

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
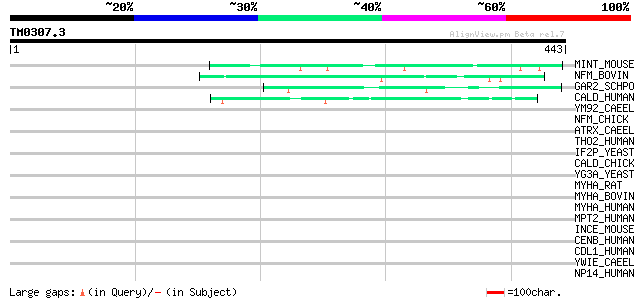
Query= TM0307.3
(443 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MINT_MOUSE (Q62504) Msx2-interacting protein (SMART/HDAC1 associ... 50 1e-05
NFM_BOVIN (O77788) Neurofilament triplet M protein (160 kDa neur... 48 4e-05
GAR2_SCHPO (P41891) Protein gar2 45 5e-04
CALD_HUMAN (Q05682) Caldesmon (CDM) 44 8e-04
YM92_CAEEL (P34531) Hypothetical protein M01A8.2 in chromosome III 43 0.001
NFM_CHICK (P16053) Neurofilament triplet M protein (160 kDa neur... 43 0.001
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 42 0.004
THO2_HUMAN (Q8NI27) THO complex subunit 2 (Tho2) 41 0.007
IF2P_YEAST (P39730) Eukaryotic translation initiation factor 5B ... 41 0.007
CALD_CHICK (P12957) Caldesmon (CDM) 40 0.009
YG3A_YEAST (P53278) Hypothetical 92.7 kDa protein in ASN2-PHB1 i... 40 0.012
MYHA_RAT (Q9JLT0) Myosin heavy chain, nonmuscle type B (Cellular... 39 0.021
MYHA_BOVIN (Q27991) Myosin heavy chain, nonmuscle type B (Cellul... 39 0.021
MYHA_HUMAN (P35580) Myosin heavy chain, nonmuscle type B (Cellul... 39 0.027
MPT2_HUMAN (O60237) Protein phosphatase 1 regulatory subunit 12B... 39 0.035
INCE_MOUSE (Q9WU62) Inner centromere protein 38 0.046
CENB_HUMAN (P07199) Major centromere autoantigen B (Centromere p... 38 0.046
CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2... 38 0.046
YWIE_CAEEL (Q23525) Hypothetical protein ZK546.14 in chromosome II 38 0.060
NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130... 38 0.060
>MINT_MOUSE (Q62504) Msx2-interacting protein (SMART/HDAC1 associated
repressor protein)
Length = 3644
Score = 50.1 bits (118), Expect = 1e-05
Identities = 70/307 (22%), Positives = 117/307 (37%), Gaps = 43/307 (14%)
Query: 160 RQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEK 219
R QQKE E+ ++ +E E+++E + TP E E E +
Sbjct: 1607 RMQQKEKEKDQKPKEAEKQEEPETHPKTPEPAA-------ETKEPEPKAPVSAGLPAVTI 1659
Query: 220 VVVTNNVSSRG-----NVTNVPPPMKNHSAAAASLAEK---LNRRVVVPVQSKQRFGGEL 271
VVT +S P P A A ++E+ ++ V VPV+ ++
Sbjct: 1660 TVVTPEPASSAPEKAEEAAEAPSPAGEKPAEPAPVSEETKLVSEPVSVPVEQPRQSDVPP 1719
Query: 272 GTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSV----GTTASKQRQSVDRR 327
G +S S + A S P+ A T P V++ GTT S+ SVD +
Sbjct: 1720 GEDSRDSQDSA---------ALAPSAPQESAATDAVPCVNAEPLTPGTTVSQVESSVDPK 1770
Query: 328 DQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHP 387
+S + EEA + + P QNA E ++AN P
Sbjct: 1771 PSSPQPLSKLTQRSEEAEEGKVEKPDTTPSTEPDATQNAGVAS--EAQPPASEDVEANPP 1828
Query: 388 VLKRSNSYNEERRSKLGVE-------EVKLKKEMEEENRER------KGGVKEKCIPIKK 434
V + N+ +RSK V+ E + ++ E +RE+ G +K + +K
Sbjct: 1829 VAAKDRKTNKSKRSKTSVQAAAASVVEKPVTRKSERIDREKLKRSSSPRGEAQKLLELKM 1888
Query: 435 SSKESSR 441
+++ +R
Sbjct: 1889 EAEKITR 1895
>NFM_BOVIN (O77788) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M) (Fragment)
Length = 810
Score = 48.1 bits (113), Expect = 4e-05
Identities = 66/296 (22%), Positives = 109/296 (36%), Gaps = 27/296 (9%)
Query: 152 AKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEAL 211
A + L + +KE EE +E EE+EEE+ S G S +E GE E+ E
Sbjct: 401 ATAPELKEEEGEKEEEEGQE-EEEEEEEAAKSDQAEEGGSEKEGSSEKEEGEQEEEGETE 459
Query: 212 QNNTKTEKVV-VTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGE 270
E V P + + A EK V +K
