

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
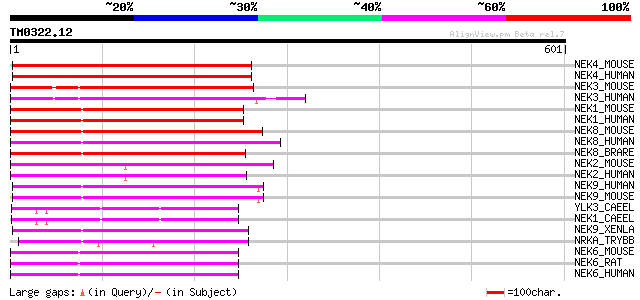
Query= TM0322.12
(601 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NEK4_MOUSE (Q9Z1J2) Serine/threonine-protein kinase Nek4 (EC 2.7... 249 2e-65
NEK4_HUMAN (P51957) Serine/threonine-protein kinase Nek4 (EC 2.7... 245 2e-64
NEK3_MOUSE (Q9R0A5) Serine/threonine-protein kinase Nek3 (EC 2.7... 223 1e-57
NEK3_HUMAN (P51956) Serine/threonine-protein kinase Nek3 (EC 2.7... 219 2e-56
NEK1_MOUSE (P51954) Serine/threonine-protein kinase Nek1 (EC 2.7... 217 6e-56
NEK1_HUMAN (Q96PY6) Serine/threonine-protein kinase Nek1 (EC 2.7... 216 1e-55
NEK8_MOUSE (Q91ZR4) Serine/threonine-protein kinase Nek8 (EC 2.7... 197 5e-50
NEK8_HUMAN (Q86SG6) Serine/threonine-protein kinase Nek8 (EC 2.7... 196 1e-49
NEK8_BRARE (Q90XC2) Serine/threonine-protein kinase Nek8 (EC 2.7... 186 2e-46
NEK2_MOUSE (O35942) Serine/threonine-protein kinase Nek2 (EC 2.7... 170 1e-41
NEK2_HUMAN (P51955) Serine/threonine-protein kinase Nek2 (EC 2.7... 170 1e-41
NEK9_HUMAN (Q8TD19) Serine/threonine-protein kinase Nek9 (EC 2.7... 167 7e-41
NEK9_MOUSE (Q8K1R7) Serine/threonine-protein kinase Nek9 (EC 2.7... 163 1e-39
YLK3_CAEEL (P41951) Putative serine/threonine-protein kinase D10... 159 2e-38
NEK1_CAEEL (P84199) Serine/threonine protein kinase D1044.8 (EC ... 159 2e-38
NEK9_XENLA (Q7ZZC8) Serine/threonine-protein kinase Nek9 (EC 2.7... 158 4e-38
NRKA_TRYBB (Q08942) Putative serine/threonine-protein kinase A (... 152 3e-36
NEK6_MOUSE (Q9ES70) Serine/threonine-protein kinase Nek6 (EC 2.7... 151 5e-36
NEK6_RAT (P59895) Serine/threonine-protein kinase Nek6 (EC 2.7.1... 150 9e-36
NEK6_HUMAN (Q9HC98) Serine/threonine-protein kinase Nek6 (EC 2.7... 150 9e-36
>NEK4_MOUSE (Q9Z1J2) Serine/threonine-protein kinase Nek4 (EC
2.7.1.37) (NimA-related protein kinase 4)
(Serine/threonine-protein kinase 2)
Length = 792
Score = 249 bits (635), Expect = 2e-65
Identities = 114/260 (43%), Positives = 177/260 (67%), Gaps = 1/260 (0%)
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFI 63
Y + +G+GS+G LV+H+ + K YV+KK+ L S R RR+A QE +L+S++++P I
Sbjct: 6 YCYMRVVGRGSYGEVTLVKHRRDGKQYVIKKLNLRNASSRERRAAEQEAQLLSQLKHPNI 65
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
V YK+SW + I++G+CE GDL +K+ G PE ++ +W VQ+ MAL YLH
Sbjct: 66 VTYKESWEGGDGLLYIVMGFCEGGDLYRKLKEQKGQLLPESQVVEWFVQIAMALQYLHEK 125
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTS-DDLASSIVGTPSYMCPELLADIPYGS 182
HILHRD+K N+FLT+ I++GD G+A++L + D+AS+++GTP YM PEL ++ PY
Sbjct: 126 HILHRDLKTQNVFLTRTNIIKVGDLGIARVLENHGDMASTLIGTPYYMSPELFSNKPYNY 185
Query: 183 KSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRKN 242
KSD+W+LGCC+YEMA LK AF A D+ +L+ +I + + P+P +YS+ L+++ML +
Sbjct: 186 KSDVWALGCCVYEMATLKHAFNAKDMNSLVYRIIEGKLPPMPKVYSTELAELIRTMLSRR 245
Query: 243 PELRPPASELLNHPHLQPYI 262
PE RP +L P+++ +I
Sbjct: 246 PEERPSVRSILRQPYIKHHI 265
>NEK4_HUMAN (P51957) Serine/threonine-protein kinase Nek4 (EC
2.7.1.37) (NimA-related protein kinase 4)
(Serine/threonine-protein kinase 2)
(Serine/threonine-protein kinase NRK2)
Length = 841
Score = 245 bits (626), Expect = 2e-64
Identities = 116/260 (44%), Positives = 173/260 (65%), Gaps = 1/260 (0%)
