

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
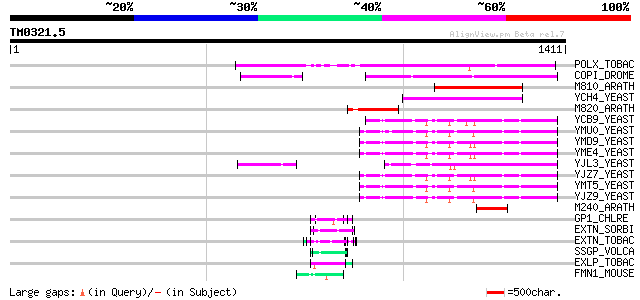
Query= TM0321.5
(1411 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 404 e-112
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 337 1e-91
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 239 4e-62
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 165 9e-40
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 121 2e-26
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 115 1e-24
YMU0_YEAST (Q04670) Transposon Ty1 protein B 107 2e-22
YMD9_YEAST (Q03434) Transposon Ty1 protein B 107 2e-22
YME4_YEAST (Q04711) Transposon Ty1 protein B 107 3e-22
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 107 3e-22
YJZ7_YEAST (P47098) Transposon Ty1 protein B 104 2e-21
YMT5_YEAST (Q04214) Transposon Ty1 protein B 103 4e-21
YJZ9_YEAST (P47100) Transposon Ty1 protein B 103 4e-21
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 79 7e-14
GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor (H... 69 9e-11
EXTN_SORBI (P24152) Extensin precursor (Proline-rich glycoprotein) 55 1e-06
EXTN_TOBAC (P13983) Extensin precursor (Cell wall hydroxyproline... 55 1e-06
SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor ... 55 2e-06
EXLP_TOBAC (Q03211) Pistil-specific extensin-like protein precur... 54 2e-06
FMN1_MOUSE (Q05860) Formin 1 isoforms I/II/III (Limb deformity p... 54 4e-06
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 404 bits (1039), Expect = e-112
Identities = 275/829 (33%), Positives = 406/829 (48%), Gaps = 64/829 (7%)
Query: 575 KSISRSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGL 634
+ R +K +SDNG E+T+ + +S G+ H + P T NG E+ +R ++E
Sbjct: 538 RETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVR 597
Query: 635 AMLYHSHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFP 694
+ML + +P +W +A TA Y+INR PS L+ +IP ++ + +Y++ FGCR F
Sbjct: 598 SMLRMAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGCRAFA 657
Query: 695 CLRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSS 754
+ K +S PCIF GY G++ ++P S F E A S
Sbjct: 658 HVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRESEVRTAADMSE 717
Query: 755 PHDLVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPP 814
+ +G P+ V + S+ P + S +D + P +
Sbjct: 718 ----------KVKNGIIPN---FVTIPSTSNNPTSAEST-----TDEVSEQGEQPGEVIE 759
Query: 815 SPAPPSPTQTATPPVGTQAPPASPPTTPPVVAQVPLAPI--AARPRTRSQNGIFRPNPRY 872
G Q P + P+ + RPR S+ P+ Y
Sbjct: 760 Q--------------GEQLDEGVEEVEHPTQGEEQHQPLRRSERPRVESRR---YPSTEY 802
Query: 873 ALVHAQPTGILTAFHTVTKPKGFKSAMKHP*---WLAAMEDELSALHKNCTWTLVPRPST 929
L+ +P+ K + HP + AM++E+ +L KN T+ LV P
Sbjct: 803 VLISDD-----------REPESLKEVLSHPEKNQLMKAMQEEMESLQKNGTYKLVELPKG 851
Query: 930 TNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHV 989
+ KWVF+ K D + R KARLV +GF Q G D+ FSPVVK T++R ILS
Sbjct: 852 KRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFDEIFSPVVKMTSIRTILSLA 911
Query: 990 VLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWF 1049
D ++ QLDVK AFLHG L E +YMEQP GF VC+LNK+LYGLKQAPR W+
Sbjct: 912 ASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWY 971
Query: 1050 QRLSSFLLRHGFSCSRADPSLFF-FYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNA 1108
+ SF+ + + +DP ++F + + + LL+YVDD+++ G D L+ + L+
Sbjct: 972 MKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSK 1031
Query: 1109 EFAIKYLGKLGYFLGLEIT--YTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHL 1166
F +K LG LG++I T L+L Q KY +L R M A VSTPLA L
Sbjct: 1032 SFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLERFNMKNAKPVSTPLAGHLKL 1091
Query: 1167 -------VSSGEAYFDPTHYRSLVGALQY-LTITRPDLSYAVNTVSQFLQAPTMEHFQAV 1218
+ Y S VG+L Y + TRPD+++AV VS+FL+ P EH++AV
Sbjct: 1092 SKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTRPDIAHAVGVVSRFLENPGKEHWEAV 1151
Query: 1219 KRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAK 1278
K ILRY+ GT L F S P + GY+DAD A D R+S+ GY +SW +K
Sbjct: 1152 KWILRYLRGTTGDCLCF-GGSDPILKGYTDADMAGDIDNRKSSTGYLFTFSGGAISWQSK 1210
Query: 1279 KQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVA 1338
Q VA S+ E+EY A T E++WL L EL + ++ D+QSA+ +++N +
Sbjct: 1211 LQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGLH-QKEYVVYCDSQSAIDLSKNSMY 1269
Query: 1339 HKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEI 1387
H KHID+ H++ E+V L V + T+ AD+ TK +PR FE+
Sbjct: 1270 HARTKHIDVRYHWIREMVDDESLKVLKISTNENPADMLTKVVPRNKFEL 1318
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 337 bits (865), Expect = 1e-91
Identities = 196/497 (39%), Positives = 284/497 (56%), Gaps = 12/497 (2%)
Query: 904 WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQ 963
W A+ EL+A N TWT+ RP N+V +WVF K++ G R KARLVA+GFTQ
Sbjct: 906 WEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQ 965
Query: 964 IPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFV 1023
DY TF+PV + ++ R ILS V+ + ++HQ+DVK AFL+G L E +YM P G
Sbjct: 966 KYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGI- 1024
Query: 1024 DPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKG--HTTLY 1081
+VC+LNKA+YGLKQA R WF+ L F S D ++ KG + +Y
Sbjct: 1025 -SCNSDNVCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIY 1083
Query: 1082 LLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYA 1141
+L+YVDD+++ D + + F L +F + L ++ +F+G+ I D ++L Q+ Y
Sbjct: 1084 VLLYVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYV 1143
Query: 1142 HDLLSRAMMLEASHVSTPLAA--GSHLVSSGEAYFDPTHYRSLVGALQYLTI-TRPDLSY 1198
+LS+ M + VSTPL + L++S E P RSL+G L Y+ + TRPDL+
Sbjct: 1144 KKILSKFNMENCNAVSTPLPSKINYELLNSDEDCNTPC--RSLIGCLMYIMLCTRPDLTT 1201
Query: 1199 AVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSS--PAVLGYSDADWARCTD 1256
AVN +S++ E +Q +KR+LRY+ GT L F++ + ++GY D+DWA
Sbjct: 1202 AVNILSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSEI 1261
Query: 1257 TRRSTYGYAIFLGD-NLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVR 1315
R+ST GY + D NL+ W+ K+Q +VA SS E+EY A+ E +WL LL + ++
Sbjct: 1262 DRKSTTGYLFKMFDFNLICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFLLTSINIK 1321
Query: 1316 LSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADI 1375
L + DNQ + +A NP HK AKHID+ HF E V + + + ++PT QLADI
Sbjct: 1322 LENPIKIYEDNQGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTENQLADI 1381
Query: 1376 FTKALPRPLFEIFRSKL 1392
FTK LP F R KL
Sbjct: 1382 FTKPLPAARFVELRDKL 1398
Score = 75.5 bits (184), Expect = 1e-12
Identities = 45/159 (28%), Positives = 77/159 (48%), Gaps = 5/159 (3%)
Query: 587 DNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHSHVPTRY 646
DNG E+ +N++ G+ + + PHT NG E+ R + E M+ + + +
Sbjct: 550 DNGREYLSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSF 609
Query: 647 WVDAFSTAVYIINRVPSKVL--SNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPYMNNKF 704
W +A TA Y+INR+PS+ L S++ P+++ + P + FG V+ ++ KF
Sbjct: 610 WGEAVLTATYLINRIPSRALVDSSKTPYEMWHNKKPYLKHLRVFGATVYVHIK-NKQGKF 668
Query: 705 SPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDE 743
+S IF GY + GFK ++ V+ DE
