

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
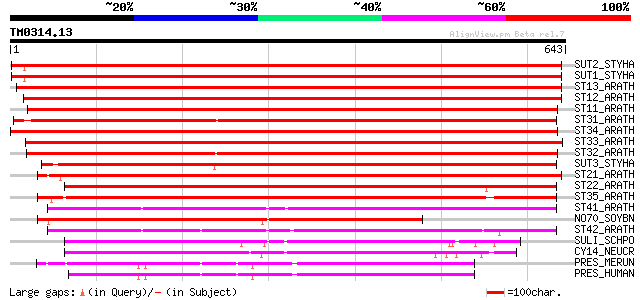
Query= TM0314.13
(643 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SUT2_STYHA (P53392) High affinity sulphate transporter 2 1014 0.0
SUT1_STYHA (P53391) High affinity sulphate transporter 1 1009 0.0
ST13_ARATH (Q9FEP7) Sulfate transporter 1.3 911 0.0
ST12_ARATH (Q9MAX3) Sulfate transporter 1.2 888 0.0
ST11_ARATH (Q9SAY1) Sulfate transporter 1.1 (High-affinity sulfa... 867 0.0
ST31_ARATH (Q9SV13) Sulfate transporter 3.1 (AST12) (AtST1) 667 0.0
ST34_ARATH (Q9LW86) Probable sulfate transporter 3.4 641 0.0
ST33_ARATH (Q9SXS2) Probable sulfate transporter 3.3 (AST91) 627 e-179
ST32_ARATH (O04289) Sulfate transporter 3.2 (AST77) 627 e-179
SUT3_STYHA (P53393) Low affinity sulphate transporter 3 610 e-174
ST21_ARATH (O04722) Sulfate transporter 2.1 (AST68) 604 e-172
ST22_ARATH (P92946) Sulfate transporter 2.2 (AST56) (AtH14) 591 e-168
ST35_ARATH (Q94LW6) Probable sulfate transporter 3.5 521 e-147
ST41_ARATH (Q9FY46) Sulfate transporter 4.1, chloroplast precurs... 389 e-107
NO70_SOYBN (Q02920) Early nodulin 70 389 e-107
ST42_ARATH (Q8GYH8) Probable sulfate transporter 4.2 379 e-104
SULI_SCHPO (Q9URY8) Probable sulfate permease C869.05c 245 3e-64
CY14_NEUCR (P23622) Sulfate permease II 224 5e-58
PRES_MERUN (Q9JKQ2) Prestin 211 5e-54
PRES_HUMAN (P58743) Prestin 211 5e-54
>SUT2_STYHA (P53392) High affinity sulphate transporter 2
Length = 662
Score = 1014 bits (2621), Expect = 0.0
Identities = 511/643 (79%), Positives = 577/643 (89%), Gaps = 6/643 (0%)
Query: 3 DTGSAPSSRRHHG------LPHIHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSR 56
+T + SSRRH G LP++HKVG PKQTL+ EIKHS ETFF D P FK QS
Sbjct: 16 ETRTNSSSRRHGGGDDTPSLPYMHKVGAPPKQTLFQEIKHSFNETFFPDKPFGNFKDQSG 75
Query: 57 KRKLVLGMQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALY 116
RK VLG+Q +FPI+EWGR Y+L+KF+GDFIAGLTIASLCIPQD+AYAKLANL+P + LY
Sbjct: 76 SRKFVLGLQYIFPILEWGRHYDLKKFRGDFIAGLTIASLCIPQDLAYAKLANLDPWYGLY 135
Query: 117 TSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPDYLRLAYTATFFAGIT 176
+SFVAPLVYAFMGTSRDIAIGPVAVVSLLLGT+L++EI+++K+ DYLRLA+TATFFAG+T
Sbjct: 136 SSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTLLSNEISNTKSHDYLRLAFTATFFAGVT 195
Query: 177 QLALGFFRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVW 236
Q+ LG RLGFLIDFLSHAAIVGFM GAAITI LQQLKGLLG+K FTK +DIVSVM SVW
Sbjct: 196 QMLLGVCRLGFLIDFLSHAAIVGFMAGAAITIGLQQLKGLLGIKDFTKNSDIVSVMHSVW 255
Query: 237 KPVEHGWNWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRAD 296
V HGWNWETI+IG+SFL+F+LIT YIAKKNKKLFWV+AI+PMI V+VSTF VYITRAD
Sbjct: 256 SNVHHGWNWETILIGLSFLIFLLITKYIAKKNKKLFWVSAISPMICVIVSTFFVYITRAD 315
Query: 297 KKGVAIVKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDY 356
K+GV IVKHIK GVNP+SA+EIFF G+Y GAGV++GVV+G+VALTEA+AI RTFAAMKDY
Sbjct: 316 KRGVTIVKHIKSGVNPSSANEIFFHGKYLGAGVRVGVVAGLVALTEAMAIGRTFAAMKDY 375
Query: 357 SIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLV 416
SIDGNKEMVAMGTMNIVGSLTS YV TGSFSRSAVNYMAGCKTAVSNIVM++V+LLTLLV
Sbjct: 376 SIDGNKEMVAMGTMNIVGSLTSCYVTTGSFSRSAVNYMAGCKTAVSNIVMAIVVLLTLLV 435
Query: 417 ITPLFKYTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGL 476
ITPLFKYTPNAVLASIIIAAV+NLV+++A +LLWKIDKFDFVACMGAFFGVIFKSVEIGL
Sbjct: 436 ITPLFKYTPNAVLASIIIAAVVNLVNIEAMVLLWKIDKFDFVACMGAFFGVIFKSVEIGL 495
Query: 477 LIAVAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSN 536
LIAVAISFAKILLQVTRPRTA+LGKL GT VYRNI QYPKA QIPGMLIIRVDSAIYFSN
Sbjct: 496 LIAVAISFAKILLQVTRPRTAVLGKLPGTSVYRNIQQYPKAEQIPGMLIIRVDSAIYFSN 555
Query: 537 SNYIKDRILKWVTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREV 596
SNYIK+RIL+W+ DE QRT SE P IQ LIVEMSPVTDIDTSGIHA E+L+K+L+KREV
Sbjct: 556 SNYIKERILRWLIDEGAQRTESELPEIQHLIVEMSPVTDIDTSGIHAFEELYKTLQKREV 615
Query: 597 QLLLANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAVATFGPK 639
QL+LANPGP+VIEKLHAS L+++IGEDKIF +V DAVAT+GPK
Sbjct: 616 QLMLANPGPVVIEKLHASNLAELIGEDKIFLTVADAVATYGPK 658
>SUT1_STYHA (P53391) High affinity sulphate transporter 1
Length = 667
Score = 1009 bits (2610), Expect = 0.0
Identities = 511/648 (78%), Positives = 580/648 (88%), Gaps = 11/648 (1%)
Query: 3 DTGSAPSSRRHHG---------LPHIHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKG 53
+T S SS RH G LP++HKVGT PKQTL+ EIKHS ETFF D P +FK
Sbjct: 16 ETRSNSSSHRHGGGGGGDDTTSLPYMHKVGTPPKQTLFQEIKHSFNETFFPDKPFGKFKD 75
Query: 54 QSRKRKLVLGMQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEH 113
QS RKL LG+Q +FPI+EWGR Y+L+KF+GDFIAGLTIASLCIPQD+AYAKLANL+P +
Sbjct: 76 QSGFRKLELGLQYIFPILEWGRHYDLKKFRGDFIAGLTIASLCIPQDLAYAKLANLDPWY 135
Query: 114 ALYTSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPDYLRLAYTATFFA 173
LY+SFVAPLVYAFMGTSRDIAIGPVAVVSLLLGT+L++EI+++K+ DYLRLA+TATFFA
Sbjct: 136 GLYSSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTLLSNEISNTKSHDYLRLAFTATFFA 195
Query: 174 GITQLALGFFRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKT--FTKKTDIVSV 231
G+TQ+ LG RLGFLIDFLSHAAIVGFM GAAITI LQQLKGLLG+ FTKKTDI+SV
Sbjct: 196 GVTQMLLGVCRLGFLIDFLSHAAIVGFMAGAAITIGLQQLKGLLGISNNNFTKKTDIISV 255
Query: 232 MRSVWKPVEHGWNWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVY 291
MRSVW V HGWNWETI+IG+SFL+F+LIT YIAKKNKKLFWV+AI+PMISV+VSTF VY
Sbjct: 256 MRSVWTHVHHGWNWETILIGLSFLIFLLITKYIAKKNKKLFWVSAISPMISVIVSTFFVY 315
Query: 292 ITRADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFA 351
ITRADK+GV+IVKHIK GVNP+SA+EIFF G+Y GAGV++GVV+G+VALTEA+AI RTFA
Sbjct: 316 ITRADKRGVSIVKHIKSGVNPSSANEIFFHGKYLGAGVRVGVVAGLVALTEAIAIGRTFA 375
Query: 352 AMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLL 411
AMKDY++DGNKEMVAMGTMNIVGSL+S YV TGSFSRSAVNYMAGCKTAVSNIVMS+V+L
Sbjct: 376 AMKDYALDGNKEMVAMGTMNIVGSLSSCYVTTGSFSRSAVNYMAGCKTAVSNIVMSIVVL 435
Query: 412 LTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMGAFFGVIFKS 471
LTLLVITPLFKYTPNAVLASIIIAAV+NLV+++A +LLWKIDKFDFVACMGAFFGVIFKS
Sbjct: 436 LTLLVITPLFKYTPNAVLASIIIAAVVNLVNIEAMVLLWKIDKFDFVACMGAFFGVIFKS 495
Query: 472 VEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSA 531
