

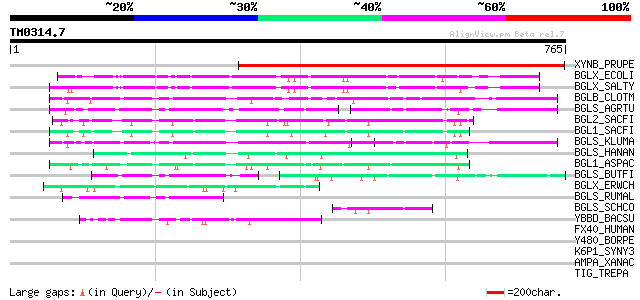
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0314.7
(765 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
XYNB_PRUPE (P83344) Putative beta-D-xylosidase (EC 3.2.1.-) (PpA... 469 e-132
BGLX_ECOLI (P33363) Periplasmic beta-glucosidase precursor (EC 3... 225 4e-58
BGLX_SALTY (Q56078) Periplasmic beta-glucosidase precursor (EC 3... 224 9e-58
BGLB_CLOTM (P14002) Thermostable beta-glucosidase B (EC 3.2.1.21... 176 3e-43
BGLS_AGRTU (P27034) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)... 147 1e-34
BGL2_SACFI (P22507) Beta-glucosidase 2 precursor (EC 3.2.1.21) (... 132 4e-30
BGL1_SACFI (P22506) Beta-glucosidase 1 precursor (EC 3.2.1.21) (... 128 5e-29
BGLS_KLUMA (P07337) Beta-glucosidase precursor (EC 3.2.1.21) (Ge... 124 9e-28
BGLS_HANAN (P06835) Beta-glucosidase precursor (EC 3.2.1.21) (Ge... 112 5e-24
BGL1_ASPAC (P48825) Beta-glucosidase 1 precursor (EC 3.2.1.21) (... 105 4e-22
BGLS_BUTFI (P16084) Beta-glucosidase A (EC 3.2.1.21) (Gentiobias... 81 9e-15
BGLX_ERWCH (Q46684) Periplasmic beta-glucosidase/beta-xylosidase... 75 5e-13
BGLS_RUMAL (P15885) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)... 75 6e-13
BGLS_SCHCO (P29091) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)... 65 5e-10
YBBD_BACSU (P40406) Hypothetical lipoprotein ybbD precursor (ORF1) 53 3e-06
FX40_HUMAN (Q9UH90) F-box only protein 40 (Muscle disease-relate... 34 1.7
Y480_BORPE (Q7VSE8) Hypothetical UPF0191 membrane protein BP0480 33 3.7
K6P1_SYNY3 (P72830) 6-phosphofructokinase 1 (EC 2.7.1.11) (Phosp... 32 4.8
AMPA_XANAC (Q8PGR0) Probable cytosol aminopeptidase (EC 3.4.11.1... 32 4.8
TIG_TREPA (O83519) Trigger factor (TF) 32 6.3
>XYNB_PRUPE (P83344) Putative beta-D-xylosidase (EC 3.2.1.-)
(PpAz152) (Fragment)
Length = 461
Score = 469 bits (1208), Expect = e-132
Identities = 238/453 (52%), Positives = 314/453 (68%), Gaps = 4/453 (0%)
Query: 316 AKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPY 375
A +I AGLDLDCG +L +TE AV++GL+ + IN A++N MRLG FDG PS Y
Sbjct: 1 ADAIKAGLDLDCGPFLAIHTEAAVRRGLVSQLEINWALANTMTVQMRLGMFDGEPSAHQY 60
Query: 376 GGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIG 435
G LGP+DVCT +Q+LA EAARQGIVLL+N SLPLS + +++AVIGPN++ T MIG
Sbjct: 61 GNLGPRDVCTPAHQQLALEAARQGIVLLENRGRSLPLSTRRHRTVAVIGPNSDVTVTMIG 120
Query: 436 NYEGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQLDDATKIAA-SSDATVIVVGASL 494
NY G+ C Y +PLQG+ T + GC DVHC QL A + AA +DATV+V+G
Sbjct: 121 NYAGVACGYTTPLQGIGRYTRTIHQAGCTDVHCNGNQLFGAAEAAARQADATVLVMGLDQ 180
Query: 495 AIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWV 554
+IEAE +DR ++LPG QQ LVS VA ASRGP ILV+MSGG +DV+FAK + +I++I+WV
Sbjct: 181 SIEAEFVDRAGLLLPGHQQELVSRVARASRGPTILVLMSGGPIDVTFAKNDPRISAIIWV 240
Query: 555 GYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRF 614
GYPG+AGG AIA+V+FG NP G+LPMTWYP++YV ++PMT+M MRADP+ GYPGRTYRF
Sbjct: 241 GYPGQAGGTAIANVLFGTANPGGKLPMTWYPQNYVTHLPMTDMAMRADPARGYPGRTYRF 300
Query: 615 YKGETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSE--CKSLDVADEHCQNMA 672
Y G VF FG G+ Y+ H + P LV VPL +S K++ V+ C ++
Sbjct: 301 YIGPVVFPFGLGLSYTTFAHNLAHGPTLVSVPLTSLKATANSTMLSKTVRVSHPDCNALS 360
Query: 673 -FDIHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVC 731
D+H+ VKN G M +HT+L+F +PP A K L+G K+H+A SE +VR V VC
Sbjct: 361 PLDVHVDVKNTGSMDGTHTLLVFTSPPDGKWASSKQLMGFHKIHIATGSEKRVRIAVHVC 420
Query: 732 KDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVR 764
K LSVVD G RR+PLG+H L +G+L + +S++
Sbjct: 421 KHLSVVDRFGIRRIPLGEHKLQIGDLSHHVSLQ 453
>BGLX_ECOLI (P33363) Periplasmic beta-glucosidase precursor (EC
3.2.1.21) (Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 765
Score = 225 bits (573), Expect = 4e-58
Identities = 203/707 (28%), Positives = 329/707 (45%), Gaps = 107/707 (15%)
Query: 66 QEKIGNLGDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAAS 125
++ I + D +++ RL IP + + + LHG + FP+ + A+S
Sbjct: 86 RQDIRAMQDQVMELSRLKIPLFFAY-DVLHGQRTV---------------FPISLGLASS 129
Query: 126 FNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDPLLSSKY 185
FN + +G+V + EA + GL +T W+P +++ RDPRWGR E GED L+S
Sbjct: 130 FNLDAVKTVGRVSAYEAA---DDGL-NMT-WAPMVDVSRDPRWGRASEGFGEDTYLTSTM 184
Query: 186 AAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDTFQPP 245
V+ +Q D + V KH+ AY +G + Y +S Q L + + PP
Sbjct: 185 GKTMVEAMQGKSPAD--RYSVMTSVKHFAAYGAV--EGGKEYN-TVDMSPQRLFNDYMPP 239
Query: 246 FKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQ 305
+K+ +D +VM + N +NG P +D LLK V+R QW G VSD +++ L K
Sbjct: 240 YKAG-LDAGSGAVMVALNSLNGTPATSDSWLLKDVLRDQWGFKGITVSDHGAIKELIK-- 296
Query: 306 HYTKT-PEEAAAKSILAGLDLDCGS-YLGKYTEGAVKQGLLDEASINNAISNNFATLMRL 363
H T PE+A ++ +G+++ Y KY G +K G + A +++A + +
Sbjct: 297 HGTAADPEDAVRVALKSGINMSMSDEYYSKYLPGLIKSGKVTMAELDDAARHVLNVKYDM 356
Query: 364 GFFDGNPSTLPYGGLGPKD---VCTSENQEL----AREAARQGIVLLKNSPGSLPLSAKS 416
G F+ PY LGPK+ V T+ L ARE AR+ +VLLKN +LPL K