Sbjct: 460 AEGEGEEAAAEAKEEKKMEEKAEEVAPKEELAAEAKVEKPEKAKSPVAKSPTTKSPTAKS 519
Query: 271 LGTESATSAAPSRYSALLQLIACGS--------SGPEFKARTQQEPRVSSVGTTASKQRQ 322
+S + +P+ S + S PE K+ T + P S + +
Sbjct: 520 PEAKSPEAKSPTAKSPTAKSPVAKSPTAKSPEAKSPEAKSPTAKSPTAKSPAAKSPAPKS 579
Query: 323 SVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMES- 381
V+ KA E+ E++E V E+P+ + AE+KE + E +
Sbjct: 580 PVEEVKPKA-EAGAEKGEQKEKVEEEKKEAKESPK-----EEKAEKKEEKPKDVPEKKKA 633
Query: 382 ---MKANHPVLK-------RSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKE 427
+KA PV + + + E + + E+ K K+++EE + +GG+KE
Sbjct: 634 ESPVKAESPVKEEVPAKPVKVSPEKEAKEEEKPQEKEKEKEKVEEVGGKEEGGLKE 689
Score = 38.1 bits (87), Expect = 0.046
Identities = 65/284 (22%), Positives = 105/284 (36%), Gaps = 50/284 (17%)
Query: 162 QQKEAEEYEEYEEQEEE----KEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKT 217
+++EAEE EE EE EEE K+ T P + G EE G++E+ E
Sbjct: 374 KEEEAEEKEEKEEAEEEVVAAKKSPVKATAPELKEEEGEKEEEEGQEEEEEE-------- 425
Query: 218 EKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGELGTESAT 277
E+ ++ G+ + E ++K+ E E A
Sbjct: 426 EEAAKSDQAEEGGSEKEGSSEKEEGEQEEEGETEAEGEGEEAAAEAKEEKKME---EKAE 482
Query: 278 SAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCE 337
AP +L A KA++ V+ TT S +S + + +A + +
Sbjct: 483 EVAPKE-----ELAAEAKVEKPEKAKSP----VAKSPTTKSPTAKSPEAKSPEAKSPTAK 533
Query: 338 EEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNE 397
+ VA + +++P E +S +A P K + +
Sbjct: 534 SPTAKSPVAK--SPTAKSP---------------------EAKSPEAKSPTAKSPTAKSP 570
Query: 398 ERRS---KLGVEEVKLKKEMEEENRERKGGVKEKCIPIKKSSKE 438
+S K VEEVK K E E E+K V+E+ K+S KE
Sbjct: 571 AAKSPAPKSPVEEVKPKAEAGAEKGEQKEKVEEEKKEAKESPKE 614
>GAR2_SCHPO (P41891) Protein gar2
Length = 500
Score = 44.7 bits (104), Expect = 5e-04
Identities = 53/245 (21%), Positives = 97/245 (38%), Gaps = 50/245 (20%)
Query: 203 EDEDRHEALQNNTKTEKVV--VTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVP 260
E + A++ +K++K+ ++ + + T+V P A AS E + V
Sbjct: 15 ETASKKGAIEKPSKSKKITKEAAKEIAKQSSKTDVSPKKSKKEAKRASSPEPSKKSVKKQ 74
Query: 261 VQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQ 320
+SK++ +ES +S++ S SS E ++ + + SS +++ +
Sbjct: 75 KKSKKKEESSSESESESSSSESE-----------SSSSESESSSSESESSSSESSSSESE 123
Query: 321 RQSVDRRDQKA-----FMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGS 375
+ + + ++K S E EEEEEAV + E+KE S S
Sbjct: 124 EEVIVKTEEKKESSSESSSSSESEEEEEAVVKI-----------------EEKKESSSDS 166
Query: 376 LVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEKCIPIKKS 435
S S + E S+ E + ++E+ E+ E+K G E + S
Sbjct: 167 ---------------SSESSSSESESESSSSESEEEEEVVEKTEEKKEGSSESSSDSESS 211
Query: 436 SKESS 440
S SS
Sbjct: 212 SDSSS 216
Score = 32.3 bits (72), Expect = 2.5
Identities = 43/223 (19%), Positives = 85/223 (37%), Gaps = 23/223 (10%)
Query: 142 EPPETNYGYNAKSKVLSNRQQQKEAE-EYEEYEEQEEEKEFTSSTTTPHSRCSRG----- 195
EP + + KSK + E+E E E E E +SS + S S
Sbjct: 65 EPSKKSVKKQKKSKKKEESSSESESESSSSESESSSSESESSSSESESSSSESSSSESEE 124
Query: 196 ---VSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEK 252
V TEE E + + + E+ VV + + + S + +S +E
Sbjct: 125 EVIVKTEEKKESSSESSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESESESSSSES 184
Query: 253 LNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSS 312
VV +++ G +ES++ + S S+ + SS E ++ ++ E
Sbjct: 185 EEEEEVVEKTEEKKEG---SSESSSDSESSSDSSSESGDSDSSSDSESESSSEDE----- 236
