

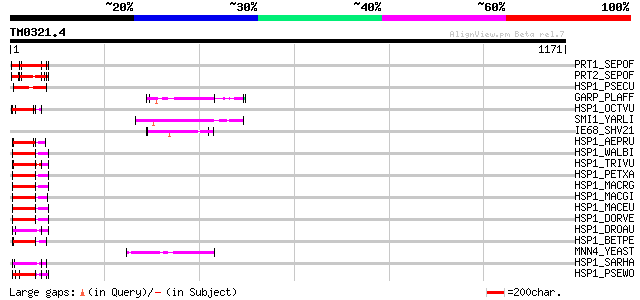
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0321.4
(1171 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PRT1_SEPOF (P80001) Spermatid-specific protein T1 [Contains: Spe... 61 2e-08
PRT2_SEPOF (P80002) Spermatid-specific protein T2 [Contains: Spe... 60 3e-08
HSP1_PSECU (P42145) Sperm protamine P1 57 2e-07
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 56 5e-07
HSP1_OCTVU (P83214) Sperm protamine P1 (Po1) [Contains: Sperm pr... 55 1e-06
SMI1_YARLI (Q6CDX0) KNR4/SMI1 homolog 53 4e-06
IE68_SHV21 (Q01042) Immediate-early protein 52 7e-06
HSP1_AEPRU (Q9GLP9) Sperm protamine P1 52 9e-06
HSP1_WALBI (P67843) Sperm protamine P1 52 1e-05
HSP1_TRIVU (P42152) Sperm protamine P1 52 1e-05
HSP1_PETXA (Q9GLQ7) Sperm protamine P1 52 1e-05
HSP1_MACRG (P67842) Sperm protamine P1 52 1e-05
HSP1_MACGI (P42139) Sperm protamine P1 52 1e-05
HSP1_MACEU (P42138) Sperm protamine P1 52 1e-05
HSP1_DORVE (P67841) Sperm protamine P1 52 1e-05
HSP1_DROAU (P42132) Sperm protamine P1 51 2e-05
HSP1_BETPE (Q9GLQ0) Sperm protamine P1 51 2e-05
MNN4_YEAST (P36044) MNN4 protein 50 3e-05
HSP1_SARHA (P62486) Sperm protamine P1 50 3e-05
HSP1_PSEWO (Q71VG4) Sperm protamine P1 50 3e-05
>PRT1_SEPOF (P80001) Spermatid-specific protein T1 [Contains:
Sperm protamine SP1]
Length = 78
Score = 60.8 bits (146), Expect = 2e-08
Identities = 39/64 (60%), Positives = 41/64 (63%), Gaps = 4/64 (6%)
Query: 4 EGGRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRG 63
+GGRRR RR RRRRRR RRR R RR RRRRRRRR+R RRR RR S RR
Sbjct: 19 KGGRRRRRRSRRRRRRSRRRSRSPYRRRYRRRRRRRRRR----SRRRRYRRRRSYSRRRY 74
Query: 64 PRRR 67
RRR
Sbjct: 75 RRRR 78
Score = 49.7 bits (117), Expect = 5e-05
Identities = 31/57 (54%), Positives = 35/57 (61%), Gaps = 2/57 (3%)
Query: 26 RRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLRR 82
RRR RR RRRRRR R+R R P RRR +RR RR RRR + RRR +R RR
Sbjct: 22 RRRRRRSRRRRRRSRRRSRSPYRRRYRRRRRRR--RRRSRRRRYRRRRSYSRRRYRR 76
Score = 49.7 bits (117), Expect = 5e-05
Identities = 30/55 (54%), Positives = 35/55 (63%)
Query: 22 RRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
RRRRRR RRRRR RRR R R+ RRR +RR + RR RRR++ RRR R
Sbjct: 22 RRRRRRSRRRRRRSRRRSRSPYRRRYRRRRRRRRRRSRRRRYRRRRSYSRRRYRR 76
Score = 43.1 bits (100), Expect = 0.004
Identities = 27/59 (45%), Positives = 34/59 (56%), Gaps = 7/59 (11%)
Query: 32 RRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLRRTWTVTRKK 90
RRRRRR RR RRR RR S + RR RRR RRR+ R +R RR + +R++
Sbjct: 22 RRRRRRSRR-------RRRRSRRRSRSPYRRRYRRRRRRRRRRSRRRRYRRRRSYSRRR 73
>PRT2_SEPOF (P80002) Spermatid-specific protein T2 [Contains:
Sperm protamine SP2]
Length = 77
Score = 60.5 bits (145), Expect = 3e-08
Identities = 39/64 (60%), Positives = 40/64 (61%), Gaps = 5/64 (7%)
Query: 4 EGGRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRG 63
+GGRRR RR RRRRR RRRR R RR RRRRRRRR R RRR RR S RR
Sbjct: 19 KGGRRRRRRSRRRRRSRRRRSRSPYRRRYRRRRRRRRSR-----RRRRYRRRRSYSRRRY 73
Query: 64 PRRR 67
RRR
Sbjct: 74 RRRR 77
Score = 50.1 bits (118), Expect = 4e-05
Identities = 31/57 (54%), Positives = 35/57 (61%), Gaps = 3/57 (5%)
Query: 26 RRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLRR 82
RRR RR RRRRR RR+R R P RRR +RR RR RRR + RRR +R RR
Sbjct: 22 RRRRRRSRRRRRSRRRRSRSPYRRRYRRRRRR---RRSRRRRRYRRRRSYSRRRYRR 75
Score = 48.9 bits (115), Expect = 8e-05
Identities = 30/56 (53%), Positives = 33/56 (58%)
Query: 18 RRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRR 73
RRRRRR RRRR RRRR R R+R R+ RRR RR RR RR + RRR
Sbjct: 22 RRRRRRSRRRRRSRRRRSRSPYRRRYRRRRRRRRSRRRRRYRRRRSYSRRRYRRRR 77
Score = 47.8 bits (112), Expect = 2e-04
Identities = 31/55 (56%), Positives = 35/55 (63%), Gaps = 1/55 (1%)
Query: 22 RRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
RRRRRR RRRR RRRR R R+ RRR +RR S RR RRR++ RRR R
Sbjct: 22 RRRRRRSRRRRRSRRRRSRSPYRRRYRRR-RRRRRSRRRRRYRRRRSYSRRRYRR 75
>HSP1_PSECU (P42145) Sperm protamine P1
Length = 68
Score = 57.4 bits (137), Expect = 2e-07
Identities = 37/69 (53%), Positives = 42/69 (60%), Gaps = 6/69 (8%)
Query: 8 RR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRR 67
R R R R R RRRRRRRR R R RRRR RR R+ RRR RR + + RRG RRR
Sbjct: 4 RHSRSRSRSRYRRRRRRRRSRYRGRRRRYRRSRR------RRRRGRRRGNCLGRRGYRRR 57
Query: 68 NHLRRRQPR 76
+ RRR+ R
Sbjct: 58 RYSRRRRRR 66
Score = 42.4 bits (98), Expect = 0.007
Identities = 26/40 (65%), Positives = 27/40 (67%), Gaps = 3/40 (7%)
Query: 6 GRRR*RRRRRRRRRRRRRRRR---RR*RRRRRRRRRRRQR 42
GRRR RR RRRRRR RRR RR RRRR RRRR+R
Sbjct: 27 GRRRRYRRSRRRRRRGRRRGNCLGRRGYRRRRYSRRRRRR 66
Score = 42.4 bits (98), Expect = 0.007
Identities = 26/40 (65%), Positives = 27/40 (67%), Gaps = 3/40 (7%)
Query: 5 GGRRR*RRRRRRRRRRRRRRR---RRR*RRRRRRRRRRRQ 41
G RRR RR RRRRRR RRR RR RRRR RRRRR+
Sbjct: 27 GRRRRYRRSRRRRRRGRRRGNCLGRRGYRRRRYSRRRRRR 66
Score = 39.7 bits (91), Expect = 0.048
Identities = 28/60 (46%), Positives = 32/60 (52%)