Sbjct: 357 GLFND-----PYSHLGPKESDPVDTNAESRLHRKEAREVARESLVLLKNRLETLPL--KK 409
Query: 417 IKSLAVIGPNANATRVMIGNYE--GIPCKYISPLQGLTALVPTS----YVPGCP------ 464
++AV+GP A++ R ++G++ G+ + ++ L G+ V + Y G
Sbjct: 410 SATIAVVGPLADSKRDVMGSWSAAGVADQSVTVLTGIKNAVGENGKVLYAKGANVTSDKG 469
Query: 465 --------------DVHCASAQLDDATKIAASSDATVIVVGASLAIEAESLDRVNIVLPG 510
D +D+A + A SD V VVG + + E+ R +I +P
Sbjct: 470 IIDFLNQYEEAVKVDPRSPQEMIDEAVQTAKQSDVVVAVVGEAQGMAHEASSRTDITIPQ 529
Query: 511 QQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIF 570
Q+ L++ + A+ P++LV+M+G ++ K + + +IL + G GG AIADV+F
Sbjct: 530 SQRDLIAAL-KATGKPLVLVLMNG--RPLALVKEDQQADAILETWFAGTEGGNAIADVLF 586
Query: 571 GFYNPSGRLPMTWYPESYVENVPM------TNMNMRADPSTGYPGRTYRFYKGETVFSFG 624
G YNPSG+LPM+ +P S V +P+ T AD Y R + G ++ FG
Sbjct: 587 GDYNPSGKLPMS-FPRS-VGQIPVYYSHLNTGRPYNADKPNKYTSRYFDEANG-ALYPFG 643
Query: 625 DGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECKSLDVADEHCQNMAFDIHLRVKNMGK 684
G+ Y+ VK L+ + R + + ++V N GK
Sbjct: 644 YGLSYTTFTVSDVK--------LSAPTMKRDGKVTA---------------SVQVTNTGK 680
Query: 685 MSSSHTVLLFFTP-PAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDV 730
+ V ++ A + P K L G EK+ L V F +D+
Sbjct: 681 REGATVVQMYLQDVTASMSRPVKQLKGFEKITLKPGETQTVSFPIDI 727
>BGLX_SALTY (Q56078) Periplasmic beta-glucosidase precursor (EC
3.2.1.21) (Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase) (T-cell inhibitor)
Length = 765
Score = 224 bits (570), Expect = 9e-58
Identities = 213/738 (28%), Positives = 345/738 (45%), Gaps = 111/738 (15%)
Query: 56 VADLVKRLTLQEKIGNLGDSAVDV-------------GRLG-----IPRYE--WWSEALH 95
V DL+K++T+ EKIG L +V G++G + R + + +
Sbjct: 38 VTDLLKKMTVDEKIGQLRLISVGPDNPKEAIREMIKDGQVGAIFNTVTRQDIRQMQDQVM 97
Query: 96 GVSNIGPGTRFS-SVVPGA-TSFPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGL 153
+S + F+ VV G T FP+ + A+SFN + +G+V + EA + GL +
Sbjct: 98 ALSRLKIPLFFAYDVVHGQRTVFPISLGLASSFNLDAVRTVGRVSAYEAA---DDGL-NM 153
Query: 154 TYWSPNINIFRDPRWGRGQETPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHY 213
T W+P +++ RDPRWGR E GED L+S VK +Q D + V KH+
Sbjct: 154 T-WAPMVDVSRDPRWGRASEGFGEDTYLTSIMGETMVKAMQGKSPAD--RYSVMTSVKHF 210
Query: 214 TAYDVDNWKGVQRYTFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCAD 273
AY +G + Y +S Q L + + PP+K+ +D +VM + N +NG P +D
Sbjct: 211 AAYGAV--EGGKEYN-TVDMSSQRLFNDYMPPYKAG-LDAGSGAVMVALNSLNGTPATSD 266
Query: 274 PDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKT-PEEAAAKSILAGLDLD-CGSYL 331
LLK V+R +W G VSD +++ L K H T PE+A ++ AG+D+ Y
Sbjct: 267 SWLLKDVLRDEWGFKGITVSDHGAIKELIK--HGTAADPEDAVRVALKAGVDMSMADEYY 324
Query: 332 GKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKD---VCTSEN 388
KY G +K G + A +++A + +G F+ PY LGPK+ V T+
Sbjct: 325 SKYLPGLIKSGKVTMAELDDATRHVLNVKYDMGLFND-----PYSHLGPKESDPVDTNAE 379
Query: 389 QEL----AREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNYE--GIPC 442
L ARE AR+ +VLLKN +LPL K ++AV+GP A++ R ++G++ G+
Sbjct: 380 SRLHRKEAREVARESVVLLKNRLETLPL--KKSGTIAVVGPLADSQRDVMGSWSAAGVAN 437
Query: 443 KYISPLQGLTALVPTS----YVPGCP--------------------DVHCASAQLDDATK 478
+ ++ L G+ V Y G D A +D+A +
Sbjct: 438 QSVTVLAGIQNAVGDGAKILYAKGANITNDKGIVDFLNLYEEAVKIDPRSPQAMIDEAVQ 497
Query: 479 IAASSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMD 538
A +D V VVG S + E+ R NI +P Q+ L++ + A+ P++LV+M+ G
Sbjct: 498 AAKQADVVVAVVGESQGMAHEASSRTNITIPQSQRDLITAL-KATGKPLVLVLMN--GRP 554
Query: 539 VSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPM--TN 596
++ K + + +IL + G GG AIADV+FG YNPSG+LP++ +P S V +P+ ++
Sbjct: 555 LALVKEDQQADAILETWFAGTEGGNAIADVLFGDYNPSGKLPIS-FPRS-VGQIPVYYSH 612
Query: 597 MNMRADPSTGYPGRTYRFYKGET---VFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVC 653
+N + P + Y E ++ FG G+ Y+ + V L+ +
Sbjct: 613 LNTGRPYNPEKPNKYTSRYFDEANGPLYPFGYGLSYTTF--------TVSDVTLSSPTMQ 664
Query: 654 RSSECKSLDVADEHCQNMAFDIHLRVKNMGKMSSSHTVLLFFTP-PAVHNAPQKHLLGIE 712
R + + + V N GK + + ++ A + P K L G E
Sbjct: 665 RDGKVTA---------------SVEVTNTGKREGATVIQMYLQDVTASMSRPVKQLKGFE 709
Query: 713 KVHLAGKSEAQVRFMVDV 730
K+ L V F +D+
Sbjct: 710 KITLKPGERKTVSFPIDI 727
>BGLB_CLOTM (P14002) Thermostable beta-glucosidase B (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 754
Score = 176 bits (445), Expect = 3e-43
Identities = 192/731 (26%), Positives = 315/731 (42%), Gaps = 113/731 (15%)
Query: 56 VADLVKRLTLQEKIGNLGDSAVD------VGRLGIPRYEWWSEALHGVSNIGPGTRFSSV 109
+ ++K++TL+EK G S +D V RLGIP ++ HG+ + +
Sbjct: 5 IKKIIKQMTLEEKAGLC--SGLDFWHTKPVERLGIPSI-MMTDGPHGLRKQREDAEIADI 61
Query: 110 ---VPGATSFPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDP 166
VP AT FP A S++ L + +G + E +A L G P NI R P
Sbjct: 62 NNSVP-ATCFPSAAGLACSWDRELVERVGAALGEECQAENVSILLG-----PGANIKRSP 115
Query: 167 RWGRGQETPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQR 226
GR E EDP LSS+ AA ++KG+Q V AC KH+ A + ++ +R
Sbjct: 116 LCGRNFEYFPEDPYLSSELAASHIKGVQSQG--------VGACLKHFAANNQEH----RR 163
