

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.13
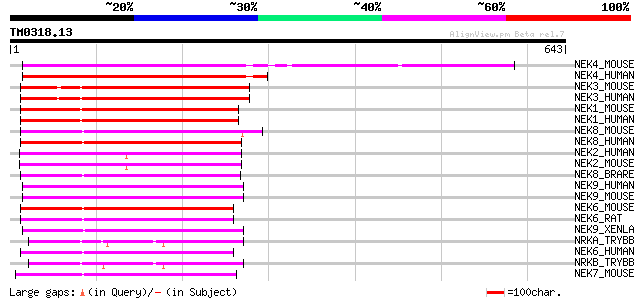
(643 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NEK4_MOUSE (Q9Z1J2) Serine/threonine-protein kinase Nek4 (EC 2.7... 248 3e-65
NEK4_HUMAN (P51957) Serine/threonine-protein kinase Nek4 (EC 2.7... 238 4e-62
NEK3_MOUSE (Q9R0A5) Serine/threonine-protein kinase Nek3 (EC 2.7... 225 2e-58
NEK3_HUMAN (P51956) Serine/threonine-protein kinase Nek3 (EC 2.7... 223 2e-57
NEK1_MOUSE (P51954) Serine/threonine-protein kinase Nek1 (EC 2.7... 207 7e-53
NEK1_HUMAN (Q96PY6) Serine/threonine-protein kinase Nek1 (EC 2.7... 206 1e-52
NEK8_MOUSE (Q91ZR4) Serine/threonine-protein kinase Nek8 (EC 2.7... 187 1e-46
NEK8_HUMAN (Q86SG6) Serine/threonine-protein kinase Nek8 (EC 2.7... 185 3e-46
NEK2_HUMAN (P51955) Serine/threonine-protein kinase Nek2 (EC 2.7... 177 6e-44
NEK2_MOUSE (O35942) Serine/threonine-protein kinase Nek2 (EC 2.7... 175 3e-43
NEK8_BRARE (Q90XC2) Serine/threonine-protein kinase Nek8 (EC 2.7... 171 4e-42
NEK9_HUMAN (Q8TD19) Serine/threonine-protein kinase Nek9 (EC 2.7... 171 7e-42
NEK9_MOUSE (Q8K1R7) Serine/threonine-protein kinase Nek9 (EC 2.7... 164 5e-40
NEK6_MOUSE (Q9ES70) Serine/threonine-protein kinase Nek6 (EC 2.7... 164 9e-40
NEK6_RAT (P59895) Serine/threonine-protein kinase Nek6 (EC 2.7.1... 163 1e-39
NEK9_XENLA (Q7ZZC8) Serine/threonine-protein kinase Nek9 (EC 2.7... 161 6e-39
NRKA_TRYBB (Q08942) Putative serine/threonine-protein kinase A (... 159 2e-38
NEK6_HUMAN (Q9HC98) Serine/threonine-protein kinase Nek6 (EC 2.7... 159 2e-38
NRKB_TRYBB (Q03428) Putative serine/threonine-protein kinase B (... 152 3e-36
NEK7_MOUSE (Q9ES74) Serine/threonine-protein kinase Nek7 (EC 2.7... 151 4e-36
>NEK4_MOUSE (Q9Z1J2) Serine/threonine-protein kinase Nek4 (EC
2.7.1.37) (NimA-related protein kinase 4)
(Serine/threonine-protein kinase 2)
Length = 792
Score = 248 bits (633), Expect = 3e-65
Identities = 162/577 (28%), Positives = 272/577 (47%), Gaps = 29/577 (5%)
Query: 16 YEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYI 75
Y + +GRG++G LV H+ + K+YV+KK+ L + + +R A QE L+++L +P I
Sbjct: 6 YCYMRVVGRGSYGEVTLVKHRRDGKQYVIKKLNLRNASSRERRAAEQEAQLLSQLKHPNI 65
Query: 76 VDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSN 135
V YK++W + + IV G+CEGGD+ +K+ +G PE +V +W Q+ +A+ YLH
Sbjct: 66 VTYKESWEGGDGLLYIVMGFCEGGDLYRKLKEQKGQLLPESQVVEWFVQIAMALQYLHEK 125
Query: 136 RVIHRDLKCSNIFLTKDNNIRLGDFGLAKRL-NAEDLTSSVVGTPNYMCPEILADIPYGY 194
++HRDLK N+FLT+ N I++GD G+A+ L N D+ S+++GTP YM PE+ ++ PY Y
Sbjct: 126 HILHRDLKTQNVFLTRTNIIKVGDLGIARVLENHGDMASTLIGTPYYMSPELFSNKPYNY 185
Query: 195 KSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSMLRKN 254
KSD+W+LGCC++E+A + AF A DM L+ +I + P+P VYS+ L ++I++ML +
Sbjct: 186 KSDVWALGCCVYEMATLKHAFNAKDMNSLVYRIIEGKLPPMPKVYSTELAELIRTMLSRR 245
Query: 255 PEHRPTAAELLRHPHLQPYVLRCQNASSNFLPIYPIVNSKDKTKKSYKTSGGKDHRDKEA 314
PE RP+ +LR P+++ ++ S FL SK+ K D R K
Sbjct: 246 PEERPSVRSILRQPYIKHHI-------SLFLEATKAKTSKNNVKNC-------DSRAKPV 291
Query: 315 GLAYHLERVHPIEGNGDVHTSNPPNNGEVTVSTRTGDNL--ENKMVDFTSYIGESSTSIS 372
+ E N DV P ++ + D + K VD +S
Sbjct: 292 AAVVSRKE----ESNTDVIHYQPRSSEGSALHVMGEDKCLSQEKPVDIGPLRSPASLEGH 347
Query: 373 GSKDGSTTSESSVCSMCKEDFKSRPAQEKGNHGNGITSKCRQDSVH--EEQALAAELFQK 430
K + S ++ + + PA+ + + G+ + + V +E + Q+
Sbjct: 348 TGKQDMNNTGESCATISRINIDILPAERRDSANAGVVQESQPQHVDAADEVDSQCSISQE 407
