

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0327.19
(490 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
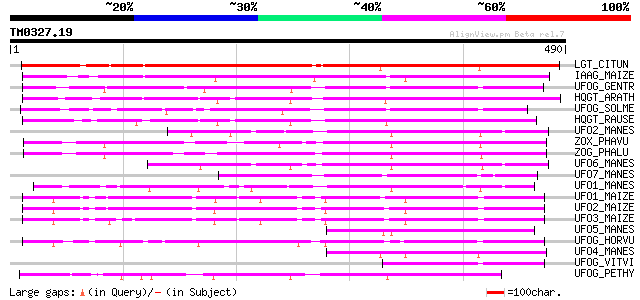
Score E
Sequences producing significant alignments: (bits) Value
LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.21... 395 e-109
IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (E... 265 3e-70
UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 188 3e-47
HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (E... 186 9e-47
UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 170 9e-42
HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.2... 169 1e-41
UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC 2.4.1... 164 4e-40
ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40) (Ze... 163 8e-40
ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203) (... 162 2e-39
UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC 2.4.1... 157 6e-38
UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC 2.4.1... 155 2e-37
UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC 2.4.1... 154 4e-37
UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 153 8e-37
UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 148 4e-35
UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 145 2e-34
UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC 2.4.1... 140 6e-33
UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 140 1e-32
UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC 2.4.1... 127 7e-29
UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 120 8e-27
UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.9... 115 2e-25
>LGT_CITUN (Q9MB73) Limonoid UDP-glucosyltransferase (EC 2.4.1.210)
(Limonoid glucosyltransferase) (Limonoid GTase) (LGTase)
Length = 511
Score = 395 bits (1015), Expect = e-109
Identities = 205/480 (42%), Positives = 303/480 (62%), Gaps = 16/480 (3%)
Query: 11 LHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTN 70
+HV+LV+F GH+NPLLRLG+ L ++G +TL T E ++ K+ + T PT
Sbjct: 7 VHVLLVSFPGHGHVNPLLRLGRLLASKGFFLTLTTPESFGKQMRKAGNFTYE-----PTP 61
Query: 71 FTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACII 130
I FF DG D + D + D YMA +E +G + +IK + ++C+I
Sbjct: 62 VGDGFIRFEFFEDGWDED-DPRREDLDQYMAQLELIGKQVIPKIIKKS-AEEYRPVSCLI 119
Query: 131 NNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPL 190
NNPF+PWV+DVA +P A LW+Q CA +A YYH+++ FP+ + PE++VQLP +PL
Sbjct: 120 NNPFIPWVSDVAESLGLPSAMLWVQSCACFAAYYHYFHGLVPFPSEKEPEIDVQLPCMPL 179
Query: 191 LKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIP 250
LK ++PSF+ P+ P+ L + + +++ K +L ++FYELEKE+ID MA+ P+ P
Sbjct: 180 LKHDEMPSFLHPSTPYPFLRRAILGQYENLGKPFCILLDTFYELEKEIIDYMAKICPIKP 239
Query: 251 VGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLESI 310
VGPL D + KP D C++WL+ +PPSSV+YISFG+++ L +++E I
Sbjct: 240 VGPLFKNPKAPTLTVRD---DCMKP-DECIDWLDKKPPSSVVYISFGTVVYLKQEQVEEI 295
Query: 311 ATALKNSNCKFLWVIK---KQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIAC 367
AL NS FLWV+K + G + LP GF E+ ++G VV W PQ KVL HP++AC
Sbjct: 296 GYALLNSGISFLWVMKPPPEDSGVKIVDLPDGFLEKVGDKGKVVQWSPQEKVLAHPSVAC 355
Query: 368 FLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRL--EQDSDGFVETGE 425
F+THCGWNS +E++A+G P+I +PQW DQ T+A + DVF+ G+RL + + + E
Sbjct: 356 FVTHCGWNSTMESLASGVPVITFPQWGDQVTDAMYLCDVFKTGLRLCRGEAENRIISRDE 415
Query: 426 LERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEILATRLDVMESS 485
+E+ + E +GPK+ L+ NA + K+ A EAVADGGSSDRNIQ FVDE+ T ++++ SS
Sbjct: 416 VEKCLLEATAGPKAVALEENALKWKKEAEEAVADGGSSDRNIQAFVDEVRRTSVEIITSS 475
>IAAG_MAIZE (Q41819) Indole-3-acetate beta-glucosyltransferase (EC
2.4.1.121) (IAA-Glu synthetase) ((Uridine
5'-diphosphate-glucose:indol-3-ylacetyl)-beta-D-glucosyl
transferase)
Length = 471
Score = 265 bits (676), Expect = 3e-70
Identities = 162/483 (33%), Positives = 238/483 (48%), Gaps = 38/483 (7%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNF 71
HV++V F QGH+NP+++ K L ++G+ TL TT + A D P
Sbjct: 4 HVLVVPFPGQGHMNPMVQFAKRLASKGVATTLVTTRFIQR---------TADVDAHPAM- 53
Query: 72 TTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACIIN 131
V SDG D + +Y+ +L++L++ +++ C++
Sbjct: 54 ------VEAISDGHDEGGFASAAGVAEYLEKQAAAASASLASLVEAR-ASSADAFTCVVY 106
Query: 132 NPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPE----------L 181
+ + WV VA +P Q CA+ A+YYHF P + L