Query: 227 YTFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWK 286
T + +V ++ L + + F++ V VMC+YN++NG+ + LL V++ +W
Sbjct: 164 MTVDTIVDERTLREIYFASFENAVKKARPWVVMCAYNKLNGEYCSENRYLLTEVLKNEWM 223
Query: 287 LNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLG---KYTEGAVKQGL 343
+G++VSD +V + AGLDL+ + G K AVK G
Sbjct: 224 HDGFVVSDWGAV--------------NDRVSGLDAGLDLEMPTSHGITDKKIVEAVKSGK 269
Query: 344 LDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLL 403
L E +N A+ ++ + Y + LAR+AA + +VLL
Sbjct: 270 LSENILNRAVERILKVIIMA--LENKKENAQYE--------QDAHHRLARQAAAESMVLL 319
Query: 404 KNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGC 463
KN LPL K ++A+IG R Y+G +I+P + L + G
Sbjct: 320 KNEDDVLPL--KKSGTIALIGAFVKKPR-----YQGSGSSHITPTR-LDDIYEEIKKAGA 371
Query: 464 PDVHCASAQ-------------LDDATKIAASSDATVIVVGASLAIEAESLDRVNIVLPG 510
V+ ++ +++A K A+SSD V+ G E+E DR ++ +P
Sbjct: 372 DKVNLVYSEGYRLENDGIDEELINEAKKAASSSDVAVVFAGLPDEYESEGFDRTHMSIPE 431
Query: 511 QQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIF 570
Q L+ VA + +++V+++G +++ + DK+ S+L G+A G + +
Sbjct: 432 NQNRLIEAVAEV-QSNIVVVLLNGSPVEMPWI---DKVKSVLEAYLGGQALGGRWR-MCY 486
Query: 571 GFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGR--TYRFY--KG-ETVFSFGD 625
+ G+L T +P N N D G YR+Y KG E +F FG
Sbjct: 487 SVKSIVGKLAET-FPVKLSHNPSYLNFPGEDDRVEYKEGLFVGYRYYDTKGIEPLFPFGH 545
Query: 626 GIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECKSLDVADEHCQNMAFDIHLRVKNMGKM 685
G+ Y+ E+ + + DV+D N ++ ++VKN+GKM
Sbjct: 546 GLSYTKFEYSDISVDK-------------------KDVSD----NSIINVSVKVKNVGKM 582
Query: 686 SSSHTVLLFFTP-PAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVVDELGNRR 744
+ V L+ + P+K L G EKV L E V F +D ++ +
Sbjct: 583 AGKEIVQLYVKDVKSSVRRPEKELKGFEKVFLNPGEEKTVTFTLDKRAFAYYNTQIKDWH 642
Query: 745 VPLGQHLLHVG 755
V G+ L+ +G
Sbjct: 643 VESGEFLILIG 653
>BGLS_AGRTU (P27034) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)
(Cellobiase) (Beta-D-glucoside glucohydrolase)
Length = 818
Score = 147 bits (371), Expect = 1e-34
Identities = 123/408 (30%), Positives = 199/408 (48%), Gaps = 55/408 (13%)
Query: 56 VADLVKRLTLQEKIGNLGDS----AVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVP 111
+ D++ ++TL+E++ L + V + RLG+P+ + ++ +G G S+V
Sbjct: 2 IDDILDKMTLEEQVSLLSGADFWTTVAIERLGVPKIKV-TDGPNGARGGG------SLVG 54
Query: 112 GATS--FPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWG 169
G S FP+ I A+++ L + G + +A++ +P +NI R G
Sbjct: 55 GVKSACFPVAIALGATWDPELIERAGVALGGQAKSK-----GASVLLAPTVNIHRSGLNG 109
Query: 170 RGQETPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTF 229
R E EDP L++ A Y+ G+Q VAA KH+ A N ++R T
Sbjct: 110 RNFECYSEDPALTAACAVAYINGVQSQG--------VAATIKHFVA----NESEIERQTM 157
Query: 230 NAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNG 289
++ V ++ L + + PPF+ V V +VM SYN++NG T +P LL V+R +W +G
Sbjct: 158 SSDVDERTLREIYLPPFEEAVKKAGVKAVMSSYNKLNGTYTSENPWLLTKVLREEWGFDG 217
Query: 290 YIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDC-GSY--LGKYTEGAVKQGLLDE 346
++SD + H T A++I AGLDL+ G + G+ AV++G +
Sbjct: 218 VVMSD-------WFGSHST-------AETINAGLDLEMPGPWRDRGEKLVAAVREGKVKA 263
Query: 347 ASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNS 406
++ + L R+G F+ P L + E++ L R+ +G VLLKN
Sbjct: 264 ETVRASARRILLLLERVGAFEKAPD------LAEHALDLPEDRALIRQLGAEGAVLLKND 317
Query: 407 PGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKY-ISPLQGLTA 453
G LPL+ S +AVIGPNA + RVM G I Y +SPL+G+ A
Sbjct: 318 -GVLPLAKSSFDQIAVIGPNAASARVMGGGSARIAAHYTVSPLEGIRA 364
Score = 89.0 bits (219), Expect = 4e-17
Identities = 85/301 (28%), Positives = 140/301 (46%), Gaps = 53/301 (17%)
Query: 471 AQLDDATKIAASSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILV 530
A + +A + A SD +++VG + E LD ++ LPG+Q+ L+ VA + V++V
Sbjct: 531 AGIAEAVETARKSDIVLLLVGREGEWDTEGLDLPDMRLPGRQEELIEAVAETNPN-VVVV 589
Query: 531 IMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVE 590
+ +GG +++ + K+ ++L + YPG+ G A+ADV+FG P+GRLP T +P++ +
Sbjct: 590 LQTGGPIEMPWL---GKVRAVLQMWYPGQELGNALADVLFGDVEPAGRLPQT-FPKALTD 645
Query: 591 NVPMTNMNMRADPSTGYPGRT--YRFYKG-------------ETVFSFGDGIGYSNVEHK 635
N +T+ DPS YPG+ R+ +G E +F FG G+GY+
Sbjct: 646 NSAITD-----DPSI-YPGQDGHVRYAEGIFVGYRHHDTREIEPLFPFGFGLGYTRFTW- 698
Query: 636 IVKAPQLVFVPLAEDHVCRSSECKSLDVADEHCQNMAFDIHLRVKNMGKMSSSHTVLLF- 694
APQL + D + + V N+G + S V L+
Sbjct: 699 --GAPQLSGTEMGAD---------------------GLTVTVDVTNIGDRAGSDVVQLYV 735
Query: 695 FTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVVD-ELGNRRVPLGQHLLH 753
+P A P K L K+ LA + + +DL+ D E G R G++ L
Sbjct: 736 HSPNARVERPFKELRAFAKLKLAPGATGTAVLKI-APRDLAYFDVEAGRFRADAGKYELI 794
Query: 754 V 754
V
Sbjct: 795 V 795
>BGL2_SACFI (P22507) Beta-glucosidase 2 precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 880
Score = 132 bits (331), Expect = 4e-30
Identities = 167/685 (24%), Positives = 277/685 (40%), Gaps = 167/685 (24%)
Query: 59 LVKRLTLQEKIG-------NLGDSAVDVGRLGIPRYEWWSEALHGVSNI----GP-GTRF 106
