

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
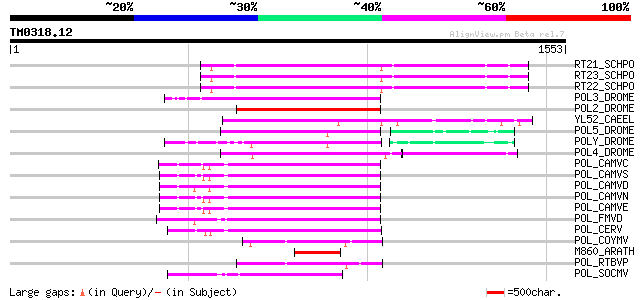
Query= TM0318.12
(1553 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 449 e-125
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 448 e-125
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 448 e-125
POL3_DROME (P04323) Retrovirus-related Pol polyprotein from tran... 376 e-103
POL2_DROME (P20825) Retrovirus-related Pol polyprotein from tran... 350 2e-95
YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III 319 4e-86
POL5_DROME (Q8I7P9) Retrovirus-related Pol polyprotein from tran... 308 8e-83
POLY_DROME (P10401) Retrovirus-related Pol polyprotein from tran... 292 4e-78
POL4_DROME (P10394) Retrovirus-related Pol polyprotein from tran... 280 2e-74
POL_CAMVC (P03555) Enzymatic polyprotein [Contains: Aspartic pro... 234 1e-60
POL_CAMVS (P03554) Enzymatic polyprotein [Contains: Aspartic pro... 234 2e-60
POL_CAMVD (P03556) Enzymatic polyprotein [Contains: Aspartic pro... 233 3e-60
POL_CAMVN (Q00962) Enzymatic polyprotein [Contains: Aspartic pro... 231 1e-59
POL_CAMVE (Q02964) Enzymatic polyprotein [Contains: Aspartic pro... 230 3e-59
POL_FMVD (P09523) Enzymatic polyprotein [Contains: Aspartic prot... 210 3e-53
POL_CERV (P05400) Enzymatic polyprotein [Contains: Aspartic prot... 209 5e-53
POL_COYMV (P19199) Putative polyprotein [Contains: Coat protein;... 179 5e-44
M860_ARATH (P92523) Hypothetical mitochondrial protein AtMg00860... 147 2e-34
POL_RTBVP (P27502) Polyprotein (P194 protein) [Contains: Coat pr... 140 2e-32
POL_SOCMV (P15629) Enzymatic polyprotein [Contains: Aspartic pro... 130 3e-29
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 449 bits (1156), Expect = e-125
Identities = 296/954 (31%), Positives = 483/954 (50%), Gaps = 46/954 (4%)
Query: 533 TLKFKLGNKKVVLKGEPELLKKRASMNALV------KAIQQQGEGYVLQYQMLEKEVAKA 586
T+K + + +K E ++KK + A+ I+ + L + K
Sbjct: 312 TIKLNISLNGISIKTEFLVVKKFSHPAAISFTTLYDNNIEISSSKHTLSQMNKVSNIVKE 371
Query: 587 EETPELLKAVLSDFPALFAAPTELPPQRRHDHAIHL-KEGASIPNIRPYRYPHYQKNEIE 645
E P++ K D A P + + + L +E +P IR Y P + +
Sbjct: 372 PELPDIYKE-FKDITAETNTEKLPKPIKGLEFEVELTQENYRLP-IRNYPLPPGKMQAMN 429
Query: 646 KLVAEMLNAGIIRPSISPYSSPVILVRKKDGSWRFCVDYRALNKITVPNKFPIPVIDELL 705
+ + L +GIIR S + + PV+ V KK+G+ R VDY+ LNK PN +P+P+I++LL
Sbjct: 430 DEINQGLKSGIIRESKAINACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPLPLIEQLL 489
Query: 706 DEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLM 765
+I + +F+K+DLKS YH I ++ D K AFR G +E+LVMP+G++ AP+ FQ +
Sbjct: 490 AKIQGSTIFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISTAPAHFQYFI 549
Query: 766 NEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIE 825
N +L + + DDILI+S + EHV H+ VLQ + +L IN K F + Q++
Sbjct: 550 NTILGEAKESHVVCYMDDILIHSKSESEHVKHVKDVLQKLKNANLIINQAKCEFHQSQVK 609
Query: 826 YLGHVLSAGTVAADPKKLEAMWLWPVPKDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQL 885
++G+ +S + ++ + W PK+ K LR FLG Y R+F+ ++ PL L
Sbjct: 610 FIGYHISEKGFTPCQENIDKVLQWKQPKNRKELRQFLGSVNYLRKFIPKTSQLTHPLNNL 669
Query: 886 LKKD-SFQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDASGTGLGAVLTQEGK- 943
LKKD ++W P+ QA E +K L P L DFSK +LETDAS +GAVL+Q+
Sbjct: 670 LKKDVRWKWTPTQTQAIENIKQCLVSPPVLRHFDFSKKILLETDASDVAVGAVLSQKHDD 729
Query: 944 ----PLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLG--RHFIIRTDQKSL--K 995
P+ ++SA +S SV ++E++A++++++ WRHYL F I TD ++L +
Sbjct: 730 DKYYPVGYYSAKMSKAQLNYSVSDKEMLAIIKSLKHWRHYLESTIEPFKILTDHRNLIGR 789
Query: 996 FLTEQQVVGEGQFKWISKLSGYDFEIQYKPGKDNSAADAMSR--------------RSSY 1041
E + + +W L ++FEI Y+PG N ADA+SR S
Sbjct: 790 ITNESEPENKRLARWQLFLQDFNFEINYRPGSANHIADALSRIVDETEPIPKDSEDNSIN 849
Query: 1042 CALSVLKVNDFE-EWQTEMLQDPKLQAIVHDLIVDPEAHKGYTLKDHRLF-YKGKLVLSK 1099
+ +DF+ + TE D KL ++++ D + LKD L K +++L
Sbjct: 850 FVNQISITDDFKNQVVTEYTNDTKLLNLLNN--EDKRVEENIQLKDGLLINSKDQILLPN 907
Query: 1100 QSSRIPILCKEFHASLLGGHSGFFRTYRRLAAVVYWEGMKSDIRDFVAECDTCQRNKFDN 1159
+ + K++H H G + W+G++ I+++V C TCQ NK N
Sbjct: 908 DTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQINKSRN 967
Query: 1160 LSPAGLLQPLPVPTQVWTDVSMDFIGGLPKSNGKDTILVVVDRLTKYAHFIPLRHPFTAP 1219
P G LQP+P + W +SMDFI LP+S+G + + VVVDR +K A +P TA
Sbjct: 968 HKPYGPLQPIPPSERPWESLSMDFITALPESSGYNALFVVVDRFSKMAILVPCTKSITAE 1027
Query: 1220 EVAALFLHEVVRLHGFPTTIVSDRDSLFLSSFWKELFRVSGTQLKFSSAYHPQTDGQTEV 1279
+ A +F V+ G P I++D D +F S WK+ +KFS Y PQTDGQTE
Sbjct: 1028 QTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTER 1087
Query: 1280 VNRCLEVYLRCLTGSRPRKWVTCLPWAEFWFNSNYNRSAKMSPFQALYGREPPVLLQGTT 1339
N+ +E LRC+ + P WV + + +N+ + + +M+PF+ ++ P L
Sbjct: 1088 TNQTVEKLLRCVCSTHPNTWVDHISLVQQSYNNAIHSATQMTPFEIVHRYSP--ALSPLE 1145
Query: 1340 IPSKIAAVNDLQVGRDELLSDLRANLLKSQDMMRAYANKKRRDV-DYQIGDEVFLKLQPY 1398
+PS ++ ++ ++ +L + M+ Y + K +++ ++Q GD V +K
Sbjct: 1146 LPSFSDKTDENSQETIQVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVK---- 1201
Query: 1399 RRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAYRLELPAHSR--VHPVFHVSLLK 1450
R ++ + KL+P + GP+ ++ K G Y L+LP + FHVS L+
Sbjct: 1202 RTKTGFLHKSNKLAPSFAGPFYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLE 1255
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 448 bits (1152), Expect = e-125
Identities = 296/954 (31%), Positives = 484/954 (50%), Gaps = 46/954 (4%)
Query: 533 TLKFKLGNKKVVLKGEPELLKKRASMNALV------KAIQQQGEGYVLQYQMLEKEVAKA 586
T+K + + +K E ++KK + A+ I+ + L + K
Sbjct: 312 TIKLNISLNGISIKTEFLVVKKFSHPAAISFTTLYDNNIEISSSKHTLSQMNKVSNIVKE 371
Query: 587 EETPELLKAVLSDFPALFAAPTELPPQRRHDHAIHL-KEGASIPNIRPYRYPHYQKNEIE 645
E P++ K D A P + + + L +E +P IR Y P + +
Sbjct: 372 PELPDIYKE-FKDITAETNTEKLPKPIKGLEFEVELTQENYRLP-IRNYPLPPGKMQAMN 429
Query: 646 KLVAEMLNAGIIRPSISPYSSPVILVRKKDGSWRFCVDYRALNKITVPNKFPIPVIDELL 705
+ + L +GIIR S + + PV+ V KK+G+ R VDY+ LNK PN +P+P+I++LL
Sbjct: 430 DEINQGLKSGIIRESKAINACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPLPLIEQLL 489
Query: 706 DEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLM 765
+I + +F+K+DLKS YH I ++ D K AFR G +E+LVMP+G++ AP+ FQ +
Sbjct: 490 AKIQGSTIFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISIAPAHFQYFI 549
Query: 766 NEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIE 825
N +L V + + D+ILI+S + EHV H+ VLQ + +L IN K F + Q++
Sbjct: 550 NTILGEVKESHVVCYMDNILIHSKSESEHVKHVKDVLQKLKNANLIINQAKCEFHQSQVK 609
Query: 826 YLGHVLSAGTVAADPKKLEAMWLWPVPKDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQL 885
++G+ +S + ++ + W PK+ K LR FLG Y R+F+ ++ PL L
Sbjct: 610 FIGYHISEKGFTPCQENIDKVLQWKQPKNRKELRQFLGSVNYLRKFIPKTSQLTHPLNNL 669
Query: 886 LKKD-SFQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDASGTGLGAVLTQEGK- 943
LKKD ++W P+ QA E +K L P L DFSK +LETDAS +GAVL+Q+
Sbjct: 670 LKKDVRWKWTPTQTQAIENIKQCLVSPPVLRHFDFSKKILLETDASDVAVGAVLSQKHDD 729
Query: 944 ----PLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLG--RHFIIRTDQKSL--K 995