Query: 23 RRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLRR 82
R R R R R R RRRRR+R + RR + R S RRG RR N L RR R +R R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRRSRYRGRRRRYRRSRRRRRRGRRRGNCLGRRGYRRRRYSR 61
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 56.2 bits (134), Expect = 5e-07
Identities = 52/199 (26%), Positives = 93/199 (46%), Gaps = 34/199 (17%)
Query: 295 VEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAEKEN 354
++K KV +E+ +S+++++ IG V + R K+ +I L+ ++ V+KE +K+
Sbjct: 495 MDKSKV-EEKNLSIQEQLIGTIGRVNVVPRRDNHKKKMAKIEEAELQKQKHVDKEEDKKE 553
Query: 355 QVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYHPNSDD 414
+ ++ +E V+E E+E + E+ +E+ E+ E+E + EE++E + +D
Sbjct: 554 E--SKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEED 611
Query: 415 SDDSSVVSLDDSDYEENWDWTTVLPRESLLDIPHDAPPPDLYELSLPVDTNDPGATYSDF 474
DD+ D+ D EE+ D DA D D D D
Sbjct: 612 EDDAEE---DEDDAEEDED---------------DAEEDD--------DEED-----DDE 640
Query: 475 EDEDGDSDDLESPSEGEGE 493
ED+D D D+ E E E E
Sbjct: 641 EDDDEDEDEDEEDEEEEEE 659
Score = 52.4 bits (124), Expect = 7e-06
Identities = 52/218 (23%), Positives = 87/218 (39%), Gaps = 52/218 (23%)
Query: 288 LLQGEKDVEKDKVIQEEVVSV--------EKEVEKDIGSVVLECERQVEKEAEKEIGSVV 339
L+ K EK+ IQE+++ ++ K + + E E Q +K +KE
Sbjct: 494 LMDKSKVEEKNLSIQEQLIGTIGRVNVVPRRDNHKKKMAKIEEAELQKQKHVDKE----- 548
Query: 340 LECERQVEKEAEKENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGV 399
E +++ KE ++E++ V E E+ V E+ E+E + E+ +E+ E+ E+E +
Sbjct: 549 -EDKKEESKEVQEESKEVQED---EEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEE-- 602
Query: 400 GGEEDDESYHPNSDDSDDSSVVSLDDSDYEENWDWTTVLPRESLLDIPHDAPPPDLYELS 459
EE+DE D +D D+ D EE+
Sbjct: 603 --EEEDEDEEDEDDAEEDEDDAEEDEDDAEED---------------------------- 632
Query: 460 LPVDTNDPGATYSDFEDEDGDSDDLESPSEGEGEGRAR 497
D + D EDED D +D E E E E +
Sbjct: 633 ---DDEEDDDEEDDDEDEDEDEEDEEEEEEEEEESEKK 667
Score = 50.8 bits (120), Expect = 2e-05
Identities = 37/143 (25%), Positives = 73/143 (50%), Gaps = 17/143 (11%)
Query: 289 LQGEKDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEK 348
LQ +K V+K++ +EE V++E ++ V E E +VE++ E+E E+
Sbjct: 539 LQKQKHVDKEEDKKEESKEVQEESKE-----VQEDEEEVEEDEEEE------------EE 581
Query: 349 EAEKENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESY 408
E E+E + E+ +E+ EE E E++ + A +++ E+ E + + EEDD+
Sbjct: 582 EEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEE 641
Query: 409 HPNSDDSDDSSVVSLDDSDYEEN 431
+ D+ +D ++ + EE+
Sbjct: 642 DDDEDEDEDEEDEEEEEEEEEES 664
Score = 34.3 bits (77), Expect = 2.0
Identities = 27/121 (22%), Positives = 65/121 (53%), Gaps = 9/121 (7%)
Query: 293 KDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAEK 352
+D ++D +E + +++K+ K + E E+ +EK+ +K+ + E++ +K+ +K
Sbjct: 245 EDKKEDHQQEEMLKTLDKKERKQKEKEMKEQEK-IEKKKKKQEEKEKKKQEKERKKQEKK 303
Query: 353 ENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYHPNS 412
E + ++ K+K + +E +K+ E+ K+K ++ +KEN+ + D S N+
Sbjct: 304 ERKQKEKEMKKQKKIEKERKKK-----EEKEKKK---KKHDKENEETMQQPDQTSEETNN 355
Query: 413 D 413
+
Sbjct: 356 E 356
Score = 32.7 bits (73), Expect = 5.9
Identities = 27/123 (21%), Positives = 58/123 (46%), Gaps = 12/123 (9%)
Query: 292 EKDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAE 351
+K EK+K QE+ +++ E+ ++ ++++EKE +K+ E + + +K+ +
Sbjct: 283 KKQEEKEKKKQEKERKKQEKKERKQKEKEMKKQKKIEKERKKK------EEKEKKKKKHD 336
Query: 352 KENQVVGEQAGK------EKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDD 405
KEN+ +Q + +++V V + KE E+ KE + GE +
Sbjct: 337 KENEETMQQPDQTSEETNNEIMVPLPSPLTDVTTPEEHKEGEHKEEEHKEGEHKEGEHKE 396
Query: 406 ESY 408
E +
Sbjct: 397 EEH 399
>HSP1_OCTVU (P83214) Sperm protamine P1 (Po1) [Contains: Sperm
protamine P2 (Po2) (Main protamine)]
Length = 56
Score = 55.1 bits (131), Expect = 1e-06
Identities = 33/46 (71%), Positives = 34/46 (73%), Gaps = 1/46 (2%)
Query: 5 GGRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR 50
GGRRR RRRRR R RR RRR RRR RRR RRR RRR+R R RRR
Sbjct: 11 GGRRR-RRRRRSRGRRGRRRGRRRGRRRGRRRGRRRRRRRGGRRRR 55
Score = 48.5 bits (114), Expect = 1e-04
Identities = 30/43 (69%), Positives = 31/43 (71%), Gaps = 2/43 (4%)
Query: 12 RRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
RRRRRRRR R RR RRR RRR RRR RRR R R+ RRR RR
Sbjct: 13 RRRRRRRRSRGRRGRRRGRRRGRRRGRRRGRRRR--RRRGGRR 53
Score = 47.4 bits (111), Expect = 2e-04
Identities = 34/61 (55%), Positives = 34/61 (55%), Gaps = 10/61 (16%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
RRR RRRRRRRR R RR RRR RRR RRR R R RRR RRG RR
Sbjct: 4 RRRVYSIGGRRRRRRRRSRGRRGRRRGRRRGRRRGRRRGRRRRR----------RRGGRR 53
Query: 67 R 67
R
Sbjct: 54 R 54
Score = 42.4 bits (98), Expect = 0.007
Identities = 24/36 (66%), Positives = 25/36 (68%)
Query: 3 NEGGRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRR 38
+ G R R R RRR RRR RRR RRRR RR RRRRR
Sbjct: 21 SRGRRGRRRGRRRGRRRGRRRGRRRRRRRGGRRRRR 56
Score = 41.2 bits (95), Expect = 0.017