LV ++T+ EK+ LG + G +PR+ G+ N+ GP G R
Sbjct: 66 LVSQMTIVEKVNLTTGTGWQLGPCVGNTG--SVPRF--------GIPNLCLQDGPLGVRL 115
Query: 107 SSVVPGATSFPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDP 166
+ +T +P + T A+FN LF G+ + E + S ++I P
Sbjct: 116 TDF---STGYPSGMATGATFNKDLFLQRGQALGHE-------------FNSKGVHIALGP 159
Query: 167 ---------RWGRGQETPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYD 217
R GR E G DP L AA +KGLQ+ + V AC KH+ +
Sbjct: 160 AVGPLGVKARGGRNFEAFGSDPYLQGIAAAATIKGLQENN--------VMACVKHFIGNE 211
Query: 218 VDNWKG------------VQRYTFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQV 265
D ++ + + +A + + + + + PF + G V SVMCSYN+V
Sbjct: 212 QDIYRQPSNSKVDPEYDPATKESISANIPDRAMHELYLWPFADSIRAG-VGSVMCSYNRV 270
Query: 266 NGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDL 325
N +C + ++ +++ + G++VSD + + + A S ++GLD+
Sbjct: 271 NNTYSCENSYMINHLLKEELGFQGFVVSDWAA--------------QMSGAYSAISGLDM 316
Query: 326 DC-GSYLGKYTEGAVKQGL-LDEASINNAI------------------SNNFATLMRLGF 365
G LG + G G L +A N + +N+F T RL
Sbjct: 317 SMPGELLGGWNTGKSYWGQNLTKAVYNETVPIERLDDMATRILAALYATNSFPTKDRLPN 376
Query: 366 FDGNPSTLPYGG------LGPK-------DVCTSENQELAREAARQGIVLLKNSPGSLPL 412
F +T YG P D ++ A + A + IVLLKN +LP+
Sbjct: 377 FSSF-TTKEYGNEFFVDKTSPVVKVNHFVDPSNDFTEDTALKVAEESIVLLKNEKNTLPI 435
Query: 413 SAKSIKSLAVIGPNANATRVMIGNYE--------------------GIPCKYISPLQGLT 452
S ++ L + G A YE G P ++P + ++
Sbjct: 436 SPNKVRKLLLSGIAAGPDPK---GYECSDQSCVDGALFEGWGSGSVGYPKYQVTPFEEIS 492
Query: 453 ALVPTSYVPGCPDVHCASAQLDDATKIAASSDATVIVVGAS-----LAIEAESLDRVNIV 507
A + + D S L + +A+ + +++VV A L I+ D+ N+
Sbjct: 493 ANARKNKMQF--DYIRESFDLTQVSTVASDAHMSIVVVSAVSGEGYLIIDGNRGDKNNVT 550
Query: 508 LPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIAD 567
L L+ VA V+ VI S G +DV + +T+I+W G G+ G AIA+
Sbjct: 551 LWHNSDNLIKAVAENCANTVV-VITSTGQVDVESFADHPNVTAIVWAGPLGDRSGTAIAN 609
Query: 568 VIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRT----------YRFYKG 617
++FG NPSG LP T +S + +P+ N P G P YR+++
Sbjct: 610 ILFGNANPSGHLPFT-VAKSNDDYIPIVTYN----PPNGEPEDNTLAEHDLLVDYRYFEE 664
Query: 618 ETV---FSFGDGIGYSNVEHKIVKA 639
+ + ++FG G+ Y+ E+K+ A
Sbjct: 665 KNIEPRYAFGYGLSYN--EYKVSNA 687
>BGL1_SACFI (P22506) Beta-glucosidase 1 precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 876
Score = 128 bits (322), Expect = 5e-29
Identities = 161/676 (23%), Positives = 272/676 (39%), Gaps = 153/676 (22%)
Query: 55 RVADLVKRLTLQEKIGNLGDSAVD----VGRLG-IPRYEWWSEALHGVSNI----GP-GT 104
R +V ++T+ EK+ + VG G +PR+ G+ N+ GP G
Sbjct: 60 RAKAIVGQMTIVEKVNLTTGTGWQLDPCVGNTGSVPRF--------GIPNLCLQDGPLGV 111
Query: 105 RFSSVVPGATSFPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFR 164
RF+ V G +P + T A+FN LF G+ + E + S ++I
Sbjct: 112 RFADFVTG---YPSGLATGATFNKDLFLQRGQALGHE-------------FNSKGVHIAL 155
Query: 165 DP---------RWGRGQETPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTA 215
P R GR E G DP L AA +KGLQ+ + V AC KH+
Sbjct: 156 GPAVGPLGVKARGGRNFEAFGSDPYLQGTAAAATIKGLQENN--------VMACVKHFIG 207
Query: 216 YDVDNWKG----------VQRYTFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQV 265
+ + ++ + +A + + + + PF V G V SVMCSYN+V
Sbjct: 208 NEQEKYRQPDDINPATNQTTKEAISANIPDRAMHALYLWPFADSVRAG-VGSVMCSYNRV 266
Query: 266 NGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDL 325
N C + ++ +++ + G++VSD + + + S ++GLD+
Sbjct: 267 NNTYACENSYMMNHLLKEELGFQGFVVSDWGA--------------QLSGVYSAISGLDM 312
Query: 326 DC-GSYLGKYTEGAVKQGL-LDEASINNAI------------------SNNFATLMRLGF 365
G G + G G L +A N + +N+F T L
Sbjct: 313 SMPGEVYGGWNTGTSFWGQNLTKAIYNETVPIERLDDMATRILAALYATNSFPTEDHLPN 372
Query: 366 FDGNPSTLPYGGLGPKDVCTSE-------------NQELAREAARQGIVLLKNSPGSLPL 412
F + +T YG D T ++ A + A + IVLLKN +LP+
Sbjct: 373 FS-SWTTKEYGNKYYADNTTEIVKVNYNVDPSNDFTEDTALKVAEESIVLLKNENNTLPI 431
Query: 413 SAKSIKSLAVIGPNANATRVMI-----------------GNYEGIPCKYISPLQGLTALV 455
S + K L + G A + G P ++P + ++ L
Sbjct: 432 SPEKAKRLLLSGIAAGPDPIGYQCEDQSCTNGALFQGWGSGSVGSPKYQVTPFEEISYLA 491
Query: 456 PTSYVPGCPDVHCASAQLDDATKIAASSDATVIVVGAS-----LAIEAESLDRVNIVLPG 510
+ + D S L TK+A+ + +++VV A+ + ++ DR N+ L
Sbjct: 492 RKNKMQF--DYIRESYDLAQVTKVASDAHLSIVVVSAASGEGYITVDGNQGDRKNLTLWN 549
Query: 511 QQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIF 570
L+ VA V++V +G FA + +T+I+W G G+ G AIA+++F
Sbjct: 550 NGDKLIETVAENCANTVVVVTSTGQINFEGFAD-HPNVTAIVWAGPLGDRSGTAIANILF 608
Query: 571 GFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRT----------YRFYKGETV 620
G NPSG LP T ++ + +P+ + PS+G P YR+++ + +
Sbjct: 609 GKANPSGHLPFT-IAKTDDDYIPIETYS----PSSGEPEDNHLVENDLLVDYRYFEEKNI 663
Query: 621 ---FSFGDGIGYSNVE 633
++FG G+ Y+ E
Sbjct: 664 EPRYAFGYGLSYNEYE 679
>BGLS_KLUMA (P07337) Beta-glucosidase precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 845
Score = 124 bits (311), Expect = 9e-28
Identities = 135/464 (29%), Positives = 200/464 (43%), Gaps = 58/464 (12%)
Query: 56 VADLVKRLTLQEKIGNLGDSAVD------VGRLGIPRYEWWSEALHGVSNIGPGTRFSSV 109