Sbjct: 669 DDKSFKSIFVGYEPN--GFKLWDAVNEKFIVARDVVVDE 705
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 239 bits (610), Expect = 4e-62
Identities = 121/225 (53%), Positives = 158/225 (69%)
Query: 1080 LYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAK 1139
+YLL+YVDDI+LTGS +LL I +L++ F++K LG + YFLG++I P GLFL Q K
Sbjct: 1 MYLLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTK 60
Query: 1140 YAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEAYFDPTHYRSLVGALQYLTITRPDLSYA 1199
YA +L+ A ML+ +STPL + S Y DP+ +RS+VGALQYLT+TRPD+SYA
Sbjct: 61 YAEQILNNAGMLDCKPMSTPLPLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYA 120
Query: 1200 VNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRR 1259
VN V Q + PT+ F +KR+LRYV GT GL + S V + D+DWA CT TRR
Sbjct: 121 VNIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRR 180
Query: 1260 STYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVW 1304
ST G+ FLG N++SWSAK+QPTV+RSS E+EYRA+A TA+EL W
Sbjct: 181 STTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 165 bits (417), Expect = 9e-40
Identities = 102/308 (33%), Positives = 160/308 (51%), Gaps = 4/308 (1%)
Query: 999 LDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLR 1058
+DV AFL+ + E +Y++QPPGFV+ R P +V L +YGLKQAP W + +++ L +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 1059 HGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKL 1118
GF + L+F +Y+ VYVDD+++ P + + L +++K LGK+
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 1119 GYFLGLEITYTPDG-LFLGQAKYAHDLLSRAMMLEASHVSTPLAAGSHLVSSGEAYF-DP 1176
FLGL I + +G + L Y S + + TPL L + + D
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 1177 THYRSLVGALQYLTIT-RPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTF 1235
T Y+S+VG L + T RPD+SY V+ +S+FL+ P H ++ +R+LRY+ T+ L +
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKY 240
Query: 1236 RRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKK-QPTVARSSCESEYRA 1294
R S A+ Y DA D ST GY L ++WS+KK + + S E+EY
Sbjct: 241 RSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYIT 300
Query: 1295 MANTASEL 1302
+ T E+
Sbjct: 301 ASETVMEI 308
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 121 bits (303), Expect = 2e-26
Identities = 65/130 (50%), Positives = 85/130 (65%), Gaps = 10/130 (7%)
Query: 859 TRSQNGIFRPNPRYALVHAQPTGILTAFHTVTK-PKGFKSAMKHP*WLAAMEDELSALHK 917
TRS+ GI + NP+Y+L T T+ K PK A+K P W AM++EL AL +
Sbjct: 3 TRSKAGINKLNPKYSL---------TITTTIKKEPKSVIFALKDPGWCQAMQEELDALSR 53
Query: 918 NCTWTLVPRPSTTNVVGIKWVFRTKFHSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVV 977
N TW LVP P N++G KWVF+TK HSDGT++RLKARLVA+GF Q G + T+SPVV
Sbjct: 54 NKTWILVPPPVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVV 113
Query: 978 KATTVRLILS 987
+ T+R IL+
Sbjct: 114 RTATIRTILN 123
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 115 bits (287), Expect = 1e-24
Identities = 133/535 (24%), Positives = 229/535 (41%), Gaps = 79/535 (14%)
Query: 904 WLAAMEDELSALHKNCTWTLVPRPSTTN-----VVGIKWVFRTKFHSDGTVERLKARLVA 958
++ A E+S L K TW + V+ ++F K DGT KAR VA
Sbjct: 1265 YVEAYHKEISQLLKMNTWDTNKYYDRNDIDPKKVINSMFIFNKK--RDGTH---KARFVA 1319
Query: 959 QGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQ 1018
+G Q P S S V + LS + ND+ + QLD+ +A+L+ + E +Y+
Sbjct: 1320 RGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLDISSAYLYADIKEELYIRP 1379
Query: 1019 PP--GFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLR-------HGFSCSRADPS 1069
PP G D + RL K+LYGLKQ+ W++ + S+L+ G+SC
Sbjct: 1380 PPHLGLND-----KLLRLRKSLYGLKQSGANWYETIKSYLINCCDMQEVRGWSC------ 1428
Query: 1070 LFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYL------GKLGY-FL 1122
F T+ L +VDD+IL D + + I L ++ K + ++ Y L
Sbjct: 1429 --VFKNSQVTICL--FVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDIL 1484
Query: 1123 GLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPL--------AAG--SHLVSSGEA 1172
GLEI Y ++KY + +++ + ++ PL A G H + E
Sbjct: 1485 GLEIKYQ-------RSKYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHYIDQDEL 1537
Query: 1173 YFDPTHYR-------SLVGALQYLTIT-RPDLSYAVNTVSQFLQAPTMEHFQAVKRILRY 1224
D Y+ L+G Y+ R DL Y +NT++Q + P+ + ++++
Sbjct: 1538 EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQF 1597
Query: 1225 VCGTQHFGLTFRRTS----SPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQ 1280
+ T+ L + + ++ SDA + +S G L ++ + K
Sbjct: 1598 MWDTRDKQLIWHKNKPTKPDNKLVAISDASYGN-QPYYKSQIGNIFLLNGKVIGGKSTKA 1656
Query: 1281 PTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPL---LLSDNQSALFMAQNPV 1337
S+ E+E A+ SE + LLN L L L+ P+ LL+D++S + + ++
Sbjct: 1657 SLTCTSTTEAEIHAV----SEAIPLLNNLSHLVQELNKKPIIKGLLTDSRSTISIIKSTN 1712
Query: 1338 AHK-HAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSK 1391
K + + + V+ L V ++ T +AD+ TK LP F++ +K
Sbjct: 1713 EEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1767
Score = 41.6 bits (96), Expect = 0.016
Identities = 22/83 (26%), Positives = 39/83 (46%)
Query: 581 IKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYHS 640
+ V Q D G+E+TN +H F + G+ ++ +G E+ +R +L +L+ S
Sbjct: 726 VLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLLNDCRTLLHCS 785
Query: 641 HVPTRYWVDAFSTAVYIINRVPS 663
+P W A + I N + S
Sbjct: 786 GLPNHLWFSAVEFSTIIRNSLVS 808
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 107 bits (268), Expect = 2e-22
Identities = 136/555 (24%), Positives = 236/555 (42%), Gaps = 92/555 (16%)
Query: 889 VTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLV-----PRPSTTNVVGIKWVFRTKF 943
+T K K K+ + A E++ L K TW V+ ++F K
Sbjct: 811 ITYNKDIKEKEKY---IQAYHKEVNQLLKMKTWDTDRYYDRKEIDPKRVINSMFIFNRK- 866
Query: 944 HSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVR-----LILSHVVLNDWQLHQ 998
DGT KAR VA+G Q P T+ P +++ TV LS + N++ + Q
Sbjct: 867 -RDGTH---KARFVARGDIQHPD-----TYDPGMQSNTVHHYALMTSLSLALDNNYYITQ 917
Query: 999 LDVKNAFLHGHLTETVYMEQPP--GFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFL 1056
LD+ +A+L+ + E +Y+ PP G D + RL K+LYGLKQ+ W++ + S+L
Sbjct: 918 LDISSAYLYADIKEELYIRPPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYL 972
Query: 1057 LR-------HGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAE 1109
++ G+SC F T+ L +VDD+IL D + + I L +
Sbjct: 973 IKQCGMEEVRGWSC--------VFKNSQVTICL--FVDDMILFSKDLNANKKIITTLKKQ 1022
Query: 1110 FAIKYL------GKLGY-FLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPL-A 1161
+ K + ++ Y LGLEI Y + KY + ++ + ++ PL
Sbjct: 1023 YDTKIINLGESDNEIQYDILGLEIKYQ-------RGKYMKLGMENSLTEKIPKLNVPLNP 1075
Query: 1162 AGSHLVSSGE--AYFDPT--------------HYRSLVGALQYLTIT-RPDLSYAVNTVS 1204
G L + G+ Y D + L+G Y+ R DL Y +NT++
Sbjct: 1076 KGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLA 1135
Query: 1205 QFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTS----SPAVLGYSDADWARCTDTRRS 1260
Q + P+ + +++++ T+ L + ++ + ++ SDA + +S
Sbjct: 1136 QHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASYGN-QPYYKS 1194
Query: 1261 TYGYAIFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATP 1320
G L ++ + K S+ E+E A+ SE V LLN L L L+ P
Sbjct: 1195 QIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SESVPLLNNLSHLVQELNKKP 1250
