

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0316.4
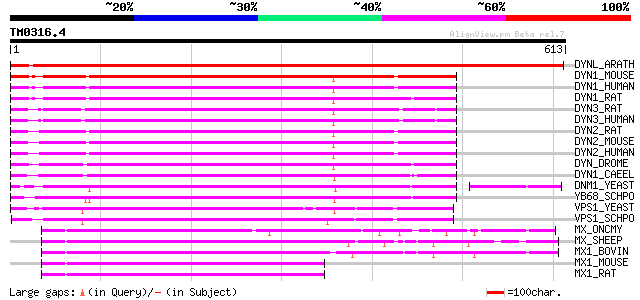
(613 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DYNL_ARATH (P42697) Dynamin-like protein 868 0.0
DYN1_MOUSE (P39053) Dynamin-1 (EC 3.6.5.5) 374 e-103
DYN1_HUMAN (Q05193) Dynamin-1 (EC 3.6.5.5) 372 e-102
DYN1_RAT (P21575) Dynamin-1 (EC 3.6.5.5) (D100) (Dynamin, brain)... 369 e-101
DYN3_RAT (Q08877) Dynamin 3 (EC 3.6.5.5) (Dynamin, testicular) (... 362 2e-99
DYN3_HUMAN (Q9UQ16) Dynamin 3 (EC 3.6.5.5) (Dynamin, testicular)... 362 2e-99
DYN2_RAT (P39052) Dynamin 2 (EC 3.6.5.5) 360 5e-99
DYN2_MOUSE (P39054) Dynamin 2 (EC 3.6.5.5) (Dynamin UDNM) 360 5e-99
DYN2_HUMAN (P50570) Dynamin 2 (EC 3.6.5.5) 359 1e-98
DYN_DROME (P27619) Dynamin (EC 3.6.5.5) (dDyn) (Shibire protein) 352 1e-96
DYN1_CAEEL (P39055) Dynamin (EC 3.6.5.5) 348 2e-95
DNM1_YEAST (P54861) Dynamin-related protein DNM1 (EC 3.6.5.5) 346 9e-95
YB68_SCHPO (Q09748) Dynamin-like protein C12C2.08 332 1e-90
VPS1_YEAST (P21576) Vacuolar sorting protein 1 317 4e-86
VPS1_SCHPO (Q9URZ5) Vacuolar sorting protein 1 309 1e-83
MX_ONCMY (Q91192) Interferon-induced GTP-binding protein Mx 202 2e-51
MX_SHEEP (P33237) Interferon-induced GTP-binding protein Mx 199 1e-50
MX1_BOVIN (P79135) Interferon-induced GTP-binding protein Mx1 198 3e-50
MX1_MOUSE (P09922) Interferon-induced GTP-binding protein Mx1 (I... 196 1e-49
MX1_RAT (P18588) Interferon-induced GTP-binding protein Mx1 196 2e-49
>DYNL_ARATH (P42697) Dynamin-like protein
Length = 610
Score = 868 bits (2242), Expect = 0.0
Identities = 416/611 (68%), Positives = 532/611 (86%), Gaps = 3/611 (0%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME+LI LVN+IQRACT LGDHG D ++ +LW++LP +AVVGGQSSGKSSVLESIVG+
Sbjct: 1 MENLISLVNKIQRACTALGDHG---DSSALPTLWDSLPAIAVVGGQSSGKSSVLESIVGK 57
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQL K++ G++EYAEFLH+PR+KFTDF+ VR+EIQDETDR TG+
Sbjct: 58 DFLPRGSGIVTRRPLVLQLQKIDDGTREYAEFLHLPRKKFTDFAAVRKEIQDETDRETGR 117
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
+K IS +PIHLSIYSPNVVNLTL+DLPGLTKVAV+GQ +SIV++IE MVR+Y+EKPN II
Sbjct: 118 SKAISSVPIHLSIYSPNVVNLTLIDLPGLTKVAVDGQSDSIVKDIENMVRSYIEKPNCII 177
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LAISPANQD+ATSDAIK++REVDP+G+RTFGVLTK+DLMDKGT+A+++LEGRS++L++PW
Sbjct: 178 LAISPANQDLATSDAIKISREVDPSGDRTFGVLTKIDLMDKGTDAVEILEGRSFKLKYPW 237
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VG+VNRSQADINKNVDMI AR++EREYF+N+ +Y HLANKMGSE+LAK+LS+HLE VI++
Sbjct: 238 VGVVNRSQADINKNVDMIAARKREREYFSNTTEYRHLANKMGSEHLAKMLSKHLERVIKS 297
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRPGG 360
RIP I SLINK++ ELE+E+ LG+PIA DAG +LY+I+E+CR F++IFKEHLDG R GG
Sbjct: 298 RIPGIQSLINKTVLELETELSRLGKPIAADAGGKLYSIMEICRLFDQIFKEHLDGVRAGG 357
Query: 361 DRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSY 420
+++Y+VFDNQLPAAL++L FD+ L++ N+RK+V+EADGYQPHLIAPEQGYRRLI+S++
Sbjct: 358 EKVYNVFDNQLPAALKRLQFDKQLAMDNIRKLVTEADGYQPHLIAPEQGYRRLIESSIVS 417
Query: 421 FRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTL 480
RGPAEASVD V+ +LK+LV KS+ ET ELK++P + + AA E+L++ RE SKK TL
Sbjct: 418 IRGPAEASVDTVHAILKDLVHKSVNETVELKQYPALRVEVTNAAIESLDKMREGSKKATL 477
Query: 481 RLVDMESSYLTVDFFRRLPQEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLR 540
+LVDME SYLTVDFFR+LPQ+VEK G P DRY + + RRI SNV SY+ +V LR
Sbjct: 478 QLVDMECSYLTVDFFRKLPQDVEKGGNPTHSIFDRYNDSYLRRIGSNVLSYVNMVCAGLR 537
Query: 541 NTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
N+IPK++V+CQVR+AK+SLL+HF+ ++G + K+LS +L+EDPA+MERR +KRLELY+
Sbjct: 538 NSIPKSIVYCQVREAKRSLLDHFFAELGTMDMKRLSSLLNEDPAIMERRSAISKRLELYR 597
Query: 601 AARDEIDSVSW 611
AA+ EID+V+W
Sbjct: 598 AAQSEIDAVAW 608
>DYN1_MOUSE (P39053) Dynamin-1 (EC 3.6.5.5)
Length = 867
Score = 374 bits (959), Expect = e-103
Identities = 211/501 (42%), Positives = 302/501 (60%), Gaps = 25/501 (4%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LI LVNR+Q A + +G + AD + LP +AVVGGQS+GKSSVLE+ VGR
Sbjct: 6 MEDLIPLVNRLQDAFSAIGQN---ADLD--------LPQIAVVGGQSAGKSSVLENFVGR 54
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQL T EYAEFLH +KFTDF VR EI+ ETDRVTG
Sbjct: 55 DFLPRGSGIVTRRPLVLQLVNSTT---EYAEFLHCKGKKFTDFEEVRLEIEAETDRVTGT 111
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K ISP+PI+L +YSP+V+NLTLVDLPG+TKV V QP I +I M+ +V K N +I
Sbjct: 112 NKGISPVPINLRVYSPHVLNLTLVDLPGMTKVPVGDQPPDIEFQIRDMLMQFVTKENCLI 171
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA+SPAN D+A SDA+K+A+EVDP G+RT GV+TKLDLMD+GT+A DVLE + L+ +
Sbjct: 172 LAVSPANSDLANSDALKIAKEVDPQGQRTIGVITKLDLMDEGTDARDVLENKLLPLRRGY 231
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DI+ D+ A ER++F + P Y HLA++MG+ YL K+L+Q L + IR
Sbjct: 232 IGVVNRSQKDIDGKKDITAALAAERKFFLSHPSYRHLADRMGTPYLQKVLNQQLTNHIRD 291
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGG---- 356
+P + + + L +E E+D D + +L++ + F F++ ++G
Sbjct: 292 TLPGLRNKLQSQLLSIEKEVDEYKNFRPDDPARKTKALLQMVQQFAVDFEKRIEGSGDQI 351
Query: 357 ----RPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRR 412
GG RI +F + P L K+ FD + + + G + L P+ +
Sbjct: 352 DTYELSGGARINRIFHERFPFELVKMEFDEKELRREISYAIKNIHGIRTGLFTPDMAFET 411
Query: 413 LIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFR 472
++ + R P + V+ V+ EL+ TK+L+++P + + +
Sbjct: 412 IVKKQVKKIREPC---LKCVDMVISELISTVRQCTKKLQQYPRLREEMERIVTTHIRERE 468
Query: 473 EESKKTTLRLVDMESSYLTVD 493
+K+ + L+D+E +Y+ +
Sbjct: 469 GRTKEQVMLLIDIELAYMNTN 489
Score = 31.2 bits (69), Expect = 8.3
Identities = 20/98 (20%), Positives = 46/98 (46%), Gaps = 5/98 (5%)
Query: 512 NVDRYAEGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKE 571
++D E I + V SY+ +V T+R+ +PK ++ + K+ + + +
Sbjct: 650 SMDPQLERQVETIRNLVDSYMAIVNKTVRDLMPKTIMHLMINNTKEFIFSELLANL-YSC 708
Query: 572 GKQLSEMLDEDPALMERRQQCAKRLELYKAARDEIDSV 609