V L+ L EKI L SAVD + RLGIP S+ +G+ GT+F
Sbjct: 6 VEQLLSELNQDEKISLL--SAVDFWHTKKIERLGIPAVRV-SDGPNGIR----GTKFFDG 58
Query: 110 VPGATSFPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWG 169
VP FP A++F+ L + GK+++ E+ A + G P N+ R P G
Sbjct: 59 VPSGC-FPNGTGLASTFDRDLLETAGKLMAKESIAKNAAVILG-----PTTNMQRGPLGG 112
Query: 170 RGQETPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTF 229
RG E+ EDP L+ + VKG+Q G+ +AA KH+ D+++ QR++
Sbjct: 113 RGFESFSEDPYLAGMATSSVVKGMQ----GEG----IAATVKHFVCNDLED----QRFSS 160
Query: 230 NAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNG 289
N++VS++ L + + PF+ V N +M +YN+VNG LL ++R +WK +G
Sbjct: 161 NSIVSERALREIYLEPFRLAVKHANPVCIMTAYNKVNGDHCSQSKKLLIDILRDEWKWDG 220
Query: 290 YIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASI 349
++SD YT A +I GLD++ T V L I
Sbjct: 221 MLMSDWFGT--------YT------TAAAIKNGLDIEFPGPTRWRTRALVSHSLNSREQI 266
Query: 350 NNAISNNFA--TLMRLGFFDGNPSTLPYGGLGPKDVC--TSENQELAREAARQGIVLLKN 405
++ L + F N GP+ T E +L RE A IVLLKN
Sbjct: 267 TTEDVDDRVRQVLKMIKFVVDNLEKTGIVENGPESTSNNTKETSDLLREIAADSIVLLKN 326
Query: 406 SPGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKY-ISPLQGLTALV--PTSYVPG 462
L++K + VIGPNA A G + Y +SP +G+ + Y G
Sbjct: 327 KNNY--LTSKERRQYHVIGPNAKAKTSSGGGSASMNSYYVVSPYEGIVNKLGKEVDYTVG 384
Query: 463 CPDVH----CASAQLDDATKIAASSDATVIVVGASLAIEAESLD 502
A + L DA K A + +A +I S +E S D
Sbjct: 385 AYSHKSIGGLAESSLIDAAKPADAENAGLIAKFYSNPVEERSED 428
Score = 87.8 bits (216), Expect = 1e-16
Identities = 81/294 (27%), Positives = 130/294 (43%), Gaps = 44/294 (14%)
Query: 472 QLDDATKIAASSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVI 531
++ +A ++AA D V+++G + E E DR N+ LP + LV V A+ VI+
Sbjct: 563 EIRNAAELAAKHDKAVLIIGLNGEWETEGYDRENMDLPKRTNELVRAVLKANPNTVIV-- 620
Query: 532 MSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVEN 591
+ G V F ++ +++ Y G G AIADV++G P+G+L ++W P +N
Sbjct: 621 -NQSGTPVEFPWL-EEANALVQAWYGGNELGNAIADVLYGDVVPNGKLSLSW-PFKLQDN 677
Query: 592 VPMTNMNMR-------ADPSTGYPGRTYRFYKGETVFSFGDGIGYSNVEHKIVKAPQLVF 644
N D GY R Y + + F FG G+ Y+ E
Sbjct: 678 PAFLNFKTEFGRVVYGEDIFVGY--RYYEKLQRKVAFPFGYGLSYTTFE----------- 724
Query: 645 VPLAEDHVCRSSECKSLDVADEHCQNMAFDIHLRVKNMG-KMSSSHTVLLFFTP-PAVHN 702
LD++D + DI + VKN G K + S V ++F+ + +
Sbjct: 725 ----------------LDISDFKVTDDKIDISVDVKNTGDKFAGSEVVQVYFSALNSKVS 768
Query: 703 APQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVV-DELGNRRVPLGQHLLHVG 755
P K L G EKVHL + V +++ +S +ELG V G++L+ VG
Sbjct: 769 RPVKELKGFEKVHLEPGEKKTVNIELELKDAISYFNEELGKWHVEAGEYLVSVG 822
>BGLS_HANAN (P06835) Beta-glucosidase precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 825
Score = 112 bits (279), Expect = 5e-24
Identities = 129/571 (22%), Positives = 226/571 (38%), Gaps = 77/571 (13%)
Query: 116 FPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETP 175
FP + ++SFN L + +E + + G Y + GRG E
Sbjct: 119 FPCGMAASSSFNKQLIYDRAVAIGSEFKGKGADAILGPVYGPMGVKA----AGGRGWEGH 174
Query: 176 GEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKG-VQRYTFNAVVS 234
G DP L A G+Q + K + +H+ D G + FN S
Sbjct: 175 GPDPYLEGVIAYLQTIGIQSQGVVSTAKHLIGNEQEHFRFAKKDKHAGKIDPGMFNTSSS 234
Query: 235 -QQDLDDT-----FQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLN 288
++DD + PF V G V+S+MCSYN++NG C + LL +++ +
Sbjct: 235 LSSEIDDRAMHEIYLWPFAEAV-RGGVSSIMCSYNKLNGSHACQNSYLLNYLLKEELGFQ 293
Query: 289 GYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDC---GSYLGKYTEGAVKQGLLD 345
G++++D ++ + + AGLD+D Y G AV G L
Sbjct: 294 GFVMTDWGALY--------------SGIDAANAGLDMDMPCEAQYFGGNLTTAVLNGTLP 339
Query: 346 EASINNAISNNFATLMRLGFFDGNPSTLPYGGLG---------------------PKDVC 384
+ +++ + + L+ G NP Y DV
Sbjct: 340 QDRLDDMATRILSALIYSGVH--NPDGPNYNAQTFLTEGHEYFKQQEGDIVVLNKHVDVR 397
Query: 385 TSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNAN---------------- 428
+ N+ +A +A +G+VLLKN +LPL + +K ++++G A
Sbjct: 398 SDINRAVALRSAVEGVVLLKNEHETLPLGREKVKRISILGQAAGDDSKGTSCSLRGCGSG 457
Query: 429 ATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPG--CPDVHCASAQLDDATKIAASSDAT 486
A G+ G +++P G+ A + D +A +D A A+ +
Sbjct: 458 AIGTGYGSGAGTFSYFVTPADGIGARAQQEKISYEFIGDSWNQAAAMDSALYADAAIEVA 517
Query: 487 VIVVGASLA-IEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKAN 545
V G + ++ D N+ L L+ +++ + +++V SG +D+ N
Sbjct: 518 NSVAGEEIGDVDGNYGDLNNLTLWHNAVPLIKNISSINNNTIVIVT-SGQQIDLEPFIDN 576
Query: 546 DKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPE--SYVENVPMTNMNMRADP 603
+ +T++++ Y G+ G +A V+FG NPSG+LP T + Y+ + ++ D
Sbjct: 577 ENVTAVIYSSYLGQDFGTVLAKVLFGDENPSGKLPFTIAKDVNDYIPVIEKVDVPDPVDK 636
Query: 604 STGYPGRTYRF---YKGETVFSFGDGIGYSN 631
T YR+ Y + FG G+ YSN
Sbjct: 637 FTESIYVDYRYFDKYNKPVRYEFGYGLSYSN 667
>BGL1_ASPAC (P48825) Beta-glucosidase 1 precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 860
Score = 105 bits (262), Expect = 4e-22
Identities = 151/656 (23%), Positives = 257/656 (39%), Gaps = 118/656 (17%)