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFI 63
Y L +GKGS+G LV+H+ + K YV+KK+ L S R RR+A QE +L+S++++P I
Sbjct: 6 YCYLRVVGKGSYGEVTLVKHRRDGKQYVIKKLNLRNASSRERRAAEQEAQLLSQLKHPNI 65
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
V YK+SW + I++G+CE GDL +K+ G PE ++ +W VQ+ MAL YLH
Sbjct: 66 VTYKESWEGGDGLLYIVMGFCEGGDLYRKLKEQKGQLLPENQVVEWFVQIAMALQYLHEK 125
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIPYGS 182
HILHRD+K N+FLT+ I++GD G+A++L + D+AS+++GTP YM PEL ++ PY
Sbjct: 126 HILHRDLKTQNVFLTRTNIIKVGDLGIARVLENHCDMASTLIGTPYYMSPELFSNKPYNY 185
Query: 183 KSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRKN 242
KSD+W+LGCC+YEMA LK AF A D+ +L+ +I + + +P YS L+++ML K
Sbjct: 186 KSDVWALGCCVYEMATLKHAFNAKDMNSLVYRIIEGKLPAMPRDYSPELAELIRTMLSKR 245
Query: 243 PELRPPASELLNHPHLQPYI 262
PE RP +L P+++ I
Sbjct: 246 PEERPSVRSILRQPYIKRQI 265
>NEK3_MOUSE (Q9R0A5) Serine/threonine-protein kinase Nek3 (EC
2.7.1.37) (NimA-related protein kinase 3)
Length = 511
Score = 223 bits (567), Expect = 1e-57
Identities = 113/265 (42%), Positives = 173/265 (64%), Gaps = 5/265 (1%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+ Y VL IG+GSFG ALLV + + + +K+IRL + +T R +E L++K+++
Sbjct: 1 MDNYTVLRVIGQGSFGRALLVLQESSNQTFAMKEIRLLKSDTQTSR---KEAVLLAKMKH 57
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P IV +K+S+ +G ++ I++ YC+ GDL IK+ G FPE+ + W +Q+ + ++++
Sbjct: 58 PNIVAFKESFEAEG-YLYIVMEYCDGGDLMQRIKQQKGNLFPEDTILNWFIQICLGVNHI 116
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIP 179
H +LHRD+K N+FLT N ++LGDFG A++L+S A + VGTP Y+ PE+ ++P
Sbjct: 117 HKRRVLHRDIKSKNVFLTHNGKVKLGDFGSARLLSSPMAFACTYVGTPYYVPPEIWENLP 176
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y +KSDIWSLGC +YE+ ALK F+A + LI KI + + PLP +YS +GLVK ML
Sbjct: 177 YNNKSDIWSLGCILYELCALKHPFQANSWKNLILKICQGPIHPLPALYSCKLQGLVKQML 236
Query: 240 RKNPELRPPASELLNHPHLQPYILK 264
++NP RP A+ LL L P + K
Sbjct: 237 KRNPSHRPSATTLLCRGSLAPLVPK 261
>NEK3_HUMAN (P51956) Serine/threonine-protein kinase Nek3 (EC
2.7.1.37) (NimA-related protein kinase 3) (HSPK 36)
Length = 506
Score = 219 bits (558), Expect = 2e-56
Identities = 123/341 (36%), Positives = 201/341 (58%), Gaps = 32/341 (9%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+ Y VL IG+GSFG ALLV+H+ +++ +K+IRL + T+ S +E L++K+++
Sbjct: 1 MDDYMVLRMIGEGSFGRALLVQHESSNQMFAMKEIRLPKSFSNTQNS-RKEAVLLAKMKH 59
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P IV +K+S+ +G + I++ YC+ GDL IK+ G FPE+ + W Q+ + ++++
Sbjct: 60 PNIVAFKESFEAEG-HLYIVMEYCDGGDLMQKIKQQKGKLFPEDMILNWFTQMCLGVNHI 118
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIP 179
H +LHRD+K NIFLT+N ++LGDFG A++L++ A + VGTP Y+ PE+ ++P
Sbjct: 119 HKKRVLHRDIKSKNIFLTQNGKVKLGDFGSARLLSNPMAFACTYVGTPYYVPPEIWENLP 178
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y +KSDIWSLGC +YE+ LK F+A + LI K+ + ++PLP+ YS + LVK M
Sbjct: 179 YNNKSDIWSLGCILYELCTLKHPFQANSWKNLILKVCQGCISPLPSHYSYELQFLVKQMF 238
Query: 240 RKNPELRPPASELLNHPHLQPYILKI------------------HLKLNSPRRSTFPFQW 281
++NP RP A+ LL+ + + K + K N+PR+ T P
Sbjct: 239 KRNPSHRPSATTLLSRGIVARLVQKCLPPEIIMEYGEEVLEEIKNSKHNTPRKKTNP--- 295
Query: 282 PESNYMRRTRFVVPDSVSTL--SDQDKCLSLSNDRSLNPSI 320