VEIGLLIAVAISFAKILLQVTRPRTA+LGKL GT VYRNI QYPKA QIPGMLIIRVDSA
Sbjct: 496 VEIGLLIAVAISFAKILLQVTRPRTAVLGKLPGTSVYRNIQQYPKAAQIPGMLIIRVDSA 555
Query: 532 IYFSNSNYIKDRILKWVTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSL 591
IYFSNSNYIK+RIL+W+ DE QRT SE P IQ LI EMSPV DIDTSGIHA E+L+K+L
Sbjct: 556 IYFSNSNYIKERILRWLIDEGAQRTESELPEIQHLITEMSPVPDIDTSGIHAFEELYKTL 615
Query: 592 KKREVQLLLANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAVATFGPK 639
+KREVQL+LANPGP+VIEKLHASKL+++IGEDKIF +V DAVAT+GPK
Sbjct: 616 QKREVQLILANPGPVVIEKLHASKLTELIGEDKIFLTVADAVATYGPK 663
>ST13_ARATH (Q9FEP7) Sulfate transporter 1.3
Length = 656
Score = 911 bits (2355), Expect = 0.0
Identities = 454/632 (71%), Positives = 540/632 (84%), Gaps = 1/632 (0%)
Query: 9 SSRRHHGLPHIHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKRKLVLGMQSVF 68
SS R P++HKV PKQ L+ E ++ KETFF DDPL FK QS+ +KL+LG+QSVF
Sbjct: 20 SSPRQANTPYVHKVEVPPKQNLFNEFMYTFKETFFHDDPLRHFKDQSKSKKLMLGIQSVF 79
Query: 69 PIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFM 128
P++EWGR YNL+ F+GD IAGLTIASLCIPQDI YAKLA+L+P++ LY+SFV PLVYA M
Sbjct: 80 PVIEWGRKYNLKLFRGDLIAGLTIASLCIPQDIGYAKLASLDPKYGLYSSFVPPLVYACM 139
Query: 129 GTSRDIAIGPVAVVSLLLGTMLTDEIADSKNP-DYLRLAYTATFFAGITQLALGFFRLGF 187
G+S+DIAIGPVAVVSLLLGT+L EI + NP +YLRLA+T+TFFAG+TQ ALGFFRLGF
Sbjct: 140 GSSKDIAIGPVAVVSLLLGTLLRAEIDPNTNPNEYLRLAFTSTFFAGVTQAALGFFRLGF 199
Query: 188 LIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWET 247
LIDFLSHAA+VGFMGGAAITIALQQLKG LG+ FTKKTDI++V+ SV HGWNW+T
Sbjct: 200 LIDFLSHAAVVGFMGGAAITIALQQLKGFLGINKFTKKTDIIAVLSSVISSAHHGWNWQT 259
Query: 248 IVIGMSFLVFILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKGVAIVKHIK 307
I+I SFL+F+LI+ +I K+NKKLFW+ AIAP++SV++STF VYITRADKKGV IVKH+
Sbjct: 260 ILISASFLIFLLISKFIGKRNKKLFWIPAIAPLVSVIISTFFVYITRADKKGVQIVKHLD 319
Query: 308 KGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAM 367
KG+NP+S I+FSG+Y G +IGVVSGMVALTEAVAI RTFAAMKDY IDGNKEMVA+
Sbjct: 320 KGLNPSSLRLIYFSGDYLLKGFRIGVVSGMVALTEAVAIGRTFAAMKDYQIDGNKEMVAL 379
Query: 368 GTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNA 427
G MN++GS+TS YV+TGSFSRSAVN+MAGC+TAVSNI+MS+V+LLTLL +TPLFKYTPNA
Sbjct: 380 GAMNVIGSMTSCYVSTGSFSRSAVNFMAGCQTAVSNIIMSIVVLLTLLFLTPLFKYTPNA 439
Query: 428 VLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKI 487
+LA+III AV+ LVDV A IL++KIDK DFVACMGAFFGVIF SVEIGLLIAV ISFAKI
Sbjct: 440 ILAAIIINAVIPLVDVNATILIFKIDKLDFVACMGAFFGVIFVSVEIGLLIAVGISFAKI 499
Query: 488 LLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSNSNYIKDRILKW 547
LLQVTRPRTAILGK+ GT VYRNI QYP+AT+IPG+L IRVDSAIYFSNSNY+++RI +W
Sbjct: 500 LLQVTRPRTAILGKIPGTSVYRNINQYPEATRIPGVLTIRVDSAIYFSNSNYVRERIQRW 559
Query: 548 VTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLLLANPGPIV 607
+TDEE A+ P IQ LI+EMSPVTDIDTSGIHALEDL+KSL+KR++QL+LANPGP V
Sbjct: 560 LTDEEEMVEAARLPRIQFLIIEMSPVTDIDTSGIHALEDLYKSLQKRDIQLVLANPGPPV 619
Query: 608 IEKLHASKLSDIIGEDKIFSSVDDAVATFGPK 639
I KLH S +D+IG DKIF +V +AV + PK
Sbjct: 620 INKLHVSHFADLIGHDKIFLTVAEAVDSCSPK 651
>ST12_ARATH (Q9MAX3) Sulfate transporter 1.2
Length = 653
Score = 888 bits (2295), Expect = 0.0
Identities = 438/624 (70%), Positives = 534/624 (85%), Gaps = 1/624 (0%)
Query: 17 PHIHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRD 76
P HKVG PKQ ++ + ++ KETFF DDPL FK Q + ++ +LG+QSVFP+ +WGR+
Sbjct: 25 PTRHKVGIPPKQNMFKDFMYTFKETFFHDDPLRDFKDQPKSKQFMLGLQSVFPVFDWGRN 84
Query: 77 YNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAI 136
Y +KF+GD I+GLTIASLCIPQDI YAKLANL+P++ LY+SFV PLVYA MG+SRDIAI
Sbjct: 85 YTFKKFRGDLISGLTIASLCIPQDIGYAKLANLDPKYGLYSSFVPPLVYACMGSSRDIAI 144
Query: 137 GPVAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQLALGFFRLGFLIDFLSHA 195
GPVAVVSLLLGT+L EI + +PD YLRLA+TATFFAGIT+ ALGFFRLGFLIDFLSHA
Sbjct: 145 GPVAVVSLLLGTLLRAEIDPNTSPDEYLRLAFTATFFAGITEAALGFFRLGFLIDFLSHA 204
Query: 196 AIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFL 255
A+VGFMGGAAITIALQQLKG LG+K FTKKTDI+SV+ SV+K HGWNW+TI+IG SFL
Sbjct: 205 AVVGFMGGAAITIALQQLKGFLGIKKFTKKTDIISVLESVFKAAHHGWNWQTILIGASFL 264
Query: 256 VFILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKGVAIVKHIKKGVNPASA 315
F+L + I KK+KKLFWV AIAP+ISV+VSTF VYITRADK+GV IVKH+ +G+NP+S
Sbjct: 265 TFLLTSKIIGKKSKKLFWVPAIAPLISVIVSTFFVYITRADKQGVQIVKHLDQGINPSSF 324
Query: 316 SEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGS 375
I+F+G+ G++IGVV+GMVALTEAVAI RTFAAMKDY IDGNKEMVA+G MN+VGS
Sbjct: 325 HLIYFTGDNLAKGIRIGVVAGMVALTEAVAIGRTFAAMKDYQIDGNKEMVALGMMNVVGS 384
Query: 376 LTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNAVLASIIIA 435
++S YVATGSFSRSAVN+MAGC+TAVSNI+MS+V+LLTLL +TPLFKYTPNA+LA+III
Sbjct: 385 MSSCYVATGSFSRSAVNFMAGCQTAVSNIIMSIVVLLTLLFLTPLFKYTPNAILAAIIIN 444
Query: 436 AVMNLVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKILLQVTRPR 495
AV+ L+D+QAAIL++K+DK DF+AC+GAFFGVIF SVEIGLLIAV+ISFAKILLQVTRPR
Sbjct: 445 AVIPLIDIQAAILIFKVDKLDFIACIGAFFGVIFVSVEIGLLIAVSISFAKILLQVTRPR 504
Query: 496 TAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSNSNYIKDRILKWVTDEEVQR 555
TA+LG + T VYRNI QYP+AT +PG+L IRVDSAIYFSNSNY+++RI +W+ +EE +
Sbjct: 505 TAVLGNIPRTSVYRNIQQYPEATMVPGVLTIRVDSAIYFSNSNYVRERIQRWLHEEEEKV 564
Query: 556 TASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLLLANPGPIVIEKLHASK 615
A+ P IQ LI+EMSPVTDIDTSGIHALEDL+KSL+KR++QL+LANPGP+VI KLH S
Sbjct: 565 KAASLPRIQFLIIEMSPVTDIDTSGIHALEDLYKSLQKRDIQLILANPGPLVIGKLHLSH 624
Query: 616 LSDIIGEDKIFSSVDDAVATFGPK 639
+D++G+D I+ +V DAV PK
Sbjct: 625 FADMLGQDNIYLTVADAVEACCPK 648
>ST11_ARATH (Q9SAY1) Sulfate transporter 1.1 (High-affinity sulfate
transporter 1) (Hst1At) (AST101)
Length = 649
Score = 867 bits (2240), Expect = 0.0
Identities = 433/615 (70%), Positives = 520/615 (84%), Gaps = 1/615 (0%)
Query: 21 KVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRDYNLQ 80
+V PK L +IK V+ETFF D PL FKGQ+ +K +LG+Q+VFPI+ W R+Y L+
Sbjct: 24 RVLAPPKAGLLKDIKSVVEETFFHDAPLRDFKGQTPAKKALLGIQAVFPIIGWAREYTLR 83
Query: 81 KFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGPVA 140
KF+GD IAGLTIASLCIPQDI YAKLAN++P++ LY+SFV PL+YA MG+SRDIAIGPVA