Query: 1321 L---LLSDNQSAL-FMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIF 1376
+ LL+D++S + + N + + + V+ L V ++ T +AD+
Sbjct: 1251 ITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVM 1310
Query: 1377 TKALPRPLFEIFRSK 1391
TK LP F++ +K
Sbjct: 1311 TKPLPIKTFKLLTNK 1325
Score = 41.2 bits (95), Expect = 0.020
Identities = 36/154 (23%), Positives = 61/154 (39%), Gaps = 2/154 (1%)
Query: 580 SIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYH 639
S+ V Q D G+E+TN +H +G+ ++ +G E+ +R +L+ L
Sbjct: 302 SVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQC 361
Query: 640 SHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPY 699
S +P W A + + N + S S + Q + PFG V
Sbjct: 362 SGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI-VNDHN 419
Query: 700 MNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLT 733
N+K PR P S + G+ + P++ T
Sbjct: 420 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 453
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 107 bits (268), Expect = 2e-22
Identities = 136/550 (24%), Positives = 232/550 (41%), Gaps = 82/550 (14%)
Query: 889 VTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVP-----RPSTTNVVGIKWVFRTKF 943
+T K K K+ + A E++ L K TW V+ ++F K
Sbjct: 811 ITYNKDIKEKEKY---IEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKK- 866
Query: 944 HSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKN 1003
DGT KAR VA+G Q P S S V + LS + N++ + QLD+ +
Sbjct: 867 -RDGTH---KARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISS 922
Query: 1004 AFLHGHLTETVYMEQPP--GFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLR--- 1058
A+L+ + E +Y+ PP G D + RL K+LYGLKQ+ W++ + S+L++
Sbjct: 923 AYLYADIKEELYIRPPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYLIKQCG 977
Query: 1059 ----HGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKY 1114
G+SC F T+ L +VDD+IL D + + I L ++ K
Sbjct: 978 MEEVRGWSC--------VFKNSQVTICL--FVDDMILFSKDLNANKKIITTLKKQYDTKI 1027
Query: 1115 L------GKLGY-FLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPL-AAGSHL 1166
+ ++ Y LGLEI Y + KY + ++ + ++ PL G L
Sbjct: 1028 INLGESDNEIQYDILGLEIKYQ-------RGKYMKLGMENSLTEKIPKLNVPLNPKGRKL 1080
Query: 1167 VSSG---------EAYFDPTHYR-------SLVGALQYLTIT-RPDLSYAVNTVSQFLQA 1209
+ G E D Y+ L+G Y+ R DL Y +NT++Q +
Sbjct: 1081 SAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILF 1140
Query: 1210 PTMEHFQAVKRILRYVCGTQHFGLTFRRTS----SPAVLGYSDADWARCTDTRRSTYGYA 1265
P+ + +++++ T+ L + + ++ SDA + +S G
Sbjct: 1141 PSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGN-QPYYKSQIGNI 1199
Query: 1266 IFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPL---L 1322
L ++ + K S+ E+E A+ SE V LLN L L L+ P+ L
Sbjct: 1200 YLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SESVPLLNNLSYLIQELNKKPIIKGL 1255
Query: 1323 LSDNQSALFMAQNPVAHK-HAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALP 1381
L+D++S + + ++ K + + + V+ L V ++ T +AD+ TK LP
Sbjct: 1256 LTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLP 1315
Query: 1382 RPLFEIFRSK 1391
F++ +K
Sbjct: 1316 IKTFKLLTNK 1325
Score = 41.2 bits (95), Expect = 0.020
Identities = 36/154 (23%), Positives = 61/154 (39%), Gaps = 2/154 (1%)
Query: 580 SIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYH 639
S+ V Q D G+E+TN +H +G+ ++ +G E+ +R +L+ L
Sbjct: 302 SVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQC 361
Query: 640 SHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPY 699
S +P W A + + N + S S + Q + PFG V
Sbjct: 362 SGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI-VNDHN 419
Query: 700 MNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLT 733
N+K PR P S + G+ + P++ T
Sbjct: 420 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 453
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 107 bits (266), Expect = 3e-22
Identities = 136/550 (24%), Positives = 230/550 (41%), Gaps = 82/550 (14%)
Query: 889 VTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVP-----RPSTTNVVGIKWVFRTKF 943
+T K K K+ + A E++ L K TW V+ ++F K
Sbjct: 811 ITYNKDIKEKEKY---IEAYHKEVNQLLKMNTWDTDKYYDRKEIDPKRVINSMFIFNRK- 866
Query: 944 HSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKN 1003
DGT KAR VA+G Q P S S V + LS + N++ + QLD+ +
Sbjct: 867 -RDGTH---KARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISS 922
Query: 1004 AFLHGHLTETVYMEQPP--GFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLR--- 1058
A+L+ + E +Y+ PP G D + RL K+LYGLKQ+ W++ + S+L++
Sbjct: 923 AYLYADIKEELYIRPPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYLIKQCG 977
Query: 1059 ----HGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKY 1114
G+SC F T+ L +VDD+IL D + + I L ++ K
Sbjct: 978 MEEVRGWSC--------VFKNSQVTICL--FVDDMILFSKDLNANKKIITTLKKQYDTKI 1027
Query: 1115 L------GKLGY-FLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPL-AAGSHL 1166
+ ++ Y LGLEI Y + KY + ++ + ++ PL G L
Sbjct: 1028 INLGESDNEIQYDILGLEIKYQ-------RGKYMKLGMENSLTEKIPKLNVPLNPKGRKL 1080
Query: 1167 VSSG---------EAYFDPTHYR-------SLVGALQYLTIT-RPDLSYAVNTVSQFLQA 1209
+ G E D Y+ L+G Y+ R DL Y +NT++Q +
Sbjct: 1081 SAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILF 1140
Query: 1210 PTMEHFQAVKRILRYVCGTQHFGLTFRRTS----SPAVLGYSDADWARCTDTRRSTYGYA 1265
P+ + +++++ T+ L + + ++ SDA + +S G
Sbjct: 1141 PSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGN-QPYYKSQIGNI 1199
Query: 1266 IFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPL---L 1322
L ++ + K S+ E+E A+ SE V LLN L L L+ P+ L
Sbjct: 1200 YLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SESVPLLNNLSHLVQELNKKPITKGL 1255
Query: 1323 LSDNQSAL-FMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALP 1381
L+D++S + + N + + + V+ L V ++ T +AD+ TK LP
Sbjct: 1256 LTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLP 1315
Query: 1382 RPLFEIFRSK 1391
F++ +K
Sbjct: 1316 IKTFKLLTNK 1325
Score = 41.2 bits (95), Expect = 0.020
Identities = 36/154 (23%), Positives = 61/154 (39%), Gaps = 2/154 (1%)
Query: 580 SIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYH 639
S+ V Q D G+E+TN +H +G+ ++ +G E+ +R +L+ L
Sbjct: 302 SVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQC 361
Query: 640 SHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPY 699
S +P W A + + N + S S + Q + PFG V
Sbjct: 362 SGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI-VNDHN 419
Query: 700 MNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLT 733
N+K PR P S + G+ + P++ T
Sbjct: 420 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 453
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 107 bits (266), Expect = 3e-22
Identities = 110/462 (23%), Positives = 196/462 (41%), Gaps = 31/462 (6%)
Query: 953 KARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTE 1012
KAR+V +G TQ P YS+ + + +++ L + + LD+ +AFL+ L E
Sbjct: 1338 KARIVCRGDTQSPD-TYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLEE 1396
Query: 1013 TVYMEQPPGFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFF 1072
+Y+ P D R V +LNKALYGLKQ+P+ W L +L G + P L
Sbjct: 1397 EIYIPHPH---DRRC---VVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGL-- 1448