R R + + ST+ +QD+ S ++ S+N ++
Sbjct: 296 ------SRIRIALGNEASTVQEEEQDRKGSHTDLESINENL 330
>NEK1_MOUSE (P51954) Serine/threonine-protein kinase Nek1 (EC
2.7.1.37) (NimA-related protein kinase 1)
Length = 774
Score = 217 bits (553), Expect = 6e-56
Identities = 105/254 (41%), Positives = 169/254 (66%), Gaps = 2/254 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME+Y L++IG+GSFG A+LV+ + + YV+K+I ++R SD+ R+ + +E+ +++ +++
Sbjct: 1 MEKYVRLQKIGEGSFGKAVLVKSTEDGRHYVIKEINISRMSDKERQESRREVAVLANMKH 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P IV+YK+S+ E G I++ YCE GDL I G F E+++ W VQ+ +AL ++
Sbjct: 61 PNIVQYKESFEENGSLY-IVMDYCEGGDLFKRINAQKGALFQEDQILDWFVQICLALKHV 119
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIP 179
H ILHRD+K NIFLTK+ ++LGDFG+A++L S +LA + +GTP Y+ PE+ + P
Sbjct: 120 HDRKILHRDIKSQNIFLTKDGTVQLGDFGIARVLNSTVELARTCIGTPYYLSPEICENKP 179
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y +KSDIW+LGC +YE+ LK AF+A +++ L+ KI P+ YS R L+ +
Sbjct: 180 YNNKSDIWALGCVLYELCTLKHAFEAGNMKNLVLKIISGSFPPVSPHYSYDLRSLLSQLF 239
Query: 240 RKNPELRPPASELL 253
++NP RP + +L
Sbjct: 240 KRNPRDRPSVNSIL 253
>NEK1_HUMAN (Q96PY6) Serine/threonine-protein kinase Nek1 (EC
2.7.1.37) (NimA-related protein kinase 1) (NY-REN-55
antigen)
Length = 1258
Score = 216 bits (551), Expect = 1e-55
Identities = 105/254 (41%), Positives = 167/254 (65%), Gaps = 2/254 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME+Y L++IG+GSFG A+LV+ + + YV+K+I ++R S + R + +E+ +++ +++
Sbjct: 1 MEKYVRLQKIGEGSFGKAILVKSTEDGRQYVIKEINISRMSSKEREESRREVAVLANMKH 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P IV+Y++S+ E G I++ YCE GDL I GV F E+++ W VQ+ +AL ++
Sbjct: 61 PNIVQYRESFEENGSLY-IVMDYCEGGDLFKRINAQKGVLFQEDQILDWFVQICLALKHV 119
Query: 121 HVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIP 179
H ILHRD+K NIFLTK+ ++LGDFG+A++L S +LA + +GTP Y+ PE+ + P
Sbjct: 120 HDRKILHRDIKSQNIFLTKDGTVQLGDFGIARVLNSTVELARTCIGTPYYLSPEICENKP 179
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y +KSDIW+LGC +YE+ LK AF+A ++ L+ KI P+ YS R LV +
Sbjct: 180 YNNKSDIWALGCVLYELCTLKHAFEAGSMKNLVLKIISGSFPPVSLHYSYDLRSLVSQLF 239
Query: 240 RKNPELRPPASELL 253
++NP RP + +L
Sbjct: 240 KRNPRDRPSVNSIL 253
>NEK8_MOUSE (Q91ZR4) Serine/threonine-protein kinase Nek8 (EC
2.7.1.37) (NimA-related protein kinase 8)
Length = 698
Score = 197 bits (502), Expect = 5e-50
Identities = 105/274 (38%), Positives = 168/274 (60%), Gaps = 2/274 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME+YE + +G+G+FG L K ++KL +LK+I + + + R++A E +++ + +
Sbjct: 1 MEKYERIRVVGRGAFGIVHLCLRKADQKLVILKQIPVEQMTKEERQAAQNECQVLKLLNH 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P ++EY ++++E + I + Y G LA+ I+K EE + + VQ+L+AL ++
Sbjct: 61 PNVIEYYENFLEDKALM-IAMEYAPGGTLAEFIQKRCNSLLEEETILHFFVQILLALHHV 119
Query: 121 HVNHILHRDVKCSNIFLTKNQDI-RLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIP 179
H + ILHRD+K NI L K++ + ++GDFG++K+L+S A ++VGTP Y+ PEL P
Sbjct: 120 HTHLILHRDLKTQNILLDKHRMVVKIGDFGISKILSSKSKAYTVVGTPCYISPELCEGKP 179
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y KSDIW+LGC +YE+A+LK AF+A ++ AL+ KI AP+ YS R LV S+L
Sbjct: 180 YNQKSDIWALGCVLYELASLKRAFEAANLPALVLKIMSGTFAPISDRYSPELRQLVLSLL 239