Query: 55 RVADLVKRLTLQEKIGNLGDSAVDVGRL-----GIPRYEWWSEALHGVSNIGPGTRFSSV 109
R +V ++TL EK+ + ++ + G+PR L + G R S
Sbjct: 46 RAVAIVSQMTLDEKVNLTTGTGWELEKCVGQTGGVPRLNIGGMCLQ---DSPLGIRDSDY 102
Query: 110 VPGATSFPMPILTAASFNSTLF----QAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRD 165
++FP + AA+++ L QA+G+ S + + AG + R
Sbjct: 103 ---NSAFPAGVNVAATWDKNLAYLRGQAMGQEFSDKGIDVQLGPAAG--------PLGRS 151
Query: 166 PRWGRGQETPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQ 225
P GR E DP L+ A +KG+Q V A KHY + ++++ V
Sbjct: 152 PDGGRNWEGFSPDPALTGVLFAETIKGIQDAG--------VVATAKHYILNEQEHFRQVA 203
Query: 226 R---YTFN------AVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDL 276
Y FN + V + + + + PF V G V ++MCSYNQ+N C +
Sbjct: 204 EAAGYGFNISDTISSNVDDKTIHEMYLWPFADAVRAG-VGAIMCSYNQINNSYGCQNSYT 262
Query: 277 LKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDL---------DC 327
L +++ + G+++SD + + S LAGLD+
Sbjct: 263 LNKLLKAELGFQGFVMSDWGA--------------HHSGVGSALAGLDMSMPGDITFDSA 308
Query: 328 GSYLG-----KYTEGAVKQGLLDEASINNAIS------------NNFATLMR--LGFFDG 368
S+ G G V Q +D+ ++ + NF++ R GF
Sbjct: 309 TSFWGTNLTIAVLNGTVPQWRVDDMAVRIMAAYYKVGRDRLYQPPNFSSWTRDEYGFKYF 368
Query: 369 NPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNA- 427
P PY + + E+ R+ VLLKN+ +LPL+ K + +A++G +A
Sbjct: 369 YPQEGPYEKVNHFVNVQRNHSEVIRKLGADSTVLLKNN-NALPLTGKE-RKVAILGEDAG 426
Query: 428 ---------------NATRVMIGNYEGIPCKY-ISPLQGLTALVPTSYVPGCPDVHCASA 471
N T M Y ++P Q + A V G +
Sbjct: 427 SNSYGANGCSDRGCDNGTLAMAWGSGTAEFPYLVTPEQAIQAEVLKH--KGSVYAITDNW 484
Query: 472 QLDDATKIAASSDATVIVVGAS-----LAIEAESLDRVNIVLPGQQQLLVSEVANASRGP 526
L +A + +++ V + ++++ DR N+ L L+ AN
Sbjct: 485 ALSQVETLAKQASVSLVFVNSDAGEGYISVDGNEGDRNNLTLWKNGDNLIKAAANNCNN- 543
Query: 527 VILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTW--Y 584
I+VI S G + V + +T+ILW G PG+ G ++ADV++G NP + P TW
Sbjct: 544 TIVVIHSVGPVLVDEWYDHPNVTAILWAGLPGQESGNSLADVLYGRVNPGAKSPFTWGKT 603
Query: 585 PESYVENVPMTNMNMRADPSTGYPGRTYRFYKG-----ET-VFSFGDGIGYSNVEH 634
E+Y + + N P + + Y+G ET ++ FG G+ Y+ +
Sbjct: 604 REAYGDYLVRELNNGNGAPQDDFSEGVFIDYRGFDKRNETPIYEFGHGLSYTTFNY 659
>BGLS_BUTFI (P16084) Beta-glucosidase A (EC 3.2.1.21) (Gentiobiase)
(Cellobiase) (Beta-D-glucoside glucohydrolase)
Length = 830
Score = 81.3 bits (199), Expect = 9e-15
Identities = 117/444 (26%), Positives = 179/444 (39%), Gaps = 68/444 (15%)
Query: 373 LPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSL---AVI------ 423
LP G G + + ++ EL+ EAA +G+VLLKN LP+ + +L V
Sbjct: 13 LPLGENGERVTASQKHIELSCEAACEGMVLLKNDRNVLPIRKGTRVALFGKGVFDYVKGG 72
Query: 424 GPNANATRVMIGN-YEGIPC----------------KYISPLQGLTALVPTSYVPGCPDV 466
G + + T I N YEG+ +Y++ L P P+
Sbjct: 73 GGSGDVTVPYIRNLYEGLSQYTSDISIYDKSVRFYQEYVADQYRLGIAPGMIKEPALPED 132
Query: 467 HCASAQLDDATKIAASS-------DATVIVVGASLAIEAESL--------DRVNIVLPGQ 511
A A T I A S D V V + EA+ L D + L
Sbjct: 133 ILADAAAYADTAIIAISRFSGEGWDRKVAGVDREIKCEAKDLVEQGNKIFDHGDFYLTNA 192
Query: 512 QQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFG 571
++ +V V + VI+V+ GG +D ++ K +D+I+S+L G GG A A ++ G
Sbjct: 193 EKKMVKMVKE-NFSSVIVVMNVGGVVDTTWFKKDDQISSVLMAWQGGIEGGLAAARILLG 251
Query: 572 FYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFY---------KGETVFS 622
NPSG+L T+ + +E+ P T D Y Y Y K + +
Sbjct: 252 KVNPSGKLSDTF--AARLEDYPSTEGFHEDDDYVDYTEDIYVGYRYFETIPGAKEKVNYP 309
Query: 623 FGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECKSLDVADEHCQNMAFDIHLRVKNM 682
FG G+ Y+ + KA FV A D V +S D+AD A + V N+
Sbjct: 310 FGYGLSYTTFLLEDYKAEP--FVASAADEVGKSDS----DLAD------AIVASVTVTNI 357
Query: 683 GKMSSSHTVLLFFT-PPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVVDELG 741
GK+ V L+++ P P K L G K L E+Q + +D++ D+LG
Sbjct: 358 GKIPGKEVVQLYYSAPQGKLGKPAKVLGGYAKTRLLQPGESQRVTIALYMEDMASYDDLG 417
Query: 742 NRRVPLGQHLLHVGNLKYPLSVRI 765
+V LL G + L +
Sbjct: 418 --KVKKAAWLLEKGEYHFFLGTSV 439
Score = 78.2 bits (191), Expect = 8e-14
Identities = 65/231 (28%), Positives = 102/231 (44%), Gaps = 32/231 (13%)
Query: 114 TSFPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQE 173
T+FP L A ++N + +G EA+ N G + +P +NI R P GR E
Sbjct: 606 TAFPCSTLLACTWNEDICYEVGVAGGEEAKEC-NFG----AWLTPAVNIHRSPLCGRNFE 660
Query: 174 TPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVV 233
EDP L+ K AA V+G+Q + + A KH+ N K R ++
Sbjct: 661 YYSEDPFLAGKQAAAMVRGIQSNN--------IIATPKHFAL----NNKESNRKGSDSRA 708
Query: 234 SQQDLDDTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVS 293
S++ + + + F+ V + + + YN VNG+ + DLL G++R +W G +VS
Sbjct: 709 SERAIREIYLKAFEIIVKEQSPGASCLQYNIVNGQRSSESHDLLTGILRDEWGFEGVVVS 768
Query: 294 DCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCG-SYLGKYTEGAVKQGL 343
D +HY K +LAG D+ G Y + E K+ L
Sbjct: 769 DWWGF-----GEHY---------KEVLAGNDIKMGCGYTEQLLEAIDKKAL 805
>BGLX_ERWCH (Q46684) Periplasmic beta-glucosidase/beta-xylosidase
precursor [Includes: Beta-glucosidase (EC 3.2.1.21)