Query: 1073 FYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKL------GYFLGLEI 1126
+ L + VYVDD ++ S+ L +FI +L + F +K G L LG+++
Sbjct: 1449 YQTEDKNLMIAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMDL 1508
Query: 1127 TY---------TPDGLFLGQAKYAHDLLSRAMMLEASHVST-PLAAGSHLVSSGEAYF-- 1174
Y T K ++ L + H+ST + ++ E F
Sbjct: 1509 VYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFRQ 1568
Query: 1175 DPTHYRSLVGALQYLT-ITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGL 1233
+ L+G L Y+ R D+ +AV V++ + P F + +I++Y+ + G+
Sbjct: 1569 GVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIGI 1628
Query: 1234 TFRR--TSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKKQPTVARSSCESE 1291
+ R V+ +DA D +S G ++ G N+ + + K SS E+E
Sbjct: 1629 HYDRDCNKDKKVIAITDASVGSEYDA-QSRIGVILWYGMNIFNVYSNKSTNRCVSSTEAE 1687
Query: 1292 YRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHF 1351
A+ ++ L L EL + ++++D++ A+ K +
Sbjct: 1688 LHAIYEGYADSETLKVTLKELGEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTWIKTEI 1747
Query: 1352 VCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLR 1393
+ E + + + + +AD+ TK + F+ F L+
Sbjct: 1748 IKEKIKEKSIKLLKITGKGNIADLLTKPVSASDFKRFIQVLK 1789
Score = 62.8 bits (151), Expect = 6e-09
Identities = 41/153 (26%), Positives = 72/153 (46%), Gaps = 5/153 (3%)
Query: 579 RSIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLY 638
R ++ SD GTEFTN+++ F S G+ H + A NGR E+ R ++ +L
Sbjct: 688 RKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITDATTLLR 747
Query: 639 HSHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFH--VAPTYANFHPFGCRVFPCL 696
S++ ++W A ++A I N + K + ++P + + V +F PFG + +
Sbjct: 748 QSNLRVKFWEYAVTSATNIRNYLEHK-STGKLPLKAISRQPVTVRLMSFLPFGEK--GII 804
Query: 697 RPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPA 729
+ + K P P I + G+K F P+
Sbjct: 805 WNHNHKKLKPSGLPSIILCKDPNSYGYKFFIPS 837
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 104 bits (259), Expect = 2e-21
Identities = 134/550 (24%), Positives = 231/550 (41%), Gaps = 82/550 (14%)
Query: 889 VTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVP-----RPSTTNVVGIKWVFRTKF 943
+T K K K+ + A E++ L K TW V+ ++F K
Sbjct: 1238 ITYNKDIKEKEKY---IEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKK- 1293
Query: 944 HSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKN 1003
DGT KAR VA+G Q P + S V + LS + N++ + QLD+ +
Sbjct: 1294 -RDGTH---KARFVARGDIQHPDTYDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISS 1349
Query: 1004 AFLHGHLTETVYMEQPP--GFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLR--- 1058
A+L+ + E +Y+ PP G D + RL K+ YGLKQ+ W++ + S+L++
Sbjct: 1350 AYLYADIKEELYIRPPPHLGMND-----KLIRLKKSHYGLKQSGANWYETIKSYLIKQCG 1404
Query: 1059 ----HGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKY 1114
G+SC F T+ L +VDD+IL D + + I L ++ K
Sbjct: 1405 MEEVRGWSC--------VFKNSQVTICL--FVDDMILFSKDLNANKKIITTLKKQYDTKI 1454
Query: 1115 L------GKLGY-FLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPL-AAGSHL 1166
+ ++ Y LGLEI Y + KY + ++ + ++ PL G L
Sbjct: 1455 INLGESDNEIQYDILGLEIKYQ-------RGKYMKLGMENSLTEKIPKLNVPLNPKGRKL 1507
Query: 1167 VSSG---------EAYFDPTHYR-------SLVGALQYLTIT-RPDLSYAVNTVSQFLQA 1209
+ G E D Y+ L+G Y+ R DL Y +NT++Q +
Sbjct: 1508 SAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILF 1567
Query: 1210 PTMEHFQAVKRILRYVCGTQHFGLTFRRTS----SPAVLGYSDADWARCTDTRRSTYGYA 1265
P+ + +++++ T+ L + + ++ SDA + +S G
Sbjct: 1568 PSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGN-QPYYKSQIGNI 1626
Query: 1266 IFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPL---L 1322
L ++ + K S+ E+E A+ SE V LLN L L L+ P+ L
Sbjct: 1627 FLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SESVPLLNNLSYLIQELNKKPIIKGL 1682
Query: 1323 LSDNQSALFMAQNPVAHK-HAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALP 1381
L+D++S + + ++ K + + + V+ L V ++ T +AD+ TK LP
Sbjct: 1683 LTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLP 1742
Query: 1382 RPLFEIFRSK 1391
F++ +K
Sbjct: 1743 IKTFKLLTNK 1752
Score = 41.2 bits (95), Expect = 0.020
Identities = 36/154 (23%), Positives = 61/154 (39%), Gaps = 2/154 (1%)
Query: 580 SIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYH 639
S+ V Q D G+E+TN +H +G+ ++ +G E+ +R +L+ L
Sbjct: 729 SVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQC 788
Query: 640 SHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPY 699
S +P W A + + N + S S + Q + PFG V
Sbjct: 789 SGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI-VNDHN 846
Query: 700 MNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLT 733
N+K PR P S + G+ + P++ T
Sbjct: 847 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 880
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 103 bits (256), Expect = 4e-21
Identities = 133/550 (24%), Positives = 232/550 (42%), Gaps = 82/550 (14%)
Query: 889 VTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVP-----RPSTTNVVGIKWVFRTKF 943
+T K K K+ + A E++ L K TW V+ ++F K
Sbjct: 811 ITYNKDIKEKEKY---IEAYHKEVNQLLKMKTWDTDKYYDRKEIDPKRVINSMFIFNRK- 866
Query: 944 HSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKN 1003
DGT KAR VA+G Q P S S V + LS + N++ + QLD+ +
Sbjct: 867 -RDGTH---KARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISS 922
Query: 1004 AFLHGHLTETVYMEQPP--GFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLR--- 1058
A+L+ + E +Y+ PP G D + RL K+LYGLKQ+ W++ + S+L++
Sbjct: 923 AYLYADIKEELYIRPPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYLIKQCG 977
Query: 1059 ----HGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKY 1114
G+SC F T+ L +VDD++L + + + I +L ++ K
Sbjct: 978 MEEVRGWSC--------VFENSQVTICL--FVDDMVLFSKNLNSNKRIIDKLKMQYDTKI 1027
Query: 1115 LG------KLGY-FLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPL-AAGSHL 1166
+ ++ Y LGLEI Y + KY + ++ + ++ PL G L
Sbjct: 1028 INLGESDEEIQYDILGLEIKYQ-------RGKYMKLGMENSLTEKIPKLNVPLNPKGRKL 1080
Query: 1167 VSSGE--AYFDPT--------------HYRSLVGALQYLTIT-RPDLSYAVNTVSQFLQA 1209
+ G+ Y D + L+G Y+ R DL Y +NT++Q +
Sbjct: 1081 SAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILF 1140
Query: 1210 PTMEHFQAVKRILRYVCGTQHFGLTFRRTS----SPAVLGYSDADWARCTDTRRSTYGYA 1265
P+ + +++++ T+ L + ++ + ++ SDA + +S G
Sbjct: 1141 PSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASYGN-QPYYKSQIGNI 1199
Query: 1266 IFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPL---L 1322
L ++ + K S+ E+E A+ SE V LLN L L L P+ L
Sbjct: 1200 YLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SESVPLLNNLSYLIQELDKKPITKGL 1255
Query: 1323 LSDNQSAL-FMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALP 1381
L+D++S + + N + + + V+ L V ++ T +AD+ TK LP
Sbjct: 1256 LTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLP 1315
Query: 1382 RPLFEIFRSK 1391
F++ +K
Sbjct: 1316 IKTFKLLTNK 1325
Score = 41.2 bits (95), Expect = 0.020
Identities = 36/154 (23%), Positives = 61/154 (39%), Gaps = 2/154 (1%)
Query: 580 SIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYH 639
S+ V Q D G+E+TN +H +G+ ++ +G E+ +R +L+ L
Sbjct: 302 SVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQC 361