Sbjct: 84 KFRGDLIAGLTIASLCIPQDIGYAKLANVDPKYGLYSSFVPPLIYAGMGSSRDIAIGPVA 143
Query: 141 VVSLLLGTMLTDEIADSKNP-DYLRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAIVG 199
VVSLL+GT+ I KNP DYLRL +TATFFAGI Q LGF RLGFLIDFLSHAA+VG
Sbjct: 144 VVSLLVGTLCQAVIDPKKNPEDYLRLVFTATFFAGIFQAGLGFLRLGFLIDFLSHAAVVG 203
Query: 200 FMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFLVFIL 259
FMGGAAITIALQQLKG LG+KTFTKKTDIVSVM SV+K EHGWNW+TIVIG SFL F+L
Sbjct: 204 FMGGAAITIALQQLKGFLGIKTFTKKTDIVSVMHSVFKNAEHGWNWQTIVIGASFLTFLL 263
Query: 260 ITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKGVAIVKHIKKGVNPASASEIF 319
+T +I K+N+KLFWV AIAP+ISV++STF V+I RADK+GV IVKHI +G+NP S +IF
Sbjct: 264 VTKFIGKRNRKLFWVPAIAPLISVIISTFFVFIFRADKQGVQIVKHIDQGINPISVHKIF 323
Query: 320 FSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSS 379
FSG+YF G++IG ++GMVALTEAVAIARTFAAMKDY IDGNKEM+A+GTMN+VGS+TS
Sbjct: 324 FSGKYFTEGIRIGGIAGMVALTEAVAIARTFAAMKDYQIDGNKEMIALGTMNVVGSMTSC 383
Query: 380 YVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAVMN 439
Y+ATGSFSRSAVN+MAG +TAVSNIVM++V+ LTL ITPLFKYTPNA+LA+III+AV+
Sbjct: 384 YIATGSFSRSAVNFMAGVETAVSNIVMAIVVALTLEFITPLFKYTPNAILAAIIISAVLG 443
Query: 440 LVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTAIL 499
L+D+ AAIL+W+IDK DF+ACMGAF GVIF SVEIGLLIAV ISFAKILLQVTRPRT +L
Sbjct: 444 LIDIDAAILIWRIDKLDFLACMGAFLGVIFISVEIGLLIAVVISFAKILLQVTRPRTTVL 503
Query: 500 GKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSNSNYIKDRILKWVTDEEVQRTASE 559
GKL + VYRN LQYP A QIPG+LIIRVDSAIYFSNSNY+++R +WV +E+
Sbjct: 504 GKLPNSNVYRNTLQYPDAAQIPGILIIRVDSAIYFSNSNYVRERASRWVREEQENAKEYG 563
Query: 560 FPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLSDI 619
P+I+ +I+EMSPVTDIDTSGIH++E+L KSL+K+E+QL+LANPGP+VIEKL+ASK +
Sbjct: 564 MPAIRFVIIEMSPVTDIDTSGIHSIEELLKSLEKQEIQLILANPGPVVIEKLYASKFVEE 623
Query: 620 IGEDKIFSSVDDAVA 634
IGE IF +V DAVA
Sbjct: 624 IGEKNIFLTVGDAVA 638
>ST31_ARATH (Q9SV13) Sulfate transporter 3.1 (AST12) (AtST1)
Length = 658
Score = 667 bits (1722), Expect = 0.0
Identities = 334/632 (52%), Positives = 463/632 (72%), Gaps = 11/632 (1%)
Query: 5 GSAPSSRRHHGLPHIHKVGTAPKQTLYLE-IKHSVKETFFFDDPLSQFKGQSRKRKLVLG 63
G+ RRHH + AP+ +L+ +++SVKET F DDP QFK Q+ RK VLG
Sbjct: 11 GAEELHRRHHTVE-------APQPQPFLKSLQYSVKETLFPDDPFRQFKNQNASRKFVLG 63
Query: 64 MQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPL 123
++ PI EW YNL+ FK D IAG+TIASL IPQ I+YAKLANL P LY+SFV PL
Sbjct: 64 LKYFLPIFEWAPRYNLKFFKSDLIAGITIASLAIPQGISYAKLANLPPILGLYSSFVPPL 123
Query: 124 VYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQLALGF 182
VYA +G+SRD+A+G VAV SLL G ML+ E+ K+P YL LA+TATFFAG+ + +LG
Sbjct: 124 VYAVLGSSRDLAVGTVAVASLLTGAMLSKEVDAEKDPKLYLHLAFTATFFAGVLEASLGI 183
Query: 183 FRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHG 242
FRLGF++DFLSHA IVGFMGGAA ++LQQLKG+ GLK FT TD++SVMRSV+ H
Sbjct: 184 FRLGFIVDFLSHATIVGFMGGAATVVSLQQLKGIFGLKHFTDSTDVISVMRSVFSQT-HE 242
Query: 243 WNWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKGVAI 302
W WE+ V+G FL F+L T Y + K K FWVAA+AP+ SV++ + VY T A++ GV +
Sbjct: 243 WRWESGVLGCGFLFFLLSTRYFSIKKPKFFWVAAMAPLTSVILGSLLVYFTHAERHGVQV 302
Query: 303 VKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNK 362
+ +KKG+NP S S++ F+ Y VK G+++G++AL E VA+ R+FA K+Y+IDGNK
Sbjct: 303 IGDLKKGLNPLSGSDLIFTSPYMSTAVKTGLITGIIALAEGVAVGRSFAMFKNYNIDGNK 362
Query: 363 EMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFK 422
EM+A G MNIVGS TS Y+ TG FSRSAVNY AGCKTA+SNIVM++ ++ TLL +TPLF
Sbjct: 363 EMIAFGMMNIVGSFTSCYLTTGPFSRSAVNYNAGCKTAMSNIVMAIAVMFTLLFLTPLFH 422
Query: 423 YTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAI 482
YTP VL++III+A++ L+D QAAI LWK+DKFDF+ CM A+ GV+F SVEIGL++AVAI
Sbjct: 423 YTPLVVLSAIIISAMLGLIDYQAAIHLWKVDKFDFLVCMSAYVGVVFGSVEIGLVVAVAI 482
Query: 483 SFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSNSNYIKD 542
S A++LL V+RP+TA+ G + + +YRN QYP + +PG+LI+ +D+ IYF+N++Y+++
Sbjct: 483 SIARLLLFVSRPKTAVKGNIPNSMIYRNTEQYPSSRTVPGILILEIDAPIYFANASYLRE 542
Query: 543 RILKWVTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLLLAN 602
RI++W+ +EE + S S+Q +I++MS V +IDTSGI + ++ K + +R ++L+L+N
Sbjct: 543 RIIRWIDEEEERVKQSGESSLQYIILDMSAVGNIDTSGISMMVEIKKVIDRRALKLVLSN 602
Query: 603 PGPIVIEKLHASK-LSDIIGEDKIFSSVDDAV 633
P V++KL SK + D +G++ +F +V +AV
Sbjct: 603 PKGEVVKKLTRSKFIGDHLGKEWMFLTVGEAV 634
>ST34_ARATH (Q9LW86) Probable sulfate transporter 3.4
Length = 653
Score = 641 bits (1653), Expect = 0.0
Identities = 310/635 (48%), Positives = 464/635 (72%), Gaps = 2/635 (0%)
Query: 1 MEDTGSAPSSRRHHGLPHIHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKRKL 60
+ED S + + IH V PK+T + ++K V + FF DDPL +F+ Q+ + ++
Sbjct: 8 VEDMASPNNGTAGETVVEIHSVCLPPKKTAFQKLKKRVGDVFFPDDPLQRFRNQTWRNRV 67
Query: 61 VLGMQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFV 120
+LG+QS+FPI WG Y+L+ + D I+GLTIASL IPQ I+YAKLANL P LY+SFV
Sbjct: 68 ILGLQSLFPIFTWGSQYDLKLLRSDVISGLTIASLAIPQGISYAKLANLPPIVGLYSSFV 127
Query: 121 APLVYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQLA 179
PL+YA +G+SR +A+GPV++ SL++G+ML++ ++ +++ YL+LA+T+TFFAG+ Q +
Sbjct: 128 PPLIYAVLGSSRHLAVGPVSIASLVMGSMLSESVSPTQDSILYLKLAFTSTFFAGVFQAS 187
Query: 180 LGFFRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPV 239
LG RLGF+IDFLS A ++GF GAA+ ++LQQLKGLLG+ FT K IV VM SV+
Sbjct: 188 LGLLRLGFMIDFLSKATLIGFTAGAAVIVSLQQLKGLLGIVHFTGKMQIVPVMSSVFNHR 247
Query: 240 EHGWNWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKG 299
W+WETIV+G+ FL +L T +I+ + KLFW++A +P+ SV++ST VY+ R+
Sbjct: 248 SE-WSWETIVMGIGFLSILLTTRHISMRKPKLFWISAASPLASVIISTLLVYLIRSKTHA 306
Query: 300 VAIVKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSID 359
++ + H+ KG+NP S + ++FSG + +K G+++G+++LTE +A+ RTFA++K+Y ++
Sbjct: 307 ISFIGHLPKGLNPPSLNMLYFSGAHLALAIKTGIITGILSLTEGIAVGRTFASLKNYQVN 366
Query: 360 GNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITP 419
GNKEM+A+G MN+ GS TS YV TGSFSRSAVNY AG KTAVSNIVM+ +L+TLL + P
Sbjct: 367 GNKEMMAIGFMNMAGSCTSCYVTTGSFSRSAVNYNAGAKTAVSNIVMASAVLVTLLFLMP 426
Query: 420 LFKYTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLIA 479