Query: 313 VGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSEN 355
K+R++ +++ ++ ++ E + + R+S N
Sbjct: 237 ------KKRKAEPASEERPAKITKPSQDSNETCTVFVGRLSWN 273
Score = 32.3 bits (72), Expect = 2.5
Identities = 28/113 (24%), Positives = 49/113 (42%), Gaps = 6/113 (5%)
Query: 113 EYVLKGSELVEGCSERFQQLNVSNKNG----IHEPPETNYGYNAKSKVLSNRQQQK--EA 166
E ++K E E SE + I E E++ +++S + + E+
Sbjct: 125 EVIVKTEEKKESSSESSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESESESSSSES 184
Query: 167 EEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQNNTKTEK 219
EE EE E+ EEK+ SS ++ S S S+E D +++++ EK
Sbjct: 185 EEEEEVVEKTEEKKEGSSESSSDSESSSDSSSESGDSDSSSDSESESSSEDEK 237
>CALD_HUMAN (Q05682) Caldesmon (CDM)
Length = 793
Score = 43.9 bits (102), Expect = 8e-04
Identities = 64/267 (23%), Positives = 107/267 (39%), Gaps = 25/267 (9%)
Query: 161 QQQKEAEE--YEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEA-LQNNTKT 217
+ + EAE Y+ ++ EEE R + E LG+ D+ E QN+
Sbjct: 17 EMRLEAERIAYQRNDDDEEEAARERRRRARQERLRQKQEEESLGQVTDQVEVNAQNSVPD 76
Query: 218 EKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLA---EKLNRRVVVPVQSKQRFGGELGTE 274
E+ T TN + +A LA E+ +R+ ++ ++ F +
Sbjct: 77 EEAKTTT--------TNTQVEGDDEAAFLERLARREERRQKRLQEALERQKEFDPTI--T 126
Query: 275 SATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMM 334
A+ + PSR ++ E K+ ++QE T +K Q D RD +
Sbjct: 127 DASLSLPSRRMQN-DTAENETTEKEEKSESRQERYEIEETETVTKSYQKNDWRDAEENKK 185
Query: 335 SCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNS 394
+E+EEEE + EN ++ EEK S + S+K N +
Sbjct: 186 EDKEKEEEEEEKPKRGSIGENQVEVM-----VEEKTTESQEETVVMSLK-NGQISSEEPK 239
Query: 395 YNEERRSKLGVEEVKLKKEMEEENRER 421
EER G +E+ ++MEEE++ER
Sbjct: 240 QEEEREQ--GSDEISHHEKMEEEDKER 264
Score = 38.5 bits (88), Expect = 0.035
Identities = 56/310 (18%), Positives = 116/310 (37%), Gaps = 44/310 (14%)
Query: 142 EPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEK---EFTSSTTTPHSRCSRGVST 198
+P T+ + S+ + N + E E EE E +E+ E T + T + + +
Sbjct: 122 DPTITDASLSLPSRRMQNDTAENETTEKEEKSESRQERYEIEETETVTKSYQKNDWRDAE 181
Query: 199 EELGEDEDRHEALQNNTKTEKV------VVTNNVSSRGNVTNVPPPMKNHSAAAASL--- 249
E ED+++ E + K + V+ ++ V +KN ++
Sbjct: 182 ENKKEDKEKEEEEEEKPKRGSIGENQVEVMVEEKTTESQEETVVMSLKNGQISSEEPKQE 241
Query: 250 ------AEKLNRRVVVPVQSKQRFGGE-----------LGTESATSAAPSRYSALLQLIA 292
+++++ + + K+R E + E A R A ++
Sbjct: 242 EEREQGSDEISHHEKMEEEDKERAEAERARLEAEERERIKAEQDKKIADER--ARIEAEE 299
Query: 293 CGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRV 352
++ + ++ R+ A+++RQ + +++A +EEE+ A R+
Sbjct: 300 KAAAQERERREAEERERMREEEKRAAEERQRIKEEEKRAAEERQRIKEEEKRAAEERQRI 359
Query: 353 SENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKK 412
E + Q A + E KA KR+ E++R+ ++E K+K
Sbjct: 360 KEEEKRAAEERQRARAE----------EEEKAKVEEQKRNKQLEEKKRA---MQETKIKG 406
Query: 413 EMEEENRERK 422
E E+ E K
Sbjct: 407 EKVEQKIEGK 416
Score = 33.5 bits (75), Expect = 1.1
Identities = 54/288 (18%), Positives = 112/288 (38%), Gaps = 24/288 (8%)
Query: 157 LSNRQQQKEAEEYEEYEEQEE-EKEFT-SSTTTPHSRCSRGVSTEELGEDEDRHEALQNN 214
L+ R+++++ E E Q+E + T +S + P R + E E E++ E+ Q
Sbjct: 100 LARREERRQKRLQEALERQKEFDPTITDASLSLPSRRMQNDTAENETTEKEEKSESRQER 159
Query: 215 TKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGELGTE 274
+ E+ + + + K EK R + Q E+ E