Query: 640 SHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPY 699
S +P W A + + N + S S + Q + PFG V
Sbjct: 362 SGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI-VNDHN 419
Query: 700 MNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLT 733
N+K PR P S + G+ + P++ T
Sbjct: 420 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 453
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 103 bits (256), Expect = 4e-21
Identities = 133/550 (24%), Positives = 232/550 (42%), Gaps = 82/550 (14%)
Query: 889 VTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVP-----RPSTTNVVGIKWVFRTKF 943
+T K K K+ + A E++ L K TW V+ ++F K
Sbjct: 1238 ITYNKDIKEKEKY---IEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKK- 1293
Query: 944 HSDGTVERLKARLVAQGFTQIPGFDYSLTFSPVVKATTVRLILSHVVLNDWQLHQLDVKN 1003
DGT KAR VA+G Q P S S V + LS + N++ + QLD+ +
Sbjct: 1294 -RDGTH---KARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISS 1349
Query: 1004 AFLHGHLTETVYMEQPP--GFVDPRFPTHVCRLNKALYGLKQAPRAWFQRLSSFLLR--- 1058
A+L+ + E +Y+ PP G D + RL K+LYGLKQ+ W++ + S+L++
Sbjct: 1350 AYLYADIKEELYIRPPPHLGMNDK-----LIRLKKSLYGLKQSGANWYETIKSYLIQQCG 1404
Query: 1059 ----HGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEFAIKY 1114
G+SC F T+ L +VDD++L + + + I +L ++ K
Sbjct: 1405 MEEVRGWSC--------VFKNSQVTICL--FVDDMVLFSKNLNSNKRIIEKLKMQYDTKI 1454
Query: 1115 LG------KLGY-FLGLEITYTPDGLFLGQAKYAHDLLSRAMMLEASHVSTPL-AAGSHL 1166
+ ++ Y LGLEI Y + KY + ++ + ++ PL G L
Sbjct: 1455 INLGESDEEIQYDILGLEIKYQ-------RGKYMKLGMENSLTEKIPKLNVPLNPKGRKL 1507
Query: 1167 VSSGE--AYFDPT--------------HYRSLVGALQYLTIT-RPDLSYAVNTVSQFLQA 1209
+ G+ Y D + L+G Y+ R DL Y +NT++Q +
Sbjct: 1508 SAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILF 1567
Query: 1210 PTMEHFQAVKRILRYVCGTQHFGLTFRRTS----SPAVLGYSDADWARCTDTRRSTYGYA 1265
P+ + +++++ T+ L + ++ + ++ SDA + +S G
Sbjct: 1568 PSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASYGN-QPYYKSQIGNI 1626
Query: 1266 IFLGDNLLSWSAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPL---L 1322
L ++ + K S+ E+E A+ SE V LLN L L L P+ L
Sbjct: 1627 YLLNGKVIGGKSTKASLTCTSTTEAEIHAI----SESVPLLNNLSYLIQELDKKPITKGL 1682
Query: 1323 LSDNQSAL-FMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALP 1381
L+D++S + + N + + + V+ L V ++ T +AD+ TK LP
Sbjct: 1683 LTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLP 1742
Query: 1382 RPLFEIFRSK 1391
F++ +K
Sbjct: 1743 IKTFKLLTNK 1752
Score = 41.2 bits (95), Expect = 0.020
Identities = 36/154 (23%), Positives = 61/154 (39%), Gaps = 2/154 (1%)
Query: 580 SIKVFQSDNGTEFTNNKVHTLFASSGVLHRFSCPHTQAQNGRVEQKHRHVLELGLAMLYH 639
S+ V Q D G+E+TN +H +G+ ++ +G E+ +R +L+ L
Sbjct: 729 SVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQC 788
Query: 640 SHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHVAPTYANFHPFGCRVFPCLRPY 699
S +P W A + + N + S S + Q + PFG V
Sbjct: 789 SGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVI-VNDHN 846
Query: 700 MNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLT 733
N+K PR P S + G+ + P++ T
Sbjct: 847 PNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKT 880
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 79.3 bits (194), Expect = 7e-14
Identities = 39/77 (50%), Positives = 50/77 (64%)
Query: 1188 YLTITRPDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYS 1247
YLTITRPDL++AVN +SQF A QAV ++L YV GT GL + TS + ++
Sbjct: 2 YLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAFA 61
Query: 1248 DADWARCTDTRRSTYGY 1264
D+DWA C DTRRS G+
Sbjct: 62 DSDWASCPDTRRSVTGF 78
>GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor
(Hydroxyproline-rich glycoprotein 1)
Length = 555
Score = 68.9 bits (167), Expect = 9e-11
Identities = 40/92 (43%), Positives = 44/92 (47%), Gaps = 5/92 (5%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
PA PPSP P +P S P S S P PVPP+ APPSPAPPSP A
Sbjct: 207 PAPPVPPSPAPP---SPPSPAPPSPPSPAPPSPSPPAPPSPVPPSPAPPSPAPPSPKPPA 263
Query: 826 TPPVGTQAPPASPPTTPPVVAQVPLAPIAARP 857
PP +PP PP PP A P+ P P
Sbjct: 264 PPP--PPSPPPPPPPRPPFPANTPMPPSPPSP 293
Score = 65.9 bits (159), Expect = 8e-10
Identities = 40/106 (37%), Positives = 50/106 (46%), Gaps = 8/106 (7%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
PA SPPSP P P + P + P+ P P PP+ APPSPAPPSP A
Sbjct: 73 PAPPSPPSPAPPSPAPPSPAPPSPAPPSPA-------PPSPAPPSPAPPSPAPPSPPSPA 125
Query: 826 TPPVGTQAPPA-SPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNP 870
P APP+ SPP+ P + P P + P S + P+P
Sbjct: 126 PPSPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSP 171
Score = 64.3 bits (155), Expect = 2e-09
Identities = 40/108 (37%), Positives = 51/108 (47%), Gaps = 4/108 (3%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDV--LPALPVPPADAPPSPAPPSPTQ 823
P+ PPSP P V +P P P S PA PVPP+ APPSPAPP P
Sbjct: 155 PSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPS 214
Query: 824 TATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNPR 871
A P + APP+ P PP + P AP + P + + P+P+
Sbjct: 215 PAPPSPPSPAPPSPPSPAPP--SPSPPAPPSPVPPSPAPPSPAPPSPK 260
Score = 60.8 bits (146), Expect = 2e-08
Identities = 36/95 (37%), Positives = 45/95 (46%), Gaps = 3/95 (3%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSC--DDSDVLPALPVPPADAPPSPAPPSPTQ 823
P S +PPSP P P P P S PA P+PP+ APPSP+PP P
Sbjct: 103 PPSPAPPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPS 162
Query: 824 TATPPVGTQAPPA-SPPTTPPVVAQVPLAPIAARP 857
+ P + APP+ +PP+ P V P P A P
Sbjct: 163 PSPPVPPSPAPPSPTPPSPSPPVPPSPAPPSPAPP 197
Score = 60.5 bits (145), Expect = 3e-08
Identities = 35/93 (37%), Positives = 42/93 (44%), Gaps = 1/93 (1%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
P S +PPSP P V +P P + S PA P PP+ APPSP+PP+P +
Sbjct: 186 PPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPPSPAPPSPSPPAPP-SP 244
Query: 826 TPPVGTQAPPASPPTTPPVVAQVPLAPIAARPR 858
PP PA P PP P P PR
Sbjct: 245 VPPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPR 277
Score = 60.5 bits (145), Expect = 3e-08
Identities = 35/92 (38%), Positives = 43/92 (46%), Gaps = 3/92 (3%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
P S +PPSP P V +P P + S P P P +PPSPAPPSP A
Sbjct: 173 PPSPTPPSPSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSPPSPA 232
Query: 826 TPPVGTQAPPASPPTTPPVVAQVPLAPIAARP 857
P + +PPA P PP A AP + +P
Sbjct: 233 PP---SPSPPAPPSPVPPSPAPPSPAPPSPKP 261
Score = 60.1 bits (144), Expect = 4e-08
Identities = 40/108 (37%), Positives = 48/108 (44%), Gaps = 3/108 (2%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPT-QT 824
P S +PPSP P +P P L S P P P PPSPAPPSPT +
Sbjct: 121 PPSPAPPSPSPPAPPSPSPPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPS 180
Query: 825 ATPPV-GTQAPPASPPTTPPVVA-QVPLAPIAARPRTRSQNGIFRPNP 870
+PPV + APP+ P PP A P P+ P S P+P
Sbjct: 181 PSPPVPPSPAPPSPAPPVPPSPAPPSPAPPVPPSPAPPSPPSPAPPSP 228
Score = 59.3 bits (142), Expect = 7e-08
Identities = 35/105 (33%), Positives = 42/105 (39%), Gaps = 4/105 (3%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
P S +PPSP P P P + P P P PP+ APPSPAPPSP +
Sbjct: 50 PPSPAPPSPAPPSPAPPSPGPPSPAPPSPPSP----APPSPAPPSPAPPSPAPPSPAPPS 105