Sbjct: 107 DSYEDWVLPVARRMGLPAVPFSTQSCAVSAVYYHFSQGRLAVPPGAAADGSDGGAGAAAL 166
Query: 182 NVQLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDS 241
+ GLP ++ +LPSF+ P+ ++ K WVL NSF ELE EV+
Sbjct: 167 SEAFLGLPEMERSELPSFVFDHGPYPTIAMQAIKQFAHAGKDDWVLFNSFEELETEVLAG 226
Query: 242 MAETYPVIPVGPLLPLSLLGVDENED----VGIEMWKPQDSCLEWLNDQPPSSVIYISFG 297
+ + +GP +PL G + G + KP+D+C +WL+ +P SV Y+SFG
Sbjct: 227 LTKYLKARAIGPCVPLPTAGRTAGANGRITYGANLVKPEDACTKWLDTKPDRSVAYVSFG 286
Query: 298 SLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRG--MVVPWCP 355
SL L + E +A L + FLWV++ D +P+ E G MVVPWCP
Sbjct: 287 SLASLGNAQKEELARGLLAAGKPFLWVVRASDEHQ---VPRYLLAEATATGAAMVVPWCP 343
Query: 356 QTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQ 415
Q VL HPA+ CF+THCGWNS LEA++ G PM+A WTDQPTNA+ V + G+R +
Sbjct: 344 QLDVLAHPAVGCFVTHCGWNSTLEALSFGVPMVAMALWTDQPTNARNVELAWGAGVRARR 403
Query: 416 DSD-GFVETGELERAIEEIVSGPKSEELKRNAA-ELKRAAREAVADGGSSDRNIQTFVDE 473
D+ G GE+ER + ++ G ++ R AA E + AR AVA GGSSDRN+ FV
Sbjct: 404 DAGAGVFLRGEVERCVRAVMDGGEAASAARKAAGEWRDRARAAVAPGGSSDRNLDEFVQF 463
Query: 474 ILA 476
+ A
Sbjct: 464 VRA 466
>UFOG_GENTR (Q96493) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 453
Score = 188 bits (477), Expect = 3e-47
Identities = 134/472 (28%), Positives = 228/472 (47%), Gaps = 44/472 (9%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAA-TTDTIPTN 70
HV ++AF H PLL L L A + +F ST+++ TT PTN
Sbjct: 6 HVAVLAFPFGTHAAPLLTLVNRLAASAPDI-----------IFSFFSTSSSITTIFSPTN 54
Query: 71 FTTNGIHVLFFS--DGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLAC 128
+ G ++ ++ DG P P + + N P N +K + ++C
Sbjct: 55 LISIGSNIKPYAVWDG-SPEGFVFSGNPREPIEYFLNAAPDNFDKAMKKAVEDTGVNISC 113
Query: 129 IINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNT-----NQFPTLENPELNV 183
++ + F+ + AD + + +P +W A ++ H Y + +F E E +
Sbjct: 114 LLTDAFLWFAADFSEKIGVPWIPVWT--AASCSLCLHVYTDEIRSRFAEFDIAEKAEKTI 171
Query: 184 Q-LPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSM 242
+PGL + DLP ++ + + L +M + K V NSF E++ + + +
Sbjct: 172 DFIPGLSAISFSDLPEELIMEDSQSIFALTLHNMGLKLHKATAVAVNSFEEIDPIITNHL 231
Query: 243 AETYP--VIPVGPLLPLSL-LGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSL 299
T ++ +GPL LS + ++NE CL+WL Q SSV+Y+SFG++
Sbjct: 232 RSTNQLNILNIGPLQTLSSSIPPEDNE------------CLKWLQTQKESSVVYLSFGTV 279
Query: 300 IVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKV 359
I ++ ++A+ L++ FLW ++ + K LP+ F + T G +V W PQ V
Sbjct: 280 INPPPNEMAALASTLESRKIPFLWSLRDEARKH---LPENFIDRTSTFGKIVSWAPQLHV 336
Query: 360 LVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDG 419
L +PAI F+THCGWNS LE+I P+I P + DQ NA++V DV+++G+ ++ G
Sbjct: 337 LENPAIGVFVTHCGWNSTLESIFCRVPVIGRPFFGDQKVNARMVEDVWKIGVGVK---GG 393
Query: 420 FVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFV 471
E R +E ++ K +E+++N LK A++AV GSS RN ++ +
Sbjct: 394 VFTEDETTRVLELVLFSDKGKEMRQNVGRLKEKAKDAVKANGSSTRNFESLL 445
>HQGT_ARATH (Q9M156) Probable hydroquinone glucosyltransferase (EC
2.4.1.218) (Arbutin synthase)
Length = 480
Score = 186 bits (473), Expect = 9e-47
Identities = 141/497 (28%), Positives = 231/497 (46%), Gaps = 47/497 (9%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLL-ARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTN 70
HV ++ GH+ PL+ K L+ GL VT V S D++P++
Sbjct: 8 HVAIIPSPGMGHLIPLVEFAKRLVHLHGLTVTF-----VIAGEGPPSKAQRTVLDSLPSS 62
Query: 71 FTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACII 130
++ F +D + + ++L L + + F+ + ++
Sbjct: 63 ISS------VFLPPVDLTDLSSSTRIESRISLTVTRSNPELRKVFDS-FVEGGRLPTALV 115
Query: 131 NNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELN--VQLPGL 188
+ F DVA EF +P + + + + H ++ + E EL + LPG
Sbjct: 116 VDLFGTDAFDVAVEFHVPPYIFYPTTANVLSFFLHL-PKLDETVSCEFRELTEPLMLPGC 174
Query: 189 PLLKPQDLPSFILPTNPF-GALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETY- 246
+ +D F+ P K L K K+ + +L N+F+ELE I ++ E
Sbjct: 175 VPVAGKD---FLDPAQDRKDDAYKWLLHNTKRYKEAEGILVNTFFELEPNAIKALQEPGL 231
Query: 247 ---PVIPVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLS 303
PV PVGPL+ + + E+ CL+WL++QP SV+Y+SFGS L+
Sbjct: 232 DKPPVYPVGPLVNIGKQEAKQTEE---------SECLKWLDNQPLGSVLYVSFGSGGTLT 282
Query: 304 AKKLESIATALKNSNCKFLWVIKKQDG------------KDSLP-LPQGFKEETKNRGMV 350
++L +A L +S +FLWVI+ G D L LP GF E TK RG V
Sbjct: 283 CEQLNELALGLADSEQRFLWVIRSPSGIANSSYFDSHSQTDPLTFLPPGFLERTKKRGFV 342
Query: 351 VP-WCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRV 409