Query: 431 LELVDLNTGISEVEDAFSKEGFNFAEAPREDAKFEESRKSTTSNASSSCTDKDKSINEER 490
E + NT S+ + D + E K + D+D+
Sbjct: 408 KERLQGNTKSSDQPGNLLPR----RSSDGGDGEGSELVKPLYPSNKDQKPDQDQVTGIIE 463
Query: 491 SSLIVIPVKVEHETESRNSLKKSENPVAFTEGSHMNCLS--SDSSSKLPVKDNETAKENI 548
+ + P H + S SL + + +H S + +L K +I
Sbjct: 464 NQDSIHPRSQPHSSMSEPSLSRQRRQKKREQTAHSGTKSQFQELPPRLLPSYPGIGKVDI 523
Query: 549 ICTTQKDENLVGVDQTPSDTTMDDGDETMRDSPCQQR 585
I T Q D N G +++ + +D P R
Sbjct: 524 IATQQNDGNQGGPVAGCVNSSRTSSTASAKDRPLSAR 560
>NEK4_HUMAN (P51957) Serine/threonine-protein kinase Nek4 (EC
2.7.1.37) (NimA-related protein kinase 4)
(Serine/threonine-protein kinase 2)
(Serine/threonine-protein kinase NRK2)
Length = 841
Score = 238 bits (607), Expect = 4e-62
Identities = 112/284 (39%), Positives = 179/284 (62%), Gaps = 8/284 (2%)
Query: 16 YEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYI 75
Y + +G+G++G LV H+ + K+YV+KK+ L + + +R A QE L+++L +P I
Sbjct: 6 YCYLRVVGKGSYGEVTLVKHRRDGKQYVIKKLNLRNASSRERRAAEQEAQLLSQLKHPNI 65
Query: 76 VDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSN 135
V YK++W + + IV G+CEGGD+ +K+ +G PE +V +W Q+ +A+ YLH
Sbjct: 66 VTYKESWEGGDGLLYIVMGFCEGGDLYRKLKEQKGQLLPENQVVEWFVQIAMALQYLHEK 125
Query: 136 RVIHRDLKCSNIFLTKDNNIRLGDFGLAKRL-NAEDLTSSVVGTPNYMCPEILADIPYGY 194
++HRDLK N+FLT+ N I++GD G+A+ L N D+ S+++GTP YM PE+ ++ PY Y
Sbjct: 126 HILHRDLKTQNVFLTRTNIIKVGDLGIARVLENHCDMASTLIGTPYYMSPELFSNKPYNY 185
Query: 195 KSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSMLRKN 254
KSD+W+LGCC++E+A + AF A DM L+ +I + +P YS L ++I++ML K
Sbjct: 186 KSDVWALGCCVYEMATLKHAFNAKDMNSLVYRIIEGKLPAMPRDYSPELAELIRTMLSKR 245
Query: 255 PEHRPTAAELLRHPHLQPYVLRCQNASSNFLPIYPIVNSKDKTK 298
PE RP+ +LR P+++ + S FL I SK+ K
Sbjct: 246 PEERPSVRSILRQPYIKRQI-------SFFLEATKIKTSKNNIK 282
>NEK3_MOUSE (Q9R0A5) Serine/threonine-protein kinase Nek3 (EC
2.7.1.37) (NimA-related protein kinase 3)
Length = 511
Score = 225 bits (574), Expect = 2e-58
Identities = 111/266 (41%), Positives = 173/266 (64%), Gaps = 5/266 (1%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNN 72
M++Y V+ IG+G+FG A LVL +S + + +K+IRL K + T+ +E L+AK+ +
Sbjct: 1 MDNYTVLRVIGQGSFGRALLVLQESSNQTFAMKEIRLLKSDTQ---TSRKEAVLLAKMKH 57
Query: 73 PYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYL 132
P IV +K+++ E E + IV YC+GGD+ + IK+ +G+ FPE+ + W Q+ + V+++
Sbjct: 58 PNIVAFKESF-EAEGYLYIVMEYCDGGDLMQRIKQQKGNLFPEDTILNWFIQICLGVNHI 116
Query: 133 HSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAE-DLTSSVVGTPNYMCPEILADIP 191
H RV+HRD+K N+FLT + ++LGDFG A+ L++ + VGTP Y+ PEI ++P
Sbjct: 117 HKRRVLHRDIKSKNVFLTHNGKVKLGDFGSARLLSSPMAFACTYVGTPYYVPPEIWENLP 176
Query: 192 YGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSML 251
Y KSD+WSLGC ++E+ A + F+A LI KI + I PLP +YS L+ ++K ML
Sbjct: 177 YNNKSDIWSLGCILYELCALKHPFQANSWKNLILKICQGPIHPLPALYSCKLQGLVKQML 236
Query: 252 RKNPEHRPTAAELLRHPHLQPYVLRC 277
++NP HRP+A LL L P V +C
Sbjct: 237 KRNPSHRPSATTLLCRGSLAPLVPKC 262
>NEK3_HUMAN (P51956) Serine/threonine-protein kinase Nek3 (EC
2.7.1.37) (NimA-related protein kinase 3) (HSPK 36)
Length = 506
Score = 223 bits (567), Expect = 2e-57
Identities = 110/266 (41%), Positives = 168/266 (62%), Gaps = 3/266 (1%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNN 72
M+DY V+ IG G+FG A LV H+S + + +K+IRL K + + +E L+AK+ +