Query: 826 TPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNP 870
P P +PP+ P P P P S P+P
Sbjct: 106 PAPPSPAPPSPAPPSPPSPAPPSPSPPAPPSPSPPSPAPPLPPSP 150
Score = 56.2 bits (134), Expect = 6e-07
Identities = 41/111 (36%), Positives = 52/111 (45%), Gaps = 10/111 (9%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSP---APPSPT 822
P S +PPSP P P + P + P+ PA P PP+ APPSP APPSP+
Sbjct: 83 PPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPS----PAPPSPPSPAPPSPSPPAPPSPS 138
Query: 823 --QTATPPVGTQAPPA-SPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNP 870
A P + APP+ SPP P VP +P P S + P+P
Sbjct: 139 PPSPAPPLPPSPAPPSPSPPVPPSPSPPVPPSPAPPSPTPPSPSPPVPPSP 189
Score = 54.7 bits (130), Expect = 2e-06
Identities = 34/95 (35%), Positives = 42/95 (43%), Gaps = 3/95 (3%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVP--PADAPPSPAPPSPTQ 823
P+ +PPSP P P + P P S P P P PA+ P P+PPSP
Sbjct: 236 PSPPAPPSPVPPSPAPPSPAPPSPKPPAPPPPPSPPPPPPPRPPFPANTPMPPSPPSPPP 295
Query: 824 TATPPVGTQAPPASPPT-TPPVVAQVPLAPIAARP 857
+ PP P SPP+ PP A VP +P P
Sbjct: 296 SPAPPTPPTPPSPSPPSPVPPSPAPVPPSPAPPSP 330
Score = 53.1 bits (126), Expect = 5e-06
Identities = 37/107 (34%), Positives = 49/107 (45%), Gaps = 7/107 (6%)
Query: 766 PASGSPPSPHPVVLVAPE--SSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQ 823
PA PPSP P P ++ P+ S PS S P P PP+ +PPSP PPSP
Sbjct: 262 PAPPPPPSPPPPPPPRPPFPANTPMPP-SPPSPPPSPAPPTPPTPPSPSPPSPVPPSPAP 320
Query: 824 TATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNP 870
P APP+ P+ PP A +P + + S + P+P
Sbjct: 321 VPPSP----APPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPSPSPSP 363
Score = 51.6 bits (122), Expect = 1e-05
Identities = 31/83 (37%), Positives = 39/83 (46%), Gaps = 3/83 (3%)
Query: 778 VLVAPESSLPIGSLSCP-SCDDSDVLPALPVPPADAPPSPAPPS--PTQTATPPVGTQAP 834
V A +P G +CP S P P PP+ APPSPAPPS P A P + AP
Sbjct: 24 VASANAQCVPGGIFNCPPSPAPPSPAPPSPAPPSPAPPSPAPPSPGPPSPAPPSPPSPAP 83
Query: 835 PASPPTTPPVVAQVPLAPIAARP 857
P+ P +P + P +P P
Sbjct: 84 PSPAPPSPAPPSPAPPSPAPPSP 106
Score = 51.2 bits (121), Expect = 2e-05
Identities = 37/100 (37%), Positives = 44/100 (44%), Gaps = 17/100 (17%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPP-SPAPPS---- 820
P S +PPSP P AP S +P PS P P PPA PP SP PP
Sbjct: 228 PPSPAPPSPSPP---APPSPVP------PSPAPPSPAPPSPKPPAPPPPPSPPPPPPPRP 278
Query: 821 --PTQTATPPVGTQAPPA-SPPTTPPVVAQVPLAPIAARP 857
P T PP PP+ +PPT P + P +P+ P
Sbjct: 279 PFPANTPMPPSPPSPPPSPAPPTPPTPPSPSPPSPVPPSP 318
Score = 49.7 bits (117), Expect = 6e-05
Identities = 37/105 (35%), Positives = 44/105 (41%), Gaps = 23/105 (21%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
P S +PPSP AP S P PS P PP+ APPSP PPSP
Sbjct: 40 PPSPAPPSP------APPSPAP------PS----------PAPPSPAPPSPGPPSPA-PP 76
Query: 826 TPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNP 870
+PP PA P PP A AP + P + + P+P
Sbjct: 77 SPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSPAPPSP 121
Score = 49.3 bits (116), Expect = 7e-05
Identities = 28/87 (32%), Positives = 39/87 (44%), Gaps = 3/87 (3%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAP---PSPAPPSPT 822
P SPPSP P + P S P +P P PP+ AP PSPAPP+P+
Sbjct: 285 PMPPSPPSPPPSPAPPTPPTPPSPSPPSPVPPSPAPVPPSPAPPSPAPSPPPSPAPPTPS 344
Query: 823 QTATPPVGTQAPPASPPTTPPVVAQVP 849
+ +P P+ P+ P + +P
Sbjct: 345 PSPSPSPSPSPSPSPSPSPSPSPSPIP 371
Score = 41.6 bits (96), Expect = 0.016
Identities = 30/86 (34%), Positives = 38/86 (43%), Gaps = 7/86 (8%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAP---PSPT 822
P S SPPSP P AP P PS S PA P P PSP+P PSP+
Sbjct: 305 PPSPSPPSPVPPS-PAPVPPSPAPPSPAPSPPPS---PAPPTPSPSPSPSPSPSPSPSPS 360
Query: 823 QTATPPVGTQAPPASPPTTPPVVAQV 848
+ +P P+ P+ PV ++
Sbjct: 361 PSPSPSPSPIPSPSPKPSPSPVAVKL 386
Score = 37.7 bits (86), Expect = 0.22
Identities = 33/102 (32%), Positives = 42/102 (40%), Gaps = 23/102 (22%)
Query: 747 PFAGSHSSPHDLVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALP 806
P S S P + S P S +PPSP P +P S PA P
Sbjct: 303 PTPPSPSPPSPVPPSPAPVPPSPAPPSPAP----SPPPS-----------------PAPP 341
Query: 807 VPPADAPPSPAP-PSPTQTATP-PVGTQAPPASPPTTPPVVA 846
P PSP+P PSP+ + +P P + P SP +P VA
Sbjct: 342 TPSPSPSPSPSPSPSPSPSPSPSPSPSPIPSPSPKPSPSPVA 383
>EXTN_SORBI (P24152) Extensin precursor (Proline-rich glycoprotein)
Length = 283
Score = 55.5 bits (132), Expect = 1e-06
Identities = 35/101 (34%), Positives = 42/101 (40%), Gaps = 4/101 (3%)
Query: 772 PSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAP-PSPTQTATPPVG 830
PSP P A P P S V P P P A PP+ P P P T P
Sbjct: 76 PSPKPTPPPATPKPTPPTYTPSPK-PKSPVYP--PPPKASTPPTYTPSPKPPATKPPTYP 132
Query: 831 TQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNPR 871
T PPA+ P TPPV P P+ P + ++ PNP+
Sbjct: 133 TPKPPATKPPTPPVYTPSPKPPVTKPPTPKPTPPVYTPNPK 173
Score = 45.8 bits (107), Expect = 8e-04
Identities = 34/114 (29%), Positives = 47/114 (40%), Gaps = 7/114 (6%)
Query: 765 EPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPP--SPAPPSPT 822
+P + PP+P PV +P+ P+ P P P PP PP +P+P PT
Sbjct: 135 KPPATKPPTP-PVYTPSPKP--PVTKPPTPKPTPPVYTPN-PKPPVTKPPTHTPSPKPPT 190
Query: 823 QTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNPRYALVH 876
TPPV T + P P +PP P P P + + P+ Y H
Sbjct: 191 SKPTPPVYTPS-PKPPKPSPPTYTPTPKPPATKPPTSTPTHPKPTPHTPYPQAH 243
Score = 45.1 bits (105), Expect = 0.001
Identities = 30/100 (30%), Positives = 39/100 (39%), Gaps = 9/100 (9%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSP--TQ 823
P +P P +P+ P+ P P PPA PP+ P P T+
Sbjct: 82 PPPATPKPTPPTYTPSPKPKSPVYPPP-PKASTPPTYTPSPKPPATKPPTYPTPKPPATK 140
Query: 824 TATPPVGTQAP------PASPPTTPPVVAQVPLAPIAARP 857
TPPV T +P P +P TPPV P P+ P
Sbjct: 141 PPTPPVYTPSPKPPVTKPPTPKPTPPVYTPNPKPPVTKPP 180
Score = 42.4 bits (98), Expect = 0.009
Identities = 43/144 (29%), Positives = 55/144 (37%), Gaps = 20/144 (13%)
Query: 770 SPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPAL-------PVPPADAPPSPA--PPS 820
SP PV P++S P P+ S PA P PPA PP+P PS
Sbjct: 97 SPKPKSPVYPPPPKASTP------PTYTPSPKPPATKPPTYPTPKPPATKPPTPPVYTPS 150
Query: 821 PTQTAT-PPVGTQAPPASPPTTPPVVAQVPL-APIAARPRTRSQNGIFRPNPRYALVHAQ 878
P T PP PP P P V + P P P ++ ++ P+P+ +
Sbjct: 151 PKPPVTKPPTPKPTPPVYTPNPKPPVTKPPTHTPSPKPPTSKPTPPVYTPSPK-PPKPSP 209
Query: 879 PTGILTAFHTVTKPKGFKSAMKHP 902
PT T TKP S HP
Sbjct: 210 PTYTPTPKPPATKPP--TSTPTHP 231
Score = 40.8 bits (94), Expect = 0.026
Identities = 30/92 (32%), Positives = 39/92 (41%), Gaps = 4/92 (4%)
Query: 753 SSPHDLVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADA 812
S P V + +P SPP+ P P + P + + P P P
Sbjct: 191 SKPTPPVYTPSPKPPKPSPPTYTPTPK-PPATKPPTSTPTHPKPTPHTPYPQAHPPTYKP 249
Query: 813 PPSPAPPSPT-QTATPPVGTQAPPASPPTTPP 843
P P+PP+PT T TPPV P+SPP PP
Sbjct: 250 APKPSPPAPTPPTYTPPV--SHTPSSPPPPPP 279
Score = 37.7 bits (86), Expect = 0.22
Identities = 32/90 (35%), Positives = 40/90 (43%), Gaps = 13/90 (14%)
Query: 770 SPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAP--PSPAPPSPTQTATP 827
S P+P PV +P+ P S P+ + PA PP P P P P +P A P
Sbjct: 191 SKPTP-PVYTPSPKPPKP----SPPTYTPTPKPPATK-PPTSTPTHPKPTPHTPYPQAHP 244