Identities = 30/65 (46%), Positives = 35/65 (53%), Gaps = 9/65 (13%)
Query: 18 RRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRG 77
R RRR RRRRRRRR R +R R+ RRR +RR RRR RRR+ G
Sbjct: 1 RLSRRRVYSIGGRRRRRRRRSRGRRGRRRGRRRGRRR---------GRRRGRRRRRRRGG 51
Query: 78 KRLRR 82
+R RR
Sbjct: 52 RRRRR 56
>SMI1_YARLI (Q6CDX0) KNR4/SMI1 homolog
Length = 713
Score = 53.1 bits (126), Expect = 4e-06
Identities = 59/243 (24%), Positives = 103/243 (42%), Gaps = 26/243 (10%)
Query: 266 LDAVDSDDDVAIV-DRLDVICGKLLQGEKDVEKDKVIQ---------EEVVSVEKEVEKD 315
LD VD ++D A + D+ G L+ + V++ K EE+ K E+D
Sbjct: 460 LDIVDLNEDTATIKDKGKATMGSALR-KTIVDESKTAHTIVEPLEESEEIKGKGKGKEED 518
Query: 316 IGSVVLECERQVE---KEAEKEIGSVVLECERQVEKEAEKENQVVGEQAGKEKVVVEEAE 372
+ + + + K+A KE E ++ EK AE++ + + A + +EA+
Sbjct: 519 VNEAAEKAKVEATEKTKKAAKEAADKEAELKKAAEKAAEEKAKAEKKAAEAREKEEKEAK 578
Query: 373 KENQVVGEQAGKEKVV--VEQAEKENQGVGGEEDDESYHPNSDDSDDSSVVSLDDSDYEE 430
+ E+ KE+V +AE+E + E ++ + ++D S V + + EE
Sbjct: 579 AAAKAKEEELKKEEVAKAAAKAEEEQKATAAAEAAKAEAKRAAEADASKKVEAEKAAAEE 638
Query: 431 NWDWTTVLPRESLLDIPHDAPPPDLYELSLPVDTNDPGATYSDFEDEDGDSDDLESPSEG 490
+ +ES + DL EL +D + A +D E +D D DD E
Sbjct: 639 S--------KESKAESEESKVERDLEELK--IDEENGNAEEADEEADDDDEDDEEEGDSK 688
Query: 491 EGE 493
EGE
Sbjct: 689 EGE 691
Score = 35.4 bits (80), Expect = 0.91
Identities = 26/127 (20%), Positives = 52/127 (40%), Gaps = 3/127 (2%)
Query: 296 EKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAEKENQ 355
+++K + + E+E++K+ V + + E+E + + + E + EA+ +
Sbjct: 572 KEEKEAKAAAKAKEEELKKE---EVAKAAAKAEEEQKATAAAEAAKAEAKRAAEADASKK 628
Query: 356 VVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYHPNSDDS 415
V E+A E+ +AE E V + K+ E E ++DDE D
Sbjct: 629 VEAEKAAAEESKESKAESEESKVERDLEELKIDEENGNAEEADEEADDDDEDDEEEGDSK 688
Query: 416 DDSSVVS 422
+ S
Sbjct: 689 EGEETKS 695
Score = 32.3 bits (72), Expect = 7.7
Identities = 32/140 (22%), Positives = 57/140 (39%), Gaps = 6/140 (4%)
Query: 292 EKDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAE 351
EK+ + +EE + E EV K E + EA K E + + EAE
Sbjct: 574 EKEAKAAAKAKEEELKKE-EVAKAAAKAEEEQKATAAAEAAKAEAKRAAEADASKKVEAE 632
Query: 352 KENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYHPN 411
K E++ + K EE++ E + + +E E+A++E +EDDE +
Sbjct: 633 K---AAAEESKESKAESEESKVERDLEELKIDEENGNAEEADEEAD--DDDEDDEEEGDS 687
Query: 412 SDDSDDSSVVSLDDSDYEEN 431
+ + S + ++N
Sbjct: 688 KEGEETKSTTASKSKSKKKN 707
>IE68_SHV21 (Q01042) Immediate-early protein
Length = 407
Score = 52.4 bits (124), Expect = 7e-06
Identities = 37/139 (26%), Positives = 69/139 (49%), Gaps = 3/139 (2%)
Query: 292 EKDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAE 351
E++ E+ + +EE E+E E++ E + E+E +E + E E E E E
Sbjct: 98 EEEAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEE 157
Query: 352 KENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYHPN 411
E + E+A +E EEAE+E + E+A +E E+AE+E + EE+ E
Sbjct: 158 AEEEEAEEEAEEEAEEAEEAEEEAEEEAEEA-EEAEEAEEAEEEAE--EAEEEAEEAEEE 214
Query: 412 SDDSDDSSVVSLDDSDYEE 430
+++++++ + + EE
Sbjct: 215 AEEAEEAEEAEEAEEEAEE 233
Score = 48.5 bits (114), Expect = 1e-04
Identities = 37/141 (26%), Positives = 68/141 (47%), Gaps = 3/141 (2%)
Query: 290 QGEKDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKE 349
+GE E ++ EE + E+E E+ E E E EAE+E E E + +E
Sbjct: 89 EGEGREEAEEEEAEEKEAEEEEAEE--AEEEAEEEEAEEAEAEEEEAEEE-EAEEEEAEE 145
Query: 350 AEKENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYH 409
AE+E E+ +E+ EEAE+E + E + + E+AE+ + EE+ E
Sbjct: 146 AEEEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEEEAEEAE 205
Query: 410 PNSDDSDDSSVVSLDDSDYEE 430
++++++ + + + + EE
Sbjct: 206 EEAEEAEEEAEEAEEAEEAEE 226
Score = 48.5 bits (114), Expect = 1e-04
Identities = 36/134 (26%), Positives = 66/134 (48%), Gaps = 5/134 (3%)
Query: 290 QGEKDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVE-- 347
+ E++ E+++ + E E E E+ E E + +EAE+E E E + E
Sbjct: 113 EAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEEAEEEAE 172
Query: 348 --KEAEKENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDD 405
+EAE+E + E+A +E EEAE+E + E+A + + E+AE+ + EE+
Sbjct: 173 EAEEAEEEAEEEAEEA-EEAEEAEEAEEEAEEAEEEAEEAEEEAEEAEEAEEAEEAEEEA 231
Query: 406 ESYHPNSDDSDDSS 419
E +++ S+
Sbjct: 232 EEAEEEEEEAGPST 245
Score = 45.8 bits (107), Expect = 7e-04
Identities = 36/145 (24%), Positives = 69/145 (46%), Gaps = 8/145 (5%)
Query: 290 QGEKDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKE-----IGSVVLECER 344
QG+ D+ Q++ E++ +++ E ER+ E+E E E G E E
Sbjct: 43 QGDDDINTTH--QQQAALTEEQRREEVEEEGEERERRGEEEREGEGGEEGEGREEAEEEE 100
Query: 345 QVEKEAEKENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEED 404
EKEAE+E E+ +E+ EEAE E + E+ +E+ E E+E + E +
Sbjct: 101 AEEKEAEEEEAEEAEEEAEEE-EAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAE 159
Query: 405 DESYHPNSDDSDDSSVVSLDDSDYE 429
+E +++ + + + ++++ E
Sbjct: 160 EEEAEEEAEEEAEEAEEAEEEAEEE 184