Sbjct: 1 MDDYMVLRMIGEGSFGRALLVQHESSNQMFAMKEIRLPKSFSN-TQNSRKEAVLLAKMKH 59
Query: 73 PYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYL 132
P IV +K+++ E E + IV YC+GGD+ + IK+ +G FPE+ + W TQ+ + V+++
Sbjct: 60 PNIVAFKESF-EAEGHLYIVMEYCDGGDLMQKIKQQKGKLFPEDMILNWFTQMCLGVNHI 118
Query: 133 HSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRL-NAEDLTSSVVGTPNYMCPEILADIP 191
H RV+HRD+K NIFLT++ ++LGDFG A+ L N + VGTP Y+ PEI ++P
Sbjct: 119 HKKRVLHRDIKSKNIFLTQNGKVKLGDFGSARLLSNPMAFACTYVGTPYYVPPEIWENLP 178
Query: 192 YGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSML 251
Y KSD+WSLGC ++E+ + F+A LI K+ + ISPLP YS L+ ++K M
Sbjct: 179 YNNKSDIWSLGCILYELCTLKHPFQANSWKNLILKVCQGCISPLPSHYSYELQFLVKQMF 238
Query: 252 RKNPEHRPTAAELLRHPHLQPYVLRC 277
++NP HRP+A LL + V +C
Sbjct: 239 KRNPSHRPSATTLLSRGIVARLVQKC 264
>NEK1_MOUSE (P51954) Serine/threonine-protein kinase Nek1 (EC
2.7.1.37) (NimA-related protein kinase 1)
Length = 774
Score = 207 bits (527), Expect = 7e-53
Identities = 94/254 (37%), Positives = 164/254 (64%), Gaps = 2/254 (0%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNN 72
ME Y +++IG G+FG A LV + + YV+K+I +++ ++K ++ + +E+ ++A + +
Sbjct: 1 MEKYVRLQKIGEGSFGKAVLVKSTEDGRHYVIKEINISRMSDKERQESRREVAVLANMKH 60
Query: 73 PYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYL 132
P IV YK+++ E+ + IV YCEGGD+ + I +G+ F E+++ W Q+ +A+ ++
Sbjct: 61 PNIVQYKESF-EENGSLYIVMDYCEGGDLFKRINAQKGALFQEDQILDWFVQICLALKHV 119
Query: 133 HSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAE-DLTSSVVGTPNYMCPEILADIP 191
H +++HRD+K NIFLTKD ++LGDFG+A+ LN+ +L + +GTP Y+ PEI + P
Sbjct: 120 HDRKILHRDIKSQNIFLTKDGTVQLGDFGIARVLNSTVELARTCIGTPYYLSPEICENKP 179
Query: 192 YGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSML 251
Y KSD+W+LGC ++E+ + AF A +M L+ KI S P+ YS L+ ++ +
Sbjct: 180 YNNKSDIWALGCVLYELCTLKHAFEAGNMKNLVLKIISGSFPPVSPHYSYDLRSLLSQLF 239
Query: 252 RKNPEHRPTAAELL 265
++NP RP+ +L
Sbjct: 240 KRNPRDRPSVNSIL 253
>NEK1_HUMAN (Q96PY6) Serine/threonine-protein kinase Nek1 (EC
2.7.1.37) (NimA-related protein kinase 1) (NY-REN-55
antigen)
Length = 1258
Score = 206 bits (525), Expect = 1e-52
Identities = 93/254 (36%), Positives = 162/254 (63%), Gaps = 2/254 (0%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNN 72
ME Y +++IG G+FG A LV + ++YV+K+I +++ + K + + +E+ ++A + +
Sbjct: 1 MEKYVRLQKIGEGSFGKAILVKSTEDGRQYVIKEINISRMSSKEREESRREVAVLANMKH 60
Query: 73 PYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYL 132
P IV Y++++ E+ + IV YCEGGD+ + I +G F E+++ W Q+ +A+ ++
Sbjct: 61 PNIVQYRESF-EENGSLYIVMDYCEGGDLFKRINAQKGVLFQEDQILDWFVQICLALKHV 119
Query: 133 HSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAE-DLTSSVVGTPNYMCPEILADIP 191
H +++HRD+K NIFLTKD ++LGDFG+A+ LN+ +L + +GTP Y+ PEI + P
Sbjct: 120 HDRKILHRDIKSQNIFLTKDGTVQLGDFGIARVLNSTVELARTCIGTPYYLSPEICENKP 179
Query: 192 YGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSML 251
Y KSD+W+LGC ++E+ + AF A M L+ KI S P+ + YS L+ ++ +
Sbjct: 180 YNNKSDIWALGCVLYELCTLKHAFEAGSMKNLVLKIISGSFPPVSLHYSYDLRSLVSQLF 239
Query: 252 RKNPEHRPTAAELL 265
++NP RP+ +L
Sbjct: 240 KRNPRDRPSVNSIL 253
>NEK8_MOUSE (Q91ZR4) Serine/threonine-protein kinase Nek8 (EC
2.7.1.37) (NimA-related protein kinase 8)
Length = 698
Score = 187 bits (474), Expect = 1e-46
Identities = 98/290 (33%), Positives = 169/290 (57%), Gaps = 11/290 (3%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNN 72