+P W PQ +VL HP+ FLTHCGWNS LE++ +G P+IA+P + +Q NA L+S+ R
Sbjct: 343 IPFWAPQAQVLAHPSTGGFLTHCGWNSTLESVVSGIPLIAWPLYAEQKMNAVLLSEDIRA 402
Query: 410 GIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQT 469
+R DG V E+ R ++ ++ G + + ++ ELK AA + D G+S + +
Sbjct: 403 ALRPRAGDDGLVRREEVARVVKGLMEGEEGKGVRNKMKELKEAACRVLKDDGTSTKALSL 462
Query: 470 FVDEILATRLDVMESSN 486
+ A + ++ ++ N
Sbjct: 463 VALKWKAHKKELEQNGN 479
>UFOG_SOLME (Q43641) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
Length = 433
Score = 170 bits (430), Expect = 9e-42
Identities = 125/455 (27%), Positives = 214/455 (46%), Gaps = 46/455 (10%)
Query: 10 ELHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPT 69
+LH+ +AF H PLL L + + T+ + F +SS+ ++ +P
Sbjct: 5 QLHIAFLAFPFGTHATPLLTLVQKISPFLPSSTIFS-------FFNTSSSNSSIFSKVPN 57
Query: 70 NFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENL-GPINLSTLIKTHFINNSKKLAC 128
I + DG+ D TP A+ + + +S + + K +C
Sbjct: 58 Q---ENIKIYNVWDGVKEGND----TPFGLEAIKLFIQSTLLISKITEEAEEETGVKFSC 110
Query: 129 IINNPFVPW--VADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLP 186
I ++ F+ W + + + P W A+ H Y + + N E ++++P
Sbjct: 111 IFSDAFL-WCFLVKLPKKMNAPGVAYWTGGSCSLAV--HLYTDLIR----SNKETSLKIP 163
Query: 187 GLP-LLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVI---DSM 242
G L D+P + + G +S +L +M ++ K V+ NSF EL+++ + D
Sbjct: 164 GFSSTLSINDIPPEVTAEDLEGPMSSMLYNMALNLHKADAVVLNSFQELDRDPLINKDLQ 223
Query: 243 AETYPVIPVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVL 302
V +GPL+ S +DE+ C++WL+ Q SV+Y+SFG++ L
Sbjct: 224 KNLQKVFNIGPLVLQSSRKLDES------------GCIQWLDKQKEKSVVYLSFGTVTTL 271
Query: 303 SAKKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVH 362
++ SIA AL+ F+W ++ K+ LP+GF E TK G +V W PQ ++L H
Sbjct: 272 PPNEIGSIAEALETKKTPFIWSLRNNGVKN---LPKGFLERTKEFGKIVSWAPQLEILAH 328
Query: 363 PAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVE 422
++ F+THCGWNS+LE I+ G PMI P + DQ N+++V V+ +G+++E G
Sbjct: 329 KSVGVFVTHCGWNSILEGISFGVPMICRPFFGDQKLNSRMVESVWEIGLQIE---GGIFT 385
Query: 423 TGELERAIEEIVSGPKSEELKRNAAELKRAAREAV 457
+ A++ + K + L+ N LK A EAV
Sbjct: 386 KSGIISALDTFFNEEKGKILRENVEGLKEKALEAV 420
>HQGT_RAUSE (Q9AR73) Hydroquinone glucosyltransferase (EC 2.4.1.218)
(Arbutin synthase)
Length = 470
Score = 169 bits (428), Expect = 1e-41
Identities = 136/481 (28%), Positives = 232/481 (47%), Gaps = 59/481 (12%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLAR-GLQVT-LATTELVYHRVFKSSSTTAATTDTIPT 69
H+ +V GH+ PL+ K L+ R VT + T+ + KS D +P
Sbjct: 6 HIAMVPTPGMGHLIPLVEFAKRLVLRHNFGVTFIIPTDGPLPKAQKSF------LDALPA 59
Query: 70 NFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPIN--LSTLIKTHFINNSKKLA 127
+VL D P ++ + + +L + + TL+ T KLA
Sbjct: 60 GVN----YVLLPPVSFDDLPADVRIETRICLTITRSLPFVRDAVKTLLAT------TKLA 109
Query: 128 CIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELN--VQL 185
++ + F DVA EFK+ + ++++H +Q + E ++ +Q+
Sbjct: 110 ALVVDLFGTDAFDVAIEFKVSPYIFYPTTAMCLSLFFHL-PKLDQMVSCEYRDVPEPLQI 168
Query: 186 PGLPLLKPQDLPSFILPTNPF-GALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAE 244
PG + +D F+ P K L K + + ++ N+F +LE + ++ E
Sbjct: 169 PGCIPIHGKD---FLDPAQDRKNDAYKCLLHQAKRYRLAEGIMVNTFNDLEPGPLKALQE 225
Query: 245 TY----PVIPVGPLLPL-SLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSL 299
PV P+GPL+ S VD+ E CL+WL+DQP SV++ISFGS
Sbjct: 226 EDQGKPPVYPIGPLIRADSSSKVDDCE------------CLKWLDDQPRGSVLFISFGSG 273
Query: 300 IVLSAKKLESIATALKNSNCKFLWVIKKQDGK-------------DSLP-LPQGFKEETK 345
+S + +A L+ S +FLWV++ + K D+L LP+GF E TK
Sbjct: 274 GAVSHNQFIELALGLEMSEQRFLWVVRSPNDKIANATYFSIQNQNDALAYLPEGFLERTK 333
Query: 346 NRGMVVP-WCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVS 404
R ++VP W PQT++L H + FLTHCGWNS+LE++ G P+IA+P + +Q NA +++
Sbjct: 334 GRCLLVPSWAPQTEILSHGSTGGFLTHCGWNSILESVVNGVPLIAWPLYAEQKMNAVMLT 393
Query: 405 DVFRVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSD 464
+ +V +R + +G + E+ A++ ++ G + ++ + +LK AA A++D GSS
Sbjct: 394 EGLKVALRPKAGENGLIGRVEIANAVKGLMEGEEGKKFRSTMKDLKDAASRALSDDGSST 453
Query: 465 R 465
+
Sbjct: 454 K 454
>UFO2_MANES (Q40285) Flavonol 3-O-glucosyltransferase 2 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
2) (Fragment)
Length = 346
Score = 164 bits (416), Expect = 4e-40
Identities = 107/356 (30%), Positives = 185/356 (51%), Gaps = 38/356 (10%)
Query: 140 DVAAEFKIPCACLWIQPCAL--YAIYYHFYNNTNQFPTLENPELNVQLPGLPLLKP---Q 194