Query: 828 PVGTQAPPASPPT-TPPV----VAQVPLAP 852
P AP SPP TPP V+ P +P
Sbjct: 245 PTYKPAPKPSPPAPTPPTYTPPVSHTPSSP 274
Score = 37.4 bits (85), Expect = 0.29
Identities = 34/107 (31%), Positives = 41/107 (37%), Gaps = 19/107 (17%)
Query: 765 EPASGSPPSPHPVVLV-----APESSLPIGSLSCPSCDDSDVLPAL----PVPPADAP-- 813
+P PP+P P V P + P P S P + P PP +P
Sbjct: 152 KPPVTKPPTPKPTPPVYTPNPKPPVTKPPTHTPSPKPPTSKPTPPVYTPSPKPPKPSPPT 211
Query: 814 --PSPAPPS---PTQTAT---PPVGTQAPPASPPTTPPVVAQVPLAP 852
P+P PP+ PT T T P T P A PPT P P AP
Sbjct: 212 YTPTPKPPATKPPTSTPTHPKPTPHTPYPQAHPPTYKPAPKPSPPAP 258
Score = 34.7 bits (78), Expect = 1.9
Identities = 25/84 (29%), Positives = 33/84 (38%), Gaps = 18/84 (21%)
Query: 806 PVPPADAPPSPAPPS---------------PT-QTATPPVGTQAPPASPPTTPPVV--AQ 847
P PPA P PP+ PT T TP PPA+P TPP +
Sbjct: 39 PKPPAKGPKPEKPPTKGHGHKPEKPPKEHKPTPPTYTPSPKPTPPPATPKPTPPTYTPSP 98
Query: 848 VPLAPIAARPRTRSQNGIFRPNPR 871
P +P+ P S + P+P+
Sbjct: 99 KPKSPVYPPPPKASTPPTYTPSPK 122
>EXTN_TOBAC (P13983) Extensin precursor (Cell wall
hydroxyproline-rich glycoprotein)
Length = 620
Score = 55.1 bits (131), Expect = 1e-06
Identities = 40/110 (36%), Positives = 49/110 (44%), Gaps = 6/110 (5%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCP--SCDDSDVLPALPVPPADAPPSPAPPSPTQ 823
P S SPP P + P SS P S S P + + S P PP APP+ +PP PT
Sbjct: 358 PPSYSPPPP-TYLPPPPPSSPPPPSFSPPPPTYEQSPPPPPAYSPPLPAPPTYSPPPPTY 416
Query: 824 TATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNPRYA 873
+ PP Q PP P +PP A P P P + + P P YA
Sbjct: 417 SPPPPTYAQPPPLPPTYSPPPPAYSPPPPPTYSPPPPTYS---PPPPAYA 463
Score = 50.1 bits (118), Expect = 4e-05
Identities = 34/92 (36%), Positives = 40/92 (42%), Gaps = 8/92 (8%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPAL--PVPPADAPPSP---APPS 820
P + SPP P P P P S P+ LP P PPA +PP P +PP
Sbjct: 396 PPAYSPPLPAPPTYSPPP---PTYSPPPPTYAQPPPLPPTYSPPPPAYSPPPPPTYSPPP 452
Query: 821 PTQTATPPVGTQAPPASPPTTPPVVAQVPLAP 852
PT + PP Q PP P +PP A P P
Sbjct: 453 PTYSPPPPAYAQPPPPPPTYSPPPPAYSPPPP 484
Score = 48.5 bits (114), Expect = 1e-04
Identities = 42/122 (34%), Positives = 50/122 (40%), Gaps = 19/122 (15%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPAL-PVPPADAPPSPA----PPS 820
PA PP P P P P S PS S P + P+PP +PP P PP
Sbjct: 460 PAYAQPPPPPPTYSPPP----PAYSPPPPSPIYSPPPPQVQPLPPTFSPPPPRRIHLPPP 515
Query: 821 PTQTATPPVGTQAPPASPPT-TPPVVAQVPLAPIAA-RPRTRSQNGIFRPNPRYALVHAQ 878
P + PP T P SPPT +PP Q+ P +PRT P P Y +
Sbjct: 516 PHRQPRPPTPTYGQPPSPPTFSPPPPRQIHSPPPPHWQPRT--------PTPTYGQPPSP 567
Query: 879 PT 880
PT
Sbjct: 568 PT 569
Score = 48.1 bits (113), Expect = 2e-04
Identities = 45/132 (34%), Positives = 53/132 (40%), Gaps = 19/132 (14%)
Query: 754 SPHDLVVSTFYEPASGSPP--SPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPAD 811
SP TF P PP SP P P + LP+ S S P PV
Sbjct: 308 SPPPTPTPTFSPP----PPAYSPPPTYSPPPPTYLPLPSSPIYS-------PPPPVYSPP 356
Query: 812 APPSPAPPSPTQTATPPVGTQAPPA-SPPTTPPVVAQVPLAPIAARPRTRSQNGIFRPNP 870
PPS +PP PT PP + PP+ SPP PP Q P P A P + P P
Sbjct: 357 PPPSYSPPPPTYLPPPPPSSPPPPSFSPP--PPTYEQSPPPPPAYSPPLPAPPTYSPPPP 414
Query: 871 RYA---LVHAQP 879
Y+ +AQP
Sbjct: 415 TYSPPPPTYAQP 426
Score = 48.1 bits (113), Expect = 2e-04
Identities = 43/140 (30%), Positives = 53/140 (37%), Gaps = 16/140 (11%)
Query: 747 PFAGSHSSPHDLVVSTFYEPASGSPPSPH---PVVLVAPESSLPIGSLSCPSCDDSDVLP 803
P +G H+ P + P SPPS H P A PI S PS
Sbjct: 150 PPSGGHTPPRGQHPPSHRRP---SPPSRHGHPPPPTYAQPPPTPIYS---PSPQVQPPPT 203
Query: 804 ALPVPPADAPPSPAPPSPTQTATPPVGTQAPPA---SPPTTPPVVAQVPLAPIAARPRTR 860
P PP P+P+PPS PP APP +PPT P PL + PR +
Sbjct: 204 YSPPPPTHVQPTPSPPSRGHQPQPPTHRHAPPTHRHAPPTHQP----SPLRHLPPSPRRQ 259
Query: 861 SQNGIFRPNPRYALVHAQPT 880
Q + P P QP+
Sbjct: 260 PQPPTYSPPPPAYAQSPQPS 279
Score = 47.4 bits (111), Expect = 3e-04
Identities = 32/93 (34%), Positives = 41/93 (43%), Gaps = 8/93 (8%)
Query: 764 YEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPA---PPS 820
YE + PP+ P + P S P + S P P+PP +PP PA PP
Sbjct: 389 YEQSPPPPPAYSPPLPAPPTYSPPPPTYSPPP---PTYAQPPPLPPTYSPPPPAYSPPPP 445
Query: 821 PTQTATPPVGTQAPP--ASPPTTPPVVAQVPLA 851
PT + PP + PP A PP PP + P A
Sbjct: 446 PTYSPPPPTYSPPPPAYAQPPPPPPTYSPPPPA 478
Score = 47.0 bits (110), Expect = 4e-04
Identities = 31/93 (33%), Positives = 43/93 (45%), Gaps = 4/93 (4%)
Query: 765 EPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQT 824
+P + SPP P +P+ S P S P+ P PP PSP PP+PT T
Sbjct: 261 QPPTYSPPPP--AYAQSPQPS-PTYSPPPPTYSPPPPSPIYSPPPPAYSPSP-PPTPTPT 316
Query: 825 ATPPVGTQAPPASPPTTPPVVAQVPLAPIAARP 857
+PP +PP + PP +P +PI + P
Sbjct: 317 FSPPPPAYSPPPTYSPPPPTYLPLPSSPIYSPP 349
Score = 45.4 bits (106), Expect = 0.001
Identities = 35/104 (33%), Positives = 41/104 (38%), Gaps = 10/104 (9%)
Query: 766 PASGSPP----SPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPA---- 817
P + SPP SP P P P S P+ P PP +PP PA
Sbjct: 406 PPTYSPPPPTYSPPPPTYAQPPPLPPTYSPPPPAYSPPPPPTYSPPPPTYSPPPPAYAQP 465
Query: 818 -PPSPTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTR 860
PP PT + PP PP SP +PP PL P + P R
Sbjct: 466 PPPPPTYSPPPP-AYSPPPPSPIYSPPPPQVQPLPPTFSPPPPR 508
Score = 45.4 bits (106), Expect = 0.001
Identities = 31/96 (32%), Positives = 42/96 (43%), Gaps = 7/96 (7%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPA----PPSP 821
P + SPP P P+ P + P P+ S PA PP +PP P P SP
Sbjct: 286 PPTYSPPPPSPIYSPPPPAYSP-SPPPTPTPTFSPPPPAYSPPPTYSPPPPTYLPLPSSP 344
Query: 822 TQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARP 857
+ PPV +PP P +PP +P P ++ P
Sbjct: 345 IYSPPPPV--YSPPPPPSYSPPPPTYLPPPPPSSPP 378
Score = 43.5 bits (101), Expect = 0.004
Identities = 26/72 (36%), Positives = 33/72 (45%), Gaps = 4/72 (5%)
Query: 806 PVPPADAPPSPA---PPSPTQTATPPVGTQA-PPASPPTTPPVVAQVPLAPIAARPRTRS 861
P PP +PP PA P P+ T +PP T + PP SP +PP A P P P
Sbjct: 260 PQPPTYSPPPPAYAQSPQPSPTYSPPPPTYSPPPPSPIYSPPPPAYSPSPPPTPTPTFSP 319
Query: 862 QNGIFRPNPRYA 873
+ P P Y+
Sbjct: 320 PPPAYSPPPTYS 331
Score = 39.3 bits (90), Expect = 0.077
Identities = 30/87 (34%), Positives = 34/87 (38%), Gaps = 12/87 (13%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPAL---PVPPADAPPSP----AP 818
P G PPSP +P I S P P P PP + P P +P
Sbjct: 525 PTYGQPPSPPTF---SPPPPRQIHSPPPPHWQPRTPTPTYGQPPSPPTFSAPPPRQIHSP 581
Query: 819 PSPTQTATPPVGTQAPPASPPTT--PP 843
P P + PP T P SPPTT PP
Sbjct: 582 PPPHRQPRPPTPTYGQPPSPPTTYSPP 608
Score = 37.4 bits (85), Expect = 0.29
Identities = 42/138 (30%), Positives = 50/138 (35%), Gaps = 46/138 (33%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAP---PSPA--PPS 820
P+ G P P PV+ S P S P PP+ P PS PPS
Sbjct: 101 PSRGFNPPPSPVI----SPSHPPPSYGAP-------------PPSHGPGHLPSHGQRPPS 143
Query: 821 PTQTATPPVGTQAPP----------ASPPT-----TPPVVAQVPLAPIAARPRTRSQNGI 865
P+ PP G PP SPP+ PP AQ P PI S +
Sbjct: 144 PSHGHAPPSGGHTPPRGQHPPSHRRPSPPSRHGHPPPPTYAQPPPTPI------YSPSPQ 197
Query: 866 FRPNPRYA---LVHAQPT 880
+P P Y+ H QPT
Sbjct: 198 VQPPPTYSPPPPTHVQPT 215
Score = 35.8 bits (81), Expect = 0.85
Identities = 21/56 (37%), Positives = 27/56 (47%), Gaps = 4/56 (7%)
Query: 806 PVPPADAPPSPA--PPSPTQTATPPVGTQAPPASPPTT--PPVVAQVPLAPIAARP 857
P P PPSP P PT + PP Q+P SP + PP + P +PI + P
