

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
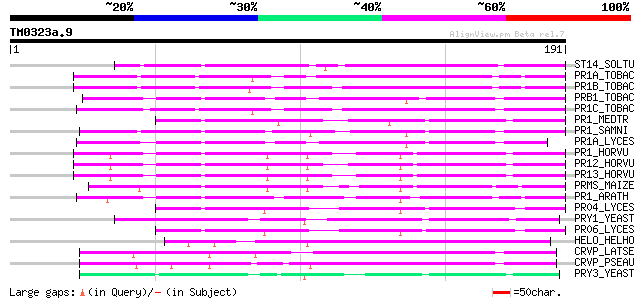
Query= TM0323a.9
(191 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ST14_SOLTU (Q41495) STS14 protein precursor 108 1e-23
PR1A_TOBAC (P08299) Pathogenesis-related protein 1A precursor (P... 78 1e-14
PR1B_TOBAC (P07053) Pathogenesis-related protein 1B precursor (P... 74 3e-13
PRB1_TOBAC (P11670) Basic form of pathogenesis-related protein 1... 71 1e-12
PR1C_TOBAC (P09042) Pathogenesis-related protein 1C precursor (P... 71 1e-12
PR1_MEDTR (Q40374) Pathogenesis-related protein PR-1 precursor 71 2e-12
PR1_SAMNI (Q41359) Pathogenesis-related protein PR-1 type precursor 67 3e-11
PR1A_LYCES (Q08697) Pathogenesis-related protein 1A1 precursor (... 66 4e-11
PR1_HORVU (Q05968) Pathogenesis-related protein 1 precursor 66 6e-11
PR12_HORVU (P35792) Pathogenesis-related protein PRB1-2 precursor 65 7e-11
PR13_HORVU (P35793) Pathogenesis-related protein PRB1-3 precurso... 65 1e-10
PRMS_MAIZE (Q00008) Pathogenesis-related protein PRMS precursor 64 2e-10
PR1_ARATH (P33154) Pathogenesis-related protein 1 precursor (PR-1) 60 4e-09
PR04_LYCES (Q04108) Pathogenesis-related leaf protein 4 precurso... 59 9e-09
PRY1_YEAST (P47032) PRY1 protein precursor (Pathogen related in ... 54 2e-07
PR06_LYCES (P04284) Pathogenesis-related leaf protein 6 precurso... 54 2e-07
HELO_HELHO (Q91055) Helothermine precursor (HLTx) 52 8e-07
CRVP_LATSE (Q8JI38) Latisemin precursor 51 1e-06
CRVP_PSEAU (Q8AVA4) Pseudechetoxin precursor (PsTx) 49 5e-06
PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3) 49 7e-06
>ST14_SOLTU (Q41495) STS14 protein precursor
Length = 214
Score = 108 bits (269), Expect = 1e-23
Identities = 66/156 (42%), Positives = 86/156 (54%), Gaps = 9/156 (5%)
Query: 37 SSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCTPGIF 96
+S ATP + A+ +L HN R E G+ L WS LAK+ SL R++RDK C+
Sbjct: 66 TSAATPPSR-AAQEFLDAHNKARSEVGVGP-LTWSPMLAKETSLLVRYQRDKQNCSFANL 123
Query: 97 DKKRYGVCCNQ-WLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQIVWRK 155
+YG NQ W TV + PR + WV ++ +Y N+ + CG YTQIVW+K
Sbjct: 124 SNGKYGG--NQLWASGTV--VTPRMAVDSWVAEKKFYNYENNSCTGDDKCGVYTQIVWKK 179
Query: 156 TKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
+ LGCAQ C G L VCFY PPGNV GE+P
Sbjct: 180 SIELGCAQRTCYE--GPATLTVCFYNPPGNVIGEKP 213
>PR1A_TOBAC (P08299) Pathogenesis-related protein 1A precursor
(PR-1A)
Length = 168
Score = 78.2 bits (191), Expect = 1e-14
Identities = 55/170 (32%), Positives = 84/170 (49%), Gaps = 13/170 (7%)
Query: 23 PFFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYA 82
P F ++ L L LV S + A + + YL HN R + G+ E L W D++A A YA
Sbjct: 10 PSFLLVSTLLLFLVISHSCRAQ-NSQQDYLDAHNTARADVGV-EPLTWDDQVAAYAQNYA 67
Query: 83 -RFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPA 141
+ D L + +YG E + M + + WV ++ YY+ +++