D+A EF IP + + +Y ++ F +E + + +L L+ P +
Sbjct: 7 DLADEFGIPSYIFFASGGGFLGFMLYVQKIHDEENFNPIEFKDSDTELIVPSLVNPFPTR 66
Query: 195 DLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIPVGPL 254
LPS IL FG L + K ++ K ++ N+F ELE I+S + P+ VGP+
Sbjct: 67 ILPSSILNKERFGQLLAIA----KKFRQAKGIIVNTFLELESRAIESF-KVPPLYHVGPI 121
Query: 255 LPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLESIATAL 314
L + G + + ++ ++WL+DQP SV+++ FGS+ S +L+ IA AL
Sbjct: 122 LDVKSDGRNTHPEI-----------MQWLDDQPEGSVVFLCFGSMGSFSEDQLKEIAYAL 170
Query: 315 KNSNCKFLWVIKKQDGKDSLP-----------LPQGFKEETKNRGMVVPWCPQTKVLVHP 363
+NS +FLW I++ D + LP+GF E T G V+ W PQ VL HP
Sbjct: 171 ENSGHRFLWSIRRPPPPDKIASPTDYEDPRDVLPEGFLERTVAVGKVIGWAPQVAVLAHP 230
Query: 364 AIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLE----QDSDG 419
AI F++HCGWNS+LE++ G P+ +P + +Q NA + +G+ ++ ++S
Sbjct: 231 AIGGFVSHCGWNSVLESLWFGVPIATWPMYAEQQFNAFEMVVELGLGVEIDMGYRKESGI 290
Query: 420 FVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEIL 475
V + ++ERAI +++ S+E ++ E++ ++ A+ DGGSS ++ F+ + +
Sbjct: 291 IVNSDKIERAIRKLME--NSDEKRKKVKEMREKSKMALIDGGSSFISLGDFIKDAM 344
>ZOX_PHAVU (P56725) Zeatin O-xylosyltransferase (EC 2.4.2.40)
(Zeatin O-beta-D-xylosyltransferase)
Length = 454
Score = 163 bits (413), Expect = 8e-40
Identities = 124/485 (25%), Positives = 212/485 (43%), Gaps = 66/485 (13%)
Query: 13 VMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNFT 72
V+L+ F QGH+NP L+L + A+ + V T T + +
Sbjct: 11 VLLLPFPVQGHLNPFLQLSHLIAAQNIAVHYVGT------------VTHIRQAKLRYHNA 58
Query: 73 TNGIHVLFFS--DGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACII 130
T+ IH F + P P+ P + E + +++ K +I
Sbjct: 59 TSNIHFHAFEVPPYVSPPPNPEDDFPSHLIPSFEASAHLREPVGKLLQSLSSQAKRVVLI 118
Query: 131 NNPFVPWVADVAAEFK-IPCACLWIQPCALYAIYYHFYNNTNQF-PTLENPEL-NVQLPG 187
N+ + VA AA F + C + + N F + P L + P
Sbjct: 119 NDSLMASVAQDAANFSNVERYCFQV---------FSALNTAGDFWEQMGKPPLADFHFPD 169
Query: 188 LPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEK-------EVID 240
+P L+ G +S D L + + Y + E+++
Sbjct: 170 IPSLQ--------------GCISAQFTDFLTAQNEFRKFNNGDIYNTSRVIEGPYVELLE 215
Query: 241 SMAETYPVIPVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLI 300
V +GP PL+ V++ + +G C+EWL+ Q PSSVIY+SFG+
Sbjct: 216 RFNGGKEVWALGPFTPLA---VEKKDSIGFS-----HPCMEWLDKQEPSSVIYVSFGTTT 267
Query: 301 VLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLP--------LPQGFKEETKNRGMVV- 351
L ++++ +AT L+ S KF+WV++ D D LP+GF+E + G+VV
Sbjct: 268 ALRDEQIQELATGLEQSKQKFIWVLRDADKGDIFDGSEAKRYELPEGFEERVEGMGLVVR 327
Query: 352 PWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGI 411
W PQ ++L H + F++HCGWNS LE++ G PM + +DQP NA LV+DV +VG+
Sbjct: 328 DWAPQMEILSHSSTGGFMSHCGWNSCLESLTRGVPMATWAMHSDQPRNAVLVTDVLKVGL 387
Query: 412 RLE--QDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQT 469
++ + V +E A+ ++ + +E+++ A +LK ++ +GG S + +
Sbjct: 388 IVKDWEQRKSLVSASVIENAVRRLMETKEGDEIRKRAVKLKDEIHRSMDEGGVSRMEMAS 447
Query: 470 FVDEI 474
F+ I
Sbjct: 448 FIAHI 452
>ZOG_PHALU (Q9ZSK5) Zeatin O-glucosyltransferase (EC 2.4.1.203)
(Trans-zeatin O-beta-D-glucosyltransferase)
Length = 459
Score = 162 bits (409), Expect = 2e-39
Identities = 125/477 (26%), Positives = 212/477 (44%), Gaps = 50/477 (10%)
Query: 13 VMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNFT 72
V+L+ F AQGH+N L L + ++A+ + V T T T+ N
Sbjct: 16 VLLIPFPAQGHLNQFLHLSRLIVAQNIPVHYVGT------------VTHIRQATLRYNNP 63
Query: 73 TNGIHVLFFS--DGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACII 130
T+ IH F + P P+ P + E + +++ K +I
Sbjct: 64 TSNIHFHAFQVPPFVSPPPNPEDDFPSHLIPSFEASAHLREPVGKLLQSLSSQAKRVVVI 123
Query: 131 NNPFVPWVADVAAEFKIPCACLWIQPCALYAIY-YHFYNNTNQFPTLENPELNVQLPGLP 189
N+ + VA AA I Y + + +N + F + G P
Sbjct: 124 NDSLMASVAQDAAN---------ISNVENYTFHSFSAFNTSGDF---------WEEMGKP 165
Query: 190 LLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLK-WVLANSFYELEKEVIDSMAETYPV 248
+ P F A K + +K + N+ +E ++ +
Sbjct: 166 PVGDFHFPEFPSLEGCIAAQFKGFRTAQYEFRKFNNGDIYNTSRVIEGPYVELLELFNGG 225
Query: 249 IPVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLE 308
V L P + L V++ + +G + C+EWL+ Q PSSVIYISFG+ L ++++
Sbjct: 226 KKVWALGPFNPLAVEKKDSIGF-----RHPCMEWLDKQEPSSVIYISFGTTTALRDEQIQ 280
Query: 309 SIATALKNSNCKFLWVIKKQDGKDSLP--------LPQGFKEETKNRGMVV-PWCPQTKV 359
IAT L+ S KF+WV+++ D D LP+GF+E + G+VV W PQ ++