(Gentiobiase) (Cellobiase); Beta-xylosidase (EC
3.2.1.37) (1,4-beta-D-xylan xylohydrolase) (Xylan
1,4-beta-xylosidase)]
Length = 654
Score = 75.5 bits (184), Expect = 5e-13
Identities = 119/464 (25%), Positives = 182/464 (38%), Gaps = 99/464 (21%)
Query: 47 DKSLPVADRVADLVKRLTLQEKIG---------------------------NLGDSAVD- 78
D LP A+R ADLV R+TL EK G + D V+
Sbjct: 60 DWRLPAAERAADLVSRMTLAEKAGVMMHGSAPTAGSVTGAGTQYDLNAAKTMIADRYVNS 119
Query: 79 -VGRLGIPRYEWWSEALHGVSNIGPGTR--------------FSSVVPGATS------FP 117
+ RL +E + + + TR F S+V + S +P
Sbjct: 120 FITRLSGDNPAQMAEENNKLQQLAEATRLGIPLTISTDPRSSFQSLVGVSVSVGKFSKWP 179
Query: 118 MPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGLTY-WSPNINIFRDPRWGRGQETPG 176
+ AA + L + +V E RA+ G+T SP ++ +PRW R T G
Sbjct: 180 ETLGLAAIGDEELVRRFADIVRQEYRAV------GITEALSPQADLATEPRWPRIDGTFG 233
Query: 177 EDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDV--DNWKGVQRYTFNAVVS 234
EDP L+ K GYV G+Q +G N V + KH+ Y D W Y A
Sbjct: 234 EDPDLTKKMVRGYVTGMQNGKNG-LNAQSVISIVKHWVGYGAAKDGWDSHNVYGKYAQFR 292
Query: 235 QQDLDDTFQPPFKSCVIDGNVASVMCSYNQV-----NGKP-----TCADPDLLKGVIRGQ 284
Q +L P + + + A +M +Y+ + +GKP + LL ++RGQ
Sbjct: 293 QNNLQWHIDP--FTGAFEAHAAGIMPTYSILRNASWHGKPIEQVGAGFNRFLLTDLLRGQ 350
Query: 285 WKLNGYIVS------DCDSVEVLFKDQHYTK-------------TPEEAAAKSILAGLDL 325
+ +G I+S DC + L + K TP E K++ AG+D
Sbjct: 351 YGFDGVILSDWLITNDCKG-DCLTGVKPGEKPVPRGMPWGVEKLTPAERFVKAVNAGVDQ 409
Query: 326 DCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKD--V 383
G AV+ G L EA ++ +++ + G F+ PY + V
Sbjct: 410 FGGVTDSALLVQAVQDGKLTEARLDTSVNRILKQKFQTGLFE-----RPYVNATQANDIV 464
Query: 384 CTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNA 427
++ Q+LA + + +VLL+N+ LPL S L I NA
Sbjct: 465 GRADWQQLADDTQARSLVLLQNN-NLLPLRKGSRVWLHGIAANA 507
>BGLS_RUMAL (P15885) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)
(Cellobiase) (Beta-D-glucoside glucohydrolase)
Length = 947
Score = 75.1 bits (183), Expect = 6e-13
Identities = 66/222 (29%), Positives = 96/222 (42%), Gaps = 26/222 (11%)
Query: 73 GDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASFNSTLFQ 132
G A + LGIP G + GP V A S P L AA+FN +L
Sbjct: 501 GGVAKHLEELGIPA---------GCCSDGPSGMRLDVGTKAFSLPNGTLIAATFNKSLIT 551
Query: 133 AIGKVVSTEARAMYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDPLLSSKYAAGYVKG 192
+ + E RA L G P +NI R P GR E EDP L+ AA ++G
Sbjct: 552 ELFTYLGLEMRANKVDCLLG-----PGMNIHRHPLNGRNFEYFSEDPFLTGTMAAAELEG 606
Query: 193 LQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDTFQPPFKSCVID 252
L + + V KH+ A N + R+ ++V S++ L + + F+ V
Sbjct: 607 L--------HSVGVEGTIKHFCA----NNQETNRHFIDSVASERALREIYLKGFEIAVRK 654
Query: 253 GNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSD 294
SVM +Y +VNG T DL ++R QW +G+ ++D
Sbjct: 655 SKARSVMTTYGKVNGLWTAGSFDLNTMILRKQWGFDGFTMTD 696
Score = 44.3 bits (103), Expect = 0.001
Identities = 59/278 (21%), Positives = 110/278 (39%), Gaps = 40/278 (14%)
Query: 473 LDDAT--KIAASSDATVIVVGASLAIEAESLDRVN--IVLPGQQQLLVSEVANASRGPVI 528
LDD+ + A SSD + ++G + E ++ + ++ G++ +L N S+ ++
Sbjct: 113 LDDSLVKRAAESSDTAICIIGRTAGEEQDNSCKAGSYLLTDGEKAILRKVRDNFSK--MV 170
Query: 529 LVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMT--WYPE 586
+++ G +D+ F ++++V G GG A V+ G +P G+LP T +
Sbjct: 171 ILLNVGNIIDMGFIDEFSP-DAVMYVWQGGMTGGTGTARVLLGEVSPCGKLPDTIAYDIT 229
Query: 587 SYVENVPMTNMNMRADPSTGYPGRTY--RFYKGETVFSFGDGIGYSNVEHKIVKAPQLVF 644
Y + N ++ + G Y F K F FG G+ Y+ E
Sbjct: 230 DYPSDKNFHNRDVDIYAEDIFVGYRYFDTFAKDRVRFPFGYGLSYTQFE----------- 278
Query: 645 VPLAEDHVCRSSECKSLDVADEHCQNMAFDIHLRVKNMGKMSSSHTVLLFFTPPAVH-NA 703
S+E + D I +VKN+G + V ++ P
Sbjct: 279 ---------ISAEGRKTD--------DGVVITAKVKNIGSAAGKEVVQVYLEAPNCKLGK 321
Query: 704 PQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVVDELG 741
+ L G EK + +E Q + +D++ D+ G
Sbjct: 322 AARVLCGFEKTKVLAPNEEQTLTIEVTERDIASYDDSG 359
>BGLS_SCHCO (P29091) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)
(Cellobiase) (Beta-D-glucoside glucohydrolase)
(Fragment)
Length = 192
Score = 65.5 bits (158), Expect = 5e-10
Identities = 41/148 (27%), Positives = 76/148 (50%), Gaps = 18/148 (12%)
Query: 445 ISPLQGLTALVPTSYVPGCPDVHCASAQLDD-----ATKIAASSDATVIVVGAS-----L 494
I+PL +TA D ++ L D A +IAA++D ++ + + L
Sbjct: 6 ITPLDAITARAQE-------DGTTVTSSLSDSDTARAAQIAAAADVAIVFISSDSGEGYL 58
Query: 495 AIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWV 554
+E + DR +++ LV VA+A+ ++ V G + ++ + + + +++W
Sbjct: 59 TVEGNAGDRNDLLAWHDGDALVQAVADANENTIVAVNTVGAIITEAWIE-HPNVKAVVWS 117
Query: 555 GYPGEAGGAAIADVIFGFYNPSGRLPMT 582
G PG+ G ++AD+++G YNPSGRLP T
Sbjct: 118 GLPGQEAGNSVADILYGAYNPSGRLPYT 145
>YBBD_BACSU (P40406) Hypothetical lipoprotein ybbD precursor (ORF1)
Length = 642
Score = 53.1 bits (126), Expect = 3e-06
Identities = 82/370 (22%), Positives = 155/370 (41%), Gaps = 69/370 (18%)
Query: 97 VSNIGPGTRFSSVVPGATSFPMPILTAASFNSTLFQAIGKVVSTEARAMYNVGLAGL-TY 155
V+ +G GT F PG + L AA +Q G ++ E A+ G+ T