Sbjct: 68 SQLAADCNL----VHSHGQYG---ENLAEGSGDFMTAAKAVEMWVDEKQYYDHDSNTCAQ 120
Query: 142 NYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR + +GCA+ +C + GG ++ C Y PPGN GE P
Sbjct: 121 GQVCGHYTQVVWRNSVRVGCARVQC--NNGGYVVS-CNYDPPGNYRGESP 167
>PR1B_TOBAC (P07053) Pathogenesis-related protein 1B precursor
(PR-1B)
Length = 168
Score = 73.6 bits (179), Expect = 3e-13
Identities = 53/170 (31%), Positives = 85/170 (49%), Gaps = 13/170 (7%)
Query: 23 PFFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLY- 81
P F ++ L L L+ S ++ A + + YL HN R + G+ E L W + +A A Y
Sbjct: 10 PSFFLVSTLLLFLIISHSSHAQ-NSQQDYLDAHNTARADVGV-EPLTWDNGVAAYAQNYV 67
Query: 82 ARFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPA 141
++ D L + +YG Q D M + + WV ++ YY+ +++
Sbjct: 68 SQLAADCNL----VHSHGQYGENLAQGSGD---FMTAAKAVEMWVDEKQYYDHDSNTCAQ 120
Query: 142 NYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR + +GCA+ +C + GG ++ C Y PPGNV G+ P
Sbjct: 121 GQVCGHYTQVVWRNSVRVGCARVKC--NNGGYVVS-CNYDPPGNVIGQSP 167
>PRB1_TOBAC (P11670) Basic form of pathogenesis-related protein 1
precursor (PRP 1)
Length = 177
Score = 71.2 bits (173), Expect = 1e-12
Identities = 55/167 (32%), Positives = 78/167 (45%), Gaps = 18/167 (10%)
Query: 26 AVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFR 85
A + A+L+ SS+A + D YL HN RR+ G+ + W ++LA A YA R
Sbjct: 9 ACFITFAILIHSSKAQNSPQD----YLNPHNAARRQVGVGP-MTWDNRLAAYAQNYANQR 63
Query: 86 RDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGN-NSCVPANYP 144
I YG + L ++ ++ WV ++ +Y+ N NSCV
Sbjct: 64 IGD---CGMIHSHGPYG----ENLAAAFPQLNAAGAVKMWVDEKRFYDYNSNSCVGG--V 114
Query: 145 CGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR + LGCA+ G C Y PPGN G+RP
Sbjct: 115 CGHYTQVVWRNSVRLGCARVRSN---NGWFFITCNYDPPGNFIGQRP 158
>PR1C_TOBAC (P09042) Pathogenesis-related protein 1C precursor
(PR-1C)
Length = 168
Score = 71.2 bits (173), Expect = 1e-12
Identities = 51/169 (30%), Positives = 77/169 (45%), Gaps = 14/169 (8%)
Query: 24 FFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYA- 82
FF V L L++S + + YL HN R + G+ E L W D++A A YA
Sbjct: 12 FFLVSTLLLFLIISHSCHAQNSQ--QDYLDAHNTARADVGV-EPLTWDDQVAAYAQNYAS 68
Query: 83 RFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPAN 142
+ D L + +YG D + + + WV ++ YY +++
Sbjct: 69 QLAADCNL----VHSHGQYGENLAWGSGD---FLTAAKAVEMWVNEKQYYAHDSNTCAQG 121
Query: 143 YPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR + +GCA+ +C G + C Y PPGNV G+ P
Sbjct: 122 QVCGHYTQVVWRNSVRVGCARVQCN---NGGYIVSCNYDPPGNVIGKSP 167
>PR1_MEDTR (Q40374) Pathogenesis-related protein PR-1 precursor
Length = 173
Score = 70.9 bits (172), Expect = 2e-12
Identities = 49/144 (34%), Positives = 64/144 (44%), Gaps = 15/144 (10%)
Query: 51 YLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLC--TPGIFDKKRYGVCCNQW 108
+L N R G+ L W DKL A YA RR+ + G + + + W
Sbjct: 41 FLIPQNIARAAVGLRP-LVWDDKLTHYAQWYANQRRNDCALEHSNGPYGENIFWGSGVGW 99