LF YTPN +LA+II+ AV+ L+D QAA LWK+DKFDF C+ +FFGV+F SV +GL IA
Sbjct: 427 LFYYTPNVILAAIILTAVIGLIDYQAAYKLWKVDKFDFFTCLCSFFGVLFVSVPLGLAIA 486
Query: 480 VAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSNSNY 539
VA+S KILL VTRP T+ G + GT++Y+++ +Y +A++IPG LI+ ++S IYF+NS Y
Sbjct: 487 VAVSVIKILLHVTRPNTSEFGNIPGTQIYQSLGRYREASRIPGFLILAIESPIYFANSTY 546
Query: 540 IKDRILKWVTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLL 599
++DRIL+W +EE + + +++ +I++M+ V+ IDTSG+ A+ +L + L+K+ +QL+
Sbjct: 547 LQDRILRWAREEENRIKENNGTTLKCIILDMTAVSAIDTSGLEAVFELRRRLEKQSLQLV 606
Query: 600 LANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAVA 634
L NP V+EKLH SK+ + +G ++ +V +AVA
Sbjct: 607 LVNPVGTVMEKLHKSKIIEALGLSGLYLTVGEAVA 641
>ST33_ARATH (Q9SXS2) Probable sulfate transporter 3.3 (AST91)
Length = 631
Score = 627 bits (1618), Expect = e-179
Identities = 307/624 (49%), Positives = 455/624 (72%), Gaps = 3/624 (0%)
Query: 19 IHKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRDYN 78
+HKV P ++ ++K +KETFF DDPL QF+GQ + KL+ Q +FPI++W +Y+
Sbjct: 3 VHKVVAPPHKSTVAKLKTKLKETFFPDDPLRQFRGQPNRTKLIRAAQYIFPILQWCPEYS 62
Query: 79 LQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGP 138
K D ++GLTIASL IPQ I+YAKLANL P LY+SFV PLVYA +G+SRD+A+GP
Sbjct: 63 FSLLKSDVVSGLTIASLAIPQGISYAKLANLPPIVGLYSSFVPPLVYAVLGSSRDLAVGP 122
Query: 139 VAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAI 197
V++ SL+LG+ML +++ +P +L+LA+++TFFAG+ Q +LG RLGF+IDFLS A +
Sbjct: 123 VSIASLILGSMLRQQVSPVDDPVLFLQLAFSSTFFAGLFQASLGILRLGFIIDFLSKATL 182
Query: 198 VGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFLVF 257
+GFMGGAAI ++LQQLKGLLG+ FTK +V V+ SV++ W+W+TIV+G+ FL+F
Sbjct: 183 IGFMGGAAIIVSLQQLKGLLGITHFTKHMSVVPVLSSVFQHTNE-WSWQTIVMGVCFLLF 241
Query: 258 ILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKGVAIVKHIKKGVNPASASE 317
+L T +++ K KLFWV+A AP++SV+VST V++ RA++ G++++ + +G+NP S +
Sbjct: 242 LLSTRHLSMKKPKLFWVSAGAPLLSVIVSTLLVFVFRAERHGISVIGKLPEGLNPPSWNM 301
Query: 318 IFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLT 377
+ F G + K G+V+G+V+LTE +A+ RTFAA+K+Y +DGNKEM+A+G MN+VGS T
Sbjct: 302 LQFHGSHLALVAKTGLVTGIVSLTEGIAVGRTFAALKNYHVDGNKEMIAIGLMNVVGSAT 361
Query: 378 SSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAV 437
S YV TG+FSRSAVN AG KTAVSNIVMS+ +++TLL + PLF+YTPN VL +II+ AV
Sbjct: 362 SCYVTTGAFSRSAVNNNAGAKTAVSNIVMSVTVMVTLLFLMPLFEYTPNVVLGAIIVTAV 421
Query: 438 MNLVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTA 497
+ L+D+ AA +WKIDKFDF+ + AFFGVIF SV+ GL IAV +S KIL+QVTRP+
Sbjct: 422 IGLIDLPAACHIWKIDKFDFLVMLCAFFGVIFLSVQNGLAIAVGLSLFKILMQVTRPKMV 481
Query: 498 ILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSNSNYIKDRILKWVTDEEVQRTA 557
I+G + GT +YR++ Y +A +IPG L++ ++S + F+NSNY+ +R +W+ + E +
Sbjct: 482 IMGNIPGTDIYRDLHHYKEAQRIPGFLVLSIESPVNFANSNYLTERTSRWIEECEEEEAQ 541
Query: 558 SEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLLLANPGPIVIEKL-HASKL 616
+ S+Q LI+EMS V+ +DT+G+ ++L K+ K++++L+ NP V+EKL A +
Sbjct: 542 EKHSSLQFLILEMSAVSGVDTNGVSFFKELKKTTAKKDIELVFVNPLSEVVEKLQRADEQ 601
Query: 617 SDIIGEDKIFSSVDDAVATFGPKG 640
+ + + +F +V +AVA+ KG
Sbjct: 602 KEFMRPEFLFLTVAEAVASLSLKG 625
>ST32_ARATH (O04289) Sulfate transporter 3.2 (AST77)
Length = 646
Score = 627 bits (1616), Expect = e-179
Identities = 309/617 (50%), Positives = 442/617 (71%), Gaps = 3/617 (0%)
Query: 20 HKVGTAPKQTLYLEIKHSVKETFFFDDPLSQFKGQSR-KRKLVLGMQSVFPIVEWGRDYN 78
H+V P Q +K+++ E F DDP + + +S+ +K+ LG++ VFPI+EW R Y+
Sbjct: 10 HQVEIPPPQPFLKSLKNTLNEILFADDPFRRIRNESKTSKKIELGLRHVFPILEWARGYS 69
Query: 79 LQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGP 138
L+ K D I+G+TIASL IPQ I+YA+LANL P LY+S V PLVYA MG+SRD+A+G
Sbjct: 70 LEYLKSDVISGITIASLAIPQGISYAQLANLPPILGLYSSLVPPLVYAIMGSSRDLAVGT 129
Query: 139 VAVVSLLLGTMLTDEIADSKNPD-YLRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAI 197
VAV SLL ML E+ NP YL LA+TATFFAG+ Q LG RLGF+++ LSHAAI
Sbjct: 130 VAVASLLTAAMLGKEVNAVVNPKLYLHLAFTATFFAGLMQTCLGLLRLGFVVEILSHAAI 189
Query: 198 VGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFLVF 257
VGFMGGAA + LQQLKGLLGL FT TDIV+V+RS++ H W WE+ V+G FL+F
Sbjct: 190 VGFMGGAATVVCLQQLKGLLGLHHFTHSTDIVTVLRSIFSQ-SHMWRWESGVLGCCFLIF 248
Query: 258 ILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKGVAIVKHIKKGVNPASASE 317
+L T YI+KK KLFW++A++P++SV+ T +Y G+ + +KKG+NP S +
Sbjct: 249 LLTTKYISKKRPKLFWISAMSPLVSVIFGTIFLYFLHDQFHGIQFIGELKKGINPPSITH 308
Query: 318 IFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLT 377
+ F+ Y +K+G+++G++AL E +A+ R+FA K+Y+IDGNKEM+A G MNI+GS +
Sbjct: 309 LVFTPPYVMLALKVGIITGVIALAEGIAVGRSFAMYKNYNIDGNKEMIAFGMMNILGSFS 368
Query: 378 SSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAV 437
S Y+ TG FSRSAVNY AGCKTA+SN+VM++ + +TLL +TPLF YTP VL+SIIIAA+
Sbjct: 369 SCYLTTGPFSRSAVNYNAGCKTALSNVVMAVAVAVTLLFLTPLFFYTPLVVLSSIIIAAM 428
Query: 438 MNLVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTA 497
+ LVD +AAI LWK+DKFDF C+ A+ GV+F ++EIGL+++V IS +++L V RP+
Sbjct: 429 LGLVDYEAAIHLWKLDKFDFFVCLSAYLGVVFGTIEIGLILSVGISVMRLVLFVGRPKIY 488
Query: 498 ILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSNSNYIKDRILKWVTDEEVQRTA 557
++G + +++YRNI YP+A +LI+ +D IYF+NS Y++DRI +W+ +EE +
Sbjct: 489 VMGNIQNSEIYRNIEHYPQAITRSSLLILHIDGPIYFANSTYLRDRIGRWIDEEEDKLRT 548
Query: 558 SEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLS 617
S S+Q ++++MS V +IDTSGI LE+L K L +RE++L++ANPG V++KL S
Sbjct: 549 SGDISLQYIVLDMSAVGNIDTSGISMLEELNKILGRRELKLVIANPGAEVMKKLSKSTFI 608
Query: 618 DIIGEDKIFSSVDDAVA 634
+ IG+++I+ +V +AVA
Sbjct: 609 ESIGKERIYLTVAEAVA 625
>SUT3_STYHA (P53393) Low affinity sulphate transporter 3
Length = 644
Score = 610 bits (1573), Expect = e-174
Identities = 311/605 (51%), Positives = 437/605 (71%), Gaps = 14/605 (2%)
Query: 38 VKETFFFDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCI 97
+K+ FF S+ ++ + V + S+FPI+ W R Y+ KFK D ++GLT+ASL I
Sbjct: 33 LKDNKFFTSSSSK-----KETRAVSFLASLFPILSWIRTYSATKFKDDLLSGLTLASLSI 87
Query: 98 PQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADS 157