Score = 41.6 bits (96), Expect = 0.013
Identities = 42/144 (29%), Positives = 74/144 (51%), Gaps = 16/144 (11%)
Query: 290 QGEKDVEKDKVIQEEVVSVEKEVE---KDIGSVVLECERQVEKEAEKEIGSVVLECERQV 346
Q ++VE++ +E E+E E + G E E EKEAE+E E E +
Sbjct: 62 QRREEVEEEGEERERRGEEEREGEGGEEGEGREEAEEEEAEEKEAEEE------EAE-EA 114
Query: 347 EKEAEKENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDE 406
E+EAE+E E A +E+ EEAE+E E+A +E+ E+AE+E + EE+ E
Sbjct: 115 EEEAEEEEAEEAE-AEEEEAEEEEAEEEE---AEEAEEEE--AEEAEEEAEEEEAEEEAE 168
Query: 407 SYHPNSDDSDDSSVVSLDDSDYEE 430
++++++ + ++++ E
Sbjct: 169 EEAEEAEEAEEEAEEEAEEAEEAE 192
Score = 40.4 bits (93), Expect = 0.028
Identities = 35/142 (24%), Positives = 67/142 (46%), Gaps = 15/142 (10%)
Query: 259 EHANEISLDAVDSDDDVAIVDRLDVICGKLLQGEKDVEKDKVIQEEVVSVEKEVEKDIGS 318
E A E + +++++ A + + + + E+ E ++ +EE E E E +
Sbjct: 116 EEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEEAEEEAEEAE 175
Query: 319 VVLECERQVEKEAEKEIGSVVLECERQVEKEAEKENQVVGEQAGKEKVVVEEAEKENQVV 378
E E + E+EAE+ E + +EAE+E + E+A + + EEAE+
Sbjct: 176 ---EAEEEAEEEAEEAE-------EAEEAEEAEEEAEEAEEEAEEAEEEAEEAEE----- 220
Query: 379 GEQAGKEKVVVEQAEKENQGVG 400
E+A + + E+AE+E + G
Sbjct: 221 AEEAEEAEEEAEEAEEEEEEAG 242
Score = 39.3 bits (90), Expect = 0.063
Identities = 37/188 (19%), Positives = 77/188 (40%), Gaps = 9/188 (4%)
Query: 313 EKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAEKENQVVG-------EQAGKEK 365
+ DI + + E++ +E+ E ER+ E+E E E G E+ +EK
Sbjct: 45 DDDINTTHQQQAALTEEQRREEVEEEGEERERRGEEEREGEGGEEGEGREEAEEEEAEEK 104
Query: 366 VVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYHPNSDDSDDSSVVSLDD 425
EE +E + E+ E+ E+ E E + EE +E+ ++++++ + +
Sbjct: 105 EAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAE 164
Query: 426 SDYEENWDWTTVLPRESLLDIPHDAPPPDLYELSLPVDTNDPGATYSDFEDEDGDSDDLE 485
+ EE + E+ + + E + + A + E+E ++++ E
Sbjct: 165 EEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEEEAEEAEEEA--EEAEEEAEEAEEAE 222
Query: 486 SPSEGEGE 493
E E E
Sbjct: 223 EAEEAEEE 230
>HSP1_AEPRU (Q9GLP9) Sperm protamine P1
Length = 60
Score = 52.0 bits (123), Expect = 9e-06
Identities = 31/48 (64%), Positives = 32/48 (66%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRR R R RRRR R RRRRR RR+R R RRR RR
Sbjct: 9 RSRSRYRRRRRRRSRYRSRRRRYRGSRRRRRSRRRRRRGYSRRRYSRR 56
Score = 48.9 bits (115), Expect = 8e-05
Identities = 33/66 (50%), Positives = 36/66 (54%), Gaps = 9/66 (13%)
Query: 9 R*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRN 68
R R R R R R RRRRRRR R R RRRR R R R+ RRR RRG RR
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRRYRGSRRRRRSRRR---------RRRGYSRRR 52
Query: 69 HLRRRQ 74
+ RRR+
Sbjct: 53 YSRRRR 58
Score = 46.6 bits (109), Expect = 4e-04
Identities = 28/44 (63%), Positives = 28/44 (63%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR 50
RRR RRR R R RRRR R RR RR RRRRRR R R RRR
Sbjct: 15 RRRRRRRSRYRSRRRRYRGSRRRRRSRRRRRRGYSRRRYSRRRR 58
Score = 45.4 bits (106), Expect = 9e-04
Identities = 30/49 (61%), Positives = 32/49 (65%), Gaps = 2/49 (4%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRR--RRRRRQR*RKPPRRR*QR 53
R R RRRRRRR R R RRRR R RRRRR RRRRR R+ RR +R
Sbjct: 11 RSRYRRRRRRRSRYRSRRRRYRGSRRRRRSRRRRRRGYSRRRYSRRRRR 59
Score = 41.2 bits (95), Expect = 0.017
Identities = 29/61 (47%), Positives = 33/61 (53%), Gaps = 4/61 (6%)
Query: 19 RRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGK 78
R R R R R R RRRRRRR R R R+ R +RR S R RRR + RRR R +
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRRYRGSRRRRRS----RRRRRRGYSRRRYSRRR 57
Query: 79 R 79
R
Sbjct: 58 R 58
Score = 35.4 bits (80), Expect = 0.91
Identities = 26/58 (44%), Positives = 29/58 (49%), Gaps = 7/58 (12%)
Query: 25 RRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLRR 82
R R R R R R RRR+R R R R +R RG RRR RRR+ RG RR
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRR-------YRGSRRRRRSRRRRRRGYSRRR 52
Score = 34.7 bits (78), Expect = 1.6
Identities = 30/74 (40%), Positives = 35/74 (46%), Gaps = 16/74 (21%)
Query: 17 RRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
R R R R R R RRRRRRR R R R RRR RG RRR RR+ R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSR-----RRR----------YRGSRRRRR-SRRRRR 45
Query: 77 GKRLRRTWTVTRKK 90
RR ++ R++
Sbjct: 46 RGYSRRRYSRRRRR 59
Score = 33.1 bits (74), Expect = 4.5
Identities = 19/27 (70%), Positives = 19/27 (70%), Gaps = 3/27 (11%)
Query: 5 GGRRR*RRRRRRRR---RRRRRRRRRR 28
G RRR R RRRRRR RRR RRRRR
Sbjct: 33 GSRRRRRSRRRRRRGYSRRRYSRRRRR 59
Score = 32.7 bits (73), Expect = 5.9
Identities = 18/27 (66%), Positives = 18/27 (66%)
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRR 32
G RR RR RRRRRR RRR R RRR
Sbjct: 33 GSRRRRRSRRRRRRGYSRRRYSRRRRR 59
>HSP1_WALBI (P67843) Sperm protamine P1
Length = 61
Score = 51.6 bits (122), Expect = 1e-05
Identities = 31/48 (64%), Positives = 32/48 (66%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRR R R RRRR R RRRRR RR +R R RRR RR
