-QDGSTTFNSATVAAVGAGFLARDITFQNTAGAAKHQAVALRVGSDLSAF 126
Query: 327 YRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIA 386
YRC I YQDSLY HS RQF+ C I GTVDFI GNAA V Q+C I ++P QKN +
Sbjct: 127 YRCDILAYQDSLYVHSNRQFFINCFIAGTVDFIFGNAAVVLQDCDIHARRPGSGQKNMVT 186
Query: 387 AQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHP 446
AQ RT PNQ +G I D QP S T+LGRPWK +SRTV M+S ++ +I+P
Sbjct: 187 AQGRTDPNQNTGIVIQKSRIGATSDLQPVQSSFPTYLGRPWKEYSRTVVMQSSITNVINP 246
Query: 447 EGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNS-QAMDFTVNQFIKGD 505
GW W+ N DTL++GEYQN G G+ + RV W+G+ V+ +S +A FT FI G
Sbjct: 247 AGWFPWDGNFAL-DTLYYGEYQNTGAGAATSGRVTWKGFKVITSSTEAQGFTPGSFIAGG 305
Query: 506 LWLPSTGVSYNAGL 519
WL +T ++ GL
Sbjct: 306 SWLKATTFPFSLGL 319
>PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 374
Score = 293 bits (750), Expect = 7e-79
Identities = 143/332 (43%), Positives = 200/332 (60%), Gaps = 5/332 (1%)
Query: 191 AEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVE 250
A +LLQ P A VA DGSG Y T+ +A+ P+ N ++I +K G+YKEY++
Sbjct: 44 ATARRLLQISNAKPNA--TVALDGSGQYKTIKEALDAVPKKNTEPFIIFIKAGVYKEYID 101
Query: 251 IKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPE 310
I K N++++G+G T I+GN+S D +TF T T V+ F+A++I FENTAGPE
Sbjct: 102 IPKSMTNVVLIGEGPTKTKITGNKSV-KDGPSTFHTTTVGVNGANFVAKNIGFENTAGPE 160
Query: 311 NKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNC 370
+QAVALR +D ++ Y C I GYQD+LY H+ RQFY++C I GTVDFI GN AV QNC
Sbjct: 161 KEQAVALRVSADKAIIYNCQIDGYQDTLYVHTYRQFYRDCTITGTVDFIFGNGEAVLQNC 220
Query: 371 LIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNF 430
+ +KP ++Q + AQ RT P Q NC I PD D+ + +T+LGRPWK +
Sbjct: 221 KVIVRKPAQNQSCMVTAQGRTEPIQKGAIVLQNCEIKPDTDYFSLSPPSKTYLGRPWKEY 280
Query: 431 SRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHV-LN 489
SRT+ M+S++ + I PEGW W DT ++ EYQN+GPG+ + R+ W+G+
Sbjct: 281 SRTIIMQSYIDKFIEPEGWAPWNITNFGRDTSYYAEYQNRGPGAALDKRITWKGFQKGFT 340
Query: 490 NSQAMDFTVNQFIKGD-LWLPSTGVSYNAGLL 520
A FT +I D WL V Y AG++
Sbjct: 341 GEAAQKFTAGVYINNDENWLQKANVPYEAGMM 372
>PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE) (P65)
Length = 447
Score = 285 bits (730), Expect = 2e-76
Identities = 160/450 (35%), Positives = 240/450 (52%), Gaps = 25/450 (5%)
Query: 86 CLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFEGTNS 145
C E+ D A + + S+ +K + DL+ W++ LS+Q+TC+DGF T +
Sbjct: 4 CNEVLDYAVDGIHKSVGTLDQFDFHK---LSEYAFDLKVWLTGTLSHQQTCLDGFANTTT 60
Query: 146 NVKDHVSGGLDQVISLVKNEL-----LLQVVPDSDPSNISNSGEF------PSWVKAEDE 194
+ ++ L + L N + + +++ D S S S PSWV
Sbjct: 61 KAGETMTKVLKTSMELSSNAIDMMDAVSRILKGFDTSQYSVSRRLLSDDGIPSWVNDGHR 120
Query: 195 KLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKK 254
+LL P A VVA DGSG + T+ DA+K P N +VIHVK G+YKE V + K+
Sbjct: 121 RLLAGGNVQPNA--VVAQDGSGQFKTLTDALKTVPPKNAVPFVIHVKAGVYKETVNVAKE 178
Query: 255 KWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQA 314
+ ++GDG T +G+ +Y D I T+ TATF V+ F+A+DI FENTAG QA
Sbjct: 179 MNYVTVIGDGPTKTKFTGSLNYA-DGINTYNTATFGVNGANFMAKDIGFENTAGTGKHQA 237
Query: 315 VALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQP 374
VALR +D ++FY C + G+QD+LY SQRQFY++C I GT+DF+ G VFQNC +
Sbjct: 238 VALRVTADQAIFYNCQMDGFQDTLYVQSQRQFYRDCSISGTIDFVFGERFGVFQNCKLVC 297
Query: 375 KKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTV 434
+ P K Q+ + A R N S F + +P + ++LGRPWK +S+ V
Sbjct: 298 RLPAKGQQCLVTAGGREKQNSASALVFQSSHFTGEPALTSVTPKV-SYLGRPWKQYSKVV 356