PQ I YA LA L+P++ LYTS + P++YA MG+SR+IAIGPVAVVS+LL +++ I
Sbjct: 88 PQSIGYANLAKLDPQYGLYTSVIPPVIYALMGSSREIAIGPVAVVSMLLSSLVPKVIDPD 147
Query: 158 KNP-DYLRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGL 216
+P DY L +T T FAGI Q A G RLGFL+DFLSHAA+VGFM GAAI I LQQLKGL
Sbjct: 148 AHPNDYRNLVFTVTLFAGIFQTAFGVLRLGFLVDFLSHAALVGFMAGAAIVIGLQQLKGL 207
Query: 217 LGLKTFTKKTDIVSVMRSVW----KPVEHGWNWETI--VIGMSFLVFILITNYIAKKNKK 270
LGL FT KTD V+V++SV+ + + NW + VIG SFL+F+L +I ++NKK
Sbjct: 208 LGLTHFTTKTDAVAVLKSVYTSLHQQITSSENWSPLNFVIGCSFLIFLLAARFIGRRNKK 267
Query: 271 LFWVAAIAPMISVVVSTFCVYITRADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAGVK 330
FW+ AIAP++SV++ST V++++ DK GV I+KH++ G+NP+S ++ +G + G K
Sbjct: 268 FFWLPAIAPLLSVILSTLIVFLSKGDKHGVNIIKHVQGGLNPSSVHKLQLNGPHVGQAAK 327
Query: 331 IGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSA 390
IG++S ++ALTEA+A+ R+FA +K Y +DGNKEM+AMG MNI GSLTS YV+TGSFSR+A
Sbjct: 328 IGLISAIIALTEAIAVGRSFANIKGYHLDGNKEMLAMGCMNIAGSLTSCYVSTGSFSRTA 387
Query: 391 VNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAILLW 450
VN+ AGCKTAVSNIVM++ +LL L + T L YTP A+LASII++A+ L+D+ A +W
Sbjct: 388 VNFSAGCKTAVSNIVMAVTVLLCLELFTRLLYYTPMAILASIILSALPGLIDIGEAYHIW 447
Query: 451 KIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYRN 510
K+DKFDF+AC+GAFFGV+F S+EIGLLIA++ISFAKILLQ RP +LG++ T+ Y +
Sbjct: 448 KVDKFDFLACLGAFFGVLFVSIEIGLLIALSISFAKILLQAIRPGVEVLGRIPTTEAYCD 507
Query: 511 ILQYPKATQIPGMLIIRVDS-AIYFSNSNYIKDRILKWVTDEEVQRTASEFPS-IQSLIV 568
+ QYP A PG+L+IR+ S ++ F+N+ ++++RILKWV DEE +Q++I+
Sbjct: 508 VAQYPMAVTTPGILVIRISSGSLCFANAGFVRERILKWVEDEEQDNIEEAAKGRVQAIII 567
Query: 569 EMSPVTDIDTSGIHALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLSDIIGEDKIFSS 628
+M+ +T++DTSGI ALE+L K L R V+L + NP VI KL + D IG++++F +
Sbjct: 568 DMTDLTNVDTSGILALEELHKKLLSRGVELAMVNPRWEVIHKLKVANFVDKIGKERVFLT 627
Query: 629 VDDAV 633
V +AV
Sbjct: 628 VAEAV 632
>ST21_ARATH (O04722) Sulfate transporter 2.1 (AST68)
Length = 677
Score = 604 bits (1558), Expect = e-172
Identities = 321/615 (52%), Positives = 439/615 (71%), Gaps = 11/615 (1%)
Query: 33 EIKHSVKETFFFDDPLSQFKGQSRK---RKLVLGMQSVFPIVEWGRDYNLQKFKGDFIAG 89
E+K VK +F +FK ++ ++++ +Q++FPI W R+Y L FK D +AG
Sbjct: 65 ELKRQVKGSFL--TKAKKFKSLQKQPFPKQILSVLQAIFPIFGWCRNYKLTMFKNDLMAG 122
Query: 90 LTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTM 149
LT+ASLCIPQ I YA LA L+P++ LYTS V PL+YA MGTSR+IAIGPVAVVSLL+ +M
Sbjct: 123 LTLASLCIPQSIGYATLAKLDPQYGLYTSVVPPLIYALMGTSREIAIGPVAVVSLLISSM 182
Query: 150 LTDEIADSKNP-DYLRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAIVGFMGGAAITI 208
L I +P Y +L T TFFAGI Q + G FRLGFL+DFLSHAAIVGFMGGAAI I
Sbjct: 183 LQKLIDPETDPLGYKKLVLTTTFFAGIFQASFGLFRLGFLVDFLSHAAIVGFMGGAAIVI 242
Query: 209 ALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFLVFILITNYIAKKN 268
LQQLKGLLG+ FT TDIVSV+R+VW+ + W+ T ++G SFL FILIT +I KK
Sbjct: 243 GLQQLKGLLGITNFTTNTDIVSVLRAVWRSCQQQWSPHTFILGCSFLSFILITRFIGKKY 302
Query: 269 KKLFWVAAIAPMISVVVSTFCVYITRADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAG 328
KKLFW+ AIAP+I+VVVST V++T+AD+ GV V+HIK G+NP S ++ F+ + G
Sbjct: 303 KKLFWLPAIAPLIAVVVSTLMVFLTKADEHGVKTVRHIKGGLNPMSIQDLDFNTPHLGQI 362
Query: 329 VKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSR 388
KIG++ +VALTEA+A+ R+FA +K Y +DGNKEMVA+G MN++GS TS Y ATGSFSR
Sbjct: 363 AKIGLIIAIVALTEAIAVGRSFAGIKGYRLDGNKEMVAIGFMNVLGSFTSCYAATGSFSR 422
Query: 389 SAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAIL 448
+AVN+ AGC+TA+SNIVM++ + + L +T L YTP A+LASII++A+ L+++ AI
Sbjct: 423 TAVNFAAGCETAMSNIVMAVTVFVALECLTRLLYYTPIAILASIILSALPGLININEAIH 482
Query: 449 LWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVY 508
+WK+DKFDF+A +GAFFGV+F SVEIGLL+AV ISFAKI+L RP LG++ GT +
Sbjct: 483 IWKVDKFDFLALIGAFFGVLFASVEIGLLVAVVISFAKIILISIRPGIETLGRMPGTDTF 542
Query: 509 RNILQYPKATQIPGMLIIRVDSAIY-FSNSNYIKDRILKWVTDEEVQRT--ASEFPSIQS 565
+ QYP + PG+LI RV SA+ F+N++ I++RI+ WV +EE + ++ I
Sbjct: 543 TDTNQYPMTVKTPGVLIFRVKSALLCFANASSIEERIMGWVDEEEEEENTKSNAKRKILF 602
Query: 566 LIVEMSPVTDIDTSGIHALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLSDIIGEDKI 625
++++MS + ++DTSGI AL +L L K V+L++ NP VI KL+ +K D IG K+
Sbjct: 603 VVLDMSSLINVDTSGITALLELHNKLIKTGVELVIVNPKWQVIHKLNQAKFVDRIG-GKV 661
Query: 626 FSSVDDAV-ATFGPK 639
+ ++ +A+ A FG K
Sbjct: 662 YLTIGEALDACFGLK 676
>ST22_ARATH (P92946) Sulfate transporter 2.2 (AST56) (AtH14)
Length = 658
Score = 591 bits (1523), Expect = e-168
Identities = 301/575 (52%), Positives = 415/575 (71%), Gaps = 5/575 (0%)
Query: 64 MQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPL 123
++S FPI+ WGR Y L FK D +AGLT+ASLCIPQ I YA LA L+PE+ LYTS V PL
Sbjct: 70 LKSAFPILSWGRQYKLNLFKKDLMAGLTLASLCIPQSIGYANLAGLDPEYGLYTSVVPPL 129
Query: 124 VYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNP-DYLRLAYTATFFAGITQLALGF 182
+Y+ MGTSR++AIGPVAVVSLLL +M+ D +P Y ++ +T TFFAG Q G
Sbjct: 130 IYSTMGTSRELAIGPVAVVSLLLSSMVRDLQDPVTDPIAYRKIVFTVTFFAGAFQAIFGL 189
Query: 183 FRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHG 242
FRLGFL+DFLSHAA+VGFM GAAI I LQQLKGL GL FT KTD+VSV+ SV+ + H
Sbjct: 190 FRLGFLVDFLSHAALVGFMAGAAIVIGLQQLKGLFGLTHFTNKTDVVSVLSSVFHSLHHP 249
Query: 243 WNWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPMISVVVSTFCVYITRADKKGVAI 302
W VIG SFL+FIL+ +I K+N KLFW+ A+AP+ISVV++T VY++ A+ +GV I
Sbjct: 250 WQPLNFVIGSSFLIFILLARFIGKRNNKLFWIPAMAPLISVVLATLIVYLSNAESRGVKI 309
Query: 303 VKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNK 362
VKHIK G N S +++ F + G KIG++S ++ALTEA+A+ R+FA +K Y +DGNK
Sbjct: 310 VKHIKPGFNQLSVNQLQFKSPHLGQIAKIGLISAIIALTEAIAVGRSFATIKGYRLDGNK 369
Query: 363 EMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFK 422
EM+AMG MNI GSL+S YVATGSFSR+AVN+ AGC+T VSNIVM++ ++++L V+T
Sbjct: 370 EMMAMGFMNIAGSLSSCYVATGSFSRTAVNFSAGCETVVSNIVMAITVMISLEVLTRFLY 429
Query: 423 YTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAI 482
+TP A+LASII++A+ L+DV A+ +WK+DK DF+ + AFFGV+F SVEIGLL+AV I
Sbjct: 430 FTPTAILASIILSALPGLIDVSGALHIWKLDKLDFLVLIAAFFGVLFASVEIGLLLAVGI 489
Query: 483 SFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIY-FSNSNYIK 541