ME YE I +GRGAFG L L K+++K +LK+I + + T++ ++ A E ++ LN+
Sbjct: 1 MEKYERIRVVGRGAFGIVHLCLRKADQKLVILKQIPVEQMTKEERQAAQNECQVLKLLNH 60
Query: 73 PYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYL 132
P +++Y + ++E + + I Y GG +AE I+K S EE + + Q+L+A+ ++
Sbjct: 61 PNVIEYYENFLE-DKALMIAMEYAPGGTLAEFIQKRCNSLLEEETILHFFVQILLALHHV 119
Query: 133 HSNRVIHRDLKCSNIFLTKDNNI-RLGDFGLAKRLNAEDLTSSVVGTPNYMCPEILADIP 191
H++ ++HRDLK NI L K + ++GDFG++K L+++ +VVGTP Y+ PE+ P
Sbjct: 120 HTHLILHRDLKTQNILLDKHRMVVKIGDFGISKILSSKSKAYTVVGTPCYISPELCEGKP 179
Query: 192 YGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSML 251
Y KSD+W+LGC ++E+A+ + AF A ++ L+ KI + +P+ YS L+Q++ S+L
Sbjct: 180 YNQKSDIWALGCVLYELASLKRAFEAANLPALVLKIMSGTFAPISDRYSPELRQLVLSLL 239
Query: 252 RKNPEHRPTAAELLRHP---------HLQPYVLRCQNASSNFLPIYPIVN 292
P RP + ++ P H +R + A + P PI +
Sbjct: 240 SLEPAQRPPLSHIMAQPLCIRALLNIHTDVGSVRMRRAEKSLTPGPPIAS 289
>NEK8_HUMAN (Q86SG6) Serine/threonine-protein kinase Nek8 (EC
2.7.1.37) (NimA-related protein kinase 8) (NIMA-related
kinase 12a)
Length = 692
Score = 185 bits (470), Expect = 3e-46
Identities = 91/257 (35%), Positives = 159/257 (61%), Gaps = 2/257 (0%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNN 72
ME YE I +GRGAFG L L K+++K ++K+I + + T++ ++ A E ++ LN+
Sbjct: 1 MEKYERIRVVGRGAFGIVHLCLRKADQKLVIIKQIPVEQMTKEERQAAQNECQVLKLLNH 60
Query: 73 PYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYL 132
P +++Y + ++E + + I Y GG +AE I+K S EE + + Q+L+A+ ++
Sbjct: 61 PNVIEYYENFLE-DKALMIAMEYAPGGTLAEFIQKRCNSLLEEETILHFFVQILLALHHV 119
Query: 133 HSNRVIHRDLKCSNIFLTKDNNI-RLGDFGLAKRLNAEDLTSSVVGTPNYMCPEILADIP 191
H++ ++HRDLK NI L K + ++GDFG++K L+++ +VVGTP Y+ PE+ P
Sbjct: 120 HTHLILHRDLKTQNILLDKHRMVVKIGDFGISKILSSKSKAYTVVGTPCYISPELCEGKP 179
Query: 192 YGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSML 251
Y KSD+W+LGC ++E+A+ + AF A ++ L+ KI + +P+ YS L+Q++ S+L
Sbjct: 180 YNQKSDIWALGCVLYELASLKRAFEAANLPALVLKIMSGTFAPISDRYSPELRQLVLSLL 239
Query: 252 RKNPEHRPTAAELLRHP 268
P RP + ++ P
Sbjct: 240 SLEPAQRPPLSHIMAQP 256
>NEK2_HUMAN (P51955) Serine/threonine-protein kinase Nek2 (EC
2.7.1.37) (NimA-related protein kinase 2) (NimA-like
protein kinase 1) (HSPK 21)
Length = 445
Score = 177 bits (450), Expect = 6e-44
Identities = 103/266 (38%), Positives = 147/266 (54%), Gaps = 9/266 (3%)
Query: 12 KMEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLN 71
+ EDYEV+ IG G++G + KS+ K V K++ TE K+ E+NL+ +L
Sbjct: 4 RAEDYEVLYTIGTGSYGRCQKIRRKSDGKILVWKELDYGSMTEAEKQMLVSEVNLLRELK 63
Query: 72 NPYIVDYKDAWVEKEDR-ICIVTGYCEGGDMAENIKKA--RGSFFPEEKVCKWLTQLLIA 128
+P IV Y D +++ + + IV YCEGGD+A I K + EE V + +TQL +A
Sbjct: 64 HPNIVRYYDRIIDRTNTTLYIVMEYCEGGDLASVITKGTKERQYLDEEFVLRVMTQLTLA 123
Query: 129 VDYLH-----SNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAE-DLTSSVVGTPNYM 182
+ H + V+HRDLK +N+FL N++LGDFGLA+ LN + + VGTP YM
Sbjct: 124 LKECHRRSDGGHTVLHRDLKPANVFLDGKQNVKLGDFGLARILNHDTSFAKTFVGTPYYM 183
Query: 183 CPEILADIPYGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSST 242
PE + + Y KSD+WSLGC ++E+ A P F A L KI +P YS
Sbjct: 184 SPEQMNRMSYNEKSDIWSLGCLLYELCALMPPFTAFSQKELAGKIREGKFRRIPYRYSDE 243
Query: 243 LKQIIKSMLRKNPEHRPTAAELLRHP 268
L +II ML HRP+ E+L +P
Sbjct: 244 LNEIITRMLNLKDYHRPSVEEILENP 269
>NEK2_MOUSE (O35942) Serine/threonine-protein kinase Nek2 (EC
2.7.1.37) (NimA-related protein kinase 2)