Query: 240 RKNPELRPPASELLNHPHLQPYILKIHLKLNSPR 273
P RPP S ++ P +L IH + S R
Sbjct: 240 SLEPAQRPPLSHIMAQPLCIRALLNIHTDVGSVR 273
>NEK8_HUMAN (Q86SG6) Serine/threonine-protein kinase Nek8 (EC
2.7.1.37) (NimA-related protein kinase 8) (NIMA-related
kinase 12a)
Length = 692
Score = 196 bits (499), Expect = 1e-49
Identities = 108/294 (36%), Positives = 173/294 (58%), Gaps = 2/294 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME+YE + +G+G+FG L K ++KL ++K+I + + + R++A E +++ + +
Sbjct: 1 MEKYERIRVVGRGAFGIVHLCLRKADQKLVIIKQIPVEQMTKEERQAAQNECQVLKLLNH 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P ++EY ++++E + I + Y G LA+ I+K EE + + VQ+L+AL ++
Sbjct: 61 PNVIEYYENFLEDKALM-IAMEYAPGGTLAEFIQKRCNSLLEEETILHFFVQILLALHHV 119
Query: 121 HVNHILHRDVKCSNIFLTKNQDI-RLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIP 179
H + ILHRD+K NI L K++ + ++GDFG++K+L+S A ++VGTP Y+ PEL P
Sbjct: 120 HTHLILHRDLKTQNILLDKHRMVVKIGDFGISKILSSKSKAYTVVGTPCYISPELCEGKP 179
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y KSDIW+LGC +YE+A+LK AF+A ++ AL+ KI AP+ YS R LV S+L
Sbjct: 180 YNQKSDIWALGCVLYELASLKRAFEAANLPALVLKIMSGTFAPISDRYSPELRQLVLSLL 239
Query: 240 RKNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESNYMRRTRFV 293
P RPP S ++ P +L +H + S R SN RT V
Sbjct: 240 SLEPAQRPPLSHIMAQPLCIRALLNLHTDVGSVRMRRAEKSVAPSNTGSRTTSV 293
>NEK8_BRARE (Q90XC2) Serine/threonine-protein kinase Nek8 (EC
2.7.1.37) (NimA-related protein kinase 8)
Length = 697
Score = 186 bits (471), Expect = 2e-46
Identities = 100/256 (39%), Positives = 157/256 (61%), Gaps = 2/256 (0%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
ME+YE + +G+G+FG L R + + L +LK+I + + + R +A E +++ + +
Sbjct: 1 MEKYEKTKVVGRGAFGIVHLCRRRTDSALVILKEIPVEQMTRDERLAAQNECQVLKLLSH 60
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P I+EY ++++E + I + Y G LAD I+K E+ + VQ+L+AL ++
Sbjct: 61 PNIIEYYENFLEDKALM-IAMEYAPGGTLADYIQKRCNSLLDEDTILHSFVQILLALYHV 119
Query: 121 HVNHILHRDVKCSNIFLTKNQDI-RLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADIP 179
H ILHRD+K NI L K+Q I ++GDFG++K+L S A ++VGTP Y+ PEL P
Sbjct: 120 HNKLILHRDLKTQNILLDKHQMIVKIGDFGISKILVSKSKAYTVVGTPCYISPELCEGKP 179
Query: 180 YGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSML 239
Y KSDIW+LGC +YE+A+LK AF+A ++ AL+ KI AP+ YS R L+ +ML
Sbjct: 180 YNQKSDIWALGCVLYELASLKRAFEAANLPALVLKIMSGTFAPISDRYSPELRQLILNML 239
Query: 240 RKNPELRPPASELLNH 255
+P RP +E++ H
Sbjct: 240 NLDPSKRPQLNEIMAH 255
>NEK2_MOUSE (O35942) Serine/threonine-protein kinase Nek2 (EC
2.7.1.37) (NimA-related protein kinase 2)
Length = 443
Score = 170 bits (430), Expect = 1e-41
Identities = 102/296 (34%), Positives = 157/296 (52%), Gaps = 11/296 (3%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
+E YEVL IG GS+G +R K + K+ V K++ ++ ++ E+ L+ ++++
Sbjct: 5 VEDYEVLHSIGTGSYGRCQKIRRKSDGKILVWKELDYGSMTEVEKQMLVSEVNLLRELKH 64
Query: 61 PFIVEYKDSWVEK-GCFVCIIVGYCERGDLADAIKKANGVN--FPEEKLCKWLVQLLMAL 117
P IV Y D +++ + I++ YCE GDLA I K EE + + + QL +AL
Sbjct: 65 PNIVRYYDRIIDRTNTTLYIVMEYCEGGDLASVISKGTKDRQYLEEEFVLRVMTQLTLAL 124
Query: 118 DYLHVNH-----ILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMC 171
H +LHRD+K +N+FL +++LGDFGLA++L D A + VGTP YM