Sbjct: 281 QIATGLEQSKQKFIWVLREADKGDIFAGSEAKRYELPKGFEERVEGMGLVVRDWAPQLEI 340
Query: 360 LVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQ--DS 417
L H + F++HCGWNS LE+I G P+ +P +DQP NA LV++V +VG+ ++
Sbjct: 341 LSHSSTGGFMSHCGWNSCLESITMGVPIATWPMHSDQPRNAVLVTEVLKVGLVVKDWAQR 400
Query: 418 DGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEI 474
+ V +E + ++ + +E+++ A LK A ++ +GG S + +F+ I
Sbjct: 401 NSLVSASVVENGVRRLMETKEGDEMRQRAVRLKNAIHRSMDEGGVSHMEMGSFIAHI 457
>UFO6_MANES (Q40288) Flavonol 3-O-glucosyltransferase 6 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
6) (Fragment)
Length = 394
Score = 157 bits (397), Expect = 6e-38
Identities = 102/378 (26%), Positives = 185/378 (47%), Gaps = 38/378 (10%)
Query: 122 NSKKLACIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFY----NNTNQFPTLE 177
+ LA + + F + DVA E +P + A ++ +
Sbjct: 27 SDSSLAGFVLDMFCTSMIDVAKELGVPYYIFFTSGAAFLGFLFYVQLIHDEQDADLTQFK 86
Query: 178 NPELNVQLPGLPLLKP-QDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEK 236
+ + + +P L P + LP+ +L + F A +++ + +++ K ++ N+F ELE
Sbjct: 87 DSDAELSVPSLANSLPARVLPASMLVKDRFYAFIRII----RGLREAKGIMVNTFMELES 142
Query: 237 EVIDSMAETY----PVIPVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVI 292
++S+ + P+ PVGP+L LS ++ DVG E +EWL+DQPPSSV+
Sbjct: 143 HALNSLKDDQSKIPPIYPVGPILKLS----NQENDVGPE----GSEIIEWLDDQPPSSVV 194
Query: 293 YISFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLP-----------LPQGFK 341
++ FGS+ + + IA AL+ S +FLW +++ K + LP GF
Sbjct: 195 FLCFGSMGGFDMDQAKEIACALEQSRHRFLWSLRRPPPKGKIETSTDYENLQEILPVGFS 254
Query: 342 EETKNRGMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAK 401
E T G VV W PQ +L HPAI F++HCGWNS+LE+I P+ +P + +Q NA
Sbjct: 255 ERTAGMGKVVGWAPQVAILEHPAIGGFVSHCGWNSILESIWFSVPIATWPLYAEQQFNAF 314
Query: 402 LVSDVFRVGIRLEQD----SDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAV 457
+ + + ++ D S+ + ++ER I+ ++ E+++ E+ +R+A+
Sbjct: 315 TMVTELGLAVEIKMDYKKESEIILSADDIERGIKCVME--HHSEIRKRVKEMSDKSRKAL 372
Query: 458 ADGGSSDRNIQTFVDEIL 475
D SS + +++++
Sbjct: 373 MDDESSSFWLDRLIEDVI 390
>UFO7_MANES (Q40289) Flavonol 3-O-glucosyltransferase 7 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
7) (Fragment)
Length = 287
Score = 155 bits (392), Expect = 2e-37
Identities = 91/283 (32%), Positives = 153/283 (53%), Gaps = 17/283 (6%)
Query: 185 LPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAE 244
+PG+ ++ +DLP +L N S++L +M + + + VL NSF EL+ ++ +
Sbjct: 5 IPGMSKIQIRDLPEGVLFGNLESLFSQMLHNMGRMLPRAAAVLMNSFEELDPTIVSDLNS 64
Query: 245 TYP-VIPVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLS 303
+ ++ +GP +S + C+ WL+ Q P+SV YISFGS+
Sbjct: 65 KFNNILCIGPFNLVSPPPPVPDTY----------GCMAWLDKQKPASVAYISFGSVATPP 114
Query: 304 AKKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHP 363
+L ++A AL+ S FLW +K + LP GF + TK+ G+V+ W PQ ++L H
Sbjct: 115 PHELVALAEALEASKVPFLWSLKDHS---KVHLPNGFLDRTKSHGIVLSWAPQVEILEHA 171
Query: 364 AIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVET 423
A+ F+THCGWNS+LE+I G PMI P + DQ N ++V DV+ +G+ + D +
Sbjct: 172 ALGVFVTHCGWNSILESIVGGVPMICRPFFGDQRLNGRMVEDVWEIGLLM--DGGVLTKN 229
Query: 424 GELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRN 466
G ++ + +I+ K ++++ N LK A+ A GSS ++
Sbjct: 230 GAID-GLNQILLQGKGKKMRENIKRLKELAKGATEPKGSSSKS 271
>UFO1_MANES (Q40284) Flavonol 3-O-glucosyltransferase 1 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
1)
Length = 449
Score = 154 bits (390), Expect = 4e-37
Identities = 130/470 (27%), Positives = 221/470 (46%), Gaps = 63/470 (13%)
Query: 22 GHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNFTTNGIHVLFF 81
GH+ + K LL+R +++ +F +S T+ + + + ++ + F
Sbjct: 2 GHLVSAVETAKLLLSRCHSLSITVL------IFNNSVVTSKVHNYVDSQIASSSNRLRF- 54
Query: 82 SDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIK-THFINN--SKKLACIINNPFVPWV 138
+ D+T ++ + +L+E P +++K T F ++ S +L I + F +
Sbjct: 55 ---IYLPRDETGIS--SFSSLIEKQKPHVKESVMKITEFGSSVESPRLVGFIVDMFCTAM 109
Query: 139 ADVAAEFKIPCACLWIQPCALYAIYYH---FYNNTNQFPTLENP-ELNVQLPGLPLLKPQ 194
DVA EF +P + A H ++ N PT N + +Q+PGL
Sbjct: 110 IDVANEFGVPSYIFYTSGAAFLNFMLHVQKIHDEENFNPTEFNASDGELQVPGLV----N 165
Query: 195 DLPSFILPTNPFGALSKV----LADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVIP 250
PS +PT LSK L + + + K V+ N+F+ELE I+S + P+ P
Sbjct: 166 SFPSKAMPT---AILSKQWFPPLLENTRRYGEAKGVIINTFFELESHAIESFKDP-PIYP 221
Query: 251 VGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLESI 310