Query: 109 LEDTVKIMDPREVMRQWVKDR-FYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECG 167
+P + + WV ++ FY +NSCV CG YTQ+VW T +GCA C
Sbjct: 100 --------NPAQAVSAWVDEKQFYNYWHNSCVDGEM-CGHYTQVVWGSTTKVGCASVVCS 150
Query: 168 PDYGGVRLNVCFYYPPGNVPGERP 191
D G C Y PPGN GERP
Sbjct: 151 DDKG--TFMTCNYDPPGNYYGERP 172
>PR1_SAMNI (Q41359) Pathogenesis-related protein PR-1 type precursor
Length = 167
Score = 67.0 bits (162), Expect = 3e-11
Identities = 53/170 (31%), Positives = 74/170 (43%), Gaps = 16/170 (9%)
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
F+V + + LV Q + A + + Y+ HN R + + W + +A A YA+
Sbjct: 10 FSVALVCVVALVMVQYSVAQ-NSPQDYVDAHNAARSAVNVGP-VTWDESVAAFARQYAQS 67
Query: 85 RRDKYLCTPGIFDKKRYG--VCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGN-NSCVPA 141
R C RYG + E T R + WV +R Y N N+C P
Sbjct: 68 RAGD--CRLVHSGDPRYGENLAFGSGFELT-----GRNAVDMWVAERNDYNPNTNTCAPG 120
Query: 142 NYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
CG YTQ+VWR + +GCA+ C G C Y PPGN G+RP
Sbjct: 121 KV-CGHYTQVVWRNSVRIGCARVRCN---NGAWFITCNYSPPGNYAGQRP 166
>PR1A_LYCES (Q08697) Pathogenesis-related protein 1A1 precursor
(PR-1A1)
Length = 175
Score = 66.2 bits (160), Expect = 4e-11
Identities = 51/163 (31%), Positives = 69/163 (42%), Gaps = 18/163 (11%)
Query: 24 FFAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYAR 83
F A + + S TP +L HN RR G+ + W D LA A YA
Sbjct: 6 FVACFITFIIFHSSQAQTPREN-----FLNAHNAARRRVGVGP-MTWDDGLAAYAQNYAN 59
Query: 84 FRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGN-NSCVPAN 142
R D I YG + L ++ ++ W ++ +Y+ N N+C P
Sbjct: 60 QRADD---CGMIHSDGPYG----ENLAAAFPQLNAAGAVKMWDDEKQWYDYNSNTCAPGK 112
Query: 143 YPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGN 185
CG YTQ+VWRK+ LGCA+ C G C Y PPGN
Sbjct: 113 V-CGHYTQVVWRKSVRLGCARVRCN---SGWVFITCNYDPPGN 151
>PR1_HORVU (Q05968) Pathogenesis-related protein 1 precursor
Length = 164
Score = 65.9 bits (159), Expect = 6e-11
Identities = 54/176 (30%), Positives = 83/176 (46%), Gaps = 23/176 (13%)
Query: 23 PFFAVLVALAL---LLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDAS 79
P A+L+ALA+ ++ SQA + D Y+ HN R G+ + WS KL A
Sbjct: 4 PKLAILLALAMAAAMVNLSQAQNSPQD----YVSPHNAARSAVGVGA-VSWSTKLQAFAQ 58
Query: 80 LYARFRRD--KYLCTPGIFDKKRY-GVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GN 135
YA R + K + G + + + G W + + WV ++ Y+ G+
Sbjct: 59 NYANQRINDCKLQHSGGPYGENIFWGSAGADWKAS--------DAVNSWVSEKKDYDYGS 110
Query: 136 NSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
N+C A CG YTQ+VWR + +GCA+ C + G C Y P GN+ G++P
Sbjct: 111 NTCA-AGKVCGHYTQVVWRASTSIGCARVVCNNNRG--VFITCNYEPRGNIIGQKP 163
>PR12_HORVU (P35792) Pathogenesis-related protein PRB1-2 precursor
Length = 164
Score = 65.5 bits (158), Expect = 7e-11
Identities = 54/176 (30%), Positives = 82/176 (45%), Gaps = 23/176 (13%)
Query: 23 PFFAVLVALAL---LLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDAS 79