P+ ++SA +S SV ++E++A++++++ WRHYL F I TD ++L +
Sbjct: 730 DKYYPVGYYSAKMSKAQLNYSVSDKEMLAIIKSLKHWRHYLESTIEPFKILTDHRNLIGR 789
Query: 996 FLTEQQVVGEGQFKWISKLSGYDFEIQYKPGKDNSAADAMSR--------------RSSY 1041
E + + +W L ++FEI Y+PG N ADA+SR S
Sbjct: 790 ITNESEPENKRLARWQLFLQDFNFEINYRPGSANHIADALSRIVDETEPIPKDSEDNSIN 849
Query: 1042 CALSVLKVNDFE-EWQTEMLQDPKLQAIVHDLIVDPEAHKGYTLKDHRLF-YKGKLVLSK 1099
+ +DF+ + TE D KL ++++ D + LKD L K +++L
Sbjct: 850 FVNQISITDDFKNQVVTEYTNDTKLLNLLNN--EDKRVEENIQLKDGLLINSKDQILLPN 907
Query: 1100 QSSRIPILCKEFHASLLGGHSGFFRTYRRLAAVVYWEGMKSDIRDFVAECDTCQRNKFDN 1159
+ + K++H H G + W+G++ I+++V C TCQ NK N
Sbjct: 908 DTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQINKSRN 967
Query: 1160 LSPAGLLQPLPVPTQVWTDVSMDFIGGLPKSNGKDTILVVVDRLTKYAHFIPLRHPFTAP 1219
P G LQP+P + W +SMDFI LP+S+G + + VVVDR +K A +P TA
Sbjct: 968 HKPYGPLQPIPPSERPWESLSMDFITALPESSGYNALFVVVDRFSKMAILVPCTKSITAE 1027
Query: 1220 EVAALFLHEVVRLHGFPTTIVSDRDSLFLSSFWKELFRVSGTQLKFSSAYHPQTDGQTEV 1279
+ A +F V+ G P I++D D +F S WK+ +KFS Y PQTDGQTE
Sbjct: 1028 QTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTER 1087
Query: 1280 VNRCLEVYLRCLTGSRPRKWVTCLPWAEFWFNSNYNRSAKMSPFQALYGREPPVLLQGTT 1339
N+ +E LRC+ + P WV + + +N+ + + +M+PF+ ++ P L
Sbjct: 1088 TNQTVEKLLRCVCSTHPNTWVDHISLVQQSYNNAIHSATQMTPFEIVHRYSP--ALSPLE 1145
Query: 1340 IPSKIAAVNDLQVGRDELLSDLRANLLKSQDMMRAYANKKRRDV-DYQIGDEVFLKLQPY 1398
+PS ++ ++ ++ +L + M+ Y + K +++ ++Q GD V +K
Sbjct: 1146 LPSFSDKTDENSQETIQVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVK---- 1201
Query: 1399 RRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAYRLELPAHSR--VHPVFHVSLLK 1450
R ++ + KL+P + GP+ ++ K G Y L+LP + FHVS L+
Sbjct: 1202 RTKTGFLHKSNKLAPSFAGPFYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLE 1255
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 448 bits (1152), Expect = e-125
Identities = 296/954 (31%), Positives = 484/954 (50%), Gaps = 46/954 (4%)
Query: 533 TLKFKLGNKKVVLKGEPELLKKRASMNALV------KAIQQQGEGYVLQYQMLEKEVAKA 586
T+K + + +K E ++KK + A+ I+ + L + K
Sbjct: 312 TIKLNISLNGISIKTEFLVVKKFSHPAAISFTTLYDNNIEISSSKHTLSQMNKVSNIVKE 371
Query: 587 EETPELLKAVLSDFPALFAAPTELPPQRRHDHAIHL-KEGASIPNIRPYRYPHYQKNEIE 645
E P++ K D A P + + + L +E +P IR Y P + +
Sbjct: 372 PELPDIYKE-FKDITAETNTEKLPKPIKGLEFEVELTQENYRLP-IRNYPLPPGKMQAMN 429
Query: 646 KLVAEMLNAGIIRPSISPYSSPVILVRKKDGSWRFCVDYRALNKITVPNKFPIPVIDELL 705
+ + L +GIIR S + + PV+ V KK+G+ R VDY+ LNK PN +P+P+I++LL
Sbjct: 430 DEINQGLKSGIIRESKAINACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPLPLIEQLL 489
Query: 706 DEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLM 765
+I + +F+K+DLKS YH I ++ D K AFR G +E+LVMP+G++ AP+ FQ +
Sbjct: 490 AKIQGSTIFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISIAPAHFQYFI 549
Query: 766 NEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIE 825
N +L V + + D+ILI+S + EHV H+ VLQ + +L IN K F + Q++
Sbjct: 550 NTILGEVKESHVVCYMDNILIHSKSESEHVKHVKDVLQKLKNANLIINQAKCEFHQSQVK 609
Query: 826 YLGHVLSAGTVAADPKKLEAMWLWPVPKDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQL 885
++G+ +S + ++ + W PK+ K LR FLG Y R+F+ ++ PL L
Sbjct: 610 FIGYHISEKGFTPCQENIDKVLQWKQPKNRKELRQFLGSVNYLRKFIPKTSQLTHPLNNL 669
Query: 886 LKKD-SFQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDASGTGLGAVLTQEGK- 943
LKKD ++W P+ QA E +K L P L DFSK +LETDAS +GAVL+Q+
Sbjct: 670 LKKDVRWKWTPTQTQAIENIKQCLVSPPVLRHFDFSKKILLETDASDVAVGAVLSQKHDD 729
Query: 944 ----PLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLG--RHFIIRTDQKSL--K 995
P+ ++SA +S SV ++E++A++++++ WRHYL F I TD ++L +
Sbjct: 730 DKYYPVGYYSAKMSKAQLNYSVSDKEMLAIIKSLKHWRHYLESTIEPFKILTDHRNLIGR 789
Query: 996 FLTEQQVVGEGQFKWISKLSGYDFEIQYKPGKDNSAADAMSR--------------RSSY 1041
E + + +W L ++FEI Y+PG N ADA+SR S
Sbjct: 790 ITNESEPENKRLARWQLFLQDFNFEINYRPGSANHIADALSRIVDETEPIPKDSEDNSIN 849
Query: 1042 CALSVLKVNDFE-EWQTEMLQDPKLQAIVHDLIVDPEAHKGYTLKDHRLF-YKGKLVLSK 1099
+ +DF+ + TE D KL ++++ D + LKD L K +++L
Sbjct: 850 FVNQISITDDFKNQVVTEYTNDTKLLNLLNN--EDKRVEENIQLKDGLLINSKDQILLPN 907
Query: 1100 QSSRIPILCKEFHASLLGGHSGFFRTYRRLAAVVYWEGMKSDIRDFVAECDTCQRNKFDN 1159
+ + K++H H G + W+G++ I+++V C TCQ NK N
Sbjct: 908 DTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQINKSRN 967
Query: 1160 LSPAGLLQPLPVPTQVWTDVSMDFIGGLPKSNGKDTILVVVDRLTKYAHFIPLRHPFTAP 1219
P G LQP+P + W +SMDFI LP+S+G + + VVVDR +K A +P TA
Sbjct: 968 HKPYGPLQPIPPSERPWESLSMDFITALPESSGYNALFVVVDRFSKMAILVPCTKSITAE 1027
Query: 1220 EVAALFLHEVVRLHGFPTTIVSDRDSLFLSSFWKELFRVSGTQLKFSSAYHPQTDGQTEV 1279
+ A +F V+ G P I++D D +F S WK+ +KFS Y PQTDGQTE
Sbjct: 1028 QTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTER 1087
Query: 1280 VNRCLEVYLRCLTGSRPRKWVTCLPWAEFWFNSNYNRSAKMSPFQALYGREPPVLLQGTT 1339
N+ +E LRC+ + P WV + + +N+ + + +M+PF+ ++ P L
Sbjct: 1088 TNQTVEKLLRCVCSTHPNTWVDHISLVQQSYNNAIHSATQMTPFEIVHRYSP--ALSPLE 1145
Query: 1340 IPSKIAAVNDLQVGRDELLSDLRANLLKSQDMMRAYANKKRRDV-DYQIGDEVFLKLQPY 1398
+PS ++ ++ ++ +L + M+ Y + K +++ ++Q GD V +K
Sbjct: 1146 LPSFSDKTDENSQETIQVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVK---- 1201
Query: 1399 RRRSLAKKMNEKLSPRYYGPYPIVAKIGAVAYRLELPAHSR--VHPVFHVSLLK 1450
R ++ + KL+P + GP+ ++ K G Y L+LP + FHVS L+
Sbjct: 1202 RTKTGFLHKSNKLAPSFAGPFYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLE 1255
>POL3_DROME (P04323) Retrovirus-related Pol polyprotein from
transposon 17.6 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1058
Score = 376 bits (966), Expect = e-103
Identities = 227/617 (36%), Positives = 351/617 (56%), Gaps = 24/617 (3%)
Query: 433 KLKGTAIVVLVDCGATHNFISQSLAREMELQITETPAYTVEVGDGHVVKSKGVCKNLSVQ 492
K K + L+D G+T N S+++ +L I T + + +G ++ +K + +
Sbjct: 19 KYKENNLKCLIDTGSTVNMTSKNI---FDLPIQNTSTF-IHTSNGPLIVNKSIIIPSKIL 74
Query: 493 IQGFVVTQDFFLFGL-RGVDVVLGLEWLA-GLGDIKANFEELTLKFKLGNKKVVLKGEPE 550
F T +F L D++LG + LA I +E+TL NK +++G
Sbjct: 75 ---FPTTNEFLLHPFSENYDLLLGRKLLAEAKATISYRDQEVTL---YNNKYKLIEGIAT 128
Query: 551 LLKKR-ASMNALVKAIQQQGEGY--VLQYQMLEKEVAKAEETPELLKAVLSDFPALFAAP 607
+ ++N + + +Q +L+ + E EE L + +
Sbjct: 129 HEQSHFQNVNMIPDTMLRQPNKISPILESDLYRLEHLNNEEKQRLCALLQKYHDIQYHEG 188
Query: 608 TELPPQRRHDHAIHLKEGASIPNIRPYRYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSP 667
+L + H I+ K ++P Y YP + E+E + +MLN GIIR S SPY+SP
Sbjct: 189 DKLTFTNQTKHTINTKH--NLPLYSKYSYPQAYEQEVESQIQDMLNQGIIRTSNSPYNSP 246
Query: 668 VILVRKKDGS-----WRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSG 722
+ +V KK + +R +DYR LN+ITV ++ PIP +DE+L ++G F+ IDL G
Sbjct: 247 IWVVPKKQDASGKQKFRIVIDYRKLNEITVGDRHPIPNMDEILGKLGRCNYFTTIDLAKG 306
Query: 723 YHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFD 782
+HQI M E V KTAF T GHYE+L MPFGL NAP+TFQ MN++LRP+L K LV+ D
Sbjct: 307 FHQIEMDPESVSKTAFSTKHGHYEYLRMPFGLKNAPATFQRCMNDILRPLLNKHCLVYLD 366
Query: 783 DILIYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKK 842
DI+++S +++EH+ L V + + + +LK+ K F + + +LGHVL+ + +P+K