Sbjct: 125 KECHRRSDGGHTVLHRDLKPANVFLDSKHNVKLGDFGLARILNHDTSFAKTFVGTPYYMS 184
Query: 172 PELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAF 231
PE ++ + Y KSDIWSL C +YE+ AL P F AF+ + L KI + +P YS
Sbjct: 185 PEQMSCLSYNEKSDIWSLACLLYELCALMPPFTAFNQKELAGKIREGRFRRIPYRYSDGL 244
Query: 232 RGLVKSMLRKNPELRPPASELLNHPHLQPYILKIHLK--LNSPRRSTFPFQWPESN 285
L+ ML RP E+L P + + + + RRS P + P+S+
Sbjct: 245 NDLITRMLFLKDYHRPSVEEILESPLIADMVAEEQRRNLERRGRRSGEPSKLPDSS 300
>NEK2_HUMAN (P51955) Serine/threonine-protein kinase Nek2 (EC
2.7.1.37) (NimA-related protein kinase 2) (NimA-like
protein kinase 1) (HSPK 21)
Length = 445
Score = 170 bits (430), Expect = 1e-41
Identities = 97/264 (36%), Positives = 144/264 (53%), Gaps = 9/264 (3%)
Query: 2 EQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNP 61
E YEVL IG GS+G +R K + K+ V K++ ++ ++ E+ L+ ++++P
Sbjct: 6 EDYEVLYTIGTGSYGRCQKIRRKSDGKILVWKELDYGSMTEAEKQMLVSEVNLLRELKHP 65
Query: 62 FIVEYKDSWVEK-GCFVCIIVGYCERGDLADAIKKANGVN--FPEEKLCKWLVQLLMALD 118
IV Y D +++ + I++ YCE GDLA I K EE + + + QL +AL
Sbjct: 66 NIVRYYDRIIDRTNTTLYIVMEYCEGGDLASVITKGTKERQYLDEEFVLRVMTQLTLALK 125
Query: 119 YLHVNH-----ILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCP 172
H +LHRD+K +N+FL Q+++LGDFGLA++L D A + VGTP YM P
Sbjct: 126 ECHRRSDGGHTVLHRDLKPANVFLDGKQNVKLGDFGLARILNHDTSFAKTFVGTPYYMSP 185
Query: 173 ELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFR 232
E + + Y KSDIWSLGC +YE+ AL P F AF + L KI + +P YS
Sbjct: 186 EQMNRMSYNEKSDIWSLGCLLYELCALMPPFTAFSQKELAGKIREGKFRRIPYRYSDELN 245
Query: 233 GLVKSMLRKNPELRPPASELLNHP 256
++ ML RP E+L +P
Sbjct: 246 EIITRMLNLKDYHRPSVEEILENP 269
>NEK9_HUMAN (Q8TD19) Serine/threonine-protein kinase Nek9 (EC
2.7.1.37) (NimA-related protein kinase 9) (Nercc1
kinase) (NIMA-related kinase 8) (Nek8)
Length = 979
Score = 167 bits (423), Expect = 7e-41
Identities = 104/277 (37%), Positives = 159/277 (56%), Gaps = 7/277 (2%)
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFI 63
Y + +G+G+FG A L R + L V K++ L R S++ RR A E+ +++ +++ I
Sbjct: 52 YIPIRVLGRGAFGEATLYRRTEDDSLVVWKEVDLTRLSEKERRDALNEIVILALLQHDNI 111
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
+ Y + +++ + I + YC G+L D I + F EE + +L Q++ A+ +H
Sbjct: 112 IAYYNHFMDNTTLL-IELEYCNGGNLYDKILRQKDKLFEEEMVVWYLFQIVSAVSCIHKA 170
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIPYGS 182
ILHRD+K NIFLTK I+LGD+GLAK L S+ +A ++VGTP YM PEL + Y
Sbjct: 171 GILHRDIKTLNIFLTKANLIKLGDYGLAKKLNSEYSMAETLVGTPYYMSPELCQGVKYNF 230
Query: 183 KSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPL--PTMYSSAFRGLVKSMLR 240
KSDIW++GC I+E+ LK F A + L KI + I A + YS +V S L
Sbjct: 231 KSDIWAVGCVIFELLTLKRTFDATNPLNLCVKIVQGIRAMEVDSSQYSLELIQMVHSCLD 290
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLK---LNSPRR 274
++PE RP A ELL+ P L+ ++ K LN+P +
Sbjct: 291 QDPEQRPTADELLDRPLLRKRRREMEEKVTLLNAPTK 327
>NEK9_MOUSE (Q8K1R7) Serine/threonine-protein kinase Nek9 (EC
2.7.1.37) (NimA-related protein kinase 9)
Length = 984
Score = 163 bits (413), Expect = 1e-39
Identities = 103/277 (37%), Positives = 158/277 (56%), Gaps = 7/277 (2%)
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFI 63