G Q + +++E +RR + L +Y A ++ + +
Sbjct: 709 GDQ-NTLMEESAEQAQRRDE---MLRMYHALKEALSII 742
>DYN1_HUMAN (Q05193) Dynamin-1 (EC 3.6.5.5)
Length = 864
Score = 372 bits (955), Expect = e-102
Identities = 210/501 (41%), Positives = 302/501 (59%), Gaps = 25/501 (4%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LI LVNR+Q A + +G + AD + LP +AVVGGQS+GKSSVLE+ VGR
Sbjct: 6 MEDLIPLVNRLQDAFSAIGQN---ADLD--------LPQIAVVGGQSAGKSSVLENFVGR 54
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQL T EYAEFLH +KFTDF VR EI+ ETDRVTG
Sbjct: 55 DFLPRGSGIVTRRPLVLQLVNATT---EYAEFLHCKGKKFTDFEEVRLEIEAETDRVTGT 111
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K ISP+PI+L +YSP+V+NLTLVDLPG+TKV V QP I +I M+ +V K N +I
Sbjct: 112 NKGISPVPINLRVYSPHVLNLTLVDLPGMTKVPVGDQPPDIEFQIRDMLMQFVTKENCLI 171
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA+SPAN D+A SDA+K+A+EVDP G+RT GV+TKLDLMD+GT+A DVLE + L+ +
Sbjct: 172 LAVSPANSDLANSDALKVAKEVDPQGQRTIGVITKLDLMDEGTDARDVLENKLLPLRRGY 231
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DI+ D+ A ER++F + P Y HLA++MG+ YL K+L+Q L + IR
Sbjct: 232 IGVVNRSQKDIDGKKDITAALAAERKFFLSHPSYRHLADRMGTPYLQKVLNQQLTNHIRD 291
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGG---- 356
+P + + + L +E E++ D + +L++ + F F++ ++G
Sbjct: 292 TLPGLRNKLQSQLLSIEKEVEEYKNFRPDDPARKTKALLQMVQQFAVDFEKRIEGSGDQI 351
Query: 357 ----RPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRR 412
GG RI +F + P L K+ FD + + + G + L P+ +
Sbjct: 352 DTYELSGGARINRIFHERFPFELVKMEFDEKELRREISYAIKNIHGIRTGLFTPDMAFET 411
Query: 413 LIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFR 472
++ + R P + V+ V+ EL+ TK+L+++P + + +
Sbjct: 412 IVKKQVKKIREPC---LKCVDMVISELISTVRQCTKKLQQYPRLREEMERIVTTHIRERE 468
Query: 473 EESKKTTLRLVDMESSYLTVD 493
+K+ + L+D+E +Y+ +
Sbjct: 469 GRTKEQVMLLIDIELAYMNTN 489
Score = 31.6 bits (70), Expect = 6.4
Identities = 20/99 (20%), Positives = 45/99 (45%), Gaps = 2/99 (2%)
Query: 512 NVDRYAEGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKE 571
++D E I + V SY+ +V T+R+ +PK ++ + K+ + + +
Sbjct: 650 SMDPQLERQVETIRNLVDSYMAIVNKTVRDLMPKTIMHLMINNTKEFIFSELLANL-YSC 708
Query: 572 GKQLSEMLDEDPALMERRQQCAKRLELYKAARDEIDSVS 610
G Q + +++E +RR + + K A I +++
Sbjct: 709 GDQ-NTLMEESAEQAQRRDEMLRMYHALKEALSIIGNIN 746
>DYN1_RAT (P21575) Dynamin-1 (EC 3.6.5.5) (D100) (Dynamin, brain)
(B-dynamin)
Length = 851
Score = 369 bits (948), Expect = e-101
Identities = 210/501 (41%), Positives = 301/501 (59%), Gaps = 25/501 (4%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LI LVNR+Q A + +G + AD + LP +AVVGGQS+GKSSVLE+ VGR
Sbjct: 6 MEDLIPLVNRLQDAFSAIGQN---ADLD--------LPQIAVVGGQSAGKSSVLENFVGR 54
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQL T EYAEFLH +KFTDF VR EI+ ETDRVTG
Sbjct: 55 DFLPRGSGIVTRRPLVLQLVNSTT---EYAEFLHCKGKKFTDFEEVRLEIEAETDRVTGT 111
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K ISP+PI+L +YSP+V+NLTLVDLPG+TKV V QP I +I M+ +V K N +I
Sbjct: 112 NKGISPVPINLRVYSPHVLNLTLVDLPGMTKVPVGDQPPDIEFQIRDMLMQFVTKENCLI 171
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA+SPAN D+A SDA+K+A+EVDP G+RT GV+TKLDLMD+GT+A DVLE + L+ +
Sbjct: 172 LAVSPANSDLANSDALKIAKEVDPQGQRTIGVITKLDLMDEGTDARDVLENKLLPLRRGY 231
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DI+ D+ A ER++F + P Y HLA++MG+ YL K+L+Q L + IR
Sbjct: 232 IGVVNRSQKDIDGKKDITAALAAERKFFLSHPSYRHLADRMGTPYLQKVLNQQLTNHIRD 291
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGG---- 356
+P + + + L +E E+D D + +L++ + F F++ ++G
Sbjct: 292 TLPGLRNKLQSQLLSIEKEVDEYKNFRPDDPARKTKALLQMVQQFAVDFEKRIEGSGDQI 351
Query: 357 ----RPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRR 412
GG RI +F + P L K+ FD + + + G + L P+ +
Sbjct: 352 DTYELSGGARINRIFHERFPFELVKMEFDEKELRREISYAIKNIHGIRTGLFTPDLAFEA 411
Query: 413 LIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFR 472
+ + + P+ VD V L +RK +++L+++P + + +
Sbjct: 412 TVKKQVQKLKEPSIKCVDMVVSELTSTIRKC---SEKLQQYPRLREEMERIVTTHIRERE 468
Query: 473 EESKKTTLRLVDMESSYLTVD 493
+K+ + L+D+E +Y+ +
Sbjct: 469 GRTKEQVMLLIDIELAYMNTN 489
Score = 31.2 bits (69), Expect = 8.3
Identities = 20/98 (20%), Positives = 46/98 (46%), Gaps = 5/98 (5%)
Query: 512 NVDRYAEGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKE 571
++D E I + V SY+ +V T+R+ +PK ++ + K+ + + +
Sbjct: 650 SMDPQLERQVETIRNLVDSYMAIVNKTVRDLMPKTIMHLMINNTKEFIFSELLANL-YSC 708
Query: 572 GKQLSEMLDEDPALMERRQQCAKRLELYKAARDEIDSV 609
G Q + +++E +RR + L +Y A ++ + +
Sbjct: 709 GDQ-NTLMEESAEQAQRRDE---MLRMYHALKEALSII 742
>DYN3_RAT (Q08877) Dynamin 3 (EC 3.6.5.5) (Dynamin, testicular)
(T-dynamin)
Length = 848
Score = 362 bits (929), Expect = 2e-99
Identities = 212/502 (42%), Positives = 301/502 (59%), Gaps = 27/502 (5%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LI LVNR+Q A + LG S L E LP +AVVGGQS+GKSSVLE+ VGR
Sbjct: 6 MEELIPLVNRLQDAFSALGQ----------SCLLE-LPQIAVVGGQSAGKSSVLENFVGR 54
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQL T EYAEFLH +KFTDF VR EI+ ETDRVTG
Sbjct: 55 DFLPRGSGIVTRRPLVLQL---VTSKAEYAEFLHCKGKKFTDFDEVRHEIEAETDRVTGM 111
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K IS +PI+L +YSP+V+NLTL+DLPG+TKV V QP I +I M+ ++ + N +I
Sbjct: 112 NKGISSVPINLRVYSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIRDMIMQFITRENCLI 171
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA++PAN D+A SDA+KLA+EVDP G RT GV+TKLDLMD+GT+A DVLE + L+ +
Sbjct: 172 LAVTPANTDLANSDALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGY 231
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VG+VNRSQ DI+ D+ A ER++F + P Y H+A++MG+ +L K+L+Q L + IR
Sbjct: 232 VGVVNRSQKDIDGKKDIKAAMLAERKFFLSHPAYRHIADRMGTPHLQKVLNQQLTNHIRD 291
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGG---- 356
+P+ + + L +E E++ D + +L++ + F F++ ++G
Sbjct: 292 TLPNFRNKLQGQLLSIEHEVEAFKNFKPEDPTRKTKALLQMVQQFAVDFEKRIEGSGDQV 351