Sbjct: 9 RSRSRYRRRRRRRSRYRSRRRRYRGRRRRRSRRGRRRRGYSRRRYSRR 56
Score = 50.1 bits (118), Expect = 4e-05
Identities = 37/75 (49%), Positives = 39/75 (51%), Gaps = 16/75 (21%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R R R R R RRRRRRR R R RRRR R RR RRR RRG RR
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRRYRGRR-------RRR---------SRRGRRR 45
Query: 67 RNHLRRRQPRGKRLR 81
R + RRR R +R R
Sbjct: 46 RGYSRRRYSRRRRRR 60
Score = 38.1 bits (87), Expect = 0.14
Identities = 30/66 (45%), Positives = 31/66 (46%), Gaps = 15/66 (22%)
Query: 17 RRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
R R R R R R RRRRRRR R R R RRR RG RRR R R+ R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSR-----RRR----------YRGRRRRRSRRGRRRR 46
Query: 77 GKRLRR 82
G RR
Sbjct: 47 GYSRRR 52
Score = 34.3 bits (77), Expect = 2.0
Identities = 19/28 (67%), Positives = 19/28 (67%)
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRR 33
GRRR R RR RRRR RRR R RRRR
Sbjct: 33 GRRRRRSRRGRRRRGYSRRRYSRRRRRR 60
Score = 32.7 bits (73), Expect = 5.9
Identities = 25/70 (35%), Positives = 31/70 (43%), Gaps = 19/70 (27%)
Query: 21 RRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRL 80
R R R R R R RRRRRRR R R RRR + RR+ R +R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRS-------------------RRRRYRGRRRRRSRRG 42
Query: 81 RRTWTVTRKK 90
RR +R++
Sbjct: 43 RRRRGYSRRR 52
>HSP1_TRIVU (P42152) Sperm protamine P1
Length = 61
Score = 51.6 bits (122), Expect = 1e-05
Identities = 31/48 (64%), Positives = 33/48 (68%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R RRRRRR R R RRRR RR RRRRRR RRRR R+ RR +RR
Sbjct: 13 RYRRRRRRRRSRYRSRRRRYRRSRRRRRRGRRRRGYSRRRYSRRGRRR 60
Score = 51.6 bits (122), Expect = 1e-05
Identities = 31/48 (64%), Positives = 32/48 (66%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRRR R R RRR RR RRRRRR +R R RRR RR
Sbjct: 9 RSRSRYRRRRRRRRSRYRSRRRRYRRSRRRRRRGRRRRGYSRRRYSRR 56
Score = 50.4 bits (119), Expect = 3e-05
Identities = 33/59 (55%), Positives = 36/59 (60%), Gaps = 2/59 (3%)
Query: 8 RR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R R R R RRRRRRRR R R RRRR RR R+R R+ RRR R + RRG RR
Sbjct: 4 RHSRSRSRSRYRRRRRRRRSRYRSRRRRYRRSRRRRRRGRRRRGYSRRRYS--RRGRRR 60
Score = 48.9 bits (115), Expect = 8e-05
Identities = 35/75 (46%), Positives = 36/75 (47%), Gaps = 21/75 (28%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R R R RRRRRRRR R R R RR RR RRRR RRG RR
Sbjct: 7 RSRSRSRYRRRRRRRRSRYRSRRRRYRRSRRRR---------------------RRGRRR 45
Query: 67 RNHLRRRQPRGKRLR 81
R + RRR R R R
Sbjct: 46 RGYSRRRYSRRGRRR 60
>HSP1_PETXA (Q9GLQ7) Sperm protamine P1
Length = 61
Score = 51.6 bits (122), Expect = 1e-05
Identities = 31/48 (64%), Positives = 32/48 (66%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRR R R RRRR R RRRRR RR +R R RRR RR
Sbjct: 9 RSRSRYRRRRRRRSRYRSRRRRYRGRRRRRSRRGRRRRGYSRRRYSRR 56
Score = 47.8 bits (112), Expect = 2e-04
Identities = 36/75 (48%), Positives = 38/75 (50%), Gaps = 16/75 (21%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R R R R RRRRRRR R R RRRR R RR RRR RRG RR
Sbjct: 2 RYRHSXSRSRSRYRRRRRRRSRYRSRRRRYRGRR-------RRR---------SRRGRRR 45
Query: 67 RNHLRRRQPRGKRLR 81
R + RRR R +R R
Sbjct: 46 RGYSRRRYSRRRRRR 60
Score = 47.0 bits (110), Expect = 3e-04
Identities = 31/50 (62%), Positives = 33/50 (66%), Gaps = 2/50 (4%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRR--RRRRRRQR*RKPPRRR*QRR 54
R R RRRRRRR R R RRRR R RRRRR R RRRR R+ RR +RR
Sbjct: 11 RSRYRRRRRRRSRYRSRRRRYRGRRRRRSRRGRRRRGYSRRRYSRRRRRR 60
Score = 35.8 bits (81), Expect = 0.70
Identities = 29/66 (43%), Positives = 30/66 (44%), Gaps = 15/66 (22%)
Query: 17 RRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
R R R R R RRRRRRR R R R RRR RG RRR R R+ R
Sbjct: 2 RYRHSXSRSRSRYRRRRRRRSRYRSR-----RRR----------YRGRRRRRSRRGRRRR 46
Query: 77 GKRLRR 82
G RR
Sbjct: 47 GYSRRR 52
Score = 34.3 bits (77), Expect = 2.0
Identities = 19/28 (67%), Positives = 19/28 (67%)
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRR 33
GRRR R RR RRRR RRR R RRRR
Sbjct: 33 GRRRRRSRRGRRRRGYSRRRYSRRRRRR 60
>HSP1_MACRG (P67842) Sperm protamine P1
Length = 61
Score = 51.6 bits (122), Expect = 1e-05
Identities = 31/48 (64%), Positives = 32/48 (66%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRR R R RRRR R RRRRR RR +R R RRR RR
Sbjct: 9 RSRSRYRRRRRRRSRYRSRRRRYRGRRRRRSRRGRRRRGYSRRRYSRR 56
Score = 50.1 bits (118), Expect = 4e-05
Identities = 37/75 (49%), Positives = 39/75 (51%), Gaps = 16/75 (21%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R R R R R RRRRRRR R R RRRR R RR RRR RRG RR
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRRYRGRR-------RRR---------SRRGRRR 45
Query: 67 RNHLRRRQPRGKRLR 81
R + RRR R +R R
Sbjct: 46 RGYSRRRYSRRRRRR 60
Score = 38.1 bits (87), Expect = 0.14
Identities = 30/66 (45%), Positives = 31/66 (46%), Gaps = 15/66 (22%)