Length = 443
Score = 175 bits (444), Expect = 3e-43
Identities = 101/266 (37%), Positives = 148/266 (54%), Gaps = 9/266 (3%)
Query: 12 KMEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLN 71
++EDYEV+ IG G++G + KS+ K V K++ TE K+ E+NL+ +L
Sbjct: 4 RVEDYEVLHSIGTGSYGRCQKIRRKSDGKILVWKELDYGSMTEVEKQMLVSEVNLLRELK 63
Query: 72 NPYIVDYKDAWVEKEDR-ICIVTGYCEGGDMAENIKKARGS--FFPEEKVCKWLTQLLIA 128
+P IV Y D +++ + + IV YCEGGD+A I K + EE V + +TQL +A
Sbjct: 64 HPNIVRYYDRIIDRTNTTLYIVMEYCEGGDLASVISKGTKDRQYLEEEFVLRVMTQLTLA 123
Query: 129 VDYLH-----SNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAE-DLTSSVVGTPNYM 182
+ H + V+HRDLK +N+FL +N++LGDFGLA+ LN + + VGTP YM
Sbjct: 124 LKECHRRSDGGHTVLHRDLKPANVFLDSKHNVKLGDFGLARILNHDTSFAKTFVGTPYYM 183
Query: 183 CPEILADIPYGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSST 242
PE ++ + Y KSD+WSL C ++E+ A P F A + L KI +P YS
Sbjct: 184 SPEQMSCLSYNEKSDIWSLACLLYELCALMPPFTAFNQKELAGKIREGRFRRIPYRYSDG 243
Query: 243 LKQIIKSMLRKNPEHRPTAAELLRHP 268
L +I ML HRP+ E+L P
Sbjct: 244 LNDLITRMLFLKDYHRPSVEEILESP 269
>NEK8_BRARE (Q90XC2) Serine/threonine-protein kinase Nek8 (EC
2.7.1.37) (NimA-related protein kinase 8)
Length = 697
Score = 171 bits (434), Expect = 4e-42
Identities = 89/256 (34%), Positives = 151/256 (58%), Gaps = 2/256 (0%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNN 72
ME YE + +GRGAFG L +++ +LK+I + + T + A E ++ L++
Sbjct: 1 MEKYEKTKVVGRGAFGIVHLCRRRTDSALVILKEIPVEQMTRDERLAAQNECQVLKLLSH 60
Query: 73 PYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYL 132
P I++Y + ++E + + I Y GG +A+ I+K S E+ + Q+L+A+ ++
Sbjct: 61 PNIIEYYENFLE-DKALMIAMEYAPGGTLADYIQKRCNSLLDEDTILHSFVQILLALYHV 119
Query: 133 HSNRVIHRDLKCSNIFLTKDNNI-RLGDFGLAKRLNAEDLTSSVVGTPNYMCPEILADIP 191
H+ ++HRDLK NI L K I ++GDFG++K L ++ +VVGTP Y+ PE+ P
Sbjct: 120 HNKLILHRDLKTQNILLDKHQMIVKIGDFGISKILVSKSKAYTVVGTPCYISPELCEGKP 179
Query: 192 YGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSML 251
Y KSD+W+LGC ++E+A+ + AF A ++ L+ KI + +P+ YS L+Q+I +ML
Sbjct: 180 YNQKSDIWALGCVLYELASLKRAFEAANLPALVLKIMSGTFAPISDRYSPELRQLILNML 239
Query: 252 RKNPEHRPTAAELLRH 267
+P RP E++ H
Sbjct: 240 NLDPSKRPQLNEIMAH 255
>NEK9_HUMAN (Q8TD19) Serine/threonine-protein kinase Nek9 (EC
2.7.1.37) (NimA-related protein kinase 9) (Nercc1
kinase) (NIMA-related kinase 8) (Nek8)
Length = 979
Score = 171 bits (432), Expect = 7e-42
Identities = 98/259 (37%), Positives = 152/259 (57%), Gaps = 4/259 (1%)
Query: 16 YEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYI 75
Y I +GRGAFG A L + V K++ L + +EK +R A E+ ++A L + I
Sbjct: 52 YIPIRVLGRGAFGEATLYRRTEDDSLVVWKEVDLTRLSEKERRDALNEIVILALLQHDNI 111
Query: 76 VDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSN 135
+ Y + +++ + I YC GG++ + I + + F EE V +L Q++ AV +H
Sbjct: 112 IAYYNHFMDNTT-LLIELEYCNGGNLYDKILRQKDKLFEEEMVVWYLFQIVSAVSCIHKA 170
Query: 136 RVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAE-DLTSSVVGTPNYMCPEILADIPYGY 194
++HRD+K NIFLTK N I+LGD+GLAK+LN+E + ++VGTP YM PE+ + Y +
Sbjct: 171 GILHRDIKTLNIFLTKANLIKLGDYGLAKKLNSEYSMAETLVGTPYYMSPELCQGVKYNF 230
Query: 195 KSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRS--SISPLPIVYSSTLKQIIKSMLR 252
KSD+W++GC +FE+ + F A + L KI + ++ YS L Q++ S L
Sbjct: 231 KSDIWAVGCVIFELLTLKRTFDATNPLNLCVKIVQGIRAMEVDSSQYSLELIQMVHSCLD 290
Query: 253 KNPEHRPTAAELLRHPHLQ 271
++PE RPTA ELL P L+
Sbjct: 291 QDPEQRPTADELLDRPLLR 309