Y + +G+G+FG A L R + L V K++ L R S++ RR A E+ +++ +++ I
Sbjct: 52 YIPIRVLGRGAFGEATLYRRTEDDSLVVWKEVDLTRLSEKERRDALNEIVILALLQHDNI 111
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
+ Y + +++ + I + YC G+L D I + F EE + +L Q++ A+ +H
Sbjct: 112 IAYYNHFMDNTTLL-IELEYCNGGNLYDKILRQKDKLFEEEMVVWYLFQIVSAVSCIHKA 170
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIPYGS 182
ILHRD+K NIFLTK I+LGD+GLAK L S+ +A ++VGTP YM PEL + Y
Sbjct: 171 GILHRDIKTLNIFLTKANLIKLGDYGLAKKLNSEYSMAETLVGTPYYMSPELCQGVKYNF 230
Query: 183 KSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPL--PTMYSSAFRGLVKSMLR 240
KSDIW++GC I+E+ LK F A + L KI + I A + YS LV + L
Sbjct: 231 KSDIWAVGCVIFELLTLKRTFDATNPLNLCVKIVQGIRAMEVDSSQYSLELIQLVHACLD 290
Query: 241 KNPELRPPASELLNHPHLQPYILKIHLK---LNSPRR 274
++PE RP A LL+ P L+ ++ K LN+P +
Sbjct: 291 QDPEQRPAADALLDLPLLRTRRREMEEKVTLLNAPTK 327
>YLK3_CAEEL (P41951) Putative serine/threonine-protein kinase
D1044.3 (EC 2.7.1.37)
Length = 1576
Score = 159 bits (402), Expect = 2e-38
Identities = 97/259 (37%), Positives = 152/259 (58%), Gaps = 16/259 (6%)
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEK------KLYVLKKIRLA----RQSDRTRRSAHQEM 52
+YE+L+Q+G G+FG VR K + KL LK+I + R+SD++ E+
Sbjct: 430 EYELLDQLGAGAFGCVYTVRKKAQSHSENPAKLLALKEIFMTNLNDRESDKSFGDMISEV 489
Query: 53 ELISK-VRNPFIVEYKDSWVEKGCFVCI--IVGYCERGDLADAIKKANGVNFPEEKLCKW 109
++I + +R+P IV Y+ +VE + ++ C DL +K+ G NF E+K+
Sbjct: 490 KIIKQQLRHPNIVRYRRIFVENHRLYIVMDLIQGCSLRDLIITMKEKKG-NFEEKKIWAM 548
Query: 110 LVQLLMALDYLHVN-HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPS 168
+VQ+++AL YLH I+HRD+K +NI +T ++ + + DFGLAK + L S+ GT
Sbjct: 549 VVQMMLALRYLHKEKQIVHRDLKPNNIMMTTDERVVITDFGLAKQKGPEYLKSA-AGTII 607
Query: 169 YMCPELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYS 228
Y CPE++ ++PYG K+DIWS GCCIYEM L+P F + ++ L +I ++ PL M+S
Sbjct: 608 YSCPEIVQNLPYGEKADIWSFGCCIYEMCQLQPVFHSTNMLTLAMQIVEAKYDPLNEMWS 667
Query: 229 SAFRGLVKSMLRKNPELRP 247
R L+ S L +P RP
Sbjct: 668 DDLRFLITSCLAPDPSARP 686
>NEK1_CAEEL (P84199) Serine/threonine protein kinase D1044.8 (EC
2.7.1.37)
Length = 959
Score = 159 bits (402), Expect = 2e-38
Identities = 97/259 (37%), Positives = 152/259 (58%), Gaps = 16/259 (6%)
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEK------KLYVLKKIRLA----RQSDRTRRSAHQEM 52
+YE+L+Q+G G+FG VR K + KL LK+I + R+SD++ E+
Sbjct: 430 EYELLDQLGAGAFGCVYTVRKKAQSHSENPAKLLALKEIFMTNLNDRESDKSFGDMISEV 489
Query: 53 ELISK-VRNPFIVEYKDSWVEKGCFVCI--IVGYCERGDLADAIKKANGVNFPEEKLCKW 109
++I + +R+P IV Y+ +VE + ++ C DL +K+ G NF E+K+
Sbjct: 490 KIIKQQLRHPNIVRYRRIFVENHRLYIVMDLIQGCSLRDLIITMKEKKG-NFEEKKIWAM 548
Query: 110 LVQLLMALDYLHVN-HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPS 168
+VQ+++AL YLH I+HRD+K +NI +T ++ + + DFGLAK + L S+ GT
Sbjct: 549 VVQMMLALRYLHKEKQIVHRDLKPNNIMMTTDERVVITDFGLAKQKGPEYLKSA-AGTII 607
Query: 169 YMCPELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYS 228
Y CPE++ ++PYG K+DIWS GCCIYEM L+P F + ++ L +I ++ PL M+S
Sbjct: 608 YSCPEIVQNLPYGEKADIWSFGCCIYEMCQLQPVFHSTNMLTLAMQIVEAKYDPLNEMWS 667
Query: 229 SAFRGLVKSMLRKNPELRP 247
R L+ S L +P RP
Sbjct: 668 DDLRFLITSCLAPDPSARP 686
>NEK9_XENLA (Q7ZZC8) Serine/threonine-protein kinase Nek9 (EC