SFA+I+L RP LG+LS T ++ +I QYP A + G+L +R+ S + F+N+N+I+
Sbjct: 490 SFARIMLSSIRPSIEALGRLSKTDIFGDINQYPMANKTAGLLTLRISSPLLCFANANFIR 549
Query: 542 DRILKWVTD---EEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALEDLFKSLKKREVQL 598
DRIL V + EE ++ + +Q +I++MS V +DTSG+ ALE+L + L +++L
Sbjct: 550 DRILNSVQEIEGEENEQEVLKENGLQVVILDMSCVMGVDTSGVFALEELHQELASNDIRL 609
Query: 599 LLANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAV 633
++A+P V+ KL +KL + I + I+ +V +AV
Sbjct: 610 VIASPRWRVLHKLKRAKLDEKIKTENIYMTVGEAV 644
>ST35_ARATH (Q94LW6) Probable sulfate transporter 3.5
Length = 634
Score = 521 bits (1341), Expect = e-147
Identities = 266/604 (44%), Positives = 397/604 (65%), Gaps = 14/604 (2%)
Query: 33 EIKHSVKETFFFDDP---LSQFKGQSRKRKLVLGMQSVFPIVEWGRDYNLQKFKGDFIAG 89
+ K KETFF DDP +SQ + K K +L + PI EW Y++QK K D +AG
Sbjct: 28 KFKSKCKETFFPDDPFKPISQEPNRLLKTKKLL--EYFVPIFEWLPKYDMQKLKYDVLAG 85
Query: 90 LTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTM 149
+TI SL +PQ I+YAKLA++ P LY+SFV P VYA G+S ++A+G VA SLL+
Sbjct: 86 ITITSLAVPQGISYAKLASIPPIIGLYSSFVPPFVYAVFGSSNNLAVGTVAACSLLIAET 145
Query: 150 LTDEIADSKNPDYLRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAIVGFMGGAAITIA 209
+E+ ++ YL L +TAT G+ Q A+GF RLG L+DFLSH+ I GFMGG AI I
Sbjct: 146 FGEEMIKNEPELYLHLIFTATLITGLFQFAMGFLRLGILVDFLSHSTITGFMGGTAIIIL 205
Query: 210 LQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFLVFILITNYIAKKNK 269
LQQLKG+ GL FT KTD+VSV+ S+ W W++ + G+ FLVF+ T YI ++
Sbjct: 206 LQQLKGIFGLVHFTHKTDVVSVLHSILDNRAE-WKWQSTLAGVCFLVFLQSTRYIKQRYP 264
Query: 270 KLFWVAAIAPMISVVVSTFCVYITRADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAGV 329
KLFWV+A+ PM+ VVV Y+ + G+A V +KKG+NP S + F +Y G
Sbjct: 265 KLFWVSAMGPMVVVVVGCVVAYLVKGTAHGIATVGPLKKGLNPPSIQLLNFDSKYLGMVF 324
Query: 330 KIGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRS 389
K G+V+G++AL E +AI R+FA MK+ DGNKEM+A G MN++GS TS Y+ TG FS++
Sbjct: 325 KAGIVTGLIALAEGIAIGRSFAVMKNEQTDGNKEMIAFGLMNVIGSFTSCYLTTGPFSKT 384
Query: 390 AVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAILL 449
AVNY AG KT +SN+VM + ++L LL + PLF YTP L++II++A++ L++ + L
Sbjct: 385 AVNYNAGTKTPMSNVVMGVCMMLVLLFLAPLFSYTPLVGLSAIIMSAMLGLINYEEMYHL 444
Query: 450 WKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYR 509
+K+DKFDF+ CM AFFGV F S++ GL+I+V S + LL V RP T LG++ + ++R
Sbjct: 445 FKVDKFDFLVCMSAFFGVSFLSMDYGLIISVGFSIVRALLYVARPSTCKLGRIPNSVMFR 504
Query: 510 NILQYPKATQIPGMLIIRVDSAIYFSNSNYIKDRILKWVTDEEVQRTASEFPSIQSLIVE 569
+I QYP + ++ G +I+++ S ++F+NS Y+++RIL+W+ DE +I+ L+++
Sbjct: 505 DIEQYPASEEMLGYIILQLGSPVFFANSTYVRERILRWIRDEP--------EAIEFLLLD 556
Query: 570 MSPVTDIDTSGIHALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLSDIIGEDKIFSSV 629
+S V+ ID +G+ L ++ + L + +++++ NP V+EK+ S + IG++ +F S+
Sbjct: 557 LSGVSTIDMTGMETLLEIQRILGSKNIKMVIINPRFEVLEKMMLSHFVEKIGKEYMFLSI 616
Query: 630 DDAV 633
DDAV
Sbjct: 617 DDAV 620
>ST41_ARATH (Q9FY46) Sulfate transporter 4.1, chloroplast precursor
(AST82)
Length = 685
Score = 389 bits (1000), Expect = e-107
Identities = 223/592 (37%), Positives = 347/592 (57%), Gaps = 10/592 (1%)
Query: 44 FDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRDYNLQK-FKGDFIAGLTIASLCIPQDIA 102
FDD S + + ++ +LV + ++FP W R Y + FK D +AG+T+ + +PQ ++
Sbjct: 56 FDDIFSGWTAKIKRMRLVDWIDTLFPCFRWIRTYRWSEYFKLDLMAGITVGIMLVPQAMS 115
Query: 103 YAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPDY 162
YAKLA L P + LY+SFV VYA G+SR +AIGPVA+VSLL+ L IAD+ +
Sbjct: 116 YAKLAGLPPIYGLYSSFVPVFVYAIFGSSRQLAIGPVALVSLLVSNALGG-IADTNEELH 174
Query: 163 LRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTF 222
+ LA GI + +G RLG+LI F+SH+ I GF +AI I L Q+K LG +
Sbjct: 175 IELAILLALLVGILECIMGLLRLGWLIRFISHSVISGFTSASAIVIGLSQIKYFLGY-SI 233
Query: 223 TKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPMIS 282
+ + IV ++ S+ + + W V+G LV + + ++ K K+L ++ A AP+
Sbjct: 234 ARSSKIVPIVESIIAGADK-FQWPPFVMGSLILVILQVMKHVGKAKKELQFLRAAAPLTG 292
Query: 283 VVVSTFCVYITRADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTE 342
+V+ T + +++V I +G+ S F ++ + + VA+ E
Sbjct: 293 IVLGTTIAKVFHPPS--ISLVGEIPQGLPTFSFPRSF---DHAKTLLPTSALITGVAILE 347
Query: 343 AVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVS 402
+V IA+ AA Y +D N E+ +G NI+GSL S+Y ATGSFSRSAVN + KT +S
Sbjct: 348 SVGIAKALAAKNRYELDSNSELFGLGVANILGSLFSAYPATGSFSRSAVNNESEAKTGLS 407
Query: 403 NIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMG 462
++ +++ +LL +TP+FKY P LA+I+I+AV LVD AI LW++DK DF
Sbjct: 408 GLITGIIIGCSLLFLTPMFKYIPQCALAAIVISAVSGLVDYDEAIFLWRVDKRDFSLWTI 467
Query: 463 AFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPG 522
+F +EIG+L+ V S A ++ + P A+LG+L GT VYRNI QYP+A G
Sbjct: 468 TSTITLFFGIEIGVLVGVGFSLAFVIHESANPHIAVLGRLPGTTVYRNIKQYPEAYTYNG 527
Query: 523 MLIIRVDSAIYFSNSNYIKDRILKW-VTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGI 581
++I+R+DS IYF+N +YIKDR+ ++ V ++ E I +I+EMSPVT ID+S +
Sbjct: 528 IVIVRIDSPIYFANISYIKDRLREYEVAVDKYTNRGLEVDRINFVILEMSPVTHIDSSAV 587
Query: 582 HALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAV 633
AL++L++ K R++QL ++NP V + S + +++G++ F V DAV
Sbjct: 588 EALKELYQEYKTRDIQLAISNPNKDVHLTIARSGMVELVGKEWFFVRVHDAV 639
>NO70_SOYBN (Q02920) Early nodulin 70
Length = 485
Score = 389 bits (1000), Expect = e-107
Identities = 208/459 (45%), Positives = 310/459 (67%), Gaps = 14/459 (3%)
Query: 33 EIKHSVKETFF---FDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRDYNLQKFKGDFIAG 89
++ +VKET + S + Q ++ +Q++FPI+ ++YN QK K D +AG
Sbjct: 18 QVVDNVKETLLPHPNPNTFSYLRNQPFSKRAFALLQNLFPILASLQNYNAQKLKCDLMAG 77
Query: 90 LTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTM 149
LT+A IPQ + A LA L+PE+ LYT V PL+YA + +SR+I IGP +V SLLL +M
Sbjct: 78 LTLAIFAIPQCMGNATLARLSPEYGLYTGIVPPLIYAMLASSREIVIGPGSVDSLLLSSM 137
Query: 150 L-TDEIADSKNPDYLRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAIVGFMGGAAITI 208
+ T ++ + Y++L +T TFFAGI Q+A G FR GFL++ LS A IVGF+ AA+ I
Sbjct: 138 IQTLKVPIHDSSTYIQLVFTVTFFAGIFQVAFGLFRFGFLVEHLSQATIVGFLAAAAVGI 197