Query: 357 ----RPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRR 412
GG +I +F + P + K+ F+ + + + G + L P+ +
Sbjct: 352 DTLELSGGAKINRIFHERFPFEIVKMEFNEKELRREISYAIKNIHGIRTGLFTPDMAFEA 411
Query: 413 LIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTF-QAALATAANEALERF 471
++ + +GP+ SVD V++EL+ TK L FP + AN ER
Sbjct: 412 IVKKQIVKLKGPSLKSVD---LVMQELINTVKKCTKRLANFPRLCEETERIVANHIRER- 467
Query: 472 REESKKTTLRLVDMESSYLTVD 493
++K L L+D++ SY+ +
Sbjct: 468 EGKTKDQVLLLIDIQVSYINTN 489
Score = 34.3 bits (77), Expect = 0.98
Identities = 22/112 (19%), Positives = 49/112 (43%), Gaps = 2/112 (1%)
Query: 501 EVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLL 560
E ++ G ++D E I + V SY+ ++ +R+ IPK ++ + K +
Sbjct: 629 ENDENGQAENFSMDPQLERQVETIRNLVDSYMSIINKCIRDLIPKTIMHLMINNVKDFIN 688
Query: 561 NHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYKAARDEIDSVSWV 612
+ Q+ E + + +++E +RR + + + K A I ++ V
Sbjct: 689 SELLAQLYSSEDQ--NTLMEESVEQAQRRDEMLRMYQALKEALAIIGDINTV 738
>DYN3_HUMAN (Q9UQ16) Dynamin 3 (EC 3.6.5.5) (Dynamin, testicular)
(T-dynamin)
Length = 859
Score = 362 bits (928), Expect = 2e-99
Identities = 213/502 (42%), Positives = 302/502 (59%), Gaps = 27/502 (5%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LI LVNR+Q A + LG S L E LP +AVVGGQS+GKSSVLE+ VGR
Sbjct: 6 MEELIPLVNRLQDAFSALGQ----------SCLLE-LPQIAVVGGQSAGKSSVLENFVGR 54
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPLVLQL T EYAEFLH +KFTDF VR EI+ ETDRVTG
Sbjct: 55 DFLPRGSGIVTRRPLVLQL---VTSKAEYAEFLHCKGKKFTDFDEVRLEIEAETDRVTGM 111
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K IS IPI+L +YSP+V+NLTL+DLPG+TKV V QP I +I M+ ++ + N +I
Sbjct: 112 NKGISSIPINLRVYSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIREMIMQFITRENCLI 171
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA++PAN D+A SDA+KLA+EVDP G RT GV+TKLDLMD+GT+A DVLE + L+ +
Sbjct: 172 LAVTPANTDLANSDALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGY 231
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VG+VNRSQ DI+ D+ A ER++F + P Y H+A++MG+ +L K+L+Q L + IR
Sbjct: 232 VGVVNRSQKDIDGKKDIKAAMLAERKFFLSHPAYRHIADRMGTPHLQKVLNQQLTNHIRD 291
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGG---- 356
+P+ + + L +E E++ D + +L++ + F F++ ++G
Sbjct: 292 TLPNFRNKLQGQLLSIEHEVEAYKNFKPEDPTRKTKALLQMVQQFAVDFEKRIEGSGDQV 351
Query: 357 ----RPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRR 412
GG +I +F + P + K+ F+ + + + G + L P+ +
Sbjct: 352 DTLELSGGAKINRIFHERFPFEIVKMEFNEKELRREISYAIKNIHGIRTGLFTPDMAFEA 411
Query: 413 LIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTF-QAALATAANEALERF 471
++ + +GP+ SVD V++EL+ TK+L FP + AN ER
Sbjct: 412 IVKKQIVKLKGPSLKSVD---LVIQELINTVKKCTKKLANFPRLCEETERIVANHIRER- 467
Query: 472 REESKKTTLRLVDMESSYLTVD 493
++K L L+D++ SY+ +
Sbjct: 468 EGKTKDQVLLLIDIQVSYINTN 489
Score = 35.4 bits (80), Expect = 0.44
Identities = 22/110 (20%), Positives = 48/110 (43%), Gaps = 2/110 (1%)
Query: 501 EVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLL 560
E ++ G ++D E I + V SY+ ++ +R+ IPK ++ + K +
Sbjct: 629 ENDENGQAENFSMDPQLERQVETIRNLVDSYMSIINKCIRDLIPKTIMHLMINNVKDFIN 688
Query: 561 NHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYKAARDEIDSVS 610
+ Q+ E + + +++E +RR + + + K A I +S
Sbjct: 689 SELLAQLYSSEDQ--NTLMEESAEQAQRRDEMLRMYQALKEALGIIGDIS 736
>DYN2_RAT (P39052) Dynamin 2 (EC 3.6.5.5)
Length = 870
Score = 360 bits (925), Expect = 5e-99
Identities = 205/501 (40%), Positives = 297/501 (58%), Gaps = 25/501 (4%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LI LVN++Q A + +G S LP +AVVGGQS+GKSSVLE+ VGR
Sbjct: 6 MEELIPLVNKLQDAFSSIGQ-----------SCHLDLPQIAVVGGQSAGKSSVLENFVGR 54
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPL+LQL +T EYAEFLH +KFTDF VRQEI+ ETDRVTG
Sbjct: 55 DFLPRGSGIVTRRPLILQLIFSKT---EYAEFLHCKSKKFTDFDEVRQEIEAETDRVTGT 111
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K ISP+PI+L +YSP+V+NLTL+DLPG+TKV V QP I +I+ M+ ++ + +S+I
Sbjct: 112 NKGISPVPINLRVYSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIKDMILQFISRESSLI 171
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA++PAN D+A SDA+KLA+EVDP G RT GV+TKLDLMD+GT+A DVLE + L+ +
Sbjct: 172 LAVTPANMDLANSDALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGY 231
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DI D+ A ER++F + P Y H+A++MG+ +L K L+Q L + IR
Sbjct: 232 IGVVNRSQKDIEGRKDIRAALAAERKFFLSHPAYRHMADRMGTPHLQKTLNQQLTNHIRE 291
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGG---- 356
+P++ S + L LE E++ D + +L++ + F F++ ++G
Sbjct: 292 SLPTLRSKLQSQLLSLEKEVEEYKNFRPDDPTRKTKALLQMVQQFGVDFEKRIEGSGDQV 351
Query: 357 ----RPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRR 412
GG RI +F + P L K+ FD + + + G + L P+ +
Sbjct: 352 DTLELSGGARINRIFHERFPFELVKMEFDEKDLRREISYAIKNIHGVRTGLFTPDLAFEA 411
Query: 413 LIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFR 472
++ + + P + V+ V++EL+ T +L +P + +
Sbjct: 412 IVKKQVVKLKEPC---LKCVDLVIQELISTVRQCTSKLSSYPRLREETERIVTTYIRERE 468
Query: 473 EESKKTTLRLVDMESSYLTVD 493
+K L L+D+E SY+ +
Sbjct: 469 GRTKDQILLLIDIEQSYINTN 489
Score = 33.5 bits (75), Expect = 1.7
Identities = 27/130 (20%), Positives = 54/130 (40%), Gaps = 11/130 (8%)
Query: 486 ESSYLTVDFFRRLPQ-EVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLRNTIP 544
++S+L + Q E E T ++D E I + V SY+ ++ ++R+ +P
Sbjct: 617 KASFLRAGVYPEKDQAENEDGAQENTFSMDPQLERQVETIRNLVDSYVAIINKSIRDLMP 676
Query: 545 KAVVFCQVRQAK----QSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
K ++ + K LL + Y+ + S +++E +RR + K
Sbjct: 677 KTIMHLMINNTKAFIHHELLAYLYSSADQ------SSLMEESAEQAQRRDDMLRMYHALK 730
Query: 601 AARDEIDSVS 610
A + I +S
Sbjct: 731 EALNIIGDIS 740
>DYN2_MOUSE (P39054) Dynamin 2 (EC 3.6.5.5) (Dynamin UDNM)
Length = 870