VGP+L + G + N+++ ++WL+DQPPSSV+++ FGS S +++ I
Sbjct: 222 VGPILDVRSNGRNTNQEI-----------MQWLDDQPPSSVVFLCFGSNGSFSKDQVKEI 270
Query: 311 ATALKNSNCKFLWVIKKQDGKDSLP-----------LPQGFKEETKNRGMVVPWCPQTKV 359
A AL++S +FLW + L LP+GF E T V+ W PQ V
Sbjct: 271 ACALEDSGHRFLWSLADHRAPGFLESPSDYEDLQEVLPEGFLERTSGIEKVIGWAPQVAV 330
Query: 360 LVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLE----- 414
L HPA ++H GWNS+LE+I G P+ +P + +Q NA V +G+ +E
Sbjct: 331 LAHPATGGLVSHSGWNSILESIWFGVPVATWPMYAEQQFNA--FQMVIELGLAVEIKMDY 388
Query: 415 -QDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSS 463
DS V+ ++ER I ++ + ++ E+ +R A+ +GGSS
Sbjct: 389 RNDSGEIVKCDQIERGIRCLMK--HDSDRRKKVKEMSEKSRGALMEGGSS 436
>UFO1_MAIZE (P16166) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-McC allele)
Length = 471
Score = 153 bits (387), Expect = 8e-37
Identities = 134/481 (27%), Positives = 217/481 (44%), Gaps = 47/481 (9%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLAR----GLQVTLATTELVYHRVFKSSSTTAATTDTI 67
HV +VAF H LL + ++L A G ++ +T ++ K+SS +A +
Sbjct: 13 HVAVVAFPFSSHAAVLLSIARALAAAAAPSGATLSFLSTASSLAQLRKASSASAG--HGL 70
Query: 68 PTNFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKT-HFINNSKKL 126
P N + + DG P ++T P +E + ++ ++
Sbjct: 71 PGN-----LRFVEVPDGA-PAAEETVPVPRQMQLFMEAAEAGGVKAWLEAARAAAGGARV 124
Query: 127 ACIINNPFVPWVADVAAEFKIPCACLWIQP-CALYAIYYHFYNNTNQFPTLENPE----- 180
C++ + FV AD AA P +W CAL A H + + +
Sbjct: 125 TCVVGDAFVWPAADAAASAGAPWVPVWTAASCALLA---HIRTDALREDVGDQAANRVDG 181
Query: 181 LNVQLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDM--KKLKWVLANSFYELEK-E 237
L + PGL + +DLP ++ + F + +L + + V N+F L+ +
Sbjct: 182 LLISHPGLASYRVRDLPDGVV-SGDFNYVINLLVHRMGQCLPRSAAAVALNTFPGLDPPD 240
Query: 238 VIDSMAETYP-VIPVGPLLPLSLLGVDENEDVGIEMWKPQD--SCLEWLNDQPPSSVIYI 294
V ++AE P +P GP LL +++ D P D CL WL QP V Y+
Sbjct: 241 VTAALAEILPNCVPFGPY---HLLLAEDDADTAA----PADPHGCLAWLGRQPARGVAYV 293
Query: 295 SFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLP-LPQGFKEETKNRG--MVV 351
SFG++ +L +A L++S FLW +++ DS P LP GF + G +VV
Sbjct: 294 SFGTVACPRPDELRELAAGLEDSGAPFLWSLRE----DSWPHLPPGFLDRAAGTGSGLVV 349
Query: 352 PWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGI 411
PW PQ VL HP++ F+TH GW S+LE +++G PM P + DQ NA+ V+ V+ G
Sbjct: 350 PWAPQVAVLRHPSVGAFVTHAGWASVLEGLSSGVPMACRPFFGDQRMNARSVAHVWGFGA 409
Query: 412 RLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFV 471
E G + + + A+EE++ G + ++ A EL+ EA GG +N FV
Sbjct: 410 AFE----GAMTSAGVATAVEELLRGEEGARMRARAKELQALVAEAFGPGGECRKNFDRFV 465
Query: 472 D 472
+
Sbjct: 466 E 466
>UFO2_MAIZE (P16165) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-Mc2 allele)
Length = 471
Score = 148 bits (373), Expect = 4e-35
Identities = 131/479 (27%), Positives = 217/479 (44%), Gaps = 43/479 (8%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLAR----GLQVTLATTELVYHRVFKSSSTTAATTDTI 67
HV +VAF H LL + ++L A G ++ +T ++ K+SS +A +
Sbjct: 13 HVAVVAFPFSSHAAVLLSIARALAAAAAPSGATLSFLSTASSLAQLRKASSASAG--HGL 70
Query: 68 PTNFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKT-HFINNSKKL 126
P N + + DG P ++T P +E + ++ ++
Sbjct: 71 PGN-----LRFVEVPDGA-PAAEETVPVPRQMQLFMEAAEAGGVKAWLEAARAAAGGARV 124
Query: 127 ACIINNPFVPWVADVAAEFKIPCACLWIQP-CALYAIYYHFYNNTNQFPTLENP-----E 180
C++ + FV AD AA P +W CAL A H ++ + + E
Sbjct: 125 TCVVGDAFVWPAADAAASAGAPWVPVWTAASCALLA---HIRTDSLREDVGDQAANRVDE 181
Query: 181 LNVQLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLA-NSFYELEK-EV 238
+ PGL + +DLP ++ + +S ++ M + + + +A N+F L+ +V
Sbjct: 182 PLISHPGLASYRVRDLPDGVVSGDFNYVISLLVHRMGQCLPRSAAAVALNTFPGLDPPDV 241
Query: 239 IDSMAETYP-VIPVGPLLPLSLLGVDENEDVGIEMWKPQD--SCLEWLNDQPPSSVIYIS 295
++AE P +P GP LL +++ D P D CL WL QP V Y+S
Sbjct: 242 TAALAEILPNCVPFGPY---HLLLAEDDADTAA----PADPHGCLAWLGRQPARGVAYVS 294
Query: 296 FGSLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRG--MVVPW 353
FG++ +L +A L+ S FLW +++ LP GF + G +VVPW
Sbjct: 295 FGTVACPRPDELRELAAGLEASAAPFLWSLREDSWT---LLPPGFLDRAAGTGSGLVVPW 351
Query: 354 CPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRL 413
PQ VL HP++ F+TH GW S+LE +++G PM P + DQ NA+ V+ V+ G
Sbjct: 352 APQVAVLRHPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGDQRMNARSVAHVWGFGAAF 411
Query: 414 EQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVD 472
E G + + + A+EE++ G + ++ A EL+ EA GG +N FV+