Sbjct: 367 DIIVFSTSLDEHLQSLGLVFEKLAKANLKLQLDKCEFLKQETTFLGHVLTPDGIKPNPEK 426
Query: 843 LEAMWLWPVPKDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQLLKKDS--FQWGPSPQQA 900
+EA+ +P+P K ++ FLGLTGYYR+F+ ++ IA+P+T+ LKK+ P A
Sbjct: 427 IEAIQKYPIPTKPKEIKAFLGLTGYYRKFIPNFADIAKPMTKCLKKNMKIDTTNPEYDSA 486
Query: 901 FEALKHTLTEIPTLAVPDFSKVFVLETDASGTGLGAVLTQEGKPLAFWSATLSDRSQAKS 960
F+ LK+ ++E P L VPDF+K F L TDAS LGAVL+Q+G PL++ S TL++ S
Sbjct: 487 FKKLKYLISEDPILKVPDFTKKFTLTTDASDVALGAVLSQDGHPLSYISRTLNEHEINYS 546
Query: 961 VYERELMAVVRAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQFKWISKLSGYDFE 1020
E+EL+A+V A + +RHYLLGRHF I +D + L +L + +W KLS +DF+
Sbjct: 547 TIEKELLAIVWATKTFRHYLLGRHFEISSDHQPLSWLYRMKDPNSKLTRWRVKLSEFDFD 606
Query: 1021 IQYKPGKDNSAADAMSR 1037
I+Y GK+N ADA+SR
Sbjct: 607 IKYIKGKENCVADALSR 623
>POL2_DROME (P20825) Retrovirus-related Pol polyprotein from
transposon 297 [Contains: Protease (EC 3.4.23.-); Reverse
transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1059
Score = 350 bits (897), Expect = 2e-95
Identities = 186/412 (45%), Positives = 265/412 (64%), Gaps = 9/412 (2%)
Query: 635 RYPHYQKNEIE--KLVAEMLNAGIIRPSISPYSSPVILVRKKDGS-----WRFCVDYRAL 687
+YP Q +EIE V EMLN G+IR S SPY+SP +V KK + +R +DYR L
Sbjct: 211 QYPLAQTHEIEVENQVQEMLNQGLIRESNSPYNSPTWVVPKKPDASGANKYRVVIDYRKL 270
Query: 688 NKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEF 747
N+IT+P+++PIP +DE+L ++G + F+ IDL G+HQI M E + KTAF T GHYE+
Sbjct: 271 NEITIPDRYPIPNMDEILGKLGKCQYFTTIDLAKGFHQIEMDEESISKTAFSTKSGHYEY 330
Query: 748 LVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQ 807
L MPFGL NAP+TFQ MN +LRP+L K LV+ DDI+I+S ++ EH+ + V +
Sbjct: 331 LRMPFGLRNAPATFQRCMNNILRPLLNKHCLVYLDDIIIFSTSLTEHLNSIQLVFTKLAD 390
Query: 808 HDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWPVPKDLKSLRGFLGLTGY 867
+LK+ K F + + +LGH+++ + +P K++A+ +P+P K +R FLGLTGY
Sbjct: 391 ANLKLQLDKCEFLKKEANFLGHIVTPDGIKPNPIKVKAIVSYPIPTKDKEIRAFLGLTGY 450
Query: 868 YRRFVKDYGKIARPLTQLLKKDSFQWGPSPQ--QAFEALKHTLTEIPTLAVPDFSKVFVL 925
YR+F+ +Y IA+P+T LKK + + +AFE LK + P L +PDF K FVL
Sbjct: 451 YRKFIPNYADIAKPMTSCLKKRTKIDTQKLEYIEAFEKLKALIIRDPILQLPDFEKKFVL 510
Query: 926 ETDASGTGLGAVLTQEGKPLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLGRHF 985
TDAS LGAVL+Q G P++F S TL+D S E+EL+A+V A + +RHYLLGR F
Sbjct: 511 TTDASNLALGAVLSQNGHPISFISRTLNDHELNYSAIEKELLAIVWATKTFRHYLLGRQF 570
Query: 986 IIRTDQKSLKFLTEQQVVGEGQFKWISKLSGYDFEIQYKPGKDNSAADAMSR 1037
+I +D + L++L + G +W +LS Y F+I Y GK+NS ADA+SR
Sbjct: 571 LIASDHQPLRWLHNLKEPGAKLERWRVRLSEYQFKIDYIKGKENSVADALSR 622
>YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III
Length = 2186
Score = 319 bits (817), Expect = 4e-86
Identities = 259/950 (27%), Positives = 437/950 (45%), Gaps = 105/950 (11%)
Query: 596 VLSDFPALFA-APTELPPQRRHDHAIHLKEGASIPNIRPYRYPHYQKNEIEKLVAEMLNA 654
V+ F +FA + EL + I LKEGA +P P K EI K++ +MLN
Sbjct: 909 VIEQFQDVFAISDDELGRNSGTECVIELKEGAEPIRQKPRPIPLALKPEIRKMIQKMLNQ 968
Query: 655 GIIRPSISPYSSPVILVRKKDGSWRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVF 714
+IR S SP+SSPV+LV+KKDGS R C+DYR +NK+ N P+P I+ L + +++
Sbjct: 969 KVIRESKSPWSSPVVLVKKKDGSIRMCIDYRKVNKVVKNNAHPLPNIEATLQSLAGKKLY 1028
Query: 715 SKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLR 774
+ D+ +G+ QI + + E TAF +E+ V+PFGL +P+ FQ M E++ +L
Sbjct: 1029 TVFDMIAGFWQIPLDEKSKEITAFAIGSELFEWNVLPFGLVISPALFQGTMEEIIGDLLG 1088
Query: 775 KCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAG 834
CA V+ DD+LI S ME+H+ + + L + + +K+ K + ++EYLGH ++
Sbjct: 1089 VCAFVYVDDLLIASKDMEQHLQDVKEALTRIRKSGMKLRASKCHIAKKEVEYLGHKVTLD 1148
Query: 835 TVAADPKKLEAMWLWPVPKDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQLLK-KDSFQW 893
V K + M + P ++K L+ FLGL GYYR+F+ ++ +IA LT L+ K ++ W
Sbjct: 1149 GVETQEVKTDKMKQFSRPTNVKELQSFLGLVGYYRKFILNFAQIASSLTSLISAKVAWIW 1208
Query: 894 GPSPQQAFEALKHTLTEIPTLAVPDF------SKVFVLETDASGTGLGAVLTQEG----- 942
+ AF+ LK + + P LA PD + F++ TDAS G+GAVL QEG
Sbjct: 1209 EKEQEIAFQELKKLVCQTPVLAQPDVEAALKGDRPFMIYTDASRKGIGAVLAQEGPDGQQ 1268
Query: 943 KPLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQV 1002
P+AF S LS + + E +A++ A++R++ + G + TD K L L +
Sbjct: 1269 HPIAFASKALSPAETRYHITDLEALAMMFALRRFKTIIYGTAITVFTDHKPLISLLKGSP 1328
Query: 1003 VGEGQFKWISKLSGYDFEIQYKPGKDNSAADAMSR------------------------- 1037
+ + ++W ++ +D +I Y GK N+ ADA+SR
Sbjct: 1329 LADRLWRWSIEILEFDVKIVYLAGKANAVADALSRGGCPPNELEEEQTKELTSIVNAIQT 1388
Query: 1038 -----RSSYCALSVLKVNDFEEWQTEM--LQDPKLQAIVHDLIVDPEAHKGY------TL 1084
S C L LK D E W+ + L+ K + + ++ E Y L
Sbjct: 1389 ELPDILDSSCWLERLKGED-EGWKEVIAALEGGKTKGTFKIVGIESEISLEYYKIVGGVL 1447
Query: 1085 KDHRLFYKGKLVLSKQSSRIPILCKEFHASLLGGHSGFFRTYRRLAAVVYWEGMKSDIRD 1144
K+ + + + V+ + R P+L KE H +L GH G + +R + YW M+ + +
Sbjct: 1448 KNTEIEEQSRSVV-PEKIRTPLL-KELHEGMLAGHFGIKKMWRMVHRKFYWPQMRVCVEN 1505
Query: 1145 FVAECDTC-----QRNKFDNLSPAGLLQPLPVPTQVWTDVSMDFIGGLPKSNGKDTILVV 1199
V C C +L+P + PL + DV + G IL +
Sbjct: 1506 CVRTCAKCLCANDHSKLTSSLTPYRMTFPLEIVACDLMDVGLSV-------QGNRYILTI 1558
Query: 1200 VDRLTKYAHFIPLRHPFTAPEVAALFLHEVVRLHGFPTTIVSDRDSLFLSSFWKELFRVS 1259
+D TKY +P+ + A + P +++D+ F++ + + +
Sbjct: 1559 IDLFTKYGTAVPIPDKKAETVLKAFVERWAIGEGRIPLKLLTDQGKEFVNGLFAQFTHML 1618
Query: 1260 GTQLKFSSAYHPQTDGQTEVVNRCLEVYLRCLTGSRPRKWVTCLPWAEFWFNSNYNRSAK 1319
+ + Y+ + +G E N+ + ++ T + P +W + +A + +N+ + +
Sbjct: 1619 KIEHITTKGYNSRANGAVERFNKTIMHIMKKKT-AVPMEWDDQVVYAVYAYNNCVHENTG 1677
Query: 1320 MSPFQALYGREPPVLLQGTTIPSKIAAVNDLQVGRDELLSDLRANLLKSQDMMR------ 1373
+P ++GR+ + G S AV DE L LLK Q + +
Sbjct: 1678 ETPMFLMHGRD----VMGPLEMSGEDAVGINYADMDEYKHLLTQELLKVQKIAKEHAMRE 1733
Query: 1374 ----------AYANKKRRDVDYQIGDEVFLKLQPYRRRSLAKKMNEKLSPRYYGPYPIVA 1423
YA+KK R Q G V L++ + + K+ K S GPY +++
Sbjct: 1734 QESYKSLFDQKYASKKHRFP--QPGSRVLLEIPSEKLGAQCPKLVNKWS----GPYRVIS 1787
Query: 1424 ----------KIGAVAYRLELPAHS-RVHP-VFHVSLLKTAVGTNFQPQP 1461
+G + L++P + RV P L+ T G + +P+P
Sbjct: 1788 CSENSAEITPVLGKRKHILQIPFENLRVIPEAMPDILIVTKKGRSKKPEP 1837
>POL5_DROME (Q8I7P9) Retrovirus-related Pol polyprotein from
transposon opus [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1003
Score = 308 bits (789), Expect = 8e-83
Identities = 175/472 (37%), Positives = 274/472 (57%), Gaps = 24/472 (5%)
Query: 589 TPELLKAVLSDFPALFAAPTELPPQRRHDHAIHLKEGASIP-NIRPYRYPHYQKNEIEKL 647
T E+L ++L +FP +F P A ++ P + Y YP + E+E+
Sbjct: 84 TQEILNSLLGEFPRIFEPPLSGMSVETAVKA-EIRTNTQDPIYAKSYPYPVNMRGEVERQ 142
Query: 648 VAEMLNAGIIRPSISPYSSPVILVRKK-----DGSWRFCVDYRALNKITVPNKFPIPVID 702
+ E+L GIIRPS SPY+SP+ +V KK + +R VD++ LN +T+P+ +PIP I+
Sbjct: 143 IDELLQDGIIRPSNSPYNSPIWIVPKKPKPNGEKQYRMVVDFKRLNTVTIPDTYPIPDIN 202
Query: 703 ELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQ 762
L +G A+ F+ +DL SG+HQI MK D+ KTAF T G YEFL +PFGL NAP+ FQ
Sbjct: 203 ATLASLGNAKYFTTLDLTSGFHQIHMKESDIPKTAFSTLNGKYEFLRLPFGLKNAPAIFQ 262
Query: 763 SLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSVFGRG 822