P A+L+ALA+ ++ SQA + D Y+ HN R G+ + WS KL A
Sbjct: 4 PKLAILLALAMAAAMVNLSQAQNSPQD----YVSPHNAARSAVGVGA-VSWSTKLQAFAQ 58
Query: 80 LYARFRRD--KYLCTPGIFDKKRY-GVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GN 135
YA R + K + G + + + G W + + WV ++ Y G+
Sbjct: 59 NYANQRINDCKLQHSGGPYGENIFWGSAGADW--------KAADAVNSWVNEKKDYNYGS 110
Query: 136 NSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
N+C A CG YTQ+VWR + +GCA+ C + G C Y P GN+ G++P
Sbjct: 111 NTCA-AGKVCGHYTQVVWRASTSIGCARVVCNNNRG--VFITCNYEPRGNIVGQKP 163
>PR13_HORVU (P35793) Pathogenesis-related protein PRB1-3 precursor
(PR-1B) (HV-8)
Length = 164
Score = 64.7 bits (156), Expect = 1e-10
Identities = 53/176 (30%), Positives = 82/176 (46%), Gaps = 23/176 (13%)
Query: 23 PFFAVLVALAL---LLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDAS 79
P +L+ALA+ ++ SQA + D Y+ HN R G+ + WS KL A
Sbjct: 4 PKLVILLALAMSAAMVNLSQAQNSPQD----YVSPHNAARAAVGVGA-VSWSTKLQAFAQ 58
Query: 80 LYARFRRD--KYLCTPGIFDKKRY-GVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GN 135
YA R + K + G + + + G W + + WV ++ Y+ G+
Sbjct: 59 NYANQRINDCKLQHSGGPYGENIFWGSAGADWKAS--------DAVNSWVSEKKDYDYGS 110
Query: 136 NSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
N+C A CG YTQ+VWR + +GCA+ C + G C Y P GN+ G++P
Sbjct: 111 NTCA-AGKVCGHYTQVVWRASTSIGCARVVCNNNRG--VFITCNYEPRGNIVGQKP 163
>PRMS_MAIZE (Q00008) Pathogenesis-related protein PRMS precursor
Length = 167
Score = 63.9 bits (154), Expect = 2e-10
Identities = 56/172 (32%), Positives = 80/172 (45%), Gaps = 20/172 (11%)
Query: 28 LVALALLLVSSQATPA----TTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYAR 83
L L L LV + AT + + + YL N R G+ + WS KL + A YA
Sbjct: 7 LAVLLLWLVMAAATAVHPSYSENSPQDYLTPQNSARAAVGVGP-VTWSTKLQQFAEKYAA 65
Query: 84 FRRD--KYLCTPGIFDKKRY-GVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCV 139
R + + G + + + G W K +D +R WV ++ +Y NSC
Sbjct: 66 QRAGDCRLQHSGGPYGENIFWGSAGFDW-----KAVD---AVRSWVDEKQWYNYATNSCA 117
Query: 140 PANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
A CG YTQ+VWR T +GCA+ C D GV + +C Y P GN+ G +P
Sbjct: 118 -AGKVCGHYTQVVWRATTSIGCARVVC-RDNRGVFI-ICNYEPRGNIAGMKP 166
>PR1_ARATH (P33154) Pathogenesis-related protein 1 precursor (PR-1)
Length = 161
Score = 59.7 bits (143), Expect = 4e-09
Identities = 54/170 (31%), Positives = 76/170 (43%), Gaps = 20/170 (11%)
Query: 24 FFAVLVALA-LLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYA 82
F V VAL L++ S+A + D YL HN R G+ ++W +++A A YA
Sbjct: 9 FLIVFVALVGALVLPSKAQDSPQD----YLRVHNQARGAVGVGP-MQWDERVAAYARSYA 63
Query: 83 RFRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPA 141
R I YG D + + WV ++ Y N+C
Sbjct: 64 EQLRGNCRL---IHSGGPYGENLAWGSGDLSGV----SAVNMWVSEKANYNYAANTC--- 113
Query: 142 NYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGERP 191
N CG YTQ+VWRK+ LGCA+ C + GG ++ C Y P GN E+P