Sbjct: 412 E----GAMTSAGVAAAVEELLRGEEGAGMRARAKELQALVAEAFGPGGECRKNFDRFVE 466
>UFO3_MAIZE (P16167) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1) (Bz-W22 allele)
Length = 471
Score = 145 bits (366), Expect = 2e-34
Identities = 135/485 (27%), Positives = 213/485 (43%), Gaps = 55/485 (11%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLAR----GLQVTLATTELVYHRVFKSSSTTAATTDTI 67
HV +VAF H LL + ++L A G ++ +T ++ K+SS +A +
Sbjct: 13 HVAVVAFPFSSHAAVLLSIARALAAAAAPSGATLSFLSTASSLAQLRKASSASAG--HGL 70
Query: 68 PTNFTTNGIHVLFFSDGLD------PNPDQTQLTPDDYMALVENLGPINLSTLIKTHFIN 121
P N + + DG P P Q QL +M E G + L
Sbjct: 71 PGN-----LRFVEVPDGAPAAEESVPVPRQMQL----FMEAAEAGGV--KAWLEAARAAA 119
Query: 122 NSKKLACIINNPFVPWVADVAAEFKIPCACLWIQP-CALYAIYYHFYNNTNQFPTLENP- 179
++ C++ + FV AD AA P +W CAL A H + + +
Sbjct: 120 GGARVTCVVGDAFVWPAADAAASAGAPWVPVWTAASCALLA---HIRTDALREDVGDQAA 176
Query: 180 ----ELNVQLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDM--KKLKWVLANSFYE 233
E + PGL + +DLP ++ + F + +L + + V N+F
Sbjct: 177 NRVDEPLISHPGLASYRVRDLPDGVV-SGDFNYVINLLVHRMGQCLPRSAAAVALNTFPG 235
Query: 234 LEK-EVIDSMAETYP-VIPVGPLLPLSLLGVDENEDVGIEMWKPQD--SCLEWLNDQPPS 289
L+ +V ++AE P +P GP LL +++ D P D CL WL QP
Sbjct: 236 LDPPDVTAALAEILPNCVPFGPY---HLLLAEDDADTAA----PADPHGCLAWLGRQPAR 288
Query: 290 SVIYISFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRG- 348
V Y+SFG++ +L +A L+ S FLW +++ LP GF + G
Sbjct: 289 GVAYVSFGTVACPRPDELRELAAGLEASGAPFLWSLREDSWT---LLPPGFLDRAAGTGS 345
Query: 349 -MVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVF 407
+VVPW PQ VL HP++ F+TH GW S+LE +++G PM P + DQ NA+ V+ V+
Sbjct: 346 GLVVPWAPQVAVLRHPSVGAFVTHAGWASVLEGVSSGVPMACRPFFGDQRMNARSVAHVW 405
Query: 408 RVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNI 467
G E G + + + A+EE++ G + ++ A L+ EA GG +N
Sbjct: 406 GFGAAFE----GAMTSAGVAAAVEELLRGEEGARMRARAKVLQALVAEAFGPGGECRKNF 461
Query: 468 QTFVD 472
FV+
Sbjct: 462 DRFVE 466
>UFO5_MANES (Q40287) Flavonol 3-O-glucosyltransferase 5 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
5)
Length = 487
Score = 140 bits (354), Expect = 6e-33
Identities = 74/201 (36%), Positives = 120/201 (58%), Gaps = 17/201 (8%)
Query: 280 LEWLNDQPPSSVIYISFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQ----------- 328
L+WL+ QP SV+Y+SFGS LS +++ +A L+ S +F+WV+++
Sbjct: 262 LDWLDQQPKESVVYVSFGSGGTLSLEQMIELAWGLERSQQRFIWVVRQPTVKTGDAAFFT 321
Query: 329 --DGKDSLP--LPQGFKEETKNRGMVVP-WCPQTKVLVHPAIACFLTHCGWNSMLEAIAA 383
DG D + P+GF +N G+VVP W PQ ++ HP++ FL+HCGWNS+LE+I A
Sbjct: 322 QGDGADDMSGYFPEGFLTRIQNVGLVVPQWSPQIHIMSHPSVGVFLSHCGWNSVLESITA 381
Query: 384 GKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQ-DSDGFVETGELERAIEEIVSGPKSEEL 442
G P+IA+P + +Q NA L+++ V +R + + V+ E+ER I I+ + E+
Sbjct: 382 GVPIIAWPIYAEQRMNATLLTEELGVAVRPKNLPAKEVVKREEIERMIRRIMVDEEGSEI 441
Query: 443 KRNAAELKRAAREAVADGGSS 463
++ ELK + +A+ +GGSS
Sbjct: 442 RKRVRELKDSGEKALNEGGSS 462
>UFOG_HORVU (P14726) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Bronze-1)
Length = 455
Score = 140 bits (352), Expect = 1e-32
Identities = 129/478 (26%), Positives = 207/478 (42%), Gaps = 51/478 (10%)
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLAR---GLQVTLATTELVYHRVFKSSSTTAATTDTIP 68
H+ +VAF H L ++L A G ++ TT ++ +P
Sbjct: 7 HIAVVAFPFSSHAAVLFSFARALAAAAPAGTSLSFLTTA--------DNAAQLRKAGALP 58
Query: 69 TNFTTNGIHVLFFSDGLDPNPDQTQLTP---DDYMALVENLGP-INLSTLIKTHFINNSK 124
N + + DG+ P P D +MA E G + L +
Sbjct: 59 GN-----LRFVEVPDGVPPGETSCLSPPRRMDLFMAAAEAGGVRVGLEAACASA---GGA 110
Query: 125 KLACIINNPFVPWVADVAAEFKIPCACLWIQP-CALYAIYYH--FYNNTNQFPTLENPEL 181
+++C++ + FV W AD A+ P +W CAL A + EL
Sbjct: 111 RVSCVVGDAFV-WTADAASAAGAPWVAVWTAASCALLAHLRTDALRRDVGDQAASRADEL 169
Query: 182 NVQLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLA-NSFYELEK-EVI 239
V GL + +DLP ++ + +S ++ + + K +A N+F L+ ++I
Sbjct: 170 LVAHAGLGGYRVRDLPDGVVSGDFNYVISLLVHRQAQRLPKAATAVALNTFPGLDPPDLI 229
Query: 240 DSMAETYP-VIPVGP--LLPLSLLGVDENEDVGIEMWKPQD--SCLEWLNDQPPSSVIYI 294
++A P +P+GP LLP + D NE P D CL WL+ +P SV Y+
Sbjct: 230 AALAAELPNCLPLGPYHLLPGAEPTADTNE-------APADPHGCLAWLDRRPARSVAYV 282
Query: 295 SFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLPLPQGFKEETKNRGMVVPWC 354
SFG+ +L+ +A L+ S FLW ++ P+GF E G+VVPW
Sbjct: 283 SFGTNATARPDELQELAAGLEASGAPFLWSLRGVVAA----APRGFLERAP--GLVVPWA 336
Query: 355 PQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLE 414