Query: 435 FMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNSQAM 494
M+S + + PEG++ W + + +T + EY NKGPG+ RVKW G VL ++ A
Sbjct: 357 IMDSTIDAIFVPEGYMPW-MGSAFKETCTYYEYNNKGPGADTNLRVKWHGVKVLTSNVAA 415
Query: 495 DFTVNQFIK------GDLWLPSTGVSYNAG 518
++ +F + D W+ +GV Y+ G
Sbjct: 416 EYYPGKFFEIVNATARDTWIVKSGVPYSLG 445
>PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 238 bits (608), Expect = 2e-62
Identities = 159/488 (32%), Positives = 232/488 (46%), Gaps = 49/488 (10%)
Query: 75 NDNRLSNAISD-CLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQ 133
N N S A+ D C + A E+L+ + G +G+ L+ W++ V +YQ
Sbjct: 103 NINATSKAVLDYCKRVLMYALEDLETIVE----EMGEDLQQSGSKMDQLKQWLTGVFNYQ 158
Query: 134 ETCIDGFEGTN---------SNVKDHVSGGLD--------------QVISLVKNELLLQV 170
CID E + ++ K S +D +V + K L
Sbjct: 159 TDCIDDIEESELRKVMGEGIAHSKILSSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETP 218
Query: 171 VPDSDPSNISNSGEFPSWVKAEDEKLLQQDAPP-PPADV----------------VVAAD 213
PD D + P W +D KL+ Q P PAD VVA D
Sbjct: 219 APDRDLLEDLDQKGLPKWHSDKDRKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKD 278
Query: 214 GSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGN 273
GSG + T+ +AVK PE N R +I++K G+YKE V I KK N+ + GDG T I+ +
Sbjct: 279 GSGQFKTISEAVKACPEKNPGRCIIYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFD 338
Query: 274 RSYG-HDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIK 332
RS G TT + T V + GF+A+ I F+NTAGP QAVA R + D +V + C
Sbjct: 339 RSVGLSPGTTTSLSGTVQVESEGFMAKWIGFQNTAGPLGHQAVAFRVNGDRAVIFNCRFD 398
Query: 333 GYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTH 392
GYQD+LY ++ RQFY+ + GTVDFI G +A V QN LI +K Q N + A
Sbjct: 399 GYQDTLYVNNGRQFYRNIVVSGTVDFIFGKSATVIQNSLILCRKGSPGQTNHVTADGNEK 458
Query: 393 PNQTS-GFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLE 451
G HNC I+ D + + +++++LGRPWK F+ T + + + ++I P GW E
Sbjct: 459 GKAVKIGIVLHNCRIMADKELEADRLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNE 518
Query: 452 WETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNSQAMDFTVNQFIKGDLWLPST 511
W+ + T + E+ N+GPG+ A RV W + ++ FTV ++ W+
Sbjct: 519 WQ-GEKFHLTATYVEFNNRGPGANTAARVPW-AKMAKSAAEVERFTVANWLTPANWIQEA 576
Query: 512 GVSYNAGL 519
V GL
Sbjct: 577 NVPVQLGL 584
>PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE) (Fragment)
Length = 571
Score = 238 bits (608), Expect = 2e-62
Identities = 159/488 (32%), Positives = 233/488 (47%), Gaps = 49/488 (10%)
Query: 75 NDNRLSNAISD-CLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQ 133
N N S A+ D C + A E+L+ + G +G+ L+ W++ V +YQ
Sbjct: 90 NMNATSKAVLDYCKRVLMYALEDLETIVE----EMGEDLQQSGSKMDQLKQWLTGVFNYQ 145
Query: 134 ETCIDGFEGTN---------SNVKDHVSGGLD--------------QVISLVKNELLLQV 170
CID E + + K S +D +V + K L
Sbjct: 146 TDCIDDIEESELRKVMGEGIRHSKILSSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETP 205
Query: 171 VPDSDPSNISNSGEFPSWVKAEDEKLL-QQDAPPPPADV----------------VVAAD 213
PD D + P W +D KL+ Q P PAD VVA D
Sbjct: 206 APDRDLLEDLDQKGLPKWHSDKDRKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKD 265
Query: 214 GSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGN 273
GSG + T+ +AVK PE N R +I++K G+YKE V I KK N+ + GDG T I+ +
Sbjct: 266 GSGQFKTISEAVKACPEKNPGRCIIYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFD 325
Query: 274 RSYG-HDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIK 332
RS G TT + T V + GF+A+ I F+NTAGP QAVA R + D +V + C
Sbjct: 326 RSVGLSPGTTTSLSGTVQVESEGFMAKWIGFQNTAGPLGNQAVAFRVNGDRAVIFNCRFD 385