2.7.1.37) (NimA-related protein kinase 9) (XNek9)
(Nercc1 kinase)
Length = 944
Score = 158 bits (399), Expect = 4e-38
Identities = 95/258 (36%), Positives = 146/258 (55%), Gaps = 4/258 (1%)
Query: 4 YEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFI 63
Y + +G G++G A L R + L V K++ LAR S++ RR A E+ ++S +++ I
Sbjct: 34 YIPIRVLGHGAYGEATLYRRTEDDSLVVWKEVGLARLSEKERRDALNEIVILSLLQHDNI 93
Query: 64 VEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVN 123
+ Y + +++ + I + YC G+L D I F EE + +L Q++ A+ +H
Sbjct: 94 IAYYNHFLDSNTLL-IELEYCNGGNLFDKIVHQKAQLFQEETVLWYLFQIVSAVSCIHKA 152
Query: 124 HILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSD-DLASSIVGTPSYMCPELLADIPYGS 182
ILHRD+K NIFLTK I+LGD+GLAK L+S+ +A + VGT YM PE+ + Y
Sbjct: 153 GILHRDIKTLNIFLTKANLIKLGDYGLAKQLSSEYSMAETCVGTLYYMSPEICQGVKYSF 212
Query: 183 KSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKS--IVAPLPTMYSSAFRGLVKSMLR 240
KSDIW++GC +YE+ L F A + L KI + V T+Y+ +V + L
Sbjct: 213 KSDIWAVGCVLYELLTLTRTFDATNPLNLCVKIVQGNWAVGLDNTVYTQELIEVVHACLE 272
Query: 241 KNPELRPPASELLNHPHL 258
++PE RP A E+L P L
Sbjct: 273 QDPEKRPTADEILERPIL 290
>NRKA_TRYBB (Q08942) Putative serine/threonine-protein kinase A (EC
2.7.1.37)
Length = 431
Score = 152 bits (383), Expect = 3e-36
Identities = 83/255 (32%), Positives = 141/255 (54%), Gaps = 7/255 (2%)
Query: 10 IGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFIVEYKDS 69
+G GS+G A + + L V K + L++ S R +R A E++ ++ +P I+ Y +
Sbjct: 26 VGLGSYGEAYVAESVEDGSLCVAKVMDLSKMSQRDKRYAQSEIKCLANCNHPNIIRYIED 85
Query: 70 WVEKGCFVCIIVGYCERGDLADAIK---KANGVNFPEEKLCKWLVQLLMALDYLHVNHIL 126
E + I++ + + G+L + IK + F E + +QL +ALDY+H + +L
Sbjct: 86 HEENDRLL-IVMEFADSGNLDEQIKLRGSGDARYFQEHEALFLFLQLCLALDYIHSHKML 144
Query: 127 HRDVKCSNIFLTKNQDIRLGDFGLAKML---TSDDLASSIVGTPSYMCPELLADIPYGSK 183
HRD+K +N+ LT ++LGDFG + S +AS+ GTP Y+ PEL + Y K
Sbjct: 145 HRDIKSANVLLTSTGLVKLGDFGFSHQYEDTVSGVVASTFCGTPYYLAPELWNNKRYNKK 204
Query: 184 SDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRKNP 243
+D+WSLG +YE+ +K F A +++ L++K+ APLP +SS F+ +V +L +P
Sbjct: 205 ADVWSLGVLLYEIMGMKKPFSASNLKGLMSKVLAGTYAPLPDSFSSEFKRVVDGILVADP 264
Query: 244 ELRPPASELLNHPHL 258
RP E+ P++
Sbjct: 265 NDRPSVREIFQIPYI 279
>NEK6_MOUSE (Q9ES70) Serine/threonine-protein kinase Nek6 (EC
2.7.1.37) (NimA-related protein kinase 6)
Length = 313
Score = 151 bits (381), Expect = 5e-36
Identities = 87/254 (34%), Positives = 146/254 (57%), Gaps = 8/254 (3%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSD-RTRRSAHQEMELISKVR 59
+ +++ ++IG+G F ++K LKK+++ D + R+ +E+ L+ ++
Sbjct: 42 LADFQIEKKIGRGQFSEVYKATCLLDRKTVALKKVQIFEMMDAKARQDCVKEIGLLKQLN 101
Query: 60 NPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIK--KANGVNFPEEKLCKWLVQLLMAL 117
+P I++Y DS++E + I++ + GDL+ IK K PE + K+ VQL A+
Sbjct: 102 HPNIIKYLDSFIEDN-ELNIVLELADAGDLSQMIKYFKKQKRLIPERTVWKYFVQLCSAV 160
Query: 118 DYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLAS-SIVGTPSYMCPELLA 176
+++H ++HRD+K +N+F+T ++LGD GL + +S+ A+ S+VGTP YM PE +
Sbjct: 161 EHMHSRRVMHRDIKPANVFITATGIVKLGDLGLGRFFSSETTAAHSLVGTPYYMSPERIH 220
Query: 177 DIPYGSKSDIWSLGCCIYEMAALKPAF--KAFDIQALINKINKSIVAPLP-TMYSSAFRG 233
+ Y KSDIWSLGC +YEMAAL+ F ++ +L KI + PLP YS R