>NEK9_MOUSE (Q8K1R7) Serine/threonine-protein kinase Nek9 (EC
2.7.1.37) (NimA-related protein kinase 9)
Length = 984
Score = 164 bits (416), Expect = 5e-40
Identities = 95/259 (36%), Positives = 150/259 (57%), Gaps = 4/259 (1%)
Query: 16 YEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYI 75
Y I +GRGAFG A L + V K++ L + +EK +R A E+ ++A L + I
Sbjct: 52 YIPIRVLGRGAFGEATLYRRTEDDSLVVWKEVDLTRLSEKERRDALNEIVILALLQHDNI 111
Query: 76 VDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSN 135
+ Y + +++ + I YC GG++ + I + + F EE V +L Q++ AV +H
Sbjct: 112 IAYYNHFMDNTT-LLIELEYCNGGNLYDKILRQKDKLFEEEMVVWYLFQIVSAVSCIHKA 170
Query: 136 RVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAE-DLTSSVVGTPNYMCPEILADIPYGY 194
++HRD+K NIFLTK N I+LGD+GLAK+LN+E + ++VGTP YM PE+ + Y +
Sbjct: 171 GILHRDIKTLNIFLTKANLIKLGDYGLAKKLNSEYSMAETLVGTPYYMSPELCQGVKYNF 230
Query: 195 KSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRS--SISPLPIVYSSTLKQIIKSMLR 252
KSD+W++GC +FE+ + F A + L KI + ++ YS L Q++ + L
Sbjct: 231 KSDIWAVGCVIFELLTLKRTFDATNPLNLCVKIVQGIRAMEVDSSQYSLELIQLVHACLD 290
Query: 253 KNPEHRPTAAELLRHPHLQ 271
++PE RP A LL P L+
Sbjct: 291 QDPEQRPAADALLDLPLLR 309
>NEK6_MOUSE (Q9ES70) Serine/threonine-protein kinase Nek6 (EC
2.7.1.37) (NimA-related protein kinase 6)
Length = 313
Score = 164 bits (414), Expect = 9e-40
Identities = 89/254 (35%), Positives = 153/254 (60%), Gaps = 8/254 (3%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTE-KFKRTAHQEMNLIAKLN 71
+ D+++ ++IGRG F + ++K LKK+++ + + K ++ +E+ L+ +LN
Sbjct: 42 LADFQIEKKIGRGQFSEVYKATCLLDRKTVALKKVQIFEMMDAKARQDCVKEIGLLKQLN 101
Query: 72 NPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIK--KARGSFFPEEKVCKWLTQLLIAV 129
+P I+ Y D+++E ++ + IV + GD+++ IK K + PE V K+ QL AV
Sbjct: 102 HPNIIKYLDSFIE-DNELNIVLELADAGDLSQMIKYFKKQKRLIPERTVWKYFVQLCSAV 160
Query: 130 DYLHSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAEDLTS-SVVGTPNYMCPEILA 188
+++HS RV+HRD+K +N+F+T ++LGD GL + ++E + S+VGTP YM PE +
Sbjct: 161 EHMHSRRVMHRDIKPANVFITATGIVKLGDLGLGRFFSSETTAAHSLVGTPYYMSPERIH 220
Query: 189 DIPYGYKSDMWSLGCCMFEIAAHQPAFRAPDM--AGLINKINRSSISPLP-IVYSSTLKQ 245
+ Y +KSD+WSLGC ++E+AA Q F M L KI + PLP YS L++
Sbjct: 221 ENGYNFKSDIWSLGCLLYEMAALQSPFYGDKMNLFSLCQKIEQCDYPPLPGEHYSEKLRE 280
Query: 246 IIKSMLRKNPEHRP 259
++ + +P+HRP
Sbjct: 281 LVSMCIYPDPDHRP 294
>NEK6_RAT (P59895) Serine/threonine-protein kinase Nek6 (EC
2.7.1.37) (NimA-related protein kinase 6)
Length = 313
Score = 163 bits (412), Expect = 1e-39
Identities = 89/254 (35%), Positives = 152/254 (59%), Gaps = 8/254 (3%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTE-KFKRTAHQEMNLIAKLN 71
+ D+++ ++IGRG F + ++K LKK+++ + + K ++ +E+ L+ +LN
Sbjct: 42 LADFQIEKKIGRGQFSEVYKATCLLDRKTVALKKVQIFEMMDAKARQDCVKEIGLLKQLN 101
Query: 72 NPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIK--KARGSFFPEEKVCKWLTQLLIAV 129
+P I+ Y D+++E ++ + IV + GD+++ IK K + PE V K+ QL AV
Sbjct: 102 HPNIIKYLDSFIE-DNELNIVLELADAGDLSQMIKYFKKQKRLIPERTVWKYFVQLCSAV 160
Query: 130 DYLHSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAEDLTS-SVVGTPNYMCPEILA 188
+++HS RV+HRD+K +N+F+T ++LGD GL + ++E + S+VGTP YM PE +
Sbjct: 161 EHMHSRRVMHRDIKPANVFITATGIVKLGDLGLGRFFSSETTAAHSLVGTPYYMSPERIH 220
Query: 189 DIPYGYKSDMWSLGCCMFEIAAHQPAFRAPDM--AGLINKINRSSISPLP-IVYSSTLKQ 245
+ Y +KSD+WSLGC ++E+AA Q F M L KI + PLP YS L++