Query: 209 ALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEH--GWNWETIVIGMSFLVFILITNYIAK 266
LQQLKGL G+ F KTD+ SV++S+W ++ W+ ++IG SFL FIL T ++ K
Sbjct: 198 GLQQLKGLFGIDNFNNKTDLFSVVKSLWTSFKNQSAWHPYNLIIGFSFLCFILFTRFLGK 257
Query: 267 KNKKLFWVAAIAPMISVVVSTFCVY------ITRADKKGVAIVKHIKKG-VNPASASEIF 319
+NKKL W++ +AP++SV+ S+ Y + D K VA++ IK G +NP+S ++
Sbjct: 258 RNKKLMWLSHVAPLLSVIGSSAIAYKINFNELQVKDYK-VAVLGPIKGGSLNPSSLHQLT 316
Query: 320 FSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSS 379
F + G ++IG+ +++LT ++A+ R+FA++K +SID N+E+V++G MNIVGSLTS
Sbjct: 317 FDSQVVGHLIRIGLTIAIISLTGSIAVGRSFASLKGHSIDPNREVVSLGIMNIVGSLTSC 376
Query: 380 YVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAVMN 439
Y+A+GS SR+AVNY AG +T VS IVM++ +L++L +T L +TP A+LA+II++AV
Sbjct: 377 YIASGSLSRTAVNYNAGSETMVSIIVMALTVLMSLKFLTGLLYFTPKAILAAIILSAVPG 436
Query: 440 LVDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLI 478
L+D+ A +WK+DK DF+AC GAF GV+F SVEIGL I
Sbjct: 437 LIDLNKAREIWKVDKMDFLACTGAFLGVLFASVEIGLAI 475
>ST42_ARATH (Q8GYH8) Probable sulfate transporter 4.2
Length = 677
Score = 379 bits (974), Expect = e-104
Identities = 217/594 (36%), Positives = 345/594 (57%), Gaps = 14/594 (2%)
Query: 44 FDDPLSQFKGQSRKRKLVLGMQSVFPIVEWGRDYNL-QKFKGDFIAGLTIASLCIPQDIA 102
F+D S++ + ++ + ++FP W R Y Q FK D +AG+T+ + +PQ ++
Sbjct: 43 FNDFFSRWTAKIKRMTFFDWIDAIFPCFLWIRTYRWHQYFKLDLMAGITVGIMLVPQAMS 102
Query: 103 YAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPDY 162
YA+LA L P + LY+SFV VYA G+SR +A+GPVA+VSLL+ L+ I D Y
Sbjct: 103 YARLAGLQPIYGLYSSFVPVFVYAVFGSSRQLAVGPVALVSLLVSNALSG-IVDPSEELY 161
Query: 163 LRLAYTATFFAGITQLALGFFRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTF 222
LA GI + +GF RLG+LI F+SH+ I GF +A+ I L QLK LG
Sbjct: 162 TELAILLALMVGIFESIMGFLRLGWLIRFISHSVISGFTTASAVVIGLSQLKYFLGYSV- 220
Query: 223 TKKTDIVSVMRSVWKPVEHGWNWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPMIS 282
++ + I+ V+ S+ + + W ++G + LV +L+ ++ K K+L ++ A P+
Sbjct: 221 SRSSKIMPVIDSIIAGADQ-FKWPPFLLGCTILVILLVMKHVGKAKKELRFIRAAGPLTG 279
Query: 283 VVVSTFCVYITRADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTE 342
+ + T + + +V I +G+ S + F + + + VA+ E
Sbjct: 280 LALGTIIAKVFHPPS--ITLVGDIPQGLPKFSFPKSFDHAKLL---LPTSALITGVAILE 334
Query: 343 AVAIARTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVS 402
+V IA+ AA Y +D N E+ +G NI GSL S+Y TGSFSRSAVN + KT +S
Sbjct: 335 SVGIAKALAAKNRYELDSNSELFGLGVANIFGSLFSAYPTTGSFSRSAVNSESEAKTGLS 394
Query: 403 NIVMSMVLLLTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAILLWKIDKFDFVACMG 462
+V +++ +LL +TP+FK+ P LA+I+I+AV LVD + AI LW++DK DF
Sbjct: 395 GLVTGIIIGCSLLFLTPMFKFIPQCALAAIVISAVSGLVDYEGAIFLWRVDKRDFTLWTI 454
Query: 463 AFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPG 522
+F +EIG+LI V S A ++ + P A+LG+L GT VYRN+ QYP+A G
Sbjct: 455 TSTTTLFFGIEIGVLIGVGFSLAFVIHESANPHIAVLGRLPGTTVYRNMKQYPEAYTYNG 514
Query: 523 MLIIRVDSAIYFSNSNYIKDRILKWVTDEEVQRTASEFPSIQSL---IVEMSPVTDIDTS 579
++I+R+D+ IYF+N +YIKDR+ ++ + + + S+ P ++ + I+EMSPVT ID+S
Sbjct: 515 IVIVRIDAPIYFANISYIKDRLREY--EVAIDKHTSKGPDMERIYFVILEMSPVTYIDSS 572
Query: 580 GIHALEDLFKSLKKREVQLLLANPGPIVIEKLHASKLSDIIGEDKIFSSVDDAV 633
+ AL+DL++ K R +QL ++NP V+ L + + ++IG++ F V DAV
Sbjct: 573 AVEALKDLYEEYKTRGIQLAISNPNKEVLLTLARAGIVELIGKEWFFVRVHDAV 626
>SULI_SCHPO (Q9URY8) Probable sulfate permease C869.05c
Length = 840
Score = 245 bits (625), Expect = 3e-64
Identities = 168/588 (28%), Positives = 299/588 (50%), Gaps = 66/588 (11%)
Query: 64 MQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPL 123
++S+FPI+EW +YN D IAG+T+ + +PQ ++YAK+A L E+ LY+SFV
Sbjct: 103 LKSLFPIIEWLPNYNPYWLINDLIAGITVGCVVVPQGMSYAKVATLPSEYGLYSSFVGVA 162
Query: 124 VYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPDYLRLAYTATFFAGITQLALGFF 183
+Y F TS+D++IGPVAV+SL+ ++ + +A + ++A AG +G
Sbjct: 163 IYCFFATSKDVSIGPVAVMSLITAKVIANVMAKDETYTAPQIATCLALLAGAITCGIGLL 222
Query: 184 RLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKT-FTKKTDIVSVMRSVWKPVEHG 242
RLGF+I+F+ A+ GF G+A+ I Q+ L+G K T K + +++S+ +
Sbjct: 223 RLGFIIEFIPVPAVAGFTTGSALNILSGQVPALMGYKNKVTAKATYMVIIQSLKHLPDTT 282
Query: 243 WNWETIVIGMSFLVFI-LITNYIAKK----NKKLFWVAAIAPMISVVVSTFCVYIT---- 293
+ ++ + L F + Y+ K+ + F + + V+V T Y
Sbjct: 283 VDAAFGLVSLFILFFTKYMCQYLGKRYPRWQQAFFLTNTLRSAVVVIVGTAISYAICKHH 342
Query: 294 RADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAM 353
R+D ++I+K + +G I + + VS +V L E ++IA++F +
Sbjct: 343 RSDPP-ISIIKTVPRGFQHVGVPLI--TKKLCRDLASELPVSVIVLLLEHISIAKSFGRV 399
Query: 354 KDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLT 413
DY I ++E++AMG N++G ++Y ATGSFSRSA+ AG KT ++ I + V++L+
Sbjct: 400 NDYRIVPDQELIAMGVTNLIGIFFNAYPATGSFSRSAIKAKAGVKTPIAGIFTAAVVILS 459
Query: 414 LLVITPLFKYTPNAVLASIIIAAVMNLV-DVQAAILLWKIDKFDFVACMGAFFGVIFKSV 472
L +T F Y PNA+L+++II AV +L+ ++ IL W++ + + +F S+
Sbjct: 460 LYCLTDAFYYIPNAILSAVIIHAVTDLILPMKQTILFWRLQPLEACIFFISVIVSVFSSI 519
Query: 473 EIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVY--------RNI------------- 511
E G+ ++V ++ A +LL++ +P + LGK+ Y R+I
Sbjct: 520 ENGIYVSVCLAAALLLLRIAKPHGSFLGKIQAANKYGSDNIANVRDIYVPLEMKEENPNL 579
Query: 512 -LQYPKATQIPGMLIIRVDSAIYFSNSN---------------------YIKDRILKW-V 548
+Q P PG+ I R+ + + N++ Y+KD W V
Sbjct: 580 EIQSPP----PGVFIFRLQESFTYPNASRVSTMISRRIKDLTRRGIDNIYVKDIDRPWNV 635
Query: 549 TDEEVQRTASEF----PSIQSLIVEMSPVTDIDTSGIHALEDLFKSLK 592
+ ++ SE P +Q++I + S V ++DT+ + +L D+ K L+
Sbjct: 636 PRQRKKKENSEIEDLRPLLQAIIFDFSAVNNLDTTAVQSLIDIRKELE 683
>CY14_NEUCR (P23622) Sulfate permease II
Length = 819
Score = 224 bits (571), Expect = 5e-58
Identities = 174/602 (28%), Positives = 288/602 (46%), Gaps = 86/602 (14%)
Query: 64 MQSVFPIVEWGRDYNLQKFKGDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPL 123
++ +FP V W YNL GDFIAG+T+ + +PQ +AYAKLANL PE+ LYTSFV +
Sbjct: 59 LRELFPFVNWIFHYNLTWLLGDFIAGVTVGFVVVPQGMAYAKLANLAPEYGLYTSFVGFV 118
Query: 124 VYAFMGTSRDIAIGPVAVVSLLLGTMLTDEIADSKNPDYLRLAYTATFFAGITQLALGFF 183
+Y TS+DI IG VAV+S ++G ++ + D + D +A T F +G L LG
Sbjct: 119 LYWAFATSKDITIGAVAVMSTIVGNIIANVQKDHPDFDAGDIARTLAFISGAMLLFLGLI 178