Sbjct: 114 NGVCGHYTQVVWRKSVRLGCAKVRC--NNGGTIIS-CNYDPRGNYVNEKP 160
>PR04_LYCES (Q04108) Pathogenesis-related leaf protein 4 precursor
(P4)
Length = 159
Score = 58.5 bits (140), Expect = 9e-09
Identities = 47/143 (32%), Positives = 59/143 (40%), Gaps = 17/143 (11%)
Query: 51 YLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRR-DKYLCTPGIFDKKRYGVCCNQWL 109
YL HN R + G+ + W LA A YA R D L G + G
Sbjct: 31 YLAVHNDARAQVGVGP-MSWDANLASRAQNYANSRAGDCNLIHSGAGENLAKGG------ 83
Query: 110 EDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGP 168
R ++ WV +R Y N CV CG YTQ+VWR + LGC +A C
Sbjct: 84 ----GDFTGRAAVQLWVSERPDYNYATNQCVGGKM-CGHYTQVVWRNSVRLGCGRARCNN 138
Query: 169 DYGGVRLNVCFYYPPGNVPGERP 191
+ + C Y P GN GERP
Sbjct: 139 GWWFIS---CNYDPVGNWVGERP 158
>PRY1_YEAST (P47032) PRY1 protein precursor (Pathogen related in Sc
1)
Length = 299
Score = 54.3 bits (129), Expect = 2e-07
Identities = 46/154 (29%), Positives = 64/154 (40%), Gaps = 16/154 (10%)
Query: 37 SSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRRDKYLCTPGIF 96
+S + +D A L EHN R + L WSD LA A YA D Y C+ +
Sbjct: 152 ASSSDSDLSDFASSVLAEHNKKRALHKDTPALSWSDTLASYAQDYA----DNYDCSGTLT 207
Query: 97 DKKR-YGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQIVWRK 155
YG E+ D + W + Y+ +N +N G +TQ+VW+
Sbjct: 208 HSGGPYG-------ENLALGYDGPAAVDAWYNEISNYDFSNPGFSSN--TGHFTQVVWKS 258
Query: 156 TKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGE 189
T +GC CG +G +C Y P GN GE
Sbjct: 259 TTQVGCGIKTCGGAWGD--YVICSYDPAGNYEGE 290
>PR06_LYCES (P04284) Pathogenesis-related leaf protein 6 precursor
(P6) (Ethylene-induced protein P1) (P14) (P14A) (PR
protein)
Length = 159
Score = 54.3 bits (129), Expect = 2e-07
Identities = 45/143 (31%), Positives = 58/143 (40%), Gaps = 17/143 (11%)
Query: 51 YLFEHNFVRREKGISEHLEWSDKLAKDASLYARFRR-DKYLCTPGIFDKKRYGVCCNQWL 109
YL HN R + G+ + W LA A YA R D L G + G
Sbjct: 31 YLAVHNDARAQVGVGP-MSWDANLASRAQNYANSRAGDCNLIHSGAGENLAKGG------ 83
Query: 110 EDTVKIMDPREVMRQWVKDRFYYE-GNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGP 168
R ++ WV +R Y N CV C YTQ+VWR + LGC +A C
Sbjct: 84 ----GDFTGRAAVQLWVSERPSYNYATNQCVGGK-KCRHYTQVVWRNSVRLGCGRARCNN 138
Query: 169 DYGGVRLNVCFYYPPGNVPGERP 191
+ + C Y P GN G+RP
Sbjct: 139 GWWFIS---CNYDPVGNWIGQRP 158
>HELO_HELHO (Q91055) Helothermine precursor (HLTx)
Length = 242
Score = 52.0 bits (123), Expect = 8e-07
Identities = 43/140 (30%), Positives = 62/140 (43%), Gaps = 13/140 (9%)
Query: 54 EHNFVRR--EKGISEHLE--WSDKLAKDASLYARFRRDKYLCTPGIFDKKRY---GVCCN 106
+HN +RR E S L+ WS+K+A++A +R CT K+ GV C
Sbjct: 42 KHNNLRRIVEPTASNMLKMTWSNKIAQNA------QRSANQCTLEHTSKEERTIDGVECG 95
Query: 107 QWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAEC 166
+ L + ++ W +R Y+ N N G YTQ+VW ++ LGCA A C
Sbjct: 96 ENLFFSSAPYTWSYAIQNWFDERKYFRFNYGPTAQNVMIGHYTQVVWYRSYELGCAIAYC 155
Query: 167 GPDYGGVRLNVCFYYPPGNV 186
VC Y P GN+