+++++LR + K V+ DDI+++S + H +L VL + + +L++N +KS F
Sbjct: 263 RMIDDILREHIGKVCYVYIDDIIVFSEDYDTHWKNLRLVLASLSKANLQVNLEKSHFLDT 322
Query: 823 QIEYLGHVLSAGTVAADPKKLEAMWLWPVPKDLKSLRGFLGLTGYYRRFVKDYGKIARPL 882
Q+E+LG++++A + ADPKK+ A+ P P +K L+ FLG+T YYR+F++DY K+A+PL
Sbjct: 323 QVEFLGYIVTADGIKADPKKVRAISEMPPPTSVKELKRFLGMTSYYRKFIQDYAKVAKPL 382
Query: 883 TQLLK------------KDSFQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDAS 930
T L + K + Q+F LK L LA P F+K F L TDAS
Sbjct: 383 TNLTRGLYANIKSSQSSKVPITLDETALQSFNDLKSILCSSEILAFPCFTKPFHLTTDAS 442
Query: 931 GTGLGAVLTQE----GKPLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLGRHFI 986
+GAVL+Q+ +P+A+ S +L+ + + E+E++A++ ++ R YL G I
Sbjct: 443 NWAIGAVLSQDDQGRDRPIAYISRSLNKTEENYATIEKEMLAIIWSLDNLRAYLYGAGTI 502
Query: 987 -IRTDQKSLKFLTEQQVVGEGQFKWISKLSGYDFEIQYKPGKDNSAADAMSR 1037
+ TD + L F + +W +++ Y+ E+ YKPGK N ADA+SR
Sbjct: 503 KVYTDHQPLTFALGNRNFNAKLKRWKARIEEYNCELIYKPGKSNVVADALSR 554
Score = 55.5 bits (132), Expect = 1e-06
Identities = 89/365 (24%), Positives = 144/365 (39%), Gaps = 44/365 (12%)
Query: 1065 LQAIVHDLIVD----PEAH----KGYTLKDHRLFYKGKLV--LSKQSSRIPILCKEFHAS 1114
LQ+ + +I++ PEAH + L + L YK ++ L S +C+
Sbjct: 642 LQSCLRPVIINGVKIPEAHLQRFQSICLANF-LLYKIRITQRLVADVSGAEEICEIIEKE 700
Query: 1115 LLGGHSGFFRTYRRLAAVVYWEGMKSDIRDFVAECDTCQRNKFDNLSPAGLLQPLPVPTQ 1174
H G +L Y+ M S IR + C C+ K++ LQP P+P
Sbjct: 701 HRRAHRGPTEIRLQLLEKYYFPRMSSTIRLQTSSCQCCKLYKYERHPNKPNLQPTPIPNY 760
Query: 1175 VWTDVSMDFIGGLPKSNGKDTILVVVDRLTKYAHFIPLRHPFTAPEVAALFLHE--VVRL 1232
+ +D + K L +D+ +K+A L+ A++ L E V L
Sbjct: 761 PCEILHIDIF-----ALEKRLYLSCIDKFSKFAKLFHLQ------SKASVHLRETLVEAL 809
Query: 1233 HGF--PTTIVSDRDSLFLSSFWKELFRVSGTQLKFSSAYHPQTDGQTEVVNRC-LEVYLR 1289
H F P +VSD + L R L ++ + +GQ E + LE+Y R
Sbjct: 810 HYFTAPKVLVSDNERGLLCPTVLNYLRSLDIDLYYAPTQKSEVNGQVERFHSTFLEIY-R 868
Query: 1290 CLTGSRPR-KWVTCLPWAEFWFNSNYNRSAKMSPFQALYGREPPVLLQGTTIPSKIAAVN 1348
CL P K V + A +N++ + P + R V QG T
Sbjct: 869 CLKDELPTFKPVELVHIAVDRYNTSVHSVTNRKPADVFFDRSSRVNYQGLT--------- 919
Query: 1349 DLQVGRDELLSDLRANLLKSQDMMRAYANKKRRD--VDYQIGDEVFLKLQPYRRRSLAKK 1406
D R + L D++ L++ + + A K RD Y GDEVF+ + + + A+
Sbjct: 920 DF---RRQTLEDIK-GLIEYKQIRGNMARNKNRDEPKSYGPGDEVFVANKQIKTKEKARF 975
Query: 1407 MNEKL 1411
EK+
Sbjct: 976 RCEKV 980
>POLY_DROME (P10401) Retrovirus-related Pol polyprotein from
transposon gypsy [Contains: Reverse transcriptase (EC
2.7.7.49); Endonuclease]
Length = 1035
Score = 292 bits (748), Expect = 4e-78
Identities = 196/628 (31%), Positives = 333/628 (52%), Gaps = 54/628 (8%)
Query: 433 KLKGTAIVVLVDCGATHNFISQSLAREMELQITETPAYTVEVGDGHV-VKSKGVCKNLSV 491
+L G + +L+D A N+I +E++ + ++V G +K K + K
Sbjct: 18 RLAGRTLKMLIDTDAAKNYIRP--VKELKNVMPVASPFSVSSIHGSTEIKHKCLMKVFKH 75
Query: 492 QIQGFVVTQDFFLFGLRGVDVVLGLEWLAGLGDIKANFEELTLKFKLGNKKVVLKGEPEL 551
++ F L L D ++GL+ L G +K N E +L+++ +K+ P +
Sbjct: 76 ------ISPFFLLDSLNAFDAIIGLDLLTQAG-VKLNLAEDSLEYQGIAEKLHYFSCPSV 128
Query: 552 LKKRASMNALVKAIQQQGEGYVLQYQMLEKEVAKAEETPELLKAVLSDFPALFAAPTELP 611
+ + +++++ + +++ + K + E AV + + P
Sbjct: 129 NFTDVNDIVVPDSVKKEFKDTIIRRK---KAFSTTNEALPFNTAVTATIRTVDNEPV--- 182
Query: 612 PQRRHDHAIHLKEGASIPNIRPYRYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILV 671
+ A G S + NE+++L+ + GIIRPS SPY+SP +V
Sbjct: 183 ----YSRAYPTLMGVS----------DFVNNEVKQLLKD----GIIRPSRSPYNSPTWVV 224
Query: 672 RKK------DGSWRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQ 725
KK + + R +D+R LN+ T+P+++P+P I +L +G A+ F+ +DLKSGYHQ
Sbjct: 225 DKKGTDAFGNPNKRLVIDFRKLNEKTIPDRYPMPSIPMILANLGKAKFFTTLDLKSGYHQ 284
Query: 726 IMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDIL 785
I + D EKT+F + G YEF +PFGL NA S FQ +++VLR + K V+ DD++
Sbjct: 285 IYLAEHDREKTSFSVNGGKYEFCRLPFGLRNASSIFQRALDDVLREQIGKICYVYVDDVI 344
Query: 786 IYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEA 845
I+S +HV H+ VL+ + +++++ +K+ F + +EYLG ++S +DP+K++A
Sbjct: 345 IFSENESDHVRHIDTVLKCLIDANMRVSQEKTRFFKESVEYLGFIVSKDGTKSDPEKVKA 404
Query: 846 MWLWPVPKDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQLL------------KKDSFQW 893
+ +P P + +R FLGL YYR F+KD+ IARP+T +L KK ++
Sbjct: 405 IQEYPEPDCVYKVRSFLGLASYYRVFIKDFAAIARPITDILKGENGSVSKHMSKKIPVEF 464
Query: 894 GPSPQQAFEALKHTL-TEIPTLAVPDFSKVFVLETDASGTGLGAVLTQEGKPLAFWSATL 952
+ + AF+ L++ L +E L PDF K F L TDAS +G+GAVL+QEG+P+ S TL
Sbjct: 465 NETQRNAFQRLRNILASEDVILKYPDFKKPFDLTTDASASGIGAVLSQEGRPITMISRTL 524
Query: 953 SDRSQAKSVYERELMAVVRAVQRWRHYLLG-RHFIIRTDQKSLKFLTEQQVVGEGQFKWI 1011
Q + EREL+A+V A+ + +++L G R I TD + L F + +W
Sbjct: 525 KQPEQNYATNERELLAIVWALGKLQNFLYGSREINIFTDHQPLTFAVADRNTNAKIKRWK 584
Query: 1012 SKLSGYDFEIQYKPGKDNSAADAMSRRS 1039
S + ++ ++ YKPGK+N ADA+SR++
Sbjct: 585 SYIDQHNAKVFYKPGKENFVADALSRQN 612
Score = 55.5 bits (132), Expect = 1e-06
Identities = 73/353 (20%), Positives = 136/353 (37%), Gaps = 51/353 (14%)
Query: 1063 PKLQAIVHDLIVDPEAHKGYTLKDHRLFYKGKLVLSKQSSRIPILCKEFHASLLGGHSGF 1122
P L + HDLI AH + R L ++ ++ +I I+ E + + H
Sbjct: 704 PTLASFQHDLI----AH--FPATQFRHCKNVVLDITDKNEQIEIVTAEHNRA----HRAA 753
Query: 1123 FRTYRRLAAVVYWEGMKSDIRDFVAECDTCQRNKFDNLSPAGLLQPLPVPTQVWTDVSMD 1182
+++ Y+ M S ++ VA C C + K+D L P+P+ V +D
Sbjct: 754 QENIKQVLRDYYFPKMGSLAKEVVANCRVCTQAKYDRHPKKQELGETPIPSYTGEMVHID 813
Query: 1183 FIGGLPKSNGKDTILVVVDRLTKYAHFIPLRHPFTAPEVAALFLHEVVRLHGFPTTIVSD 1242
S + L +D+ +KYA P+ T ++ A L +++ L T+ D
Sbjct: 814 IF-----STDRKLFLTCIDKFSKYAIVQPVVSR-TIVDITAPLL-QIINLFPNIKTVYCD 866
Query: 1243 RDSLFLSSFWKELFRVS-GTQLKFSSAYHPQTDGQTEVVNRCLEVYLRCLT-GSRPRKWV 1300
+ F S + + S G + + H ++GQ E + L RCL + V
Sbjct: 867 NEPAFNSETVTSMLKNSFGIDIVNAPPLHSSSNGQVERFHSTLAEIARCLKLDKKTNDTV 926
Query: 1301 TCLPWAEFWFNSNYNRSAKMSPFQALYGREPPVLLQGTTIPSKIAAVNDLQVGRDELLSD 1360
+ A +N + + P + ++ G E +
Sbjct: 927 ELILRATIEYNKTVHSVTRERPIEVVHP------------------------GAHERCLE 962
Query: 1361 LRANLLKSQDMMRAYANKKRRDVDYQIGDEVFLKLQPYRRRSLAKKMNEKLSP 1413
++A L+K+Q N R++ +++G+ VF+K K++ KL+P
Sbjct: 963 IKARLVKAQQDSIGRNNPSRQNRVFEVGERVFVKNN--------KRLGNKLTP 1007
>POL4_DROME (P10394) Retrovirus-related Pol polyprotein from
transposon 412 [Contains: Protease (EC 3.4.23.-); Reverse
transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1237
Score = 280 bits (717), Expect = 2e-74
Identities = 178/549 (32%), Positives = 288/549 (52%), Gaps = 44/549 (8%)
Query: 590 PELLKAVL----SDFPALFAAPTE-LPPQRRHDHAIHLKEGASIPNIRPYRYPHYQKNEI 644
PEL K+ L S++ +FA +E + + + LK+ + + YR PH Q EI
Sbjct: 272 PELFKSQLENICSEYIDIFALESEPITVNNLYKQQLRLKDDEPVYT-KNYRSPHSQVEEI 330
Query: 645 EKLVAEMLNAGIIRPSISPYSSPVILVRKKDG------SWRFCVDYRALNKITVPNKFPI 698
+ V +++ I+ PS+S Y+SP++LV KK WR +DYR +NK + +KFP+
Sbjct: 331 QAQVQKLIKDKIVEPSVSQYNSPLLLVPKKSSPNSDKKKWRLVIDYRQINKKLLADKFPL 390
Query: 699 PVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAP 758
P ID++LD++G A+ FS +DL SG+HQI + + T+F T G Y F +PFGL AP
Sbjct: 391 PRIDDILDQLGRAKYFSCLDLMSGFHQIELDEGSRDITSFSTSNGSYRFTRLPFGLKIAP 450