Query: 184 RLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWK--PVEH 241
R GF+++F+ AI FM G+AI+IA Q+ L+G+ + + V+ + K P H
Sbjct: 179 RFGFIVEFIPIVAISAFMTGSAISIAAGQVSTLMGIPNINSREETYKVIINTLKGLPNTH 238
Query: 242 --GWNWETIVIGMSFLVFIL--ITNYIAKKNKKLFWVAAIAPMISVVVSTFCV------Y 291
T + G+ F+ + + ++ + F+V+ + M+ +++ V +
Sbjct: 239 LDAAMGLTALFGLYFIRWFCTQMGKRYPRQQRAWFFVSTLR-MVFIIILYILVSWLVNRH 297
Query: 292 ITRADKKGVAIVKHIKKGVNPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFA 351
+ K I+ H+ G A + E A + +V L E +AI+++F
Sbjct: 298 VKDPKKAHFKILGHVPSGFQHKGAPRL--DNEILSAISGDIPTTILVLLIEHIAISKSFG 355
Query: 352 AMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLL 411
+ +Y I+ ++E+VA+G N++G Y ATGSFSR+A+ AG +T ++ I ++++L
Sbjct: 356 RVNNYIINPSQELVAIGFTNLLGPFLGGYPATGSFSRTAIKAKAGVRTPLAGIFTAVLVL 415
Query: 412 LTLLVITPLFKYTPNAVLASIIIAAVMNLVDVQAAIL-LWKIDKFDFVACMGAFFGVIFK 470
L L +T +F Y PN+ LA++II AV +L+ + W + V F IF
Sbjct: 416 LALYALTSVFFYIPNSALAAMIIHAVGDLITPPREVYKFWLTSPLEVVIFFAGVFVSIFT 475
Query: 471 SVEIGLLIAVAISFAKILLQV--------------TRPRTAILG-KLSG----------- 504
S+E G+ + VA S A +L ++ T PR + G K SG
Sbjct: 476 SIENGIYVTVAASGAVLLWRIAKSPGKFLGQTEIYTAPRELVRGSKDSGLTQSLLQKSEH 535
Query: 505 ----TKVYRNILQYPK---ATQIPGMLIIRVDSAIYFSNSNYIKDRIL------------ 545
+ R+ L P+ +T PG+ + R + + NS D +
Sbjct: 536 HTAFLSLDRDDLSNPELQISTPWPGIFVYRFGEGLNYVNSAKHLDNLTIHVFKHTRRTEL 595
Query: 546 --------------------KWVTDEEVQRTASEFPSIQSLIVEMSPVTDIDTSGIHALE 585
++TDE V R P+++++I++ S V ID + AL+
Sbjct: 596 NKFEKLGDRPWNDPGPRRGQAFLTDELVSR-----PTLRAIILDFSAVNCIDVTAAQALQ 650
Query: 586 DL 587
DL
Sbjct: 651 DL 652
>PRES_MERUN (Q9JKQ2) Prestin
Length = 744
Score = 211 bits (537), Expect = 5e-54
Identities = 152/531 (28%), Positives = 270/531 (50%), Gaps = 36/531 (6%)
Query: 32 LEIKHSVKETFFFDDPLSQ-FKGQSRKRKLVLGMQSVFPIVEWGRDYNLQKFK-GDFIAG 89
L +K V E+ D L Q F +K + ++ M PI +W Y +++ GD ++G
Sbjct: 32 LHVKDKVSESI--GDKLKQAFTCTPKKIRNIIYM--FLPITKWLPAYKFKEYVLGDLVSG 87
Query: 90 LTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAFMGTSRDIAIGPVAVVSLLLG-- 147
++ L +PQ +A+A LA + P LY+SF ++Y F GTSR I+IGP AV+SL++G
Sbjct: 88 ISTGVLQLPQGLAFAMLAAVPPVFGLYSSFYPVIMYCFFGTSRHISIGPFAVISLMIGGV 147
Query: 148 --TMLTDEIA-----------DSKNPDYLRLAYTATFFAGITQLALGFFRLGFLIDFLSH 194
++ D+I ++++ +++A + T +GI Q LG R GF+ +L+
Sbjct: 148 AVRLVPDDIVIPGGVNATNGTEARDALRVKVAMSVTLLSGIIQFCLGVCRFGFVAIYLTE 207
Query: 195 AAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVMRSVWKPVEHGWNWETIVIGMSF 254
+ GF AA+ + LK L G+KT + + I SV+ S +++ N +G+
Sbjct: 208 PLVRGFTTAAAVHVFTSMLKYLFGVKT-KRYSGIFSVVYSTVAVLQNVKNLNVCSLGVGL 266
Query: 255 LVFILITNYIAKKNKKLFWVAAIAPM----ISVVVST-FCVYITRADKKGVAIVKHIKKG 309
+VF L+ K+ + F AP+ +VV+ T + V +V + G
Sbjct: 267 MVFGLLLG--GKEFNERFKEKLPAPIPLEFFAVVMGTGISAGFNLHESYSVDVVGTLPLG 324
Query: 310 V-NPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAIARTFAAMKDYSIDGNKEMVAMG 368
+ PA+ F Y A + +V + +++A+T A Y +DGN+E++A+G
Sbjct: 325 LLPPANPDTSLFHLVYVDA-----IAIAIVGFSVTISMAKTLANKHGYQVDGNQELIALG 379
Query: 369 TMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVMSMVLLLTLLVITPLFKYTPNAV 428
N +GSL ++ + S SRS V G KT ++ + S+++LL +L LF+ P AV
Sbjct: 380 ICNSIGSLFQTFSISCSLSRSLVQEGTGGKTQLAGCLASLMILLVILATGFLFESLPQAV 439
Query: 429 LASIIIAAVMNL-VDVQAAILLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKI 487
L++I+I + + + W+ K + + F +F ++ GL+ AV I+ +
Sbjct: 440 LSAIVIVNLKGMFMQFSDLPFFWRTSKIELTIWLTTFVSSLFLGLDYGLITAVIIALLTV 499
Query: 488 LLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLIIRVDSAIYFSNSN 538
+ + P +LG+L T VY +I Y + +IPG+ I ++++ IY++NS+
Sbjct: 500 IYRTQSPSYKVLGQLPDTDVYIDIDAYEEVKEIPGIKIFQINAPIYYANSD 550
>PRES_HUMAN (P58743) Prestin
Length = 744
Score = 211 bits (537), Expect = 5e-54
Identities = 143/493 (29%), Positives = 255/493 (51%), Gaps = 31/493 (6%)
Query: 69 PIVEWGRDYNLQKFK-GDFIAGLTIASLCIPQDIAYAKLANLNPEHALYTSFVAPLVYAF 127
PI +W Y +++ GD ++G++ L +PQ +A+A LA + P LY+SF ++Y F
Sbjct: 66 PITKWLPAYKFKEYVLGDLVSGISTGVLQLPQGLAFAMLAAVPPIFGLYSSFYPVIMYCF 125
Query: 128 MGTSRDIAIGPVAVVSLLLG----TMLTDEIA-----------DSKNPDYLRLAYTATFF 172
+GTSR I+IGP AV+SL++G ++ D+I ++++ +++A + T
Sbjct: 126 LGTSRHISIGPFAVISLMIGGVAVRLVPDDIVIPGGVNATNGTEARDALRVKVAMSVTLL 185
Query: 173 AGITQLALGFFRLGFLIDFLSHAAIVGFMGGAAITIALQQLKGLLGLKTFTKKTDIVSVM 232
+GI Q LG R GF+ +L+ + GF AA+ + LK L G+KT + + I SV+
Sbjct: 186 SGIIQFCLGVCRFGFVAIYLTEPLVRGFTTAAAVHVFTSMLKYLFGVKT-KRYSGIFSVV 244
Query: 233 RSVWKPVEHGWNWETIVIGMSFLVFILITNYIAKKNKKLFWVAAIAPM----ISVVVST- 287
S +++ N +G+ +VF L+ K+ + F AP+ +VV+ T
Sbjct: 245 YSTVAVLQNVKNLNVCSLGVGLMVFGLLLG--GKEFNERFKEKLPAPIPLEFFAVVMGTG 302
Query: 288 FCVYITRADKKGVAIVKHIKKGV-NPASASEIFFSGEYFGAGVKIGVVSGMVALTEAVAI 346
+ V +V + G+ PA+ F Y A + +V + +++
Sbjct: 303 ISAGFNLKESYNVDVVGTLPLGLLPPANPDTSLFHLVYVDA-----IAIAIVGFSVTISM 357
Query: 347 ARTFAAMKDYSIDGNKEMVAMGTMNIVGSLTSSYVATGSFSRSAVNYMAGCKTAVSNIVM 406
A+T A Y +DGN+E++A+G N +GSL ++ + S SRS V G KT ++ +
Sbjct: 358 AKTLANKHGYQVDGNQELIALGLCNSIGSLFQTFSISCSLSRSLVQEGTGGKTQLAGCLA 417
Query: 407 SMVLLLTLLVITPLFKYTPNAVLASIIIAAVMNL-VDVQAAILLWKIDKFDFVACMGAFF 465
S+++LL +L LF+ P AVL++I+I + + + W+ K + + F
Sbjct: 418 SLMILLVILATGFLFESLPQAVLSAIVIVNLKGMFMQFSDLPFFWRTSKIELTIWLTTFV 477
Query: 466 GVIFKSVEIGLLIAVAISFAKILLQVTRPRTAILGKLSGTKVYRNILQYPKATQIPGMLI 525
+F ++ GL+ AV I+ ++ + P +LGKL T VY +I Y + +IPG+ I
Sbjct: 478 SSLFLGLDYGLITAVIIALLTVIYRTQSPSYKVLGKLPETDVYIDIDAYEEVKEIPGIKI 537
Query: 526 IRVDSAIYFSNSN 538
++++ IY++NS+
Sbjct: 538 FQINAPIYYANSD 550
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.138 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,386,166
Number of Sequences: 164201
Number of extensions: 2820588
Number of successful extensions: 7200
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 7014
Number of HSP's gapped (non-prelim): 90
length of query: 643
length of database: 59,974,054
effective HSP length: 117
effective length of query: 526
effective length of database: 40,762,537
effective search space: 21441094462
effective search space used: 21441094462
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0314.13