Sbjct: 221 ENGYNFKSDIWSLGCLLYEMAALQSPFYGDKMNLFSLCQKIEQCDYPPLPGEHYSEKLRE 280
Query: 234 LVKSMLRKNPELRP 247
LV + +P+ RP
Sbjct: 281 LVSMCIYPDPDHRP 294
>NEK6_RAT (P59895) Serine/threonine-protein kinase Nek6 (EC
2.7.1.37) (NimA-related protein kinase 6)
Length = 313
Score = 150 bits (379), Expect = 9e-36
Identities = 87/254 (34%), Positives = 145/254 (56%), Gaps = 8/254 (3%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSD-RTRRSAHQEMELISKVR 59
+ +++ ++IG+G F ++K LKK+++ D + R+ +E+ L+ ++
Sbjct: 42 LADFQIEKKIGRGQFSEVYKATCLLDRKTVALKKVQIFEMMDAKARQDCVKEIGLLKQLN 101
Query: 60 NPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIK--KANGVNFPEEKLCKWLVQLLMAL 117
+P I++Y DS++E + I++ + GDL+ IK K PE + K+ VQL A+
Sbjct: 102 HPNIIKYLDSFIEDN-ELNIVLELADAGDLSQMIKYFKKQKRLIPERTVWKYFVQLCSAV 160
Query: 118 DYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLAS-SIVGTPSYMCPELLA 176
+++H ++HRD+K +N+F+T ++LGD GL + +S+ A+ S+VGTP YM PE +
Sbjct: 161 EHMHSRRVMHRDIKPANVFITATGIVKLGDLGLGRFFSSETTAAHSLVGTPYYMSPERIH 220
Query: 177 DIPYGSKSDIWSLGCCIYEMAALKPAF--KAFDIQALINKINKSIVAPLP-TMYSSAFRG 233
+ Y KSDIWSLGC +YEMAAL+ F ++ +L KI + PLP YS R
Sbjct: 221 ENGYNFKSDIWSLGCLLYEMAALQSPFYGDKMNLFSLCQKIEQCDYPPLPGEHYSEKLRE 280
Query: 234 LVKSMLRKNPELRP 247
LV + +P RP
Sbjct: 281 LVSMCIYPDPNHRP 294
>NEK6_HUMAN (Q9HC98) Serine/threonine-protein kinase Nek6 (EC
2.7.1.37) (NimA-related protein kinase 6) (Protein
kinase SID6-1512)
Length = 313
Score = 150 bits (379), Expect = 9e-36
Identities = 87/254 (34%), Positives = 145/254 (56%), Gaps = 8/254 (3%)
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSD-RTRRSAHQEMELISKVR 59
+ +++ ++IG+G F ++K LKK+++ D + R+ +E+ L+ ++
Sbjct: 42 LADFQIEKKIGRGQFSEVYKATCLLDRKTVALKKVQIFEMMDAKARQDCVKEIGLLKQLN 101
Query: 60 NPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIK--KANGVNFPEEKLCKWLVQLLMAL 117
+P I++Y DS++E + I++ + GDL+ IK K PE + K+ VQL A+
Sbjct: 102 HPNIIKYLDSFIEDN-ELNIVLELADAGDLSQMIKYFKKQKRLIPERTVWKYFVQLCSAV 160
Query: 118 DYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLAS-SIVGTPSYMCPELLA 176
+++H ++HRD+K +N+F+T ++LGD GL + +S+ A+ S+VGTP YM PE +
Sbjct: 161 EHMHSRRVMHRDIKPANVFITATGVVKLGDLGLGRFFSSETTAAHSLVGTPYYMSPERIH 220
Query: 177 DIPYGSKSDIWSLGCCIYEMAALKPAF--KAFDIQALINKINKSIVAPLP-TMYSSAFRG 233
+ Y KSDIWSLGC +YEMAAL+ F ++ +L KI + PLP YS R
Sbjct: 221 ENGYNFKSDIWSLGCLLYEMAALQSPFYGDKMNLFSLCQKIEQCDYPPLPGEHYSEKLRE 280
Query: 234 LVKSMLRKNPELRP 247
LV + +P RP
Sbjct: 281 LVSMCICPDPHQRP 294
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,073,146
Number of Sequences: 164201
Number of extensions: 2967911
Number of successful extensions: 12356
Number of sequences better than 10.0: 1693
Number of HSP's better than 10.0 without gapping: 1414
Number of HSP's successfully gapped in prelim test: 280
Number of HSP's that attempted gapping in prelim test: 7822
Number of HSP's gapped (non-prelim): 2080
length of query: 601
length of database: 59,974,054
effective HSP length: 116
effective length of query: 485
effective length of database: 40,926,738
effective search space: 19849467930
effective search space used: 19849467930
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0322.12