Sbjct: 129 VTRLGEGTNF----PGNMA-----LGAARSRINAYQT-GSIIGKELSAL------GINTD 172
Query: 156 WSPNINIFRDP-RWGRGQETPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYT 214
+SP ++I +P G + + L+S+ +KGLQ+ D +A+ KH+
Sbjct: 173 FSPVVDINNNPDNPVIGVRSFSSNRELTSRLGLYTMKGLQRQD--------IASALKHFP 224
Query: 215 AY---DVDNWKGVQRYTFNAVVSQQDLDDTFQPPFKSCVIDGNVASVMCSYNQ------- 264
+ DVD+ G+ + Q+ L + PF+ ID VM ++ Q
Sbjct: 225 GHGDTDVDSHYGLPLVSHG----QERLREVELYPFQKA-IDAGADMVMTAHVQFPAFDDT 279
Query: 265 -----VNGK----PTCADPDLLKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAA 315
++G P ++ G++R + NG IV+D +++ + H+ + EEA
Sbjct: 280 TYKSKLDGSDILVPATLSKKVMTGLLRQEMGFNGVIVTDALNMKAIA--DHFGQ--EEAV 335
Query: 316 AKSILAGLDLDCG-------------SYLGKYTEGAVKQGLLDEASINNAISNNFATLMR 362
++ AG+D+ + + + + AVK G + E INN++ + ++
Sbjct: 336 VMAVKAGVDIALMPASVTSLKEEQKFARVIQALKEAVKNGDIPEQQINNSVERIISLKIK 395
Query: 363 LGFFDG--NPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSL 420
G + + ST K V + ++ + ++ A + + +LKN +LP K +
Sbjct: 396 RGMYPARNSDSTKEKIAKAKKIVGSKQHLKAEKKLAEKAVTVLKNEQHTLPFKPKKGSRI 455
Query: 421 AVIGPNANAT 430
++ P T
Sbjct: 456 LIVAPYEEQT 465
>FX40_HUMAN (Q9UH90) F-box only protein 40 (Muscle disease-related
protein)
Length = 709
Score = 33.9 bits (76), Expect = 1.7
Identities = 42/182 (23%), Positives = 70/182 (38%), Gaps = 30/182 (16%)
Query: 172 QETPGEDPLLSSKYAAGYVKGLQQTDDGDSNKLKVAACCKHYTAYDVDNWKGVQRYTFNA 231
++ P E+ A GL DG +LK A K Y Y V N + + +
Sbjct: 266 KKEPQENQKQQDVRTAMETTGLAPWQDGVLERLKTAVDAKDYNMYLVHNGRMLIHFGQMP 325
Query: 232 VVSQQDLD--------------DTFQPPFKSCVIDGNVASVMCSYNQVNGKP-------- 269
+ ++ D TF+ P C + M S K
Sbjct: 326 ACTPKERDFVYGKLEAQEVKTVYTFKVPVSYCGKRARLGDAMLSCKPSEHKAVDTSDLGI 385
Query: 270 TCAD---PDLLKGVIRG--QWKLNGYIVSDCDSVEVLFKD---QHYTKTPEEAAAKSILA 321
T D DL+K ++ + +L G+++S+ S++ LF D Q Y PE+ ++ ++LA
Sbjct: 386 TVGDLPKSDLIKTTLQCALERELKGHVISESRSIDGLFMDFATQTYNFEPEQFSSGTVLA 445
Query: 322 GL 323
L
Sbjct: 446 DL 447
>Y480_BORPE (Q7VSE8) Hypothetical UPF0191 membrane protein BP0480
Length = 206
Score = 32.7 bits (73), Expect = 3.7
Identities = 29/102 (28%), Positives = 43/102 (41%), Gaps = 5/102 (4%)
Query: 60 VKRLTLQEKIGNL----GDSAVDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATS 115
++RLT Q + L G A G L + WW L VS + + G +
Sbjct: 67 LRRLTGQPALVRLRRMCGLFAFFYGSLHFLAWVWWDRGLDPVSMLQDVGERPFITVGFAA 126
Query: 116 FPMPILTAASFNSTLFQAIGKVVSTEARAMYNVG-LAGLTYW 156
F + AA+ + +GK T RA+Y +G LA L +W
Sbjct: 127 FVLMAALAATSTQWAMRKLGKRWQTLHRAVYAIGLLAILHFW 168
>K6P1_SYNY3 (P72830) 6-phosphofructokinase 1 (EC 2.7.1.11)
(Phosphofructokinase 1) (Phosphohexokinase 1)
Length = 361
Score = 32.3 bits (72), Expect = 4.8
Identities = 19/57 (33%), Positives = 28/57 (48%), Gaps = 2/57 (3%)
Query: 485 ATVIVVGASLA--IEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDV 539
AT I +G A I E+LDR++ +++V EV G + L GGG D+
Sbjct: 147 ATEISIGFDTATNIATEALDRLHFTAASHNRVMVLEVMGRDAGHIALAAGIGGGADI 203
>AMPA_XANAC (Q8PGR0) Probable cytosol aminopeptidase (EC 3.4.11.1)
(Leucine aminopeptidase) (LAP) (Leucyl aminopeptidase)
Length = 490
Score = 32.3 bits (72), Expect = 4.8
Identities = 41/174 (23%), Positives = 67/174 (37%), Gaps = 33/174 (18%)
Query: 373 LPYGGLGPKDVCTSENQELAREAA---RQGIVLLKNSP-------GSLPLSAKSIKSLAV 422
L G +G + +E AR+AA RQ + + ++ G + + +LA+
Sbjct: 102 LKTGAVGTALLTLTELTVKARDAAWNIRQAVTVSDHAAYRYTATLGKKKVDETGLTTLAI 161
Query: 423 IGPNANATRVMIGNYEGIPCKYISPLQGLTALVPTSYVPGCPDVHCASAQLDDATKIAAS 482
G +A A V + EG+ ++ L L P +C A L
Sbjct: 162 AGDDARALAVGVATAEGV--EFARELGNL------------PPNYCTPAYL--------- 198
Query: 483 SDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGG 536
+D G EAE LD + G LL +A+R +I++ +GGG
Sbjct: 199 ADTAAAFAGKFRGAEAEILDEAQMEALGMGSLLSVARGSANRPRLIVLKWNGGG 252
>TIG_TREPA (O83519) Trigger factor (TF)
Length = 442
Score = 32.0 bits (71), Expect = 6.3
Identities = 31/140 (22%), Positives = 52/140 (37%), Gaps = 4/140 (2%)
Query: 391 LAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQG 450
L +A +G LP+S S+K V P+ + + +I Y+ P + G
Sbjct: 72 LMEKALEEGFAQASQDSQPLPISRPSLKKKPVFDPDEDFSFAVI--YDVFPSVELRNTSG 129
Query: 451 LTALVPTSYVPGCPDVHCASAQLDDATKIAASSDA-TVIVVGASLAIEAESLDRVNIVLP 509
+ VPT V DV ++ + + A + VG ++ +D V P
Sbjct: 130 FSLSVPTVSVTE-EDVSRELTRIQERNALVTDKGADSCAEVGDIATVDYHEVDDSGAVRP 188
Query: 510 GQQQLLVSEVANASRGPVIL 529
G ++ V GP L
Sbjct: 189 GTERAGVVFTLGVEEGPFAL 208
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 93,393,604
Number of Sequences: 164201
Number of extensions: 4124347
Number of successful extensions: 8668
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 8590
Number of HSP's gapped (non-prelim): 38
length of query: 765
length of database: 59,974,054
effective HSP length: 118
effective length of query: 647
effective length of database: 40,598,336
effective search space: 26267123392
effective search space used: 26267123392
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0314.7