Sbjct: 156 PDQPTYKYYQVCQYCPGGNI 175
>CRVP_LATSE (Q8JI38) Latisemin precursor
Length = 238
Score = 51.2 bits (121), Expect = 1e-06
Identities = 49/179 (27%), Positives = 73/179 (40%), Gaps = 25/179 (13%)
Query: 25 FAVLVALALLLVSSQAT-------PATTDGAKVYLFEHNFVRREKGISEH----LEWSDK 73
F VL++LA +L S T + K + +HN +RR + +EW+
Sbjct: 4 FIVLLSLAAVLQQSSGTVDFASESSNKRENQKEIVDKHNALRRSVRPTARNMLQMEWNSD 63
Query: 74 LAKDASLYAR----FRRDKYLCTPGIFDKKRYGVCCNQWLEDTVKIMDPREVMRQWVKDR 129
A++A +A +L T G F C + L + + V++ W +
Sbjct: 64 AAQNAQRWADRCSFAHSPSHLRTVGKFS-------CGENLFMSSQPYAWSRVIQSWYDEN 116
Query: 130 FYYEGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPG 188
+ + P G YTQIVW K+ LGCA A C L VC Y P GN+ G
Sbjct: 117 KNFIYDVGANPPGSVIGHYTQIVWYKSHLLGCAAARCS---SSKYLYVCQYCPAGNIIG 172
>CRVP_PSEAU (Q8AVA4) Pseudechetoxin precursor (PsTx)
Length = 238
Score = 49.3 bits (116), Expect = 5e-06
Identities = 47/176 (26%), Positives = 79/176 (44%), Gaps = 19/176 (10%)
Query: 25 FAVLVALALLLVSSQATP---ATTDGAKVYLFE----HNFVRREKGISEH----LEWSDK 73
F VL++LA +L S T + + K Y E HN +RR + ++W+ +
Sbjct: 4 FIVLLSLAAVLQQSSGTADFASESSNKKNYQKEIVDKHNALRRSVKPTARNMLQMKWNSR 63
Query: 74 LAKDASLYARFRRDKYLCTPGIFDKKRYG-VCCNQWLEDTVKIMDPREVMRQWVKDRFYY 132
A++A +A R + +P +K+ G + C + + + + V++ W + +
Sbjct: 64 AAQNAKRWAN--RCTFAHSPP--NKRTVGKLRCGENIFMSSQPFPWSGVVQAWYDEIKNF 119
Query: 133 EGNNSCVPANYPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPG 188
P G YTQ+VW K+ +GCA A+C L VC Y P GN+ G
Sbjct: 120 VYGIGAKPPGSVIGHYTQVVWYKSYLIGCASAKCS---SSKYLYVCQYCPAGNIRG 172
>PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3)
Length = 881
Score = 48.9 bits (115), Expect = 7e-06
Identities = 50/167 (29%), Positives = 66/167 (38%), Gaps = 19/167 (11%)
Query: 25 FAVLVALALLLVSSQATPATTDGAKVYLFEHNFVRREKGISEHLEWSDKLAKDASLYARF 84
F + V L L+ T + V L EHN R + L WSD LA A YA
Sbjct: 4 FPISVLLGCLVAVKAQTTFPNFESDV-LNEHNKFRALHVDTAPLTWSDTLATYAQNYA-- 60
Query: 85 RRDKYLCTPGIFDKKR--YGVCCNQWLEDTVKIMDPREVMRQWVKDRFYYEGNNSCVPAN 142
D+Y C+ G+ YG DT + W + Y +N +
Sbjct: 61 --DQYDCS-GVLTHSDGPYGENLALGYTDTGAV-------DAWYGEISKYNYSNPGFSES 110
Query: 143 YPCGDYTQIVWRKTKYLGCAQAECGPDYGGVRLNVCFYYPPGNVPGE 189
G +TQ+VW+ T +GC CG + VC Y PPGN GE
Sbjct: 111 --TGHFTQVVWKSTAEIGCGYKYCGTTWN--NYIVCSYNPPGNYLGE 153
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.142 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,085,592
Number of Sequences: 164201
Number of extensions: 1080976
Number of successful extensions: 2432
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 2319
Number of HSP's gapped (non-prelim): 90
length of query: 191
length of database: 59,974,054
effective HSP length: 104
effective length of query: 87
effective length of database: 42,897,150
effective search space: 3732052050
effective search space used: 3732052050
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0323a.9