Sbjct: 160 YEIEETETVTKSYQKNDWRDAEENKKEDKEKEEEEEEKPKRGSIGENQV------EVMVE 213
Query: 275 SATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMM 334
T+ S+ ++ + G E + + ++E S + ++ + D+ +A
Sbjct: 214 EKTTE--SQEETVVMSLKNGQISSE-EPKQEEEREQGSDEISHHEKMEEEDKERAEAERA 270
Query: 335 SCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNS 394
E EE E A ++++ + AEEK + E E +A +R
Sbjct: 271 RLEAEERERIKAEQDKKIADERARI-----EAEEK----AAAQERERREAE----ERERM 317
Query: 395 YNEERRSKLGVEEVKLKKEMEEENRERKGGVKEKCIPIKKSSKESSRR 442
EE+R+ + +K +++ E R+R +++ ++ KE +R
Sbjct: 318 REEEKRAAEERQRIKEEEKRAAEERQRIKEEEKRAAEERQRIKEEEKR 365
>YM92_CAEEL (P34531) Hypothetical protein M01A8.2 in chromosome III
Length = 937
Score = 43.1 bits (100), Expect = 0.001
Identities = 67/312 (21%), Positives = 118/312 (37%), Gaps = 44/312 (14%)
Query: 143 PPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELG 202
PP+ A SK +Q K + E E+ + ++E K S P + +
Sbjct: 359 PPKFPVKQKAPSKHQLMMEQLKASIEAEKTKPKKEIKSRVSLLPPPAPKAPQK------- 411
Query: 203 EDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSA-----AAASLAEKLNRRV 257
E+++ E + +T V+++ + PP + A E++ +
Sbjct: 412 ENKEGGEMTETPRRTITKTPLKTVNAKAKTSPTPPVERQKKERKPLYVAPPAKERVEKEK 471
Query: 258 VVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGP----EFKARTQQEPR-VSS 312
+P + + S+ PS SA S GP F Q PR SS
Sbjct: 472 KIPSKPVVSPPTTAEKKPVVSSIPSTSSA--------SKGPFPTSSFAGGKLQGPRKTSS 523
Query: 313 VGTTASKQRQSVDRRDQKAFMMSCEEEEEE-EAVAMMMNRVSENPRLMLGNLQNAEEKEY 371
TT S ++Q D+K + + EA ++MNR++E+ LGN+ EK+
Sbjct: 524 SSTTTSAKKQKNPPIDEKEKLSRLQHSTHAFEATLIVMNRINEDNERKLGNISEQYEKKV 583
Query: 372 FS-GSLVEM------------ESMK-ANHPVLKRS----NSYNEERRSKLGVEEVKLKKE 413
G L +M E MK +N V++ S + +++ + + ++
Sbjct: 584 SELGDLKKMLDEARKKFEEDVEQMKNSNQQVIRNHANAVESLQKTHETQIAEKNKEFERN 643
Query: 414 MEEENRERKGGV 425
EEE R+ V
Sbjct: 644 FEEERARREAEV 655
>NFM_CHICK (P16053) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M)
Length = 857
Score = 43.1 bits (100), Expect = 0.001
Identities = 67/332 (20%), Positives = 119/332 (35%), Gaps = 36/332 (10%)
Query: 119 SELVEGCSERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEA-----EEYEEYE 173
++L E QL + + +H+ E + +K+ +Q KEA EE EY
Sbjct: 258 TDLTTALKEIRAQLECQSDHNMHQAEE--WFKCRYAKLTEAAEQNKEAIRSAKEEIAEYR 315
Query: 174 EQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHE-----------ALQNNTKTEKVVV 222
Q + K + R ++ +L + E+RH L+N + K +
Sbjct: 316 RQLQSKSIELESV----RGTKESLERQLSDIEERHNNDLTTYQDTIHQLENELRGTKWEM 371
Query: 223 TNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGELGTESATSAAPS 282
++ ++ NV + AA L E R F G + T PS
Sbjct: 372 ARHLREYQDLLNVKMALDIEIAAYRKLLEGEETRF-------SAFSGSITGPIFTHRQPS 424
Query: 283 RYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEEEEEE 342
A ++ P+ K + + + ++ + D A M+ + +EEE
Sbjct: 425 VTIASTKIQKTKIEPPKLKVQHKFVEEIIEETKVEDEKSEMEDALSAIAEEMAAKAQEEE 484
Query: 343 EAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSK 402
+ E + + A E+E E E +A +S++ E K
Sbjct: 485 QEEEKAEEEAVEEEAVSEKAAEQAAEEE-------EKEEEEAEEEEAAKSDAAEEGGSKK 537
Query: 403 LGVEEVKLKKEMEEENRERKGGVKEKCIPIKK 434
+EE + +E EEE E KG +E ++K
Sbjct: 538 EEIEEKEEGEEAEEEEAEAKGKAEEAGAKVEK 569
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog
(X-linked nuclear protein-1)
Length = 1359
Score = 41.6 bits (96), Expect = 0.004
Identities = 56/293 (19%), Positives = 106/293 (36%), Gaps = 15/293 (5%)