Score = 360 bits (925), Expect = 5e-99
Identities = 205/501 (40%), Positives = 297/501 (58%), Gaps = 25/501 (4%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LI LVN++Q A + +G S LP +AVVGGQS+GKSSVLE+ VGR
Sbjct: 6 MEELIPLVNKLQDAFSSIGQ-----------SCHLDLPQIAVVGGQSAGKSSVLENFVGR 54
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPL+LQL +T EYAEFLH +KFTDF VRQEI+ ETDRVTG
Sbjct: 55 DFLPRGSGIVTRRPLILQLIFSKT---EYAEFLHCKSKKFTDFDEVRQEIEAETDRVTGT 111
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K ISP+PI+L +YSP+V+NLTL+DLPG+TKV V QP I +I+ M+ ++ + +S+I
Sbjct: 112 NKGISPVPINLRVYSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIKDMILQFISRESSLI 171
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA++PAN D+A SDA+KLA+EVDP G RT GV+TKLDLMD+GT+A DVLE + L+ +
Sbjct: 172 LAVTPANMDLANSDALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGY 231
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DI D+ A ER++F + P Y H+A++MG+ +L K L+Q L + IR
Sbjct: 232 IGVVNRSQKDIEGKKDIRAALAAERKFFLSHPAYRHMADRMGTPHLQKTLNQQLTNHIRE 291
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGG---- 356
+P++ S + L LE E++ D + +L++ + F F++ ++G
Sbjct: 292 SLPTLRSKLQSQLLSLEKEVEEYKNFRPDDPTRKTKALLQMVQQFGVDFEKRIEGSGDQV 351
Query: 357 ----RPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRR 412
GG RI +F + P L K+ FD + + + G + L P+ +
Sbjct: 352 DTLELSGGARINRIFHERFPFELVKMEFDEKDLRREISYAIKNIHGVRTGLFTPDLAFEA 411
Query: 413 LIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFR 472
++ + + P + V+ V++EL+ T +L +P + +
Sbjct: 412 IVKKQVVKLKEPC---LKCVDLVIQELISTVRQCTSKLSSYPRLREETERIVTTYIRERE 468
Query: 473 EESKKTTLRLVDMESSYLTVD 493
+K L L+D+E SY+ +
Sbjct: 469 GRTKDQILLLIDIEQSYINTN 489
Score = 33.5 bits (75), Expect = 1.7
Identities = 27/130 (20%), Positives = 54/130 (40%), Gaps = 11/130 (8%)
Query: 486 ESSYLTVDFFRRLPQ-EVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLRNTIP 544
++S+L + Q E E T ++D E I + V SY+ ++ ++R+ +P
Sbjct: 617 KASFLRAGVYPEKDQAENEDGAQENTFSMDPQLERQVETIRNLVDSYVAIINKSIRDLMP 676
Query: 545 KAVVFCQVRQAK----QSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
K ++ + K LL + Y+ + S +++E +RR + K
Sbjct: 677 KTIMHLMINNTKAFIHHELLAYLYSSADQ------SSLMEESAEQAQRRDDMLRMYHALK 730
Query: 601 AARDEIDSVS 610
A + I +S
Sbjct: 731 EALNIIGDIS 740
>DYN2_HUMAN (P50570) Dynamin 2 (EC 3.6.5.5)
Length = 870
Score = 359 bits (921), Expect = 1e-98
Identities = 204/501 (40%), Positives = 297/501 (58%), Gaps = 25/501 (4%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LI LVN++Q A + +G S LP +AVVGGQS+GKSSVLE+ VGR
Sbjct: 6 MEELIPLVNKLQDAFSSIGQ-----------SCHLDLPQIAVVGGQSAGKSSVLENFVGR 54
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPL+LQL +T E+AEFLH +KFTDF VRQEI+ ETDRVTG
Sbjct: 55 DFLPRGSGIVTRRPLILQLIFSKT---EHAEFLHCKSKKFTDFDEVRQEIEAETDRVTGT 111
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K ISP+PI+L +YSP+V+NLTL+DLPG+TKV V QP I +I+ M+ ++ + +S+I
Sbjct: 112 NKGISPVPINLRVYSPHVLNLTLIDLPGITKVPVGDQPPDIEYQIKDMILQFISRESSLI 171
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA++PAN D+A SDA+KLA+EVDP G RT GV+TKLDLMD+GT+A DVLE + L+ +
Sbjct: 172 LAVTPANMDLANSDALKLAKEVDPQGLRTIGVITKLDLMDEGTDARDVLENKLLPLRRGY 231
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DI D+ A ER++F + P Y H+A++MG+ +L K L+Q L + IR
Sbjct: 232 IGVVNRSQKDIEGKKDIRAALAAERKFFLSHPAYRHMADRMGTPHLQKTLNQQLTNHIRE 291
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGG---- 356
+P++ S + L LE E++ D + +L++ + F F++ ++G
Sbjct: 292 SLPALRSKLQSQLLSLEKEVEEYKNFRPDDPTRKTKALLQMVQQFGVDFEKRIEGSGDQV 351
Query: 357 ----RPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRR 412
GG RI +F + P L K+ FD + + + G + L P+ +
Sbjct: 352 DTLELSGGARINRIFHERFPFELVKMEFDEKDLRREISYAIKNIHGVRTGLFTPDLAFEA 411
Query: 413 LIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERFR 472
++ + + P + V+ V++EL+ T +L +P + +
Sbjct: 412 IVKKQVVKLKEPC---LKCVDLVIQELINTVRQCTSKLSSYPRLREETERIVTTYIRERE 468
Query: 473 EESKKTTLRLVDMESSYLTVD 493
+K L L+D+E SY+ +
Sbjct: 469 GRTKDQILLLIDIEQSYINTN 489
Score = 33.1 bits (74), Expect = 2.2
Identities = 27/130 (20%), Positives = 54/130 (40%), Gaps = 11/130 (8%)
Query: 486 ESSYLTVDFFRRLPQ-EVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLRNTIP 544
++S+L + Q E E T ++D E I + V SY+ ++ ++R+ +P
Sbjct: 617 KASFLRAGVYPEKDQAENEDGAQENTFSMDPQLERQVETIRNLVDSYVAIINKSIRDLMP 676
Query: 545 KAVVFCQVRQAK----QSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYK 600
K ++ + K LL + Y+ + S +++E +RR + K
Sbjct: 677 KTIMHLMINNTKAFIHHELLAYLYSSADQ------SSLMEESADQAQRRDDMLRMYHALK 730
Query: 601 AARDEIDSVS 610
A + I +S
Sbjct: 731 EALNIIGDIS 740
>DYN_DROME (P27619) Dynamin (EC 3.6.5.5) (dDyn) (Shibire protein)
Length = 877
Score = 352 bits (904), Expect = 1e-96
Identities = 198/502 (39%), Positives = 296/502 (58%), Gaps = 26/502 (5%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
M+SLI +VN++Q A T LG H + LP +AVVGGQS+GKSSVLE+ VG+
Sbjct: 1 MDSLITIVNKLQDAFTSLGVH-----------MQLDLPQIAVVGGQSAGKSSVLENFVGK 49
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPL+LQL G EY EFLH+ +KF+ F +R+EI+DETDRVTG
Sbjct: 50 DFLPRGSGIVTRRPLILQLIN---GVTEYGEFLHIKGKKFSSFDEIRKEIEDETDRVTGS 106
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K IS IPI+L +YSP+V+NLTL+DLPGLTKVA+ QP I Q+I+ M+ ++ K +I
Sbjct: 107 NKGISNIPINLRVYSPHVLNLTLIDLPGLTKVAIGDQPVDIEQQIKQMIFQFIRKETCLI 166
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA++PAN D+A SDA+KLA+EVDP G RT GV+TKLDLMD+GT+A D+LE + L+ +
Sbjct: 167 LAVTPANTDLANSDALKLAKEVDPQGVRTIGVITKLDLMDEGTDARDILENKLLPLRRGY 226
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
+G+VNRSQ DI D+ A ER++F + P Y H+A+++G+ YL ++L+Q L + IR
Sbjct: 227 IGVVNRSQKDIEGRKDIHQALAAERKFFLSHPSYRHMADRLGTPYLQRVLNQQLTNHIRD 286
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGG---- 356
+P + + K + LE E++ DA + +L++ + + F+ ++G