PQ VL H A+ F+TH GW S++E +++G PM P + DQ NA+ V+ V+ G
Sbjct: 337 PQVGVLRHAAVGAFVTHAGWASVMEGVSSGVPMACRPFFGDQTMNARSVASVWGFGTAF- 395
Query: 415 QDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVD 472
DG + G + A+ ++ G E ++ A EL+ +A G +N FV+
Sbjct: 396 ---DGPMTRGAVANAVATLLRGEDGERMRAKAQELQAMVGKAFEPDGGCRKNFDEFVE 450
>UFO4_MANES (Q40286) Flavonol 3-O-glucosyltransferase 4 (EC
2.4.1.91) (UDP-glucose flavonoid 3-O-glucosyltransferase
4) (Fragment)
Length = 241
Score = 127 bits (319), Expect = 7e-29
Identities = 71/211 (33%), Positives = 118/211 (55%), Gaps = 16/211 (7%)
Query: 280 LEWLNDQPPSSVIYISFGSLIVLSAKKLESIATALKNSNCKFLWVIK---KQDGKDSLPL 336
L+WL+ P SVIY GS+ L++ +L + L+++N F+WVI+ K +G + L
Sbjct: 23 LKWLDLWEPGSVIYACLGSISGLTSWQLAELGLGLESTNQPFIWVIREGEKSEGLEKWIL 82
Query: 337 PQGFKEETKNRG--MVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWT 394
+G++E + R + W PQ +L HPAI F THCGWNS LE I+AG P++A P +
Sbjct: 83 EEGYEERKRKREDFWIRGWSPQVLILSHPAIGAFFTHCGWNSTLEGISAGVPIVACPLFA 142
Query: 395 DQPTNAKLVSDVFRVGIR----------LEQDSDGFVETGELERAIEEIV-SGPKSEELK 443
+Q N KLV +V +G+ LE ++ ++++AIE ++ G + EE +
Sbjct: 143 EQFYNEKLVVEVLGIGVSVGVEAAVTWGLEDKCGAVMKKEQVKKAIEIVMDKGKEGEERR 202
Query: 444 RNAAELKRAAREAVADGGSSDRNIQTFVDEI 474
R A E+ A+ + +GGSS +++ + +
Sbjct: 203 RRAREIGEMAKRTIEEGGSSYLDMEMLIQYV 233
>UFOG_VITVI (P51094) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Fragment)
Length = 154
Score = 120 bits (301), Expect = 8e-27
Identities = 60/143 (41%), Positives = 84/143 (57%), Gaps = 3/143 (2%)
Query: 330 GKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIA 389
GK S+ LP+GF E+T+ GMVVPW PQ +VL F+THCGWNS+ E++A G P+I
Sbjct: 7 GKASVHLPEGFLEKTRGYGMVVPWAPQAEVLALRQFGAFVTHCGWNSLWESVAGGVPLIC 66
Query: 390 YPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAEL 449
P + DQ N ++V DV +G+R+E G L ++I+S K ++L+ N L
Sbjct: 67 RPFFGDQRLNGRMVEDVLEIGVRIE---GGVFTKSGLMSCFDQILSQEKGKKLRENLRAL 123
Query: 450 KRAAREAVADGGSSDRNIQTFVD 472
+ A AV GSS N +T VD
Sbjct: 124 RETADRAVGPKGSSTENFKTLVD 146
>UFOG_PETHY (Q43716) Flavonol 3-O-glucosyltransferase (EC 2.4.1.91)
(UDP-glucose flavonoid 3-O-glucosyltransferase)
(Anthocyanin rhamnosyl transferase)
Length = 473
Score = 115 bits (289), Expect = 2e-25
Identities = 111/451 (24%), Positives = 201/451 (43%), Gaps = 60/451 (13%)
Query: 9 EELHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIP 68
+ LHV++ F A GHI+P ++L L + G++V+ T RV KS +A TT +P
Sbjct: 10 DALHVVMFPFFAFGHISPFVQLANKLSSYGVKVSFFTASGNASRV-KSMLNSAPTTHIVP 68
Query: 69 TNFTTNGIHVLFFSDGLDPNPDQT-QLTPD--DYMALVENLGPINLSTLI---KTHFINN 122
HV +GL P + T +LTP + + + +L + TL+ K HF+
Sbjct: 69 LTLP----HV----EGLPPGAESTAELTPASAELLKVALDLMQPQIKTLLSHLKPHFVLF 120
Query: 123 S------KKLACIINNPFVPWVADVAAEFK-IPCACLWIQPCALYAIYYHFYNNTNQFPT 175
K+A + V + VA + C ++P ++P+
Sbjct: 121 DFAQEWLPKMANGLGIKTVYYSVVVALSTAFLTCPARVLEP--------------KKYPS 166
Query: 176 LEN---PELNVQLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFY 232
LE+ P L + ++ + F+ F + + ++ +LA +
Sbjct: 167 LEDMKKPPLGFPQTSVTSVRTFEARDFLYVFKSFHNGPTLYDRIQSGLRGCSAILAKTCS 226
Query: 233 ELEKEVIDSMAETY--PVIPVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSS 290
++E I + + PV +GP++P G K ++ WLN +
Sbjct: 227 QMEGPYIKYVEAQFNKPVFLIGPVVPDPPSG------------KLEEKWATWLNKFEGGT 274
Query: 291 VIYISFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKD-----SLPLPQGFKEETK 345
VIY SFGS L+ +++ +A L+ + F V+ D + LP+GF E K
Sbjct: 275 VIYCSFGSETFLTDDQVKELALGLEQTGLPFFLVLNFPANVDVSAELNRALPEGFLERVK 334
Query: 346 NRGMVVP-WCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVS 404
++G++ W Q +L H ++ C++ H G++S++EA+ ++ PQ DQ NAKLVS
Sbjct: 335 DKGIIHSGWVQQQNILAHSSVGCYVCHAGFSSVIEALVNDCQVVMLPQKGDQILNAKLVS 394
Query: 405 DVFRVGIRL-EQDSDGFVETGELERAIEEIV 434
G+ + +D DG+ +++ A+E+++
Sbjct: 395 GDMEAGVEINRRDEDGYFGKEDIKEAVEKVM 425
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 58,055,597
Number of Sequences: 164201
Number of extensions: 2515656
Number of successful extensions: 6607
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 6437
Number of HSP's gapped (non-prelim): 126
length of query: 490
length of database: 59,974,054
effective HSP length: 114
effective length of query: 376
effective length of database: 41,255,140
effective search space: 15511932640
effective search space used: 15511932640
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0327.19