Query: 159 NRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSR--CSRGVSTEELGEDEDRHEALQNNTK 216
+R++ K E +E +E+E+ K+ S + S+ T EDED E + +K
Sbjct: 82 SRKRAKSESESDESDEEEDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDEEREQKSK 141
Query: 217 TEKVVVTNNVSSRGNVTN-----VPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGGEL 271
+ SS + + V KN + AE SK+ G
Sbjct: 142 KKSKKTKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKGLK 201
Query: 272 GTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKA 331
+ S + S ++ + S K ++ E T ++R +
Sbjct: 202 KKAKSESESESEDEKEVKK-SKKKSKKVVKKESESEDEAPEKKKTEKRKRSKTSSEESSE 260
Query: 332 FMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKR 391
S EEEEE+E+ + + L + L + EE E ++ + + ++
Sbjct: 261 SEKSDEEEEEKES----SPKPKKKKPLAVKKLSSDEESEESDVEVLPQKKKRGAVTLISD 316
Query: 392 SNSYNEERRSKLG---VEEVKLKKEMEEENRERKGGVKEKCIPIKKSSKESSR 441
S +++ E+V KK ++E+ E E I + + SK+ +
Sbjct: 317 SEDEKDQKSESEASDVEEKVSKKKAKKQESSESGSDSSEGSITVNRKSKKKEK 369
Score = 32.7 bits (73), Expect = 1.9
Identities = 60/292 (20%), Positives = 106/292 (35%), Gaps = 56/292 (19%)
Query: 153 KSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVS----TEELGEDEDRH 208
KSK + +K AE EE +E E+ P + +G+ +E E ED
Sbjct: 167 KSKKNKEKSVKKRAETSEESDEDEK----------PSKKSKKGLKKKAKSESESESEDEK 216
Query: 209 EALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFG 268
E ++ K++KVV + S P K + + + S+
Sbjct: 217 EVKKSKKKSKKVVKKESESE----DEAPEKKKTEKRKRSKTSSE--------ESSESEKS 264
Query: 269 GELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRD 328
E E +S P + L K + +E S V K+++
Sbjct: 265 DEEEEEKESSPKPKKKKPLAVK----------KLSSDEESEESDVEVLPQKKKRGA---- 310
Query: 329 QKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPV 388
++S E+E+++ + V E Q + E SGS +S + + V
Sbjct: 311 --VTLISDSEDEKDQKSESEASDVEEKVSKKKAKKQESSE----SGS----DSSEGSITV 360
Query: 389 LKRSNSYNEERRSKLGV--EEVKLKKEMEEENRERKGGVKEKCIPIKKSSKE 438
++S + + K G+ + KL+KE + R KE+ ++K KE
Sbjct: 361 NRKSKKKEKPEKKKKGIIMDSSKLQKETIDAERAE----KERRKRLEKKQKE 408
>THO2_HUMAN (Q8NI27) THO complex subunit 2 (Tho2)
Length = 1478
Score = 40.8 bits (94), Expect = 0.007
Identities = 57/313 (18%), Positives = 125/313 (39%), Gaps = 35/313 (11%)
Query: 131 QLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHS 190
+ V K+G +P E + K++ R + + E+ E+++++E+ K+ TT P++
Sbjct: 1176 EARVLGKDGKEKPKEERPNKDEKARETKERTPKSDKEK-EKFKKEEKAKDEKFKTTVPNA 1234
Query: 191 RCSRGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLA 250
S+ E ++ R + K+++ V + P P
Sbjct: 1235 E-SKSTQEREREKEPSRERDIAKEMKSKENVKGGEKTPVSGSLKSPVP------------ 1281
Query: 251 EKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQL------IACGSSGPEFKART 304
R +P +++ ++ T + S + + +L++L + + P
Sbjct: 1282 -----RSDIPEPEREQKRRKIDTHPSPSHSSTVKDSLIELKESSAKLYINHTPPPLSKSK 1336
Query: 305 QQEPRVSSVGTT--ASKQRQSVDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGN 362
++E + + S++R+ D +D+K + E + R EN + +
Sbjct: 1337 EREMDKKDLDKSRERSREREKKDEKDRKERKRDHSNNDREVPPDLTKRRKEENGTMGVSK 1396
Query: 363 LQNAE--EKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKL---GVEEVKLKKEM--E 415
++ E Y + E K++ K S+S+ E+ K+ G +E + KE +
Sbjct: 1397 HKSESPCESPYPNEKDKEKNKSKSSGKE-KGSDSFKSEKMDKISSGGKKESRHDKEKIEK 1455
Query: 416 EENRERKGGVKEK 428
+E R+ GG +EK
Sbjct: 1456 KEKRDSSGGKEEK 1468