Query: 759 STFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSV 818
++FQ +M + A ++ DD+++ + + + +LT+V +++LK++ +K
Sbjct: 451 NSFQRMMTIAFSGIEPSQAFLYMDDLIVIGCSEKHMLKNLTEVFGKCREYNLKLHPEKCS 510
Query: 819 FGRGQIEYLGHVLSAGTVAADPKKLEAMWLWPVPKDLKSLRGFLGLTGYYRRFVKDYGKI 878
F ++ +LGH + + D KK + + +PVP D S R F+ YYRRF+K++
Sbjct: 511 FFMHEVTFLGHKCTDKGILPDDKKYDVIQNYPVPHDADSARRFVAFCNYYRRFIKNFADY 570
Query: 879 ARPLTQLLKKD-SFQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDASGTGLGAV 937
+R +T+L KK+ F+W Q+AF LK L L PDFSK F + TDAS GAV
Sbjct: 571 SRHITRLCKKNVPFEWTDECQKAFIHLKSQLINPTLLQYPDFSKEFCITTDASKQACGAV 630
Query: 938 LTQEGK----PLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLGRHFIIRTDQKS 993
LTQ P+A+ S + KS E+EL A+ A+ +R Y+ G+HF ++TD +
Sbjct: 631 LTQNHNGHQLPVAYASRAFTKGESNKSTTEQELAAIHWAIIHFRPYIYGKHFTVKTDHRP 690
Query: 994 LKFLTEQQVVGEGQFKWISKLSGYDFEIQYKPGKDNSAADAMSR----RSSYCALSVLKV 1049
L +L + +L Y+F ++Y GKDN ADA+SR ++LKV
Sbjct: 691 LTYLFSMVNPSSKLTRIRLELEEYNFTVEYLKGKDNHVADALSRITIKELKDITGNILKV 750
Query: 1050 N----------------DFEEWQTEMLQDPKLQAIVHDLIVDPEAHKGYTLKDHR---LF 1090
D ++ E+ +P V+++I + E K TL+ + LF
Sbjct: 751 TTRFQSRQKSCAGKEQLDLQKQTKEIASEPN----VYEVITNDEVRKVVTLQLNDSICLF 806
Query: 1091 YKGKLVLSK 1099
GK ++++
Sbjct: 807 KHGKKIIAR 815
Score = 100 bits (248), Expect = 4e-20
Identities = 76/326 (23%), Positives = 147/326 (44%), Gaps = 7/326 (2%)
Query: 1097 LSKQSSRIPILCKEFHASLLGGHSGFFRTYRRLAAVVYWEGMKSDIRDFVAECDTCQRNK 1156
++ + + IL + GGH+G +T ++ YW+ M I+++V +C CQ+ K
Sbjct: 887 INNEKEKEAILSTLHDDPIQGGHTGITKTLAKVKRHYYWKNMSKYIKEYVRKCQKCQKAK 946
Query: 1157 FDNLSPAGLLQPLPVPTQVWTDVSMDFIGGLPKS-NGKDTILVVVDRLTKYAHFIPLRHP 1215
+ + P + V +D IG LPKS NG + + ++ LTKY IP+ +
Sbjct: 947 TTKHTKTPMTIT-ETPEHAFDRVVVDTIGPLPKSENGNEYAVTLICDLTKYLVAIPIANK 1005
Query: 1216 FTAPEVAALFLHEVVRLHGFPTTIVSDRDSLFLSSFWKELFRVSGTQLKFSSAYHPQTDG 1275
+A VA + +G T ++D + + +S +L + + S+A+H QT G
Sbjct: 1006 -SAKTVAKAIFESFILKYGPMKTFITDMGTEYKNSIITDLCKYLKIKNITSTAHHHQTVG 1064
Query: 1276 QTEVVNRCLEVYLRCLTGSRPRKWVTCLPWAEFWFNSNYNRSAKMSPFQALYGREPPVLL 1335
E +R L Y+R + W L + + FN+ + P++ ++GR +
Sbjct: 1065 VVERSHRTLNEYIRSYISTDKTDWDVWLQYFVYCFNTTQSMVHNYCPYELVFGRTSNLPK 1124
Query: 1336 QGTTIPSKIAAVNDLQVGRDELLSDLRANLLKSQDMMRAYANKKRRDVDYQIGDEVFLKL 1395
+ S I + ++ E L +++ ++ A+ K + + D ++ D ++L
Sbjct: 1125 HFNKLHS-IEPIYNIDDYAKESKYRLEVAYARARKLLEAHKEKNKENYDLKVKD---IEL 1180
Query: 1396 QPYRRRSLAKKMNEKLSPRYYGPYPI 1421
+ + L ++ KL +Y GPY I
Sbjct: 1181 EVGDKVLLRNEVGHKLDFKYTGPYKI 1206
>POL_CAMVC (P03555) Enzymatic polyprotein [Contains: Aspartic protease
(EC 3.4.23.-); Endonuclease; Reverse transcriptase (EC
2.7.7.49)]
Length = 679
Score = 234 bits (597), Expect = 1e-60
Identities = 189/674 (28%), Positives = 324/674 (48%), Gaps = 65/674 (9%)
Query: 416 LNSIQGISSRKSLKVWGKL-----KGTAIVVLVDCGATHNFISQSLAREMELQITETPAY 470
+ + +++ S+ + G+L K + VD GA+ S+ + E E P
Sbjct: 12 IEQVMNVTNPNSIYIKGRLYFKGYKKIELHCFVDTGASLCIASKFVIPEEHWVNAERPIM 71
Query: 471 TVEVGDGHVVKSKGVCKNLSVQIQGFVVTQDFFLFGLRGVDVVLGLEWLAGLGDIKANFE 530
V++ DG + VCK++ + I G + G+D ++G + L + F
Sbjct: 72 -VKIADGSSITISKVCKDIDLIIAGEIFKIPTVYQQESGIDFIIGNNF-CQLYEPFIQFT 129
Query: 531 ELTLKFKLGNK-------------KVVLKGEPELLKKRA----------SMNALVKAIQQ 567
+ + K NK +V ++G E +KKR+ S N + +++
Sbjct: 130 DRVIFTK--NKSYPVHITKLTRAVRVGIEGFLESMKKRSKTQQPEPVNISTNKIENPLEE 187
Query: 568 ---QGEGYVLQYQMLEKEVAKAEETPELLKAVLSDFPALFAAPTELPPQRRHD---HAIH 621
EG L + L + ++ ELL+ V S+ P L P + +I
Sbjct: 188 IAILSEGRRLSEEKLFITQQRMQKIEELLEKVCSENP--------LDPNKTKQWMKASIK 239
Query: 622 LKEGASIPNIRPYRYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILV----RKKDGS 677
L + + ++P +Y + E +K + E+L+ +I+PS SP+ +P LV K+ G
Sbjct: 240 LSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGK 299
Query: 678 WRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTA 737
R V+Y+A+NK T+ + + +P DELL I ++FS D KSG+ Q+++ E TA
Sbjct: 300 KRMVVNYKAMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTA 359
Query: 738 FRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVH 797
F +GHYE+ V+PFGL APS FQ M+E R V RK V+ DDIL++S E+H++H
Sbjct: 360 FTCPQGHYEWNVVPFGLKQAPSIFQRHMDEAFR-VFRKFCCVYVDDILVFSNNEEDHLLH 418
Query: 798 LTQVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWP-VPKDLK 856
+ +LQ +QH + ++ KK+ + +I +LG + GT LE + +P +D K
Sbjct: 419 VAMILQKCNQHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKK 478
Query: 857 SLRGFLGLTGYYRRFVKDYGKIARPLTQLLKKD-SFQWGPSPQQAFEALKHTLTEIPTLA 915
L+ FLG+ Y ++ +I +PL LK++ ++W + +K L P L
Sbjct: 479 QLQRFLGILTYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLH 538
Query: 916 VPDFSKVFVLETDAS----GTGLGAVLTQEG--KPLAFWSATLSDRSQAKSVY--ERELM 967
P + ++ETDAS G L A+ EG L A+ S ++ ++ + ++E +
Sbjct: 539 HPLPEEKLIIETDASDDYWGGMLKAIKINEGTNTELICRYASGSFKAAERNYHSNDKETL 598
Query: 968 AVVRAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQ----FKWISKLSGYDFEIQY 1023
AV+ ++++ YL HF+IRTD K G+ + +W + LS Y F++++
Sbjct: 599 AVINTIKKFSIYLTPVHFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEH 658
Query: 1024 KPGKDNSAADAMSR 1037
G DN AD +SR
Sbjct: 659 IKGTDNHFADFLSR 672
>POL_CAMVS (P03554) Enzymatic polyprotein [Contains: Aspartic protease
(EC 3.4.23.-); Endonuclease; Reverse transcriptase (EC
2.7.7.49)]
Length = 679
Score = 234 bits (596), Expect = 2e-60
Identities = 191/671 (28%), Positives = 322/671 (47%), Gaps = 65/671 (9%)
Query: 419 IQGISSRKSLKVWGKL-----KGTAIVVLVDCGATHNFISQSLAREMELQITETPAYTVE 473
+ +++ S+ + G+L K + VD GA+ S+ + E E P V+
Sbjct: 15 VMNVTNPNSIYIKGRLYFKGYKKIELHCFVDTGASLCIASKFVIPEEHWVNAERPIM-VK 73
Query: 474 VGDGHVVKSKGVCKNLSVQIQGFVVTQDFFLFGLRGVDVVLGLEWLAGLGDIKANFEELT 533
+ DG + VCK++ + I G + G+D ++G + L + F +
Sbjct: 74 IADGSSITISKVCKDIDLIIAGEIFRIPTVYQQESGIDFIIGNNFCQ-LYEPFIQFTDRV 132
Query: 534 LKFKLGNK-------------KVVLKGEPELLKKRA----------SMNALVKAIQQ--- 567
+ K NK +V +G E +KKR+ S N + +++
Sbjct: 133 IFTK--NKSYPVHIAKLTRAVRVGTEGFLESMKKRSKTQQPEPVNISTNKIENPLEEIAI 190
Query: 568 QGEGYVLQYQMLEKEVAKAEETPELLKAVLSDFPALFAAPTELPPQRRHDH---AIHLKE 624
EG L + L + ++ ELL+ V S+ P L P + +I L +
Sbjct: 191 LSEGRRLSEEKLFITQQRMQKIEELLEKVCSENP--------LDPNKTKQWMKASIKLSD 242
Query: 625 GASIPNIRPYRYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILVR----KKDGSWRF 680
+ ++P +Y + E +K + E+L+ +I+PS SP+ +P LV K+ G R
Sbjct: 243 PSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRM 302
Query: 681 CVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRT 740
V+Y+A+NK TV + + +P DELL I ++FS D KSG+ Q+++ E TAF
Sbjct: 303 VVNYKAMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTC 362
Query: 741 HEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQ 800
+GHYE+ V+PFGL APS FQ M+E R V RK V+ DDIL++S E+H++H+
Sbjct: 363 PQGHYEWNVVPFGLKQAPSIFQRHMDEAFR-VFRKFCCVYVDDILVFSNNEEDHLLHVAM 421
Query: 801 VLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWP-VPKDLKSLR 859
+LQ +QH + ++ KK+ + +I +LG + GT LE + +P +D K L+