Query: 333 GYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTH 392
GYQD+LY ++ RQFY+ + GTVDFI G +A V QN LI +K Q N + A +
Sbjct: 386 GYQDTLYVNNGRQFYRNIVVSGTVDFINGKSATVIQNSLILCRKGSPGQTNHVTADGKQK 445
Query: 393 PNQTS-GFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLE 451
G HNC I+ D + + +++++LGRPWK F+ T + + + ++I P GW E
Sbjct: 446 GKAVKIGIVLHNCRIMADKELEADRLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNE 505
Query: 452 WETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNSQAMDFTVNQFIKGDLWLPST 511
W+ + T + E+ N+GPG+ A RV W + ++ FTV ++ W+
Sbjct: 506 WQ-GEKFHLTATYVEFNNRGPGANPAARVPW-AKMAKSAAEVERFTVANWLTPANWIQEA 563
Query: 512 GVSYNAGL 519
V+ GL
Sbjct: 564 NVTVQLGL 571
>PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 135 bits (341), Expect = 2e-31
Identities = 88/278 (31%), Positives = 133/278 (47%), Gaps = 12/278 (4%)
Query: 205 PADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGD- 263
P+ +V A G+YTT+ DA+ I +++G Y E V + ++I G
Sbjct: 24 PSGAIVVAKSGGDYTTIGDAIDALSTSTTDTQTIFIEEGTYDEQVYLPAMTGKVIIYGQT 83
Query: 264 ------GMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVAL 317
++ TI+ SY + TATF A+G ++ NT G QA+AL
Sbjct: 84 ENTDSYADNLVTITHAISYEDAGESDDLTATFRNKAVGSQVYNLNIANTCGQACHQALAL 143
Query: 318 RSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICG-NAAAVFQNCLIQPKK 376
+ +D +Y C GYQD+L A + Q Y I G VDFI G +A A FQN I+ +
Sbjct: 144 SAWADQQGYYGCNFTGYQDTLLAQTGNQLYINSYIEGAVDFIFGQHARAWFQNVDIRVVE 203
Query: 377 PLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFM 436
+I A R+ TS + + T+ + G+ +LGRPW ++R VF
Sbjct: 204 G--PTSASITANGRSSETDTSYYVINKSTVAAKEGDDVAEGTY--YLGRPWSEYARVVFQ 259
Query: 437 ESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGS 474
++ M+ +I+ GW EW T+T + + FGEY N G GS
Sbjct: 260 QTSMTNVINSLGWTEWSTSTPNTEYVTFGEYANTGAGS 297
>PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 120 bits (301), Expect = 9e-27
Identities = 83/277 (29%), Positives = 128/277 (45%), Gaps = 11/277 (3%)
Query: 205 PADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDG 264
P+ +V A G+Y T+ AV + I +++G Y E V I +++ G
Sbjct: 24 PSGAIVVAKSGGDYDTISAAVDALSTTSTETQTIFIEEGSYDEQVYIPALSGKLIVYGQT 83
Query: 265 MDVTTISGNR-SYGH-----DNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALR 318
D TT + N + H D TAT A G ++ NT G QA+A+
Sbjct: 84 EDTTTYTSNLVNITHAIALADVDNDDETATLRNYAEGSAIYNLNIANTCGQACHQALAVS 143
Query: 319 SDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICG-NAAAVFQNCLIQPKKP 377
+ + +Y C GYQD+L A + Q Y I G VDFI G +A A F C I+ +
Sbjct: 144 AYASEQGYYACQFTGYQDTLLAETGYQVYAGTYIEGAVDFIFGQHARAWFHECDIRVLEG 203
Query: 378 LKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFME 437
+I A R+ + S + H T+ S+G+ +LGRPW ++R F +
Sbjct: 204 --PSSASITANGRSSESDDSYYVIHKSTVAAADGNDVSSGTY--YLGRPWSQYARVCFQK 259
Query: 438 SFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGS 474
+ M+++I+ GW EW T+T + + F EY N G G+
Sbjct: 260 TSMTDVINHLGWTEWSTSTPNTENVTFVEYGNTGTGA 296
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,793,220
Number of Sequences: 164201
Number of extensions: 2847883
Number of successful extensions: 7036
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 6907
Number of HSP's gapped (non-prelim): 47
length of query: 521
length of database: 59,974,054
effective HSP length: 115
effective length of query: 406
effective length of database: 41,090,939
effective search space: 16682921234
effective search space used: 16682921234
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0343.3