Sbjct: 287 TLPGLRDKLQKQMLTLEKEVEEFKHFQPGDASIKTKAMLQMIQQLQSDFERTIEGSGSAL 346
Query: 357 -----RPGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYR 411
GG +I +F +L + K+ D + + + G + L P+ +
Sbjct: 347 VNTNELSGGAKINRIFHERLRFEIVKMACDEKELRREISFAIRNIHGIRVGLFTPDMAFE 406
Query: 412 RLIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERF 471
++ ++ + P VD V L +VR T ++ R+P + + +
Sbjct: 407 AIVKRQIALLKEPVIKCVDLVVQELSVVVRMC---TAKMSRYPRLREETERIITTHVRQR 463
Query: 472 REESKKTTLRLVDMESSYLTVD 493
K+ L L+D E +Y+ +
Sbjct: 464 EHSCKEQILLLIDFELAYMNTN 485
Score = 39.3 bits (90), Expect = 0.031
Identities = 28/100 (28%), Positives = 47/100 (47%), Gaps = 13/100 (13%)
Query: 514 DRYAEGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQ----SLLNHFYTQIGK 569
D E I + V SY+ +V T R+ +PKA++ + AK LL H Y
Sbjct: 643 DPQLERQVETIRNLVDSYMKIVTKTTRDMVPKAIMMLIINNAKDFINGELLAHLYA---- 698
Query: 570 KEGKQLSEMLDEDPALMERRQQCAKRLELYKAARDEIDSV 609
G Q ++M++E RR++ L +Y+A +D + +
Sbjct: 699 -SGDQ-AQMMEESAESATRREE---MLRMYRACKDALQII 733
>DYN1_CAEEL (P39055) Dynamin (EC 3.6.5.5)
Length = 830
Score = 348 bits (894), Expect = 2e-95
Identities = 197/502 (39%), Positives = 297/502 (58%), Gaps = 26/502 (5%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
M++LI ++NR+Q A + LG +S+ LP +AVVGGQS+GKSSVLE+ VG+
Sbjct: 8 MQALIPVINRVQDAFSQLG-----------TSVSFELPQIAVVGGQSAGKSSVLENFVGK 56
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGK 120
DFLPRGSGIVTRRPL+LQL + EYAEFLH +F DF VR+EI+DETDRVTG+
Sbjct: 57 DFLPRGSGIVTRRPLILQLIQ---DRNEYAEFLHKKGHRFVDFDAVRKEIEDETDRVTGQ 113
Query: 121 TKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSII 180
K ISP PI+L ++SPNV+NLTL+DLPGLTKV V QP I Q+I M+ T++ + +I
Sbjct: 114 NKGISPHPINLRVFSPNVLNLTLIDLPGLTKVPVGDQPADIEQQIRDMILTFINRETCLI 173
Query: 181 LAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPW 240
LA++PAN D+ATSDA+KLA+EVDP G RT GVLTKLDLMD+GT+A ++LE + + L+ +
Sbjct: 174 LAVTPANSDLATSDALKLAKEVDPQGLRTIGVLTKLDLMDEGTDAREILENKLFTLRRGY 233
Query: 241 VGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRA 300
VG+VNR Q DI D+ A ER++F + P Y H+A+++G+ YL L+Q L + IR
Sbjct: 234 VGVVNRGQKDIVGRKDIRAALDAERKFFISHPSYRHMADRLGTSYLQHTLNQQLTNHIRD 293
Query: 301 RIPSITSLINKSLEELESEMDHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGR--- 357
+P++ + K + +E ++ D G + +L++ F + ++G
Sbjct: 294 TLPTLRDSLQKKMFAMEKDVAEYKNYQPNDPGRKTKALLQMVTQFNADIERSIEGSSAKL 353
Query: 358 ------PGGDRIYHVFDNQLPAALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYR 411
GG RI +F + P + K+ D + ++ + G + L P+ +
Sbjct: 354 VSTNELSGGARINRLFHERFPFEIVKMEIDEKEMRKEIQYAIRNIHGIRVGLFTPDMAFE 413
Query: 412 RLIDSALSYFRGPAEASVDAVNFVLKELVRKSIAETKELKRFPTFQAALATAANEALERF 471
+ ++ + P+ VD V L ++R+ A+T + R+P + L +
Sbjct: 414 AIAKKQITRLKEPSLKCVDLVVNELANVIRQ-CADT--MARYPRLRDELERIVVSHMRER 470
Query: 472 REESKKTTLRLVDMESSYLTVD 493
+ +K+ +VD E +Y+ +
Sbjct: 471 EQIAKQQIGLIVDYELAYMNTN 492
Score = 37.7 bits (86), Expect = 0.089
Identities = 30/117 (25%), Positives = 54/117 (45%), Gaps = 13/117 (11%)
Query: 500 QEVEKAGGPATPNVDRYAEGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQA---- 555
QE E ++D E I + V SY+ ++ T+++ +PKAV+ V Q
Sbjct: 632 QEDESQQEMEDTSIDPQLERQVETIRNLVDSYMRIITKTIKDLVPKAVMHLIVNQTGEFM 691
Query: 556 KQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYKAARDEIDSVSWV 612
K LL H Y Q G + +++E ++R++ L +Y A ++ + +S V
Sbjct: 692 KDELLAHLY-QCGDTDA-----LMEESQIEAQKREE---MLRMYHACKEALRIISEV 739
>DNM1_YEAST (P54861) Dynamin-related protein DNM1 (EC 3.6.5.5)
Length = 757
Score = 346 bits (888), Expect = 9e-95
Identities = 204/540 (37%), Positives = 304/540 (55%), Gaps = 61/540 (11%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
+E LI VN++Q V+ D G LP +AVVG QSSGKSS+LE++VGR
Sbjct: 4 LEDLIPTVNKLQ---DVMYDSGIDTLD---------LPILAVVGSQSSGKSSILETLVGR 51
Query: 61 DFLPRGSGIVTRRPLVLQLHKLETGS---------------------------------- 86
DFLPRG+GIVTRRPLVLQL+ + S
Sbjct: 52 DFLPRGTGIVTRRPLVLQLNNISPNSPLIEEDDNSVNPHDEVTKISGFEAGTKPLEYRGK 111
Query: 87 -----QEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNL 141
E+ EFLH+P ++F DF +++EI++ET R+ GK K IS IPI+L ++SP+V+NL
Sbjct: 112 ERNHADEWGEFLHIPGKRFYDFDDIKREIENETARIAGKDKGISKIPINLKVFSPHVLNL 171
Query: 142 TLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLARE 201
TLVDLPG+TKV + QP I ++I+ ++ Y+ PN +ILA+SPAN D+ S+++KLARE
Sbjct: 172 TLVDLPGITKVPIGEQPPDIEKQIKNLILDYIATPNCLILAVSPANVDLVNSESLKLARE 231
Query: 202 VDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIAR 261
VDP G+RT GV+TKLDLMD GTNALD+L G+ Y L+ +VG+VNRSQ DI N + +
Sbjct: 232 VDPQGKRTIGVITKLDLMDSGTNALDILSGKMYPLKLGFVGVVNRSQQDIQLNKTVEESL 291
Query: 262 RKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRARIPSITSLINKSLEELESEMD 321
KE +YF P Y ++ K G+ YLAKLL+Q L S IR ++P I + +N + + E E+
Sbjct: 292 DKEEDYFRKHPVYRTISTKCGTRYLAKLLNQTLLSHIRDKLPDIKTKLNTLISQTEQELA 351
Query: 322 HLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGRP--------GGDRIYHVFDNQLPA 373
G A ++ +L+L F F +DG GG RIY++++N
Sbjct: 352 RYGGVGATTNESRASLVLQLMNKFSTNFISSIDGTSSDINTKELCGGARIYYIYNNVFGN 411
Query: 374 ALRKLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSYFRGPAEASVDAVN 433
+L+ + +LS+ +VR + + G +P L PE + L+ + P++ V+ V
Sbjct: 412 SLKSIDPTSNLSVLDVRTAIRNSTGPRPTLFVPELAFDLLVKPQIKLLLEPSQRCVELVY 471
Query: 434 FVLKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTLRLVDMESSYLTVD 493
L ++ K + EL R+P ++ L +E L + ++ L+D+ +Y+ +
Sbjct: 472 EELMKICHK--CGSAELARYPKLKSMLIEVISELLRERLQPTRSYVESLIDIHRAYINTN 529
Score = 46.6 bits (109), Expect = 2e-04
Identities = 29/101 (28%), Positives = 48/101 (46%), Gaps = 2/101 (1%)
Query: 509 ATPNVDRYAEGHFRRIASNVSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIG 568
A P + E I + SY ++ + + + +PKAV+ V K S+ N T++
Sbjct: 658 AEPPLTEREELECELIKRLIVSYFDIIREMIEDQVPKAVMCLLVNYCKDSVQNRLVTKLY 717