Sbjct: 221 ENGYNFKSDIWSLGCLLYEMAALQSPFYGDKMNLFSLCQKIEQCDYPPLPGEHYSEKLRE 280
Query: 246 IIKSMLRKNPEHRP 259
++ + +P HRP
Sbjct: 281 LVSMCIYPDPNHRP 294
>NEK9_XENLA (Q7ZZC8) Serine/threonine-protein kinase Nek9 (EC
2.7.1.37) (NimA-related protein kinase 9) (XNek9)
(Nercc1 kinase)
Length = 944
Score = 161 bits (407), Expect = 6e-39
Identities = 91/258 (35%), Positives = 149/258 (57%), Gaps = 4/258 (1%)
Query: 16 YEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYI 75
Y I +G GA+G A L + V K++ LA+ +EK +R A E+ +++ L + I
Sbjct: 34 YIPIRVLGHGAYGEATLYRRTEDDSLVVWKEVGLARLSEKERRDALNEIVILSLLQHDNI 93
Query: 76 VDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSN 135
+ Y + +++ + + I YC GG++ + I + F EE V +L Q++ AV +H
Sbjct: 94 IAYYNHFLDS-NTLLIELEYCNGGNLFDKIVHQKAQLFQEETVLWYLFQIVSAVSCIHKA 152
Query: 136 RVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAE-DLTSSVVGTPNYMCPEILADIPYGY 194
++HRD+K NIFLTK N I+LGD+GLAK+L++E + + VGT YM PEI + Y +
Sbjct: 153 GILHRDIKTLNIFLTKANLIKLGDYGLAKQLSSEYSMAETCVGTLYYMSPEICQGVKYSF 212
Query: 195 KSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRS--SISPLPIVYSSTLKQIIKSMLR 252
KSD+W++GC ++E+ F A + L KI + ++ VY+ L +++ + L
Sbjct: 213 KSDIWAVGCVLYELLTLTRTFDATNPLNLCVKIVQGNWAVGLDNTVYTQELIEVVHACLE 272
Query: 253 KNPEHRPTAAELLRHPHL 270
++PE RPTA E+L P L
Sbjct: 273 QDPEKRPTADEILERPIL 290
>NRKA_TRYBB (Q08942) Putative serine/threonine-protein kinase A (EC
2.7.1.37)
Length = 431
Score = 159 bits (403), Expect = 2e-38
Identities = 87/258 (33%), Positives = 151/258 (57%), Gaps = 13/258 (5%)
Query: 22 IGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYIVDYKDA 81
+G G++G A++ + V K + L+K +++ KR A E+ +A N+P I+ Y +
Sbjct: 26 VGLGSYGEAYVAESVEDGSLCVAKVMDLSKMSQRDKRYAQSEIKCLANCNHPNIIRYIED 85
Query: 82 WVEKEDRICIVTGYCEGGDMAENIKKARGS----FFPEEKVCKWLTQLLIAVDYLHSNRV 137
E+ DR+ IV + + G++ E IK RGS +F E + QL +A+DY+HS+++
Sbjct: 86 H-EENDRLLIVMEFADSGNLDEQIK-LRGSGDARYFQEHEALFLFLQLCLALDYIHSHKM 143
Query: 138 IHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAEDLTSSVV-----GTPNYMCPEILADIPY 192
+HRD+K +N+ LT ++LGDFG + + ED S VV GTP Y+ PE+ + Y
Sbjct: 144 LHRDIKSANVLLTSTGLVKLGDFGFSHQY--EDTVSGVVASTFCGTPYYLAPELWNNKRY 201
Query: 193 GYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSMLR 252
K+D+WSLG ++EI + F A ++ GL++K+ + +PLP +SS K+++ +L
Sbjct: 202 NKKADVWSLGVLLYEIMGMKKPFSASNLKGLMSKVLAGTYAPLPDSFSSEFKRVVDGILV 261
Query: 253 KNPEHRPTAAELLRHPHL 270
+P RP+ E+ + P++
Sbjct: 262 ADPNDRPSVREIFQIPYI 279
>NEK6_HUMAN (Q9HC98) Serine/threonine-protein kinase Nek6 (EC
2.7.1.37) (NimA-related protein kinase 6) (Protein
kinase SID6-1512)
Length = 313
Score = 159 bits (403), Expect = 2e-38
Identities = 88/254 (34%), Positives = 151/254 (58%), Gaps = 8/254 (3%)
Query: 13 MEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTE-KFKRTAHQEMNLIAKLN 71
+ D+++ ++IGRG F + ++K LKK+++ + + K ++ +E+ L+ +LN
Sbjct: 42 LADFQIEKKIGRGQFSEVYKATCLLDRKTVALKKVQIFEMMDAKARQDCVKEIGLLKQLN 101
Query: 72 NPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIK--KARGSFFPEEKVCKWLTQLLIAV 129
+P I+ Y D+++E ++ + IV + GD+++ IK K + PE V K+ QL AV
Sbjct: 102 HPNIIKYLDSFIE-DNELNIVLELADAGDLSQMIKYFKKQKRLIPERTVWKYFVQLCSAV 160
Query: 130 DYLHSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAEDLTS-SVVGTPNYMCPEILA 188
+++HS RV+HRD+K +N+F+T ++LGD GL + ++E + S+VGTP YM PE +
Sbjct: 161 EHMHSRRVMHRDIKPANVFITATGVVKLGDLGLGRFFSSETTAAHSLVGTPYYMSPERIH 220