Sbjct: 422 ILQKCNQHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQ 481
Query: 860 GFLGLTGYYRRFVKDYGKIARPLTQLLKKD-SFQWGPSPQQAFEALKHTLTEIPTLAVPD 918
FLG+ Y ++ +I +PL LK++ ++W + +K L P L P
Sbjct: 482 RFLGILTYASDYIPKLAQIRKPLQAKLKENVPWRWTKEDTLYMQKVKKNLQGFPPLHHPL 541
Query: 919 FSKVFVLETDAS----GTGLGAVLTQEG--KPLAFWSATLSDRSQAKSVY--ERELMAVV 970
+ ++ETDAS G L A+ EG L A+ S ++ K+ + ++E +AV+
Sbjct: 542 PEEKLIIETDASDDYWGGMLKAIKINEGTNTELICRYASGSFKAAEKNYHSNDKETLAVI 601
Query: 971 RAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQ----FKWISKLSGYDFEIQYKPG 1026
++++ YL HF+IRTD K G+ + +W + LS Y F++++ G
Sbjct: 602 NTIKKFSIYLTPVHFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKG 661
Query: 1027 KDNSAADAMSR 1037
DN AD +SR
Sbjct: 662 TDNHFADFLSR 672
>POL_CAMVD (P03556) Enzymatic polyprotein [Contains: Aspartic protease
(EC 3.4.23.-); Endonuclease; Reverse transcriptase (EC
2.7.7.49)]
Length = 674
Score = 233 bits (594), Expect = 3e-60
Identities = 189/661 (28%), Positives = 315/661 (47%), Gaps = 52/661 (7%)
Query: 419 IQGISSRKSLKVWGKL-----KGTAIVVLVDCGATHNFISQSLAREMELQITETPAYTVE 473
+ I++ S+ + G+L K + VD GA+ S+ + E E P V+
Sbjct: 17 VMNITNPNSIYIKGRLYFKGYKKIELHCFVDTGASLCIASKFVIPEEHWINAERPIM-VK 75
Query: 474 VGDGHVVKSKGVCKNLSVQIQGFVVTQDFFLFGLRGVDVVLG------LEWLAGLGDIKA 527
+ DG + VC+++ + I G + G+D ++G E D
Sbjct: 76 IADGSSITINKVCRDIDLIIAGEIFHIPTVYQQESGIDFIIGNNFCQLYEPFIQFTDRVI 135
Query: 528 NFEELTLKFKLGNK----KVVLKGEPELLKKRAS------MNALVKAIQQQGEGYVLQYQ 577
++ T + +V +G E +KKR+ +N I EG L +
Sbjct: 136 FTKDRTYPVHIAKLTRAVRVGTEGFLESMKKRSKTQQPEPVNISTNKIAILSEGRRLSEE 195
Query: 578 MLEKEVAKAEETPELLKAVLSDFPALFAAPTELPPQRRHDH---AIHLKEGASIPNIRPY 634
L + ++ ELL+ V S+ P L P + +I L + + ++P
Sbjct: 196 KLFITQQRMQKIEELLEKVCSENP--------LDPNKTKQWMKASIKLSDPSKAIKVKPM 247
Query: 635 RYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILVR----KKDGSWRFCVDYRALNKI 690
+Y + E +K + E+L+ +I+PS SP+ +P LV K+ G R V+Y+A+NK
Sbjct: 248 KYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKA 307
Query: 691 TVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVM 750
TV + + P DELL I ++FS D KSG+ Q+++ E TAF +GHYE+ V+
Sbjct: 308 TVGDAYNPPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVV 367
Query: 751 PFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHDL 810
PFGL APS FQ M+E R V RK V+ DDIL++S E+H++H+ +LQ +QH +
Sbjct: 368 PFGLKQAPSIFQRHMDEAFR-VFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGI 426
Query: 811 KINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWP-VPKDLKSLRGFLGLTGYYR 869
++ KK+ + +I +LG + GT LE + +P +D K L+ FLG+ Y
Sbjct: 427 ILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYAS 486
Query: 870 RFVKDYGKIARPLTQLLKKD-SFQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETD 928
++ +I +PL LK++ ++W + +K L P L P + ++ETD
Sbjct: 487 DYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETD 546
Query: 929 AS----GTGLGAVLTQEG--KPLAFWSATLSDRSQAKSVY--ERELMAVVRAVQRWRHYL 980
AS G L A+ EG L A+ S ++ K+ + ++E +AV+ ++++ YL
Sbjct: 547 ASDDYWGGMLKAIKINEGTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYL 606
Query: 981 LGRHFIIRTDQKSLKFLTEQQVVGEGQ----FKWISKLSGYDFEIQYKPGKDNSAADAMS 1036
HF+IRTD K G+ + +W + LS Y F++++ G DN AD +S
Sbjct: 607 TPVHFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLS 666
Query: 1037 R 1037
R
Sbjct: 667 R 667
>POL_CAMVN (Q00962) Enzymatic polyprotein [Contains: Aspartic protease
(EC 3.4.23.-); Endonuclease; Reverse transcriptase (EC
2.7.7.49)]
Length = 680
Score = 231 bits (589), Expect = 1e-59
Identities = 186/671 (27%), Positives = 319/671 (46%), Gaps = 65/671 (9%)
Query: 419 IQGISSRKSLKVWGKL-----KGTAIVVLVDCGATHNFISQSLAREMELQITETPAYTVE 473
+ +++ S+ + G+L K + VD GA+ S+ + E E P V+
Sbjct: 16 VMNVTNPNSIYIKGRLYFKGYKKIELHCFVDTGASLCIASKFVIPEEHWVNAERPIM-VK 74
Query: 474 VGDGHVVKSKGVCKNLSVQIQGFVVTQDFFLFGLRGVDVVLGLEWLAGLGDIKANFEELT 533
+ DG + VCK++ + I G + G+D ++G + L + F +
Sbjct: 75 IADGSSITISKVCKDIDLIIVGVIFKIPTVYQQESGIDFIIGNNFCQ-LYEPFIQFTDRV 133
Query: 534 LKFKLGNK-------------KVVLKGEPELLKKRA----------SMNALVKAIQQ--- 567
+ K NK +V +G E +KKR+ S N + +++
Sbjct: 134 IFTK--NKSYPVHIAKLTRAVRVGTEGFLESMKKRSKTQQPEPVNISTNKIENPLEEIAI 191
Query: 568 QGEGYVLQYQMLEKEVAKAEETPELLKAVLSDFPALFAAPTELPPQRRHDH---AIHLKE 624
EG L + L + ++T ELL+ V S+ P L P + +I L +
Sbjct: 192 LSEGRRLSEEKLFITQQRMQKTEELLEKVCSENP--------LDPNKTKQWMKASIKLSD 243
Query: 625 GASIPNIRPYRYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILVRKKD----GSWRF 680
+ ++P +Y + E +K + E+L+ +I+PS SP+ +P LV + G+ R
Sbjct: 244 PSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAENGRGNKRM 303
Query: 681 CVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRT 740
V+Y+A+NK TV + + +P DELL I ++FS D KSG+ Q+++ E TAF
Sbjct: 304 VVNYKAMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTC 363
Query: 741 HEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQ 800
+GHYE+ V+PFGL APS FQ M+E R V RK V+ DDI+++S E+H++H+
Sbjct: 364 PQGHYEWNVVPFGLKQAPSIFQRHMDEAFR-VFRKFCCVYVDDIVVFSNNEEDHLLHVAM 422
Query: 801 VLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWP-VPKDLKSLR 859
+LQ +QH + ++ KK+ + +I +LG + GT LE + +P +D K L+
Sbjct: 423 ILQKCNQHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQ 482
Query: 860 GFLGLTGYYRRFVKDYGKIARPLTQLLKKD-SFQWGPSPQQAFEALKHTLTEIPTLAVPD 918
FLG+ Y ++ + ++ +PL LK++ ++W + +K L P L P
Sbjct: 483 RFLGILTYASDYIPNLAQMRQPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPL 542
Query: 919 FSKVFVLETDAS----GTGLGAVLTQEGKP----LAFWSATLSDRSQAKSVYERELMAVV 970
+ ++ETDAS G L A+ EG + S + + ++E +AV+
Sbjct: 543 PEEKLIIETDASDDYWGGMLKAIKINEGTNTELICRYRSGSFKAAERNYHSNDKETLAVI 602
Query: 971 RAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQ----FKWISKLSGYDFEIQYKPG 1026
++++ YL HF+IRTD K G+ + +W + LS Y F++++ G
Sbjct: 603 NTIKKFSIYLTPVHFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKG 662
Query: 1027 KDNSAADAMSR 1037
DN AD +SR
Sbjct: 663 TDNHFADFLSR 673
>POL_CAMVE (Q02964) Enzymatic polyprotein [Contains: Aspartic protease
(EC 3.4.23.-); Endonuclease; Reverse transcriptase (EC
2.7.7.49)]
Length = 679
Score = 230 bits (586), Expect = 3e-59
Identities = 190/671 (28%), Positives = 321/671 (47%), Gaps = 65/671 (9%)
Query: 419 IQGISSRKSLKVWGKL-----KGTAIVVLVDCGATHNFISQSLAREMELQITETPAYTVE 473
+ +++ S+ + G+L K + VD GA+ S+ + E E P V+
Sbjct: 15 VMNVTNPNSIYIKGRLYFKGYKKIELHCFVDTGASLCIASKFVIPEEHWVNAERPIM-VK 73
Query: 474 VGDGHVVKSKGVCKNLSVQIQGFVVTQDFFLFGLRGVDVVLGLEWLAGLGDIKANFEELT 533
+ DG + VCK++ + I + G+D ++G + L + F +
Sbjct: 74 IADGSSITISKVCKDIDLIIAREIFKIPTVYQQESGIDFIIGNNFCQ-LYEPFIQFTDRV 132
Query: 534 LKFKLGNK-------------KVVLKGEPELLKKRA----------SMNAL---VKAIQQ 567
+ K NK +V +G E +KKR+ S N + +K I
Sbjct: 133 IFTK--NKSYPVHIAKLTRAVRVGTEGFLESMKKRSKTQQPEPVNISTNKIENPLKEIAI 190
Query: 568 QGEGYVLQYQMLEKEVAKAEETPELLKAVLSDFPALFAAPTELPPQRRHDH---AIHLKE 624
EG L + L + ++ ELL+ V S+ P L P + +I L +
Sbjct: 191 LSEGRRLSEEKLFITQQRMQKIEELLEKVCSENP--------LDPNKTKQWMKASIKLSD 242