Sbjct: 246 PSPLRHLPPSPRRQPQPPTYSPPPPAYAQSPQPSPTYSPPPPTYSPPPPSPIYSPP 301
Score = 34.3 bits (77), Expect = 2.5
Identities = 36/125 (28%), Positives = 48/125 (37%), Gaps = 10/125 (8%)
Query: 763 FYEPASGSPPSPHPVVLVAPES--SLPIGSLSCPSCDDSDVLPALPVPP-ADAPPSPAPP 819
+ P S P P + L P + + P PS + A P P PPS P
Sbjct: 33 YLPPPVTSQPPPSSIGLSPPSAPTTTPPSRGHVPSPRHAPPRHAYPPPSHGHLPPSVGGP 92
Query: 820 SPTQTATPPVGTQAPPASP---PTTPPVVAQVPLAPIAARPRTRSQNGIFRPNPRYALVH 876
P + PP PP SP P+ PP P P + P +G P+P + H
Sbjct: 93 PPHRGHLPPSRGFNPPPSPVISPSHPPPSYGAP--PPSHGPGHLPSHGQRPPSPSHG--H 148
Query: 877 AQPTG 881
A P+G
Sbjct: 149 APPSG 153
Score = 32.7 bits (73), Expect = 7.2
Identities = 25/82 (30%), Positives = 32/82 (38%), Gaps = 10/82 (12%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPA---DAPP----SPAP 818
P + SPP P + P P P+ P PP +PP P P
Sbjct: 533 PPTFSPPPPRQIHSPPPPHWQP--RTPTPTYGQPPSPPTFSAPPPRQIHSPPPPHRQPRP 590
Query: 819 PSPTQTATP-PVGTQAPPASPP 839
P+PT P P T +PP+ PP
Sbjct: 591 PTPTYGQPPSPPTTYSPPSPPP 612
>SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185 precursor
(SSG 185)
Length = 485
Score = 54.7 bits (130), Expect = 2e-06
Identities = 32/87 (36%), Positives = 34/87 (38%), Gaps = 2/87 (2%)
Query: 771 PPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTATPPVG 830
PPSP P P S P S PS P P PP PP P PPSP PP
Sbjct: 230 PPSPQPTASSRPPSPPP--SPRPPSPPPPSPSPPPPPPPPPPPPPPPPPSPPPPPPPPPP 287
Query: 831 TQAPPASPPTTPPVVAQVPLAPIAARP 857
PP P +PP P P+ P
Sbjct: 288 PPPPPPPPSPSPPRKPPSPSPPVPPPP 314
Score = 49.3 bits (116), Expect = 7e-05
Identities = 29/83 (34%), Positives = 31/83 (36%)
Query: 770 SPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTATPPV 829
+P P P P S P S PS S P+ P P PP P PP P PP
Sbjct: 218 NPIGPAPNNSPLPPSPQPTASSRPPSPPPSPRPPSPPPPSPSPPPPPPPPPPPPPPPPPS 277
Query: 830 GTQAPPASPPTTPPVVAQVPLAP 852
PP PP PP P P
Sbjct: 278 PPPPPPPPPPPPPPPPPPSPSPP 300
Score = 47.8 bits (112), Expect = 2e-04
Identities = 30/91 (32%), Positives = 33/91 (35%), Gaps = 6/91 (6%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
P S PPSP P P P P S P P PP PP P PSP +
Sbjct: 250 PPSPPPPSPSPPPPPPPPPPPP------PPPPPSPPPPPPPPPPPPPPPPPPSPSPPRKP 303
Query: 826 TPPVGTQAPPASPPTTPPVVAQVPLAPIAAR 856
P PP SPP+ P P +R
Sbjct: 304 PSPSPPVPPPPSPPSVLPAATGFPFCECVSR 334
Score = 33.1 bits (74), Expect = 5.5
Identities = 19/53 (35%), Positives = 24/53 (44%), Gaps = 3/53 (5%)
Query: 806 PVPPADAPP-SPAPPSPTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARP 857
P+PP+ P S PPSP + PP + PP+ P PP P P P
Sbjct: 228 PLPPSPQPTASSRPPSPPPSPRPP--SPPPPSPSPPPPPPPPPPPPPPPPPSP 278
Score = 32.3 bits (72), Expect = 9.4
Identities = 20/62 (32%), Positives = 30/62 (48%), Gaps = 3/62 (4%)
Query: 793 CPS-CDDSDVLPALPVPP-ADAPPSPAPPSPTQTATPPVGTQAPPASPPTTPPVVAQVPL 850
CP+ +V P P P + PPSP P + ++ +PP + PP+ PP +P P
Sbjct: 208 CPTGLSGPNVNPIGPAPNNSPLPPSPQPTASSRPPSPPPSPR-PPSPPPPSPSPPPPPPP 266
Query: 851 AP 852
P
Sbjct: 267 PP 268
>EXLP_TOBAC (Q03211) Pistil-specific extensin-like protein precursor
(PELP)
Length = 426
Score = 54.3 bits (129), Expect = 2e-06
Identities = 31/88 (35%), Positives = 41/88 (46%), Gaps = 2/88 (2%)
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
PA PP P PV +P + + P + P LP PP A P PSP+ A
Sbjct: 193 PAKQPPPPPPPVKAPSPSPATQPPTKQPPPPPRAKKSPLLPPPPPVAYPPVMTPSPSPAA 252
Query: 826 TPPVGTQAPPASPPTTPPVVAQVPLAPI 853
PP+ AP SPP PP++ + P P+
Sbjct: 253 EPPI--IAPFPSPPANPPLIPRRPAPPV 278
Score = 45.8 bits (107), Expect = 8e-04
Identities = 34/115 (29%), Positives = 40/115 (34%), Gaps = 14/115 (12%)
Query: 766 PASGSPP-------SPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAP 818
P G PP SP P+V P P PS D PPA P P P
Sbjct: 127 PIGGGPPVNQPKPSSPSPLVKPPPPPPSPCK----PSPPDQSAKQPPQPPPAKQPSPPPP 182
Query: 819 PSPTQTATPPVGTQAPPASPPT---TPPVVAQVPLAPIAARPRTRSQNGIFRPNP 870
P P + +P Q PP PP +P Q P PR + + P P
Sbjct: 183 PPPVKAPSPSPAKQPPPPPPPVKAPSPSPATQPPTKQPPPPPRAKKSPLLPPPPP 237
Score = 42.0 bits (97), Expect = 0.012
Identities = 32/105 (30%), Positives = 39/105 (36%), Gaps = 19/105 (18%)
Query: 765 EPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPV----------PPADAPP 814
+ ++ PP P P +P P PS P PV PP PP
Sbjct: 162 DQSAKQPPQPPPAKQPSPPPPPPPVKAPSPSPAKQPPPPPPPVKAPSPSPATQPPTKQPP 221
Query: 815 -------SPAPPSPTQTATPPVGTQAPPASPPTTPPVVAQVPLAP 852
SP P P A PPV T +P SP PP++A P P
Sbjct: 222 PPPRAKKSPLLPPPPPVAYPPVMTPSP--SPAAEPPIIAPFPSPP 264
Score = 42.0 bits (97), Expect = 0.012
Identities = 28/104 (26%), Positives = 43/104 (40%), Gaps = 9/104 (8%)
Query: 758 LVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPA 817
LV+ +F + + G P+ P + +P+ + P + VLP P+ PPSP+
Sbjct: 17 LVLGSFSKLSHGELWLELPLPFDWPPAEIPLPDIPSPFDGPTFVLPPPSPLPSPPPPSPS 76
Query: 818 PPSPTQTATPP---------VGTQAPPASPPTTPPVVAQVPLAP 852
PP P+ + PP G PP P +PL P
Sbjct: 77 PPPPSPSPPPPSTIPLIPPFTGGFLPPLPGSKLPDFAGLLPLIP 120
Score = 37.4 bits (85), Expect = 0.29
Identities = 31/106 (29%), Positives = 41/106 (38%), Gaps = 21/106 (19%)
Query: 766 PASGSPPSPHPVVLVAPESS---LPIGSLSCPSCDDSDVLPALP----VPPADA-----P 813
P S SPP P + L+ P + P+ P D + +LP +P VPP
Sbjct: 79 PPSPSPPPPSTIPLIPPFTGGFLPPLPGSKLP--DFAGLLPLIPNLPDVPPIGGGPPVNQ 136
Query: 814 PSPAPPSPTQTATPPVGTQAPPA-------SPPTTPPVVAQVPLAP 852
P P+ PSP PP + P+ PP PP P P
Sbjct: 137 PKPSSPSPLVKPPPPPPSPCKPSPPDQSAKQPPQPPPAKQPSPPPP 182
>FMN1_MOUSE (Q05860) Formin 1 isoforms I/II/III (Limb deformity
protein)
Length = 1468
Score = 53.5 bits (127), Expect = 4e-06
Identities = 45/138 (32%), Positives = 54/138 (38%), Gaps = 23/138 (16%)
Query: 730 ISLTYVSHHAQFDEFCFPFA---GSHSSPHDLVVSTFYEPASGSPPSPHPVVLVAPESSL 786
ISLT +S + PF G+ SS + PA +PP P P L+ P L
Sbjct: 836 ISLTQLSPSKDSKDIHAPFQTREGTSSSSQQKISP----PAPPTPP-PLPPPLIPPPPPL 890
Query: 787 PIGSLSCPSCDDSDVL------------PALPVPPADAPPSPAPPS---PTQTATPPVGT 831
P G P + P PVPP+D PP P PP P A P G
Sbjct: 891 PPGLGPLPPAPPIPPVCPVSPPPPPPPPPPTPVPPSDGPPPPPPPPPPLPNVLALPNSGG 950
Query: 832 QAPPASPPTTPPVVAQVP 849
PP PP PP +A P
Sbjct: 951 PPPPPPPPPPPPGLAPPP 968
Score = 33.5 bits (75), Expect = 4.2
Identities = 24/86 (27%), Positives = 30/86 (33%), Gaps = 4/86 (4%)
Query: 771 PPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTATPP-- 828
PP P P V P P P + LP PP PP P PP PP
Sbjct: 914 PPPPPPPTPVPPSDGPPPPPPPPPPLPNVLALPNSGGPPPP-PPPPPPPPGLAPPPPPGL 972
Query: 829 -VGTQAPPASPPTTPPVVAQVPLAPI 853
G + + P P + P+ P+
Sbjct: 973 SFGLSSSSSQYPRKPAIEPSCPMKPL 998
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.347 0.151 0.556
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 155,494,076
Number of Sequences: 164201
Number of extensions: 6512424
Number of successful extensions: 64518
Number of sequences better than 10.0: 880
Number of HSP's better than 10.0 without gapping: 240
Number of HSP's successfully gapped in prelim test: 714
Number of HSP's that attempted gapping in prelim test: 44592
Number of HSP's gapped (non-prelim): 6699
length of query: 1411
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1288
effective length of database: 39,777,331
effective search space: 51233202328
effective search space used: 51233202328
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0321.5