Query: 189 DIPYGYKSDMWSLGCCMFEIAAHQPAFRAPDM--AGLINKINRSSISPLP-IVYSSTLKQ 245
+ Y +KSD+WSLGC ++E+AA Q F M L KI + PLP YS L++
Sbjct: 221 ENGYNFKSDIWSLGCLLYEMAALQSPFYGDKMNLFSLCQKIEQCDYPPLPGEHYSEKLRE 280
Query: 246 IIKSMLRKNPEHRP 259
++ + +P RP
Sbjct: 281 LVSMCICPDPHQRP 294
>NRKB_TRYBB (Q03428) Putative serine/threonine-protein kinase B (EC
2.7.1.37)
Length = 431
Score = 152 bits (384), Expect = 3e-36
Identities = 82/257 (31%), Positives = 144/257 (55%), Gaps = 11/257 (4%)
Query: 22 IGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYIVDYKDA 81
+G G++G ++ + V K + L+K + + KR A E+ N+P I+ Y +
Sbjct: 26 VGLGSYGEGYVAERVEDGSLCVAKVMDLSKMSRRDKRYAQSEIKYPTNCNHPNIIRYIED 85
Query: 82 WVEKEDRICIVTGYCEGGDMAENIKK---ARGSFFPEEKVCKWLTQLLIAVDYLHSNRVI 138
E+ DR+ IV + + G++ E IK +F E + QL +A+DY+HS++++
Sbjct: 86 H-EENDRLLIVMEFADSGNLDEQIKPWGTGDARYFQEHEALFLFLQLCLALDYIHSHKML 144
Query: 139 HRDLKCSNIFLTKDNNIRLGDFGLAKRLNAEDLTSSVV-----GTPNYMCPEILADIPYG 193
HRD+K +N+ LT ++LGDFG + + ED S VV GTP Y+ PE+ ++ Y
Sbjct: 145 HRDIKSANVLLTSTGLVKLGDFGFSHQY--EDTVSGVVASTFCGTPYYLAPELWNNLRYN 202
Query: 194 YKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSMLRK 253
K+D+WSLG ++EI + F A ++ GL++K+ + +PLP +SS K+++ +L
Sbjct: 203 KKADVWSLGVLLYEIMGMKKPFSASNLKGLMSKVLAGTYAPLPDSFSSEFKRVVDGILVA 262
Query: 254 NPEHRPTAAELLRHPHL 270
+P RP+ E + P++
Sbjct: 263 DPNDRPSVRENFQIPYI 279
>NEK7_MOUSE (Q9ES74) Serine/threonine-protein kinase Nek7 (EC
2.7.1.37) (NimA-related protein kinase 7)
Length = 302
Score = 151 bits (382), Expect = 4e-36
Identities = 88/263 (33%), Positives = 149/263 (56%), Gaps = 8/263 (3%)
Query: 7 DMRSKKMEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTE-KFKRTAHQEMN 65
DM + ++ + ++IGRG F + + LKK+++ + K + +E++
Sbjct: 25 DMGYNTLANFRIEKKIGRGQFSEVYRASCLLDGVPVALKKVQIFDLMDAKARADCIKEID 84
Query: 66 LIAKLNNPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIK--KARGSFFPEEKVCKWLT 123
L+ +LN+P ++ Y +++E ++ + IV + GD++ IK K + PE V K+
Sbjct: 85 LLKQLNHPNVIKYYASFIE-DNELNIVLELADAGDLSRMIKHFKKQKRLIPERTVWKYFV 143
Query: 124 QLLIAVDYLHSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAEDLTS-SVVGTPNYM 182
QL A+D++HS RV+HRD+K +N+F+T ++LGD GL + +++ + S+VGTP YM
Sbjct: 144 QLCSALDHMHSRRVMHRDIKPANVFITATGVVKLGDLGLGRFFSSKTTAAHSLVGTPYYM 203
Query: 183 CPEILADIPYGYKSDMWSLGCCMFEIAAHQPAFRAPDM--AGLINKINRSSISPLPI-VY 239
PE + + Y +KSD+WSLGC ++E+AA Q F M L KI + PLP Y
Sbjct: 204 SPERIHENGYNFKSDIWSLGCLLYEMAALQSPFYGDKMNLYSLCKKIEQCDYPPLPSDHY 263
Query: 240 SSTLKQIIKSMLRKNPEHRPTAA 262
S L+Q++ + +PE RP A
Sbjct: 264 SEELRQLVNICINPDPEKRPDIA 286
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.129 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,294,384
Number of Sequences: 164201
Number of extensions: 3248049
Number of successful extensions: 12570
Number of sequences better than 10.0: 1752
Number of HSP's better than 10.0 without gapping: 1416
Number of HSP's successfully gapped in prelim test: 338
Number of HSP's that attempted gapping in prelim test: 7801
Number of HSP's gapped (non-prelim): 2179
length of query: 643
length of database: 59,974,054
effective HSP length: 117
effective length of query: 526
effective length of database: 40,762,537
effective search space: 21441094462
effective search space used: 21441094462
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0318.13