Query: 569 KKEGKQLSEMLDEDPALMERRQQCAKRLELYKAARDEIDSV 609
K+ E+L ED L + R+ C K L +YK A I ++
Sbjct: 718 KE--TLFEELLVEDQTLAQDRELCVKSLGVYKKAATLISNI 756
>YB68_SCHPO (Q09748) Dynamin-like protein C12C2.08
Length = 781
Score = 332 bits (852), Expect = 1e-90
Identities = 204/537 (37%), Positives = 293/537 (53%), Gaps = 57/537 (10%)
Query: 1 MESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGR 60
ME LI LVN++Q N+ S + LP++ VVG QS GKSSVLE+IVG+
Sbjct: 1 MEQLIPLVNQLQDLVY-----------NTIGSDFLDLPSIVVVGSQSCGKSSVLENIVGK 49
Query: 61 DFLPRGSGIVTRRPLVLQLHKL-------------------------ETGS--------- 86
DFLPRG+GIVTRRPL+LQL L ETGS
Sbjct: 50 DFLPRGTGIVTRRPLILQLINLKRKTKNNHDEESTSDNNSEETSAAGETGSLEGIEEDSD 109
Query: 87 --QEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNLTLV 144
++YAEFLH+P KFTD + VR EI++ET RV G K I+ +PI+L IYS V+NLTL+
Sbjct: 110 EIEDYAEFLHIPDTKFTDMNKVRAEIENETLRVAGANKGINKLPINLKIYSTRVLNLTLI 169
Query: 145 DLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDP 204
DLPGLTK+ V QP I + +++ Y+ +PNSIILA+SPAN DI S+ +KLAR VDP
Sbjct: 170 DLPGLTKIPVGDQPTDIEAQTRSLIMEYISRPNSIILAVSPANFDIVNSEGLKLARSVDP 229
Query: 205 TGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKE 264
G+RT GVLTKLDLMD+GTNA+D+L GR Y L+ +V VNRSQ+DI + M A + E
Sbjct: 230 KGKRTIGVLTKLDLMDQGTNAMDILSGRVYPLKLGFVATVNRSQSDIVSHKSMRDALQSE 289
Query: 265 REYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLG 324
R +F + P Y + ++ G+ YLAK LS L S IR R+P I + ++ + + + ++++ G
Sbjct: 290 RSFFEHHPAYRTIKDRCGTPYLAKTLSNLLVSHIRERLPDIKARLSTLISQTQQQLNNYG 349
Query: 325 RPIALDAGAQLYTILELCRAFERIFKEHLDGGR--------PGGDRIYHVFDNQLPAALR 376
D + +L+ F F +DG GG R+Y +F+N AL
Sbjct: 350 DFKLSDQSQRGIILLQAMNRFANTFIASIDGNSSNIPTKELSGGARLYSIFNNVFTTALN 409
Query: 377 KLPFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSYFRGPAEASVDAVNFVL 436
+ ++LS ++R + + G + L E + L+ L+ P V+ V L
Sbjct: 410 SIDPLQNLSTVDIRTAILNSTGSRATLFLSEMAFDILVKPQLNLLAAPCHQCVELVYEEL 469
Query: 437 KELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTLRLVDMESSYLTVD 493
++ S ++ FP Q AL ++ L + L+ ++S+Y+ +
Sbjct: 470 MKICHYS--GDSDISHFPKLQTALVETVSDLLRENLTPTYSFVESLIAIQSAYINTN 524
Score = 35.8 bits (81), Expect = 0.34
Identities = 18/82 (21%), Positives = 41/82 (49%), Gaps = 2/82 (2%)
Query: 528 VSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALME 587
++SY L + + +PK ++ V +K ++ N +++ +++ +L ED +
Sbjct: 701 ITSYFNLTRKIIIDQVPKVIMHLLVNASKDAIQNRLVSKLYRED--FFDTLLIEDENVKS 758
Query: 588 RRQQCAKRLELYKAARDEIDSV 609
R++C + L +Y A I +V
Sbjct: 759 EREKCERLLSVYNQANKIISTV 780
>VPS1_YEAST (P21576) Vacuolar sorting protein 1
Length = 704
Score = 317 bits (813), Expect = 4e-86
Identities = 203/541 (37%), Positives = 295/541 (54%), Gaps = 68/541 (12%)
Query: 2 ESLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGRD 61
E LI +N++Q A LG G S S + LP + VVG QSSGKSSVLE+IVGRD
Sbjct: 3 EHLISTINKLQDALAPLG-------GGSQSPI--DLPQITVVGSQSSGKSSVLENIVGRD 53
Query: 62 FLPRGSGIVTRRPLVLQL-----------------------------------------H 80
FLPRG+GIVTRRPLVLQL
Sbjct: 54 FLPRGTGIVTRRPLVLQLINRRPKKSEHAKVNQTANELIDLNINDDDKKKDESGKHQNEG 113
Query: 81 KLETGSQEYAEFLHMPRRKFTDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVN 140
+ E +E+ EFLH+P +KF +F +R+EI ETD+VTG IS +PI+L IYSP+V+
Sbjct: 114 QSEDNKEEWGEFLHLPGKKFYNFDEIRKEIVKETDKVTGANSGISSVPINLRIYSPHVLT 173
Query: 141 LTLVDLPGLTKVAVEGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAR 200
LTLVDLPGLTKV V QP I ++I+ M+ Y+ KPN+IIL+++ AN D+A SD +KLAR
Sbjct: 174 LTLVDLPGLTKVPVGDQPPDIERQIKDMLLKYISKPNAIILSVNAANTDLANSDGLKLAR 233
Query: 201 EVDPTGERTFGVLTKLDLMDKGTNALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIA 260
EVDP G RT GVLTK+DLMD+GT+ +D+L GR L++ ++ ++NR Q DI + A
Sbjct: 234 EVDPEGTRTIGVLTKVDLMDQGTDVIDILAGRVIPLRYGYIPVINRGQKDIEHKKTIREA 293
Query: 261 RRKEREYFANSPDYGHLANKMGSEYLAKLLSQHLESVIRARIPSITSLINKSLEELESEM 320
ER++F N P Y A+ G+ YLAK L+ L IR +P I + I +L++ ++E+
Sbjct: 294 LENERKFFENHPSYSSKAHYCGTPYLAKKLNSILLHHIRQTLPEIKAKIEATLKKYQNEL 353
Query: 321 DHLGRPIALDAGAQLYTILELCRAFERIFKEHLDGGR--------PGGDRIYHVFDNQLP 372
+LG P +D+ + + +L + F + LDG GG RI +VF
Sbjct: 354 INLG-PETMDSASSV--VLSMITDFSNEYAGILDGEAKELSSQELSGGARISYVFHETFK 410
Query: 373 AALRKL-PFDRHLSLQNVRKVVSEADGYQPHLIAPEQGYRRLIDSALSYFRGPAEASVDA 431
+ L PFD+ + ++R ++ + G P L + + L+ + F P S+
Sbjct: 411 NGVDSLDPFDQ-IKDSDIRTIMYNSSGSAPSLFVGTEAFEVLVKQQIRRFEEP---SLRL 466
Query: 432 VNFVLKELVR--KSIAETKELKRFPTFQAALATAANEALERFREESKKTTLRLVDMESSY 489
V V ELVR K I + R+P + A++ + L+ + + + ++ E +Y
Sbjct: 467 VTLVFDELVRMLKQIISQPKYSRYPALREAISNQFIQFLKDATIPTNEFVVDIIKAEQTY 526
Query: 490 L 490
+
Sbjct: 527 I 527
Score = 41.2 bits (95), Expect = 0.008
Identities = 24/83 (28%), Positives = 46/83 (54%), Gaps = 4/83 (4%)
Query: 528 VSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQ-LSEMLDEDPALM 586
+SSY +V T+ + IPKA++ + ++K + + K GKQ + E+ E+ +
Sbjct: 625 ISSYFSIVKRTIADIIPKALMLKLIVKSKTDIQK---VLLEKLYGKQDIEELTKENDITI 681
Query: 587 ERRQQCAKRLELYKAARDEIDSV 609
+RR++C K +E+ + A + SV
Sbjct: 682 QRRKECKKMVEILRNASQIVSSV 704
>VPS1_SCHPO (Q9URZ5) Vacuolar sorting protein 1
Length = 678
Score = 309 bits (792), Expect = 1e-83
Identities = 197/514 (38%), Positives = 277/514 (53%), Gaps = 41/514 (7%)
Query: 3 SLIGLVNRIQRACTVLGDHGAAADGNSFSSLWEALPTVAVVGGQSSGKSSVLESIVGRDF 62
SLI +VN++Q A + +G LP + VV QSSGKSSVLE+IVGRDF
Sbjct: 4 SLIKVVNQLQEAFSTVGVQNLID-----------LPQITVVRSQSSGKSSVLENIVGRDF 52
Query: 63 LPRGSGIVTRRPLVLQL------------------HKLETGSQEYAEFLHMPRRKFTDFS 104
LPRG+GIVTRRPLVLQL K + S E+ EFLH+P +KF +F
Sbjct: 53 LPRGTGIVTRRPLVLQLINRPSASGKNEETTTDSDGKDQNNSSEWGEFLHLPGQKFFEFE 112
Query: 105 LVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAVEGQPESIVQE 164
+R+EI ET+ TGK IS +PI+L IYSP+V+ LTLVDLPGLTKV V QP I ++