Query: 625 GASIPNIRPYRYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILVR----KKDGSWRF 680
+ ++P +Y + E +K + E+L+ +I+PS SP+ +P LV K+ G R
Sbjct: 243 PSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRM 302
Query: 681 CVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRT 740
V+Y+A+NK T+ + + +P DELL I ++FS D KSG+ Q+++ E TAF
Sbjct: 303 VVNYKAMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTC 362
Query: 741 HEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQ 800
+GHYE+ V+PFGL APS FQ M+E R V RK V+ DDIL++S E+H++H+
Sbjct: 363 PQGHYEWNVVPFGLKQAPSIFQRHMDEAFR-VFRKFCCVYVDDILVFSNNEEDHLLHVAM 421
Query: 801 VLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWP-VPKDLKSLR 859
+LQ +QH + ++ KK+ + +I +LG + GT LE + +P +D K L+
Sbjct: 422 ILQKCNQHGIILSKKKAQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQ 481
Query: 860 GFLGLTGYYRRFVKDYGKIARPLTQLLKKD-SFQWGPSPQQAFEALKHTLTEIPTLAVPD 918
FLG+ Y ++ +I +PL LK++ ++W + +K L P L P
Sbjct: 482 RFLGILTYASDYIPKLAQIRKPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPL 541
Query: 919 FSKVFVLETDAS----GTGLGAVLTQEG--KPLAFWSATLSDRSQAKSVY--ERELMAVV 970
+ ++ETDAS G L A+ EG L A+ S ++ ++ + ++E +AV+
Sbjct: 542 PEEKLIIETDASDDYWGGMLKAIKINEGTNTELICRYASGSFKAAERNYHSNDKETLAVI 601
Query: 971 RAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQ----FKWISKLSGYDFEIQYKPG 1026
++++ YL HF+IRTD K G+ + +W + LS Y F++++ G
Sbjct: 602 NTIKKFSIYLTPVHFLIRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKG 661
Query: 1027 KDNSAADAMSR 1037
DN AD +SR
Sbjct: 662 TDNHFADFLSR 672
>POL_FMVD (P09523) Enzymatic polyprotein [Contains: Aspartic protease
(EC 3.4.23.-); Endonuclease; Reverse transcriptase (EC
2.7.7.49)]
Length = 666
Score = 210 bits (534), Expect = 3e-53
Identities = 175/669 (26%), Positives = 313/669 (46%), Gaps = 58/669 (8%)
Query: 410 ELKHLSLNSIQ----GISSRKSLKVWGKL-----KGTAIVVLVDCGATHNFISQSLAREM 460
EL H LN + +++ S+ + GKL K I VD GA+ S+ + E
Sbjct: 11 ELGHFCLNKQEMLHLNVTNPNSIYIEGKLSFEGYKSFNIHCYVDTGASLCIASRYIIPE- 69
Query: 461 ELQITETPAYTVEVGDGHVVKSKGVCKNLSVQIQGFVVTQDFFLFGLRGVDVVLG----- 515
EL V++ + ++K VCKNL V+ G G+D ++G
Sbjct: 70 ELWENSPKDIQVKIANQELIKITKVCKNLKVKFAGKSFEIPTVYQQETGIDFLIGNNFCR 129
Query: 516 -----LEWLAGLGDIKAN----FEELTLKFKLGNKKVVLKGEPELLKKRASMNALVKAIQ 566
++W + N +++T F + N + + + ++ + K I
Sbjct: 130 LYNPFIQWEDRIAFHLKNEMVLIKKVTKAFSVSNPSFLENMKKDSKTEQIPGTNISKNII 189
Query: 567 QQGEGYVL---QYQMLEKEVAKAEETPELLKAVLSDFPALFAAPTELPPQRRHDHAIHLK 623
E Y L +YQ +E +LL V S+ P + ++ +I L
Sbjct: 190 NPEERYFLITEKYQKIE----------QLLDKVCSENPI-----DPIKSKQWMKASIKLI 234
Query: 624 EGASIPNIRPYRYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILVR----KKDGSWR 679
+ + ++P Y + K + E+L+ G+I PS S + SP LV ++ G R
Sbjct: 235 DPLKVIRVKPMSYSPQDREGFAKQIKELLDLGLIIPSKSQHMSPAFLVENEAERRRGKKR 294
Query: 680 FCVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFR 739
V+Y+A+N+ T+ + +P + ELL + +FS D KSG+ Q+++ E + TAF
Sbjct: 295 MVVNYKAINQATIGDSHNLPNMQELLTLLRGKSIFSSFDCKSGFWQVVLDEESQKLTAFT 354
Query: 740 THEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLT 799
+GH+++ V+PFGL APS FQ M L + C +V+ DDI+++S + +H H+
Sbjct: 355 CPQGHFQWKVVPFGLKQAPSIFQRHMQTALNGADKFC-MVYVDDIIVFSNSELDHYNHVY 413
Query: 800 QVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWPVP-KDLKSL 858
VL++++++ + ++ KK+ + +I +LG + GT LE + +P +D K L
Sbjct: 414 AVLKIVEKYGIILSKKKANLFKEKINFLGLEIDKGTHCPQNHILENIHKFPDRLEDKKHL 473
Query: 859 RGFLGLTGYYRRFVKDYGKIARPLTQLLKKD-SFQWGPSPQQAFEALKHTLTEIPTLAVP 917
+ FLG+ Y ++ +I +PL LKKD ++ W S + +K L P L +P
Sbjct: 474 QRFLGVLTYAETYIPKLAEIRKPLQVKLKKDVTWNWTQSDSDYVKKIKKNLGSFPKLYLP 533
Query: 918 DFSKVFVLETDASGTGLGAVL---TQEGKPLAFWSATLSDRSQAKSVY--ERELMAVVRA 972
++ETDAS + G VL +G L ++ S + K+ + ++EL+AV +
Sbjct: 534 KPEDHLIIETDASDSFWGGVLKARALDGVELICRYSSGSFKQAEKNYHSNDKELLAVKQV 593
Query: 973 VQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQ----FKWISKLSGYDFEIQYKPGKD 1028
+ ++ YL F +RTD K+ + + G+ + +W + S Y F++++ G
Sbjct: 594 ITKFSAYLTPVRFTVRTDNKNFTYFLRINLKGDSKQGRLVRWQNWFSKYQFDVEHLEGVK 653
Query: 1029 NSAADAMSR 1037
N AD ++R
Sbjct: 654 NVLADCLTR 662
>POL_CERV (P05400) Enzymatic polyprotein [Contains: Aspartic protease
(EC 3.4.23.-); Endonuclease; Reverse transcriptase (EC
2.7.7.49)]
Length = 659
Score = 209 bits (532), Expect = 5e-53
Identities = 166/636 (26%), Positives = 297/636 (46%), Gaps = 53/636 (8%)
Query: 443 VDCGATHNFISQSLAREMELQITETPAYTVEVGDGHVVKSKGVCKNLSVQIQGFVVTQDF 502
VD G++ S+ + E Q E P +++ +G +++ VC L +++ G
Sbjct: 33 VDTGSSLCMASKYVIPEEYWQTAEKPL-NIKIANGKIIQLTKVCSKLPIRLGGERFLIPT 91
Query: 503 FLFGLRGVDVVLGLEWLAGLGDIKANFEELT--LKFKLGNKKVVL-----------KGEP 549
G+D++LG + + + F + T + F L + V++ KG
Sbjct: 92 LFQQESGIDLLLGNNFC----QLYSPFIQYTDRIYFHLNKQSVIIGKITKAYQYGVKGFL 147
Query: 550 ELLKKRASMN--------ALVKAIQQQGEGYVLQYQMLEKEVAKAEETPELLKAVLSDFP 601
E +KK++ +N + ++G +V + + E +++K E+L+ V S+ P
Sbjct: 148 ESMKKKSKVNRPEPINITSNQHLFLEEGGNHVDE-MLYEIQISKFSAIEEMLERVSSENP 206
Query: 602 ALFAAPTELPPQRRHDH---AIHLKEGASIPNIRPYRYPHYQKNEIEKLVAEMLNAGIIR 658
+ P++ I L + ++ ++P Y + E ++ + E+L +I+
Sbjct: 207 --------IDPEKSKQWMTATIELIDPKTVVKVKPMSYSPSDREEFDRQIKELLELKVIK 258
Query: 659 PSISPYSSPVILVR----KKDGSWRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVF 714
PS S + SP LV ++ G R V+Y+A+NK T + +P DELL + +++
Sbjct: 259 PSKSTHMSPAFLVENEAERRRGKKRMVVNYKAMNKATKGDAHNLPNKDELLTLVRGKKIY 318
Query: 715 SKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLR 774
S D KSG Q+++ E TAF +GHY++ V+PFGL APS F
Sbjct: 319 SSFDCKSGLWQVLLDKESQLLTAFTCPQGHYQWNVVPFGLKQAPSIFPKTYANSHSNQYS 378
Query: 775 KCALVFFDDILIYSLT-MEEHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIEYLGHVLSA 833
K V+ DDIL++S T +EH +H+ +L+ ++ + ++ KK+ + +I +LG +
Sbjct: 379 KYCCVYVDDILVFSNTGRKEHYIHVLNILRRCEKLGIILSKKKAQLFKEKINFLGLEIDQ 438
Query: 834 GTVAADPKKLEAMWLWPVP-KDLKSLRGFLGLTGYYRRFVKDYGKIARPLTQLLKKDS-F 891
GT LE + +P +D K L+ FLG+ Y ++ I +PL LK+DS +
Sbjct: 439 GTHCPQNHILEHIHKFPDRIEDKKQLQRFLGILTYASDYIPKLASIRKPLQSKLKEDSTW 498
Query: 892 QWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDAS----GTGLGAVLTQEGKPLAF 947
W + Q +K L P L P+ + V+ETDAS G L A+ +
Sbjct: 499 TWNDTDSQYMAKIKKNLKSFPKLYHPEPNDKLVIETDASEEFWGGILKAIHNSHEYICRY 558
Query: 948 WSATLSDRSQAKSVYERELMAVVRAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQ 1007
S + + E+EL+AV+R ++++ YL F+IRTD K+ + G+ +
Sbjct: 559 ASGSFKAAERNYHSNEKELLAVIRVIKKFSIYLTPSRFLIRTDNKNFTHFVNINLKGDRK 618
Query: 1008 ----FKWISKLSGYDFEIQYKPGKDNSAADAMSRRS 1039
+W LS YDF++++ G N AD + +
Sbjct: 619 QGRLVRWQMWLSQYDFDVEHIAGTKNVFADFLQENT 654
>POL_COYMV (P19199) Putative polyprotein [Contains: Coat protein;
Protease (EC 3.4.23.-); Reverse transcriptase (EC
2.7.7.49); Ribonuclease H (EC 3.1.26.4)]
Length = 1886
Score = 179 bits (454), Expect = 5e-44
Identities = 122/421 (28%), Positives = 212/421 (49%), Gaps = 34/421 (8%)
Query: 651 MLNAGIIRPSISPYSSPVILVR-----------KKDGSWRFCVDYRALNKITVPNKFPIP 699