Query: 17 RRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
R R R R R R RRRRRRR R R R RRR RG RRR R R+ R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSR-----RRR----------YRGRRRRRSRRGRRRR 46
Query: 77 GKRLRR 82
G RR
Sbjct: 47 GYSRRR 52
Score = 34.3 bits (77), Expect = 2.0
Identities = 19/28 (67%), Positives = 19/28 (67%)
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRR 33
GRRR R RR RRRR RRR R RRRR
Sbjct: 33 GRRRRRSRRGRRRRGYSRRRYSRRRRRR 60
Score = 32.7 bits (73), Expect = 5.9
Identities = 25/70 (35%), Positives = 31/70 (43%), Gaps = 19/70 (27%)
Query: 21 RRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRL 80
R R R R R R RRRRRRR R R RRR + RR+ R +R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRS-------------------RRRRYRGRRRRRSRRG 42
Query: 81 RRTWTVTRKK 90
RR +R++
Sbjct: 43 RRRRGYSRRR 52
>HSP1_MACGI (P42139) Sperm protamine P1
Length = 60
Score = 51.6 bits (122), Expect = 1e-05
Identities = 31/48 (64%), Positives = 32/48 (66%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRR R R RRRR R RRRRR RR +R R RRR RR
Sbjct: 9 RSRSRYRRRRRRRSRYRSRRRRYRGRRRRRSRRGRRRRGYSRRRYSRR 56
Score = 48.9 bits (115), Expect = 8e-05
Identities = 36/73 (49%), Positives = 38/73 (51%), Gaps = 16/73 (21%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R R R R R RRRRRRR R R RRRR R RR RRR RRG RR
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRRYRGRR-------RRR---------SRRGRRR 45
Query: 67 RNHLRRRQPRGKR 79
R + RRR R +R
Sbjct: 46 RGYSRRRYSRRRR 58
Score = 45.1 bits (105), Expect = 0.001
Identities = 30/49 (61%), Positives = 32/49 (65%), Gaps = 2/49 (4%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRR--RRRRRRQR*RKPPRRR*QR 53
R R RRRRRRR R R RRRR R RRRRR R RRRR R+ RR +R
Sbjct: 11 RSRYRRRRRRRSRYRSRRRRYRGRRRRRSRRGRRRRGYSRRRYSRRRRR 59
Score = 38.1 bits (87), Expect = 0.14
Identities = 30/66 (45%), Positives = 31/66 (46%), Gaps = 15/66 (22%)
Query: 17 RRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
R R R R R R RRRRRRR R R R RRR RG RRR R R+ R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSR-----RRR----------YRGRRRRRSRRGRRRR 46
Query: 77 GKRLRR 82
G RR
Sbjct: 47 GYSRRR 52
Score = 32.7 bits (73), Expect = 5.9
Identities = 25/70 (35%), Positives = 31/70 (43%), Gaps = 19/70 (27%)
Query: 21 RRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRL 80
R R R R R R RRRRRRR R R RRR + RR+ R +R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRS-------------------RRRRYRGRRRRRSRRG 42
Query: 81 RRTWTVTRKK 90
RR +R++
Sbjct: 43 RRRRGYSRRR 52
Score = 32.3 bits (72), Expect = 7.7
Identities = 18/27 (66%), Positives = 18/27 (66%)
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRR 32
GRRR R RR RRRR RRR R RRR
Sbjct: 33 GRRRRRSRRGRRRRGYSRRRYSRRRRR 59
>HSP1_MACEU (P42138) Sperm protamine P1
Length = 61
Score = 51.6 bits (122), Expect = 1e-05
Identities = 31/48 (64%), Positives = 32/48 (66%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRR R R RRRR R RRRRR RR +R R RRR RR
Sbjct: 9 RSRSRYRRRRRRRSRYRSRRRRSRGRRRRRSRRGRRRRGYSRRRYSRR 56
Score = 50.4 bits (119), Expect = 3e-05
Identities = 37/75 (49%), Positives = 39/75 (51%), Gaps = 16/75 (21%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R R R R R RRRRRRR R R RRRR R RR RRR RRG RR
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRRSRGRR-------RRR---------SRRGRRR 45
Query: 67 RNHLRRRQPRGKRLR 81
R + RRR R +R R
Sbjct: 46 RGYSRRRYSRRRRRR 60
Score = 38.5 bits (88), Expect = 0.11
Identities = 30/66 (45%), Positives = 31/66 (46%), Gaps = 15/66 (22%)
Query: 17 RRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
R R R R R R RRRRRRR R R R RRR RG RRR R R+ R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSR-----RRR----------SRGRRRRRSRRGRRRR 46
Query: 77 GKRLRR 82
G RR
Sbjct: 47 GYSRRR 52
Score = 34.3 bits (77), Expect = 2.0
Identities = 19/28 (67%), Positives = 19/28 (67%)
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRR 33
GRRR R RR RRRR RRR R RRRR
Sbjct: 33 GRRRRRSRRGRRRRGYSRRRYSRRRRRR 60
>HSP1_DORVE (P67841) Sperm protamine P1
Length = 61
Score = 51.6 bits (122), Expect = 1e-05
Identities = 31/48 (64%), Positives = 32/48 (66%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRR R R RRRR R RRRRR RR +R R RRR RR
Sbjct: 9 RSRSRYRRRRRRRSRYRSRRRRYRGRRRRRSRRGRRRRGYSRRRYSRR 56
Score = 50.1 bits (118), Expect = 4e-05
Identities = 37/75 (49%), Positives = 39/75 (51%), Gaps = 16/75 (21%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R R R R R RRRRRRR R R RRRR R RR RRR RRG RR
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRRYRGRR-------RRR---------SRRGRRR 45
Query: 67 RNHLRRRQPRGKRLR 81
R + RRR R +R R
Sbjct: 46 RGYSRRRYSRRRRRR 60
Score = 38.1 bits (87), Expect = 0.14
Identities = 30/66 (45%), Positives = 31/66 (46%), Gaps = 15/66 (22%)
Query: 17 RRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
R R R R R R RRRRRRR R R R RRR RG RRR R R+ R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSR-----RRR----------YRGRRRRRSRRGRRRR 46