Sbjct: 113 KIREEIVRETEEKTGKNVGISSVPIYLRIYSPHVLTLTLVDLPGLTKVPVGDQPRDIEKQ 172
Query: 165 IETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLTKLDLMDKGTN 224
I MV Y+ K N+IILA++ AN D+A SD +KLAREVDP G RT GVLTK+DLMDKGT+
Sbjct: 173 IREMVLKYISKNNAIILAVNAANTDLANSDGLKLAREVDPEGLRTIGVLTKVDLMDKGTD 232
Query: 225 ALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYFANSPDYGHLANKMGSE 284
+D+L GR L+ +V ++NR Q DI + IA ER +F P YG A G+
Sbjct: 233 VVDILAGRVIPLRLGYVPVINRGQKDIEGKKSIRIALEAERNFFETHPSYGSKAQYCGTP 292
Query: 285 YLAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLG-RPIALDAGAQLYTILELCR 343
+LA+ L+ L IR +P I IN +L + ++E+ LG P+ ++ L I + C
Sbjct: 293 FLARKLNMILMHHIRNTLPEIKVRINAALAKYQAELHSLGDTPVGDNSSIVLNLITDFCN 352
Query: 344 AFERIF---KEHLDGGR-PGGDRIYHVFDNQLPAALRKL-PFDRHLSLQNVRKVVSEADG 398
+ + E L GG RI VF ++ + PFD + ++R ++ + G
Sbjct: 353 EYRTVVDGRSEELSATELSGGARIAFVFHEIFSNGIQAIDPFD-EVKDSDIRTILYNSSG 411
Query: 399 YQPHLIAPEQGYRRLIDSALSYFRGPAEASVDAVNFVLKELVR--KSIAETKELKRFPTF 456
P L + ++ + P S+ V+ + ELVR + + KR+P
Sbjct: 412 PSPSLFMGTAAFEVIVKQQIKRLEDP---SLKCVSLIYDELVRILNQLLQRPIFKRYPLL 468
Query: 457 QAALATAANEALERFREESKKTTLRLVDMESSYL 490
+ + + + + +V ME SY+
Sbjct: 469 KDEFYKVVIGFFRKCMQPTNTLVMDMVAMEGSYI 502
Score = 39.3 bits (90), Expect = 0.031
Identities = 18/82 (21%), Positives = 43/82 (51%), Gaps = 2/82 (2%)
Query: 528 VSSYIGLVADTLRNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALME 587
+ SY +V TL + +PK++ ++ +K+ + + Q+ K + ++L E ++
Sbjct: 599 IMSYFNIVKRTLADMVPKSISLKMIKYSKEHIQHELLEQLYKSQA--FDKLLQESEVTVQ 656
Query: 588 RRQQCAKRLELYKAARDEIDSV 609
RR++C + +E A + + +V
Sbjct: 657 RRKECEQMVESLLQASEIVSNV 678
>MX_ONCMY (Q91192) Interferon-induced GTP-binding protein Mx
Length = 621
Score = 202 bits (515), Expect = 2e-51
Identities = 170/601 (28%), Positives = 290/601 (47%), Gaps = 51/601 (8%)
Query: 36 ALPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHM 95
ALP +AV+G QSSGKSSVLE++ G LPRGSGIVTR PL L++ + + G + + + +
Sbjct: 33 ALPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLELKMKRKKEGEEWHGKISYQ 91
Query: 96 PRRK-FTDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAV 154
+ D S V ++I++ D + G IS I L I SP+V +LTL+DLPG+ +VAV
Sbjct: 92 DHEEEIEDPSDVEKKIREAQDEMAGVGVGISDDLISLEIGSPDVPDLTLIDLPGIARVAV 151
Query: 155 EGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLT 214
+GQPE+I ++I+ ++R ++ K +I L + P N DIAT++A+K+A+EVDP GERT G+LT
Sbjct: 152 KGQPENIGEQIKRLIRKFIMKQETISLVVVPCNVDIATTEALKMAQEVDPEGERTLGILT 211
Query: 215 KLDLMDKGT--NALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYFANSP 272
K DL+DKGT +D++ L ++ + R Q +I + V + A +E+ +F
Sbjct: 212 KPDLVDKGTEETVVDIVHNEVIHLTKGYMIVKCRGQKEIMERVSLTEATEREKAFF---K 268
Query: 273 DYGHLANKMGSEY-----LAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPI 327
++ HL+ + LA+ L+ L I +P + I L E +E++ G
Sbjct: 269 EHAHLSTLYDEGHATIPKLAEKLTLELVHHIEKSLPRLEEQIEAKLSETHAELERYGTGP 328
Query: 328 ALDAGAQLYTILELCRAFERIFKEHLDGGRPGGDRIYHVFDN-QLPAALRKLPFDRHLSL 386
D+ +LY +++ AF + G +VF + KL +R +
Sbjct: 329 PEDSAERLYFLIDKVTAFTQDAINLSTGEEMKSGVRLNVFSTLRKEFGKWKLHLERSGEI 388
Query: 387 QNVRKVVSEADGYQPHLIAPE-------QGYRRLIDSALSYFRGPAEASV----DAVNFV 435
N +++ E D Y+ E + + ++ + GPA + DAV V
Sbjct: 389 FN-QRIEGEVDDYEKTYRGRELPGFINYKTFEVMVKDQIKQLEGPAVKKLKEISDAVRKV 447
Query: 436 LKELVRKSIAETKELKRFPTFQAALATAANEALERFREESKKTTLR-------LVDMESS 488
L + S FP + T EA+++ E + ++ LR +V + S
Sbjct: 448 FLLLAQSSFT------GFPNLLKSAKTKI-EAIKQVNESTAESMLRTQFKMELIVYTQDS 500
Query: 489 YLTVDFFRRLPQEVEKAGGPATP------NVDRYAEGHFRRIASNVSSYIGLVADTLRNT 542
+ R +E E P T + D +A + + ++ SY + + L +
Sbjct: 501 TYSHSLCERKREEDEDQ--PLTEIRSTIFSTDNHAT--LQEMMLHLKSYYWISSQRLADQ 556
Query: 543 IPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYKAA 602
IP + + +++ L + +K+ + ++L ED + +R +L+ A
Sbjct: 557 IPMVIRYLVLQEFASQLQREMLQTLQEKD--NIEQLLKEDIDIGSKRAALQSKLKRLMKA 614
Query: 603 R 603
R
Sbjct: 615 R 615
>MX_SHEEP (P33237) Interferon-induced GTP-binding protein Mx
Length = 654
Score = 199 bits (507), Expect = 1e-50
Identities = 169/607 (27%), Positives = 298/607 (48%), Gaps = 57/607 (9%)
Query: 36 ALPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHM 95
ALP +AV+G QSSGKSSVLE++ G LPRGSGIVTR PLVL+L KLE + + +
Sbjct: 64 ALPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLVLRLKKLEKEGEWKGKVSFL 122
Query: 96 PRR-KFTDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAV 154
R + +D S V +EI + + G+ IS I L + SP+V +LTL+DLPG+T+VAV
Sbjct: 123 DREIEISDASQVEKEISEAQIAIAGEGMGISHELISLEVSSPHVPDLTLIDLPGITRVAV 182
Query: 155 EGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLT 214
QP I +I++++R Y+ + +I L + PAN DIAT++A+++A++VDP G+RT G+LT
Sbjct: 183 GNQPHDIEYQIKSLIRKYILRQETINLVVVPANVDIATTEALRMAQDVDPQGDRTIGILT 242
Query: 215 KLDLMDKGT--NALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYFANSP 272
K DL+DKGT +DV+ + L+ ++ + R Q +I + + A ++ER +F +
Sbjct: 243 KPDLVDKGTEDKVVDVVRNLVFHLKKGYMIVKCRGQQEIQHRLSLDKALQRERIFFEDHT 302
Query: 273 DYGHLANKMGSEY--LAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPIALD 330
+ L + + LA+ L+ L I +P + + I ++ + + E+ G+ I +
Sbjct: 303 HFRDLLEEGRATIPCLAERLTNELIMHICKTLPLLENQIKETHQRITEELQKYGKDIPEE 362
Query: 331 AGAQLYTILELCRAFERIFKEHLDGGRPGGDRIYHVFDNQLP-----AALRKLPFDRHLS 385
++++++E F + ++G G +F +A+ + F++
Sbjct: 363 ESEKMFSLIEKIDTFNKEIISTIEGEEHVGQYDSRLFTKVRAEFCKWSAVVEKNFEK--G 420
Query: 386 LQNVRKVVSEADG-YQPHLIAPEQGYRR---LIDSALSYFRGPAEASVDAVNFVLKELVR 441
+ +RK + + + Y+ + Y+ +I + PA VD ++ V +++R
Sbjct: 421 HEAIRKEIKQFENRYRGRELPGFVNYKTFEIIIKKQVIVLEEPA---VDMLHTV-TDIIR 476
Query: 442 KSIAETKELKRFPTFQAALATAANE----ALERFREESKKTTLRLVDMESSYLTVDFFRR 497
+ E K F F TA ++ LE+ E K L + Y +RR
Sbjct: 477 NTFTEVSG-KHFSEFFNLHRTAKSKIEDIRLEQENEAEKSIRLHFQMEQLVYCQDQVYRR 535
Query: 498 LPQEVEKA------------------GGPATPNVDRYAEGHFRRIASNVSSYIGLVADTL 539