+L +IRPS S + S +VR +K G R +Y+ LN+ T +++ +P
Sbjct: 1425 LLQMKVIRPSESKHRSTAFIVRSGTEIDPITGKEKKGKERMVFNYKLLNENTESDQYSLP 1484
Query: 700 VIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLVMPFGLTNAPS 759
I+ ++ ++G ++++SK DLKSG+ Q+ M+ E V TAF YE+LVMPFGL NAP+
Sbjct: 1485 GINTIISKVGRSKIYSKFDLKSGFWQVAMEEESVPWTAFLAGNKLYEWLVMPFGLKNAPA 1544
Query: 760 TFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHDLKINGKKSVF 819
FQ M+ V + K V+ DDIL++S T E+H HL +LQL ++ L ++ K
Sbjct: 1545 IFQRKMDNVFKGT-EKFIAVYIDDILVFSETAEQHSQHLYTMLQLCKENGLILSPTKMKI 1603
Query: 820 GRGQIEYLGHVLSAGTVAADPKKLEAMWLWPVPK--DLKSLRGFLGLTGYYRRFVKDYGK 877
G +I++LG L + P + + + K + +R +LG+ Y R +++D GK
Sbjct: 1604 GTPEIDFLGASLGCTKIKLQPHIISKICDFSDEKLATPEGMRSWLGILSYARNYIQDIGK 1663
Query: 878 IARPLTQ-LLKKDSFQWGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLETDASGTGLGA 936
+ +PL Q + + P + +K + +P L +P ++ETD TG GA
Sbjct: 1664 LVQPLRQKMAPTGDKRMNPETWKMVRQIKEKVKNLPDLQLPPKDSFIIIETDGCMTGWGA 1723
Query: 937 VL---------TQEGKPLAFWSATLSDRSQAKSVYERELMAVVRAVQRWR-HYLLGRHFI 986
V + A+ S + + KS + E+ A + + +++ +YL + I
Sbjct: 1724 VCKWKMSKHDPRSTERICAYASGSF---NPIKSTIDAEIQAAIHGLDKFKIYYLDKKELI 1780
Query: 987 IRTD-QKSLKFLTEQQVVGEGQFKWIS---KLSGYDFEIQYK--PGKDNSAADAMSRRSS 1040
IR+D + +KF + + +W++ L+G + ++ GK N ADA+SR +
Sbjct: 1781 IRSDCEAIIKFYNKTNENKPSRVRWLTFSDFLTGLGITVTFEHIDGKHNGLADALSRMIN 1840
Query: 1041 Y 1041
+
Sbjct: 1841 F 1841
>M860_ARATH (P92523) Hypothetical mitochondrial protein AtMg00860
(ORF158)
Length = 158
Score = 147 bits (371), Expect = 2e-34
Identities = 75/130 (57%), Positives = 90/130 (68%), Gaps = 2/130 (1%)
Query: 797 HLTQVLQLMDQHDLKINGKKSVFGRGQIEYLGH--VLSAGTVAADPKKLEAMWLWPVPKD 854
HL VLQ+ +QH N KK FG+ QI YLGH ++S V+ADP KLEAM WP PK+
Sbjct: 3 HLGMVLQIWEQHQFYANRKKCAFGQPQIAYLGHRHIISGEGVSADPAKLEAMVGWPEPKN 62
Query: 855 LKSLRGFLGLTGYYRRFVKDYGKIARPLTQLLKKDSFQWGPSPQQAFEALKHTLTEIPTL 914
LRGFLGLTGYYRRFVK+YGKI RPLT+LLKK+S +W AF+ALK +T +P L
Sbjct: 63 TTELRGFLGLTGYYRRFVKNYGKIVRPLTELLKKNSLKWTEMAALAFKALKGAVTTLPVL 122
Query: 915 AVPDFSKVFV 924
A+PD FV
Sbjct: 123 ALPDLKLPFV 132
>POL_RTBVP (P27502) Polyprotein (P194 protein) [Contains: Coat
protein; Protease (EC 3.4.23.-); Reverse transcriptase
(EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4)]
Length = 1675
Score = 140 bits (354), Expect = 2e-32
Identities = 113/429 (26%), Positives = 201/429 (46%), Gaps = 26/429 (6%)
Query: 636 YPHYQKNEIEKLVAEMLNAGIIRPS--ISPYSSPVILVRKKDGSW----RFCVDYRALNK 689
Y K EK + E+L+ +I+ + + + +VR R +Y+ LN
Sbjct: 1191 YTPADKEVFEKQIKELLDNKLIKKADPTCRHRTAAFIVRNHSEEVAQKPRIVYNYKRLND 1250
Query: 690 ITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVEKTAFRTHEGHYEFLV 749
+ F IP +++ I A +FSK DLK+G+H + +K + + T F EG Y + V
Sbjct: 1251 NMHTDPFNIPHKISMINLIQKANIFSKFDLKAGFHHMKLKDDFKDWTTFTCSEGLYTWNV 1310
Query: 750 MPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSLTMEEHVVHLTQVLQLMDQHD 809
PFG+ NAP FQ M E + K AL++ DDILI S +EH+ HL + +
Sbjct: 1311 CPFGIANAPCAFQRFMQESFGDL--KFALLYIDDILIASNNEKEHIEHLKIFFNRVKEVG 1368
Query: 810 LKINGKKSVFGRGQIEYLGHVLSAGTVAADPKKLEAMWLWPVPK--DLKSLRGFLGLTGY 867
++ KKS ++EYLG + G ++ P ++ + + K LK L+ +LGL Y
Sbjct: 1369 CVLSKKKSKMFLKEVEYLGVEIKEGKISLQPHIVDKIKKFDKNKLNTLKGLQAYLGLLNY 1428
Query: 868 YRRFVKDYGKIARPLTQLLKKDSFQ-WGPSPQQAFEALKHTLTEIPTLAVPDFSKVFVLE 926
R ++KD K+ PL + K+ + + ++ +++I L P + ++E
Sbjct: 1429 ARGYIKDLSKLVGPLYKKTGKNGQRIFNKEDWNIIFKIEREVSKIKPLERPKETDYIIIE 1488
Query: 927 TDASGTGLGAVLT--------QEGKPLAFWSATLSDRSQAKSVYERELMAVVRAVQRWRH 978
TDAS G GAVL ++ + +A +++ + + + E+ A+ A+ +++
Sbjct: 1489 TDASEEGWGAVLVCKPDKYSGKDTEKIAGYASGNFGEKKTWTSLDYEIEAINEALNKFQI 1548
Query: 979 YLLGRHFIIRTDQKSL-KFLTEQQVVGEGQFKWIS-----KLSGYDFEIQYKPGKDNSAA 1032
Y L + F IRTD +++ K + + + +WI GY ++ G N
Sbjct: 1549 Y-LDKDFTIRTDCEAIVKGIKTEDYKKRSKTRWIKLRDNLLKDGYKPTFEHIKGNKNFLP 1607
Query: 1033 DAMSRRSSY 1041
+ +SR +
Sbjct: 1608 NFLSREGDF 1616
>POL_SOCMV (P15629) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 692
Score = 130 bits (327), Expect = 3e-29
Identities = 127/503 (25%), Positives = 231/503 (45%), Gaps = 37/503 (7%)
Query: 443 VDCGATHNFISQSLAREMELQITETPAYTVEVGDGHVVKSKGVCKNLSVQIQGFVVTQDF 502
+D GAT F + ++ E+ + + + H ++ N+ ++I+
Sbjct: 35 IDTGATLCFGKRKISNNWEI-LKQPKEIIIADKSKHYIRE--AISNVFLKIENKEFLIPI 91
Query: 503 FLFGLRGVDVVLGLEWLAGLGDIKANFEELTLKFK-LGNKKVVLKGEPELLKKRASMNAL 561
G+D+++G +L E + L++K L N K ++L K +
Sbjct: 92 IYLHDSGLDLIIGNNFLKLYQPFIQRLETIELRWKNLNNPKESQMISTKILTKNEVLKLS 151
Query: 562 VKAIQQQGEGYVLQYQMLEKEVAKAEETPELLKAVLSDFPALFAAPTELPPQRRHDHAIH 621
+ I E Y+ ++ +E++ L+ V S+ P E + I
Sbjct: 152 FEKIHICLEKYLF-FKTIEEQ----------LEEVCSEHPL-----DETKNKNGLLIEIR 195
Query: 622 LKEGA---SIPNIRPYRYPHYQKNEIEKLVAEMLNAGIIRPSISPYSSPVILVRK----K 674
LK+ ++ N PY Q E ++ ++L G+IR S SP+S+P V K
Sbjct: 196 LKDPLQEINVTNRIPYTIRDVQ--EFKEECEDLLKKGLIRESQSPHSAPAFYVENHNEIK 253
Query: 675 DGSWRFCVDYRALNKITVPNKFPIPVIDELLDEIGPARVFSKIDLKSGYHQIMMKAEDVE 734
G R ++Y+ +N+ T+ + + +P D +L++I + FS +D KSGY+Q+ +
Sbjct: 254 RGKRRMVINYKKMNEATIGDSYKLPRKDFILEKIKGSLWFSSLDAKSGYYQLRLHENTKP 313
Query: 735 KTAFRTH-EGHYEFLVMPFGLTNAPSTFQSLMNEVLRPVLRKCALVFFDDILIYSL-TME 792
TAF + HYE+ V+ FGL APS +Q M++ L+ + C L + DDILI++ + E
Sbjct: 314 LTAFSCPPQKHYEWNVLSFGLKQAPSIYQRFMDQSLKGLEHIC-LAYIDDILIFTKGSKE 372
Query: 793 EHVVHLTQVLQLMDQHDLKINGKKSVFGRGQIEYLG-HVLSAGTVAADPKKLEAMWLWPV 851
+HV + VLQ + + + I+ KKS + +IEYLG + G + P E + +P
Sbjct: 373 QHVNDVRIVLQRIKEKGIIISKKKSKLIQQEIEYLGLKIQGNGEIDLSPHTQEKILQFPD 432
Query: 852 P-KDLKSLRGFLGLTGYYRR--FVKDYGKIARPLTQLLK-KDSFQWGPSPQQAFEALKHT 907
+D K ++ FLG Y F K+ + L + + K+ ++W + +++K
Sbjct: 433 ELEDRKQIQRFLGCINYIANEGFFKNLALERKHLQKKISVKNPWKWDTIDTKMVQSIKGK 492
Query: 908 LTEIPTLAVPDFSKVFVLETDAS 930
+ +P L ++ETDAS
Sbjct: 493 IQSLPKLYNASIQDFLIVETDAS 515
Score = 41.2 bits (95), Expect = 0.022
Identities = 24/96 (25%), Positives = 45/96 (46%), Gaps = 4/96 (4%)
Query: 949 SATLSDRSQAKSVYERELMAVVRAVQRWRHYLLGRHFIIRTDQKSLKFLTEQQVVGEGQ- 1007
S T +D + E E++A V+ +++WR LL F++RTD K + + +
Sbjct: 592 SGTFTDTETRYPIAELEVLAGVKVLEKWRIDLLQTRFLLRTDSKYFAGFCRYNIKTDYRN 651
Query: 1008 ---FKWISKLSGYDFEIQYKPGKDNSAADAMSRRSS 1040
+W +L Y ++ ++N AD ++R S
Sbjct: 652 GRLIRWQLRLQAYQPYVELIKSENNPFADTLTREWS 687
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 182,063,552
Number of Sequences: 164201
Number of extensions: 8035118
Number of successful extensions: 27714
Number of sequences better than 10.0: 329
Number of HSP's better than 10.0 without gapping: 130
Number of HSP's successfully gapped in prelim test: 206
Number of HSP's that attempted gapping in prelim test: 25079
Number of HSP's gapped (non-prelim): 1301
length of query: 1553
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1430
effective length of database: 39,777,331
effective search space: 56881583330
effective search space used: 56881583330
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0318.12