Query: 77 GKRLRR 82
G RR
Sbjct: 47 GYSRRR 52
Score = 34.3 bits (77), Expect = 2.0
Identities = 19/28 (67%), Positives = 19/28 (67%)
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRR 33
GRRR R RR RRRR RRR R RRRR
Sbjct: 33 GRRRRRSRRGRRRRGYSRRRYSRRRRRR 60
Score = 32.7 bits (73), Expect = 5.9
Identities = 25/70 (35%), Positives = 31/70 (43%), Gaps = 19/70 (27%)
Query: 21 RRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRL 80
R R R R R R RRRRRRR R R RRR + RR+ R +R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRS-------------------RRRRYRGRRRRRSRRG 42
Query: 81 RRTWTVTRKK 90
RR +R++
Sbjct: 43 RRRRGYSRRR 52
>HSP1_DROAU (P42132) Sperm protamine P1
Length = 63
Score = 51.2 bits (121), Expect = 2e-05
Identities = 35/71 (49%), Positives = 38/71 (53%), Gaps = 10/71 (14%)
Query: 12 RRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLR 71
R RR R R R R RRR RRR R RRRR +R R+ RRR RRG RR R
Sbjct: 2 RYRRHSRSRSRSRYRRRRRRRLRNRRRRYRRSRRGRRRR----------RRGSRRGYSRR 51
Query: 72 RRQPRGKRLRR 82
R Q R +R RR
Sbjct: 52 RYQSRRRRRRR 62
Score = 49.3 bits (116), Expect = 6e-05
Identities = 33/61 (54%), Positives = 36/61 (58%), Gaps = 2/61 (3%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
RR R R R R RRRRRRR R RRR RR RR R+R R+ RR RR + RR RR
Sbjct: 4 RRHSRSRSRSRYRRRRRRRLRNRRRRYRRSRRGRRRRRRGSRRGYSRRRYQS--RRRRRR 61
Query: 67 R 67
R
Sbjct: 62 R 62
Score = 35.0 bits (79), Expect = 1.2
Identities = 28/80 (35%), Positives = 32/80 (40%), Gaps = 23/80 (28%)
Query: 12 RRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLR 71
RR R R R R RRRRR R R RRRR RR R R R
Sbjct: 4 RRHSRSRSRSRYRRRRRRRLRNRRRRYRRSR-----------------------RGRRRR 40
Query: 72 RRQPRGKRLRRTWTVTRKKQ 91
RR R RR + R+++
Sbjct: 41 RRGSRRGYSRRRYQSRRRRR 60
>HSP1_BETPE (Q9GLQ0) Sperm protamine P1
Length = 61
Score = 50.8 bits (120), Expect = 2e-05
Identities = 31/48 (64%), Positives = 31/48 (64%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRR R R RRRR R RRRR RRR R R RRR RR
Sbjct: 9 RSRSRYRRRRRRRSRYRSRRRRYRGSRRRRSRRRGRRRGYSRRRYSRR 56
Score = 48.1 bits (113), Expect = 1e-04
Identities = 35/68 (51%), Positives = 38/68 (55%), Gaps = 9/68 (13%)
Query: 9 R*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRN 68
R R R R R R RRRRRRR R R RRRR R R RRR +RR RRG RR
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRRYRGSR-----RRRSRRRGR----RRGYSRRR 52
Query: 69 HLRRRQPR 76
+ RRR+ R
Sbjct: 53 YSRRRRRR 60
Score = 44.7 bits (104), Expect = 0.002
Identities = 30/50 (60%), Positives = 32/50 (64%), Gaps = 2/50 (4%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRR--RRRRRRQR*RKPPRRR*QRR 54
R R RRRRRRR R R RRRR R RRRR RR RRR R+ RR +RR
Sbjct: 11 RSRYRRRRRRRSRYRSRRRRYRGSRRRRSRRRGRRRGYSRRRYSRRRRRR 60
Score = 42.7 bits (99), Expect = 0.006
Identities = 28/59 (47%), Positives = 32/59 (53%)
Query: 23 RRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLR 81
R R R R R R RRRRR+R R RRR R + RR RRR + RRR R +R R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSRRRRYRGSRRRRSRRRGRRRGYSRRRYSRRRRRR 60
Score = 38.5 bits (88), Expect = 0.11
Identities = 30/66 (45%), Positives = 31/66 (46%), Gaps = 15/66 (22%)
Query: 17 RRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPR 76
R R R R R R RRRRRRR R R R RRR RG RRR RR + R
Sbjct: 2 RYRHSRSRSRSRYRRRRRRRSRYRSR-----RRR----------YRGSRRRRSRRRGRRR 46
Query: 77 GKRLRR 82
G RR
Sbjct: 47 GYSRRR 52
Score = 32.3 bits (72), Expect = 7.7
Identities = 18/28 (64%), Positives = 18/28 (64%)
Query: 5 GGRRR*RRRRRRRRRRRRRRRRRR*RRR 32
G RRR RRR RRR RRR RR RRR
Sbjct: 33 GSRRRRSRRRGRRRGYSRRRYSRRRRRR 60
>MNN4_YEAST (P36044) MNN4 protein
Length = 1178
Score = 50.4 bits (119), Expect = 3e-05
Identities = 44/185 (23%), Positives = 85/185 (45%), Gaps = 13/185 (7%)
Query: 247 DVKGYNLVHVFVEHANEISLDAVDSDDDVAIVDRLDVICGKLLQGEKDVEKDKVIQEEVV 306
D+ Y L +F ++++ + D I+ D KLL+ K EK K +EE
Sbjct: 993 DLNSYGL-DLFAPTLSDVNRKGIQMFDKDPIIVYEDYAYAKLLEERKRREKKKKEEEEKK 1051
Query: 307 SVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAEKENQVVGEQAGKEKV 366
E+E +K E E++ ++E EK+ +++ E++ +KE + +Q +EK
Sbjct: 1052 KKEEEEKKKKE----EEEKKKKEEEEKK--------KKEEEEKKKKEEEEKKKQEEEEKK 1099
Query: 367 VVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYHPNSDDSDDSSVVSLDDS 426
EE EK+ Q GE+ E ++ E E + EE+ + + ++D ++
Sbjct: 1100 KKEEEEKKKQEEGEKMKNEDEENKKNEDEEKKKNEEEEKKKQEEKNKKNEDEEKKKQEEE 1159
Query: 427 DYEEN 431
+ ++N
Sbjct: 1160 EKKKN 1164
Score = 38.1 bits (87), Expect = 0.14
Identities = 28/109 (25%), Positives = 55/109 (49%)
Query: 292 EKDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAE 351
+K E++K +EE +KE E+ E +++ E+E +K+ ++ E + K+ E
Sbjct: 1067 KKKEEEEKKKKEEEEKKKKEEEEKKKQEEEEKKKKEEEEKKKQEEGEKMKNEDEENKKNE 1126
Query: 352 KENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVG 400
E + E+ K+K + + E++ +Q +EK E+ EK+ Q G
Sbjct: 1127 DEEKKKNEEEEKKKQEEKNKKNEDEEKKKQEEEEKKKNEEEEKKKQEEG 1175