>IF2P_YEAST (P39730) Eukaryotic translation initiation factor 5B
(eIF-5B) (Translation initiation factor IF-2)
Length = 1002
Score = 40.8 bits (94), Expect = 0.007
Identities = 60/284 (21%), Positives = 105/284 (36%), Gaps = 29/284 (10%)
Query: 160 RQQQKEAEEY--EEYEEQEEEKEFTSSTTTPHSRCSRGV--STEELGEDEDRHEAL---- 211
++ +K + Y EE+EE + E S+T TP+ S G ++ E + EA+
Sbjct: 3 KKSKKNQQNYWDEEFEEDAAQNEEISATPTPNPESSAGADDTSREASASAEGAEAIEGDF 62
Query: 212 -----QNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQR 266
Q+ K EK V+ + + + K ++ R Q+++
Sbjct: 63 MSTLKQSKKKQEKKVIEEKKDGKPILKSKKEKEKEKKEKEKQKKKEQAARKKAQQQAQKE 122
Query: 267 FGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDR 326
EL ++ AA + +A + S G K + V + R+ ++
Sbjct: 123 KNKELNKQNVEKAAAEKAAAEK---SQKSKGESDKPSASAKKPAKKVPAGLAALRRQLEL 179
Query: 327 RDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANH 386
+ Q E EEEE R++ ++ +EKE + E KA
Sbjct: 180 KKQLEEQEKLEREEEERLEKEEEERLANEEKMKEEAKAAKKEKE-----KAKREKRKAEG 234
Query: 387 PVLKRSNSYN----EERRSKL----GVEEVKLKKEMEEENRERK 422
+L R E RR+ L V+ L K+ EEN+ +K
Sbjct: 235 KLLTRKQKEEKKLLERRRAALLSSGNVKVAGLAKKDGEENKPKK 278
>CALD_CHICK (P12957) Caldesmon (CDM)
Length = 771
Score = 40.4 bits (93), Expect = 0.009
Identities = 66/281 (23%), Positives = 112/281 (39%), Gaps = 45/281 (16%)
Query: 153 KSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEALQ 212
KS +N +Q E E +E ++ EEEK + V TEE D A++
Sbjct: 163 KSYQRNNWRQDGEEEGKKEEKDSEEEKP-------------KEVPTEENQVDV----AVE 205
Query: 213 NNTKTEKVVVTNNVSSRG-NVTN--VPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFGG 269
+T E+VV T ++ N TN + AA E R + + K+R
Sbjct: 206 KSTDKEEVVETKTLAVNAENDTNAMLEGEQSITDAADKEKEEAEKEREKLEAEEKER--- 262
Query: 270 ELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQ 329
L E AA + A E KA ++E + A+++R+ ++
Sbjct: 263 -LKAEEEKKAAEEKQKA----------EEEKKAAEERERAKAEEEKRAAEERERAKAEEE 311
Query: 330 KAFMMSCEEEE----EEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKAN 385
+ + EE E EEE A +E R AEE+ + + E +
Sbjct: 312 RK---AAEERERAKAEEERKAAEERAKAEEERKAAEERAKAEEERKAAEERAKAEKERKA 368
Query: 386 HPVLKRSNSYNEER----RSKLGVEEVKLKKEMEEENRERK 422
+R+ + E+R +++L E++K KK+MEE+ + +
Sbjct: 369 AEERERAKAEEEKRAAEEKARLEAEKLKEKKKMEEKKAQEE 409
Score = 30.4 bits (67), Expect = 9.5
Identities = 20/69 (28%), Positives = 32/69 (45%), Gaps = 3/69 (4%)
Query: 112 GEYVLKGSELVEGCSERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEE 171
G LKG+ E SE+ ++ + E E + K+L +Q+K+ EE E
Sbjct: 496 GRSNLKGAANAEAGSEKLKE---KQQEAAVELDELKKRREERRKILEEEEQKKKQEEAER 552
Query: 172 YEEQEEEKE 180
+EEEK+
Sbjct: 553 KIREEEEKK 561
Score = 30.4 bits (67), Expect = 9.5
Identities = 64/295 (21%), Positives = 105/295 (34%), Gaps = 31/295 (10%)
Query: 152 AKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRHEAL 211
A+ + + +++K A E E + EEE++ + R + EE + E+ +A
Sbjct: 285 AEERERAKAEEEKRAAEERERAKAEEERKAAEERERAKAEEER-KAAEERAKAEEERKAA 343
Query: 212 QNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAAS--LAEKLNRRVVVPVQSKQRFGG 269
+ K E+ ++ + + A AE+ R ++ K++
Sbjct: 344 EERAKAEEERKAAEERAKAEKERKAAEERERAKAEEEKRAAEEKARLEAEKLKEKKKMEE 403
Query: 270 ELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQ 329
+ E A L+ E K ++E + T SK+ Q D +D+
Sbjct: 404 KKAQEEKAQA---------NLLRKQEEDKEAKVEAKKESLPEKLQPT-SKKDQVKDNKDK 453