Q+V + P+T + ++ + + +++ +SS+I L+
Sbjct: 536 ALQQVREKEAEEEKKKKSNHYYQSEDSEPSTAEIFQHLMAYHQEVSTRISSHIPLI---- 591
Query: 540 RNTIPKAVVFCQVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELY 599
I V+ Q K+S+L ++ Q +L E ++R+ +RLE
Sbjct: 592 ---IQFFVLRTYGEQLKKSMLQLL------QDKDQYDWLLKERTDTRDKRKFLKERLERL 642
Query: 600 KAARDEI 606
AR +
Sbjct: 643 SRARQRL 649
>MX1_BOVIN (P79135) Interferon-induced GTP-binding protein Mx1
Length = 654
Score = 198 bits (504), Expect = 3e-50
Identities = 166/596 (27%), Positives = 286/596 (47%), Gaps = 35/596 (5%)
Query: 36 ALPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHM 95
ALP +AV+G QSSGKSSVLE++ G LPRGSGIVTR PLVL+L KL + + +
Sbjct: 64 ALPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLVLRLKKLGNEDEWKGKVSFL 122
Query: 96 PRR-KFTDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAV 154
+ + D S V +EI + + G+ IS I L + SP+V +LTL+DLPG+T+VAV
Sbjct: 123 DKEIEIPDASQVEKEISEAQIAIAGEGTGISHELISLEVSSPHVPDLTLIDLPGITRVAV 182
Query: 155 EGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLT 214
QP I +I++++R Y+ + +I L + PAN DIAT++A+++A+EVDP G+RT G+LT
Sbjct: 183 GNQPPDIEYQIKSLIRKYILRQETINLVVVPANVDIATTEALRMAQEVDPQGDRTIGILT 242
Query: 215 KLDLMDKGT--NALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYFANSP 272
K DL+DKGT +DV+ + L+ ++ + R Q DI + + A ++ER +F +
Sbjct: 243 KPDLVDKGTEDKVVDVVRNLVFHLKKGYMIVKCRGQQDIKHRMSLDKALQRERIFFEDHA 302
Query: 273 DYGHL--ANKMGSEYLAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPIALD 330
+ L K LA+ L+ L I +P + + I ++ + + E+ G+ I +
Sbjct: 303 HFRDLLEEGKATIPCLAERLTSELIMHICKTLPLLENQIKETHQRITEELQKYGKDIPEE 362
Query: 331 AGAQLYTILELCRAFERIFKEHLDGGRPGGDRIYHVFDNQLPAALRK--------LPFDR 382
+++ +E F + ++ G+ +D++L +R + +
Sbjct: 363 ESEKMFCPIEKIDTFNKEIISTIE-----GEEFVEQYDSRLFTKVRAEFSKWSAVVEKNF 417
Query: 383 HLSLQNVRKVVSEADG-YQPHLIAPEQGYRRLIDSALSYFRGPAEASVDAVNFVLKELVR 441
+ +RK + + + Y+ + Y+ R E +VD ++ V +++R
Sbjct: 418 EKGYEAIRKEIKQFENRYRGRELPGFVNYKTFETIIKKQVRVLEEPAVDMLHTV-TDIIR 476
Query: 442 KSIAETKELKRFPTFQAALATAANE----ALERFREESKKTTLRLVDMESSYLTVDFFRR 497
+ + K F F TA ++ LE+ E K L + Y +RR
Sbjct: 477 NTFTDVSG-KHFNEFFNLHRTAKSKIEDIRLEQENEAEKSIRLHFQMEQLVYCQDQVYRR 535
Query: 498 LPQEVEKAGGPATPN-------VDRYAEGHFRRIASNVSSYIGLVADTLRNTIPKAVVFC 550
Q+V + N + +E I ++++Y V+ + IP + F
Sbjct: 536 ALQQVREKEAEEEKNKKSNHYFQSQVSEPSTDEIFQHLTAYQQEVSTRISGHIPLIIQFF 595
Query: 551 QVRQAKQSLLNHFYTQIGKKEGKQLSEMLDEDPALMERRQQCAKRLELYKAARDEI 606
+R + L + K+ Q +L E ++R+ +RLE AR +
Sbjct: 596 VLRTYGEQLKKSMLQLLQDKD--QYDWLLKERTDTRDKRKFLKERLERLTRARQRL 649
>MX1_MOUSE (P09922) Interferon-induced GTP-binding protein Mx1
(Influenza resistance protein)
Length = 631
Score = 196 bits (498), Expect = 1e-49
Identities = 124/317 (39%), Positives = 186/317 (58%), Gaps = 6/317 (1%)
Query: 36 ALPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHM 95
ALP +AV+G QSSGKSSVLE++ G LPRGSGIVTR PLVL+L KL+ G + + +
Sbjct: 35 ALPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLVLKLRKLKEGEEWRGKVSYD 93
Query: 96 PRR-KFTDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAV 154
+ +D S V + I + + G IS I L + SPNV +LTL+DLPG+T+VAV
Sbjct: 94 DIEVELSDPSEVEEAINKGQNFIAGVGLGISDKLISLDVSSPNVPDLTLIDLPGITRVAV 153
Query: 155 EGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLT 214
QP I ++I+ +++TY++K +I L + P+N DIAT++A+ +A+EVDP G+RT GVLT
Sbjct: 154 GNQPADIGRQIKRLIKTYIQKQETINLVVVPSNVDIATTEALSMAQEVDPEGDRTIGVLT 213
Query: 215 KLDLMDKGT--NALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYFANSP 272
K DL+D+G LDV+ Y L+ ++ + R Q DI + + + A +KE+ +F +
Sbjct: 214 KPDLVDRGAEGKVLDVMRNLVYPLKKGYMIVKCRGQQDIQEQLSLTEAFQKEQVFFKDHS 273
Query: 273 DYGHLA--NKMGSEYLAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPIALD 330
+ L K LA+ L++ L S I +P + IN S + E+ G I D
Sbjct: 274 YFSILLEDGKATVPCLAERLTEELTSHICKSLPLLEDQINSSHQSASEELQKYGADIPED 333
Query: 331 AGAQLYTILELCRAFER 347
++ ++ AF R
Sbjct: 334 DRTRMSFLVNKISAFNR 350
>MX1_RAT (P18588) Interferon-induced GTP-binding protein Mx1
Length = 652
Score = 196 bits (497), Expect = 2e-49
Identities = 121/317 (38%), Positives = 187/317 (58%), Gaps = 6/317 (1%)
Query: 36 ALPTVAVVGGQSSGKSSVLESIVGRDFLPRGSGIVTRRPLVLQLHKLETGSQEYAEFLHM 95
ALP +AV+G QSSGKSSVLE++ G LPRGSGIVTR PLVL+L +L+ G + + ++
Sbjct: 60 ALPAIAVIGDQSSGKSSVLEALSGVA-LPRGSGIVTRCPLVLKLKQLKQGEKWSGKVIYK 118
Query: 96 PRR-KFTDFSLVRQEIQDETDRVTGKTKQISPIPIHLSIYSPNVVNLTLVDLPGLTKVAV 154
+ + SLV +EI + + G+ +IS I L + SP+V +LTL+DLPG+T+VAV
Sbjct: 119 DTEIEISHPSLVEREINKAQNLIAGEGLKISSDLISLEVSSPHVPDLTLIDLPGITRVAV 178
Query: 155 EGQPESIVQEIETMVRTYVEKPNSIILAISPANQDIATSDAIKLAREVDPTGERTFGVLT 214
QP I +I+ ++ Y++K +I L + P+N DIAT++A+K+A+EVDP G+RT G+LT
Sbjct: 179 GDQPADIEHKIKRLITEYIQKQETINLVVVPSNVDIATTEALKMAQEVDPQGDRTIGILT 238
Query: 215 KLDLMDKGT--NALDVLEGRSYRLQHPWVGIVNRSQADINKNVDMIIARRKEREYFANSP 272
K DL+D+GT +DV+ L+ ++ + R Q DI + + + A +KE+ +F P
Sbjct: 239 KPDLVDRGTEDKVVDVVRNLVCHLKKGYMIVKCRGQQDIQEQLSLAEALQKEQVFFKEHP 298
Query: 273 DYGHLA--NKMGSEYLAKLLSQHLESVIRARIPSITSLINKSLEELESEMDHLGRPIALD 330
+ L K LAK L+ L S I +P + + IN + + E+ G I D
Sbjct: 299 QFRVLLEDGKATVPCLAKRLTMELTSHICKSLPILENQINVNHQIASEELQKYGADIPED 358
Query: 331 AGAQLYTILELCRAFER 347
+L ++ F +
Sbjct: 359 DSKRLSFLMNKINVFNK 375
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,328,584
Number of Sequences: 164201
Number of extensions: 2938861
Number of successful extensions: 9452
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 9298
Number of HSP's gapped (non-prelim): 85
length of query: 613
length of database: 59,974,054
effective HSP length: 116
effective length of query: 497
effective length of database: 40,926,738
effective search space: 20340588786
effective search space used: 20340588786
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0316.4