Score = 36.2 bits (82), Expect = 0.54
Identities = 30/120 (25%), Positives = 54/120 (45%)
Query: 292 EKDVEKDKVIQEEVVSVEKEVEKDIGSVVLECERQVEKEAEKEIGSVVLECERQVEKEAE 351
+K E++K +EE +KE E+ E ++Q E+E +K+ + E + + E
Sbjct: 1059 KKKEEEEKKKKEEEEKKKKEEEEKKKKEEEEKKKQEEEEKKKKEEEEKKKQEEGEKMKNE 1118
Query: 352 KENQVVGEQAGKEKVVVEEAEKENQVVGEQAGKEKVVVEQAEKENQGVGGEEDDESYHPN 411
E E K+K EE +K+ + + +EK E+ EK+ ++ E H N
Sbjct: 1119 DEENKKNEDEEKKKNEEEEKKKQEEKNKKNEDEEKKKQEEEEKKKNEEEEKKKQEEGHSN 1178
>HSP1_SARHA (P62486) Sperm protamine P1
Length = 61
Score = 50.4 bits (119), Expect = 3e-05
Identities = 34/60 (56%), Positives = 35/60 (57%), Gaps = 3/60 (5%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
RRR R R R R RRRRRRR R RRR RR RR R R+ RR RR S RRG RR
Sbjct: 4 RRRSRSRSRSRYRRRRRRRSRGRRRRTYRRSRRHSRRRRGRRRGYSRRRYS---RRGRRR 60
Score = 48.1 bits (113), Expect = 1e-04
Identities = 33/66 (50%), Positives = 37/66 (56%), Gaps = 7/66 (10%)
Query: 11 RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHL 70
R RRR R R R R RRRR RR R RRRR +R R+ RRR R RRG RR +
Sbjct: 2 RYRRRSRSRSRSRYRRRRRRRSRGRRRRTYRRSRRHSRRRRGR-------RRGYSRRRYS 54
Query: 71 RRRQPR 76
RR + R
Sbjct: 55 RRGRRR 60
Score = 43.5 bits (101), Expect = 0.003
Identities = 30/60 (50%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query: 22 RRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLR 81
R RRR R R R R RRRRR+R + RRR RR+ RR RRR + RRR R R R
Sbjct: 2 RYRRRSRSRSRSRYRRRRRRR-SRGRRRRTYRRSRRHSRRRRGRRRGYSRRRYSRRGRRR 60
Score = 40.0 bits (92), Expect = 0.037
Identities = 23/38 (60%), Positives = 24/38 (62%)
Query: 3 NEGGRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRR 40
+ G RRR RR RR RRRR RRR RRR RR RRR
Sbjct: 23 SRGRRRRTYRRSRRHSRRRRGRRRGYSRRRYSRRGRRR 60
Score = 38.9 bits (89), Expect = 0.083
Identities = 22/36 (61%), Positives = 23/36 (63%)
Query: 6 GRRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQ 41
GRRR RR RR RRRR RRR RRR RR RR+
Sbjct: 25 GRRRRTYRRSRRHSRRRRGRRRGYSRRRYSRRGRRR 60
Score = 37.7 bits (86), Expect = 0.18
Identities = 30/60 (50%), Positives = 32/60 (53%), Gaps = 16/60 (26%)
Query: 20 RRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKR 79
R RRR R R R R RRRRRRR R R RRR RR+ R H RRR RG+R
Sbjct: 2 RYRRRSRSRSRSRYRRRRRRRSRGR---RRRTYRRS-----------RRHSRRR--RGRR 45
>HSP1_PSEWO (Q71VG4) Sperm protamine P1
Length = 62
Score = 50.4 bits (119), Expect = 3e-05
Identities = 30/48 (62%), Positives = 30/48 (62%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R R RRRRRRR R R RRR RR RR RRRR R R RRR RR
Sbjct: 10 RSRSRYRRRRRRRSRHRNRRRTYRRSRRHSRRRRGRRRGYSRRRYSRR 57
Score = 47.4 bits (111), Expect = 2e-04
Identities = 31/55 (56%), Positives = 32/55 (57%), Gaps = 3/55 (5%)
Query: 12 RRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRR 66
R R R RRRRRRR R R RRR RR RR R R+ RR RR S RRG RR
Sbjct: 10 RSRSRYRRRRRRRSRHRNRRRTYRRSRRHSRRRRGRRRGYSRRRYS---RRGRRR 61
Score = 45.8 bits (107), Expect = 7e-04
Identities = 30/60 (50%), Positives = 33/60 (55%)
Query: 22 RRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKRLR 81
R RR R R R R RRRRR+R R RRR RR+ RR RRR + RRR R R R
Sbjct: 2 RYRRHSRSRSRSRYRRRRRRRSRHRNRRRTYRRSRRHSRRRRGRRRGYSRRRYSRRGRRR 61
Score = 45.8 bits (107), Expect = 7e-04
Identities = 31/65 (47%), Positives = 35/65 (53%), Gaps = 7/65 (10%)
Query: 12 RRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLR 71
RR R R R R RRRRR R R R RRR +R R+ RRR R RRG RR + R
Sbjct: 4 RRHSRSRSRSRYRRRRRRRSRHRNRRRTYRRSRRHSRRRRGR-------RRGYSRRRYSR 56
Query: 72 RRQPR 76
R + R
Sbjct: 57 RGRRR 61
Score = 43.9 bits (102), Expect = 0.003
Identities = 27/48 (56%), Positives = 29/48 (60%)
Query: 7 RRR*RRRRRRRRRRRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRR 54
R R RRRRR R R RRR RR R RRRR RRR R+ RR +RR
Sbjct: 14 RYRRRRRRRSRHRNRRRTYRRSRRHSRRRRGRRRGYSRRRYSRRGRRR 61
Score = 36.2 bits (82), Expect = 0.54
Identities = 29/60 (48%), Positives = 31/60 (51%), Gaps = 15/60 (25%)
Query: 20 RRRRRRRRR*RRRRRRRRRRRQR*RKPPRRR*QRRTSSTVLRRGPRRRNHLRRRQPRGKR 79
R RR R R R R RRRRRRR R R RRR RR+ R H RRR RG+R
Sbjct: 2 RYRRHSRSRSRSRYRRRRRRRSRHRN--RRRTYRRS-----------RRHSRRR--RGRR 46
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.338 0.149 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 130,156,672
Number of Sequences: 164201
Number of extensions: 5736293
Number of successful extensions: 59640
Number of sequences better than 10.0: 778
Number of HSP's better than 10.0 without gapping: 364
Number of HSP's successfully gapped in prelim test: 447
Number of HSP's that attempted gapping in prelim test: 38554
Number of HSP's gapped (non-prelim): 6731
length of query: 1171
length of database: 59,974,054
effective HSP length: 121
effective length of query: 1050
effective length of database: 40,105,733
effective search space: 42111019650
effective search space used: 42111019650
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0321.4