Query: 330 ----KAFMMSCEEEEE--EEAVAMMMNRVSENPRLM-------LGNLQNAEEKEYFSGSL 376
K M S + + E A R P+L NL+ A E S L
Sbjct: 454 EKAPKEEMKSVWDRKRGVPEQKAQNGERELTTPKLKSTENAFGRSNLKGAANAEAGSEKL 513
Query: 377 VEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEE---ENRERKGGVKEK 428
E + A L EERR L EE K K+E E E K +KE+
Sbjct: 514 KEKQQEAAVE--LDELKKRREERRKILEEEEQKKKQEEAERKIREEEEKKRMKEE 566
>YG3A_YEAST (P53278) Hypothetical 92.7 kDa protein in ASN2-PHB1
intergenic region
Length = 816
Score = 40.0 bits (92), Expect = 0.012
Identities = 61/310 (19%), Positives = 120/310 (38%), Gaps = 41/310 (13%)
Query: 134 VSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCS 193
+S++NG+ EP T+ +KS V+ +EQ+EEK P
Sbjct: 266 ISSENGVLEPRTTDQSGGSKSGVVPT-------------DEQKEEKSDVKKVNPPSGEEK 312
Query: 194 RGVSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKL 253
+ V E E+E + + + + T+ S + + + P S A ++
Sbjct: 313 KEVEAEGDAEEETEQSSAEESAERTSTPETSEPESEEDESPIDP-----SKAPKVPFQEP 367
Query: 254 NRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSV 313
+R+ + + + T+ + +AAPS A PE +T++ +S
Sbjct: 368 SRKERTGIFALWKSPTSSSTQKSKTAAPSNPVAT-------PENPELIVKTKEHGYLSKA 420
Query: 314 GTTASKQRQSV------DRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAE 367
+ + D R ++ + +E +E + + N++ E M +
Sbjct: 421 VYDKINYDEKIHQAWLADLRAKEKDKYDAKNKEYKEKLQDLQNQIDEIENSMKAMREETS 480
Query: 368 EK-EYFSGSLV-EMESMKANH-----PVLK-RSNSYNEERRSKLGV--EEVKLKKEMEEE 417
EK E LV ++ + A H +LK N N++ + K V ++ +K E+++
Sbjct: 481 EKIEVSKNRLVKKIIDVNAEHNNKKLMILKDTENMKNQKLQEKNEVLDKQTNVKSEIDDL 540
Query: 418 NRERKGGVKE 427
N E+ KE
Sbjct: 541 NNEKTNVQKE 550
>MYHA_RAT (Q9JLT0) Myosin heavy chain, nonmuscle type B (Cellular
myosin heavy chain, type B) (Nonmuscle myosin heavy
chain-B) (NMMHC-B)
Length = 1976
Score = 39.3 bits (90), Expect = 0.021
Identities = 74/321 (23%), Positives = 126/321 (39%), Gaps = 45/321 (14%)
Query: 152 AKSKVLSN-RQQQKEAEEYE-EYEEQEEEKE--FTSSTTTPHSRCSRGVSTEELGEDEDR 207
A+ +V+ R+ Q + ++Y+ E EE ++ F S + S +L E+
Sbjct: 1639 ARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQEELAS 1698
Query: 208 HEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNH-SAAAASLAEKLNRRVVVPVQSKQR 266
E + + + E+ + + +++ + + K A A L E+L R
Sbjct: 1699 SERARRHAEQERDELADEIANSASGKSALLDEKRRLEARIAQLEEELEEEQSNMELLNDR 1758
Query: 267 FG------GELGTESAT--SAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTAS 318
F L TE A SAA +A QL E KA+ Q+ G S
Sbjct: 1759 FRKTTLQVDTLNTELAAERSAAQKSDNARQQL---ERQNKELKAKLQELE-----GAVKS 1810
Query: 319 KQRQSVDRRDQKAFMMS--CEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSL 376
K + ++ + K + E+E +E A A + R +E +L +Q +E+ +
Sbjct: 1811 KFKATISALEAKIGQLEEQLEQEAKERAAANKLVRRTEK-KLKEIFMQVEDERRHADQYK 1869
Query: 377 VEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEE---------------ENRER 421
+ME A LKR EE ++ KL++E+++ +NR R
Sbjct: 1870 EQMEKANARMKQLKRQLEEAEEEATRANASRRKLQRELDDATEANEGLSREVSTLKNRLR 1929
Query: 422 KGGVKEKCIPIKKSSKESSRR 442
+GG PI SS S RR
Sbjct: 1930 RGG------PISFSSSRSGRR 1944
Score = 33.1 bits (74), Expect = 1.5
Identities = 66/300 (22%), Positives = 122/300 (40%), Gaps = 23/300 (7%)
Query: 145 ETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEE--KEFTSSTTTPHSRCS--RGVSTEE 200
ET N S++ RQ ++E +E +E+EEE K S+ + + ++
Sbjct: 1327 ETRQKLNLSSRI---RQLEEEKNSLQEQQEEEEEARKNLEKQVLALQSQLADTKKKVDDD 1383
Query: 201 LGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNH-SAAAASLAEKLN--RRV 257
LG E EA + K + + + + + + KN L L+ R++
Sbjct: 1384 LGTIEGLEEAKKKLLKDVEAL-SQRLEEKVLAYDKLEKTKNRLQQELDDLTVDLDHQRQI 1442
Query: 258 VVPVQSKQR-FGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTT 316
V ++ KQ+ F L E SA RY+ + E KA + ++
Sbjct: 1443 VSNLEKKQKKFDQLLAEEKGISA---RYAEERDRAEAEAREKETKALSLARALEEALEAK 1499
Query: 317 ASKQRQSVDRRDQKAFMMSCEEE-----EEEEAVAMMMNRVSENPRLMLGNLQNAEEKEY 371
+RQ+ R +MS +++ E E + + E R L L++ +
Sbjct: 1500 EEFERQNKQLRADMEDLMSSKDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATE 1559
Query: 372 FSGSLVE--MESMKANHPV-LKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEK 428
+ +E M++MKA L+ + NEE++ L + +L+ E+E+E ++R V K
Sbjct: 1560 DAKLRLEVNMQAMKAQFERDLQTRDEQNEEKKRLLLKQVRELEAELEDERKQRALAVASK 1619
>MYHA_BOVIN (Q27991) Myosin heavy chain, nonmuscle type B (Cellular
myosin heavy chain, type B) (Nonmuscle myosin heavy
chain-B) (NMMHC-B)
Length = 1976
Score = 39.3 bits (90), Expect = 0.021
Identities = 74/321 (23%), Positives = 126/321 (39%), Gaps = 45/321 (14%)
Query: 152 AKSKVLSN-RQQQKEAEEYE-EYEEQEEEKE--FTSSTTTPHSRCSRGVSTEELGEDEDR 207
A+ +V+ R+ Q + ++Y+ E EE ++ F S + S +L E+
Sbjct: 1639 ARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQEELAS 1698
Query: 208 HEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNH-SAAAASLAEKLNRRVVVPVQSKQR 266
E + + + E+ + + +++ + + K A A L E+L R
Sbjct: 1699 SERARRHAEQERDELADEIANSASGKSALLDEKRRLEARIAQLEEELEEEQSNMELLNDR 1758
Query: 267 FG------GELGTESAT--SAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTAS 318
F L TE A SAA +A QL E KA+ Q+ G S
Sbjct: 1759 FRKTTLQVDTLNTELAAERSAAQKSDNARQQL---ERQNKELKAKLQELE-----GAVKS 1810
Query: 319 KQRQSVDRRDQKAFMMS--CEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSL 376
K + ++ + K + E+E +E A A + R +E +L +Q +E+ +
Sbjct: 1811 KFKATISALEAKIGQLEEQLEQEAKERAAANKLVRRTEK-KLKEIFMQVEDERRHADQYK 1869
Query: 377 VEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEE---------------ENRER 421
+ME A LKR EE ++ KL++E+++ +NR R
Sbjct: 1870 EQMEKANARMKQLKRQLEEAEEEATRANASRRKLQRELDDATEANEGLSREVSTLKNRLR 1929
Query: 422 KGGVKEKCIPIKKSSKESSRR 442
+GG PI SS S RR
Sbjct: 1930 RGG------PISFSSSRSGRR 1944
Score = 35.4 bits (80), Expect = 0.30
Identities = 64/289 (22%), Positives = 114/289 (39%), Gaps = 20/289 (6%)
Query: 157 LSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVS------TEELGEDEDRHEA 210
LS+R +Q E E E+QEEE+E S ++ ++LG E+ EA
Sbjct: 1334 LSSRIRQLEEERSSLQEQQEEEEEARRSLEKQLQALQAQLTDTKKKVDDDLGTIENLEEA 1393
Query: 211 LQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLN--RRVVVPVQSKQRFG 268
+ K +V+ + L L+ R++V ++ KQ+
Sbjct: 1394 KKKLLKDVEVLSQRLEEKALAYDKLEKTKTRLQQELDDLLVDLDHQRQIVSNLEKKQKKF 1453
Query: 269 GELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRD 328
+L E +A RY+ + E KA + ++ +RQ+ R
Sbjct: 1454 DQLLAEEKNISA--RYAEERDRAEAEAREKETKALSLARALEEALEAREEAERQNKQLRA 1511
Query: 329 QKAFMMSCEEE-----EEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSL---VEME 380
+MS +++ E E + + E R L L++ E + L V M+
Sbjct: 1512 DMEDLMSSKDDVGKNVHELEKSKRALEQQVEEMRTQLEELED-ELQATEDAKLRLEVNMQ 1570
Query: 381 SMKAN-HPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEK 428
+MKA L+ + NEE++ L + +L+ E+E+E ++R V K
Sbjct: 1571 AMKAQFERDLQTRDEQNEEKKRLLIKQVRELEAELEDERKQRALAVASK 1619
>MYHA_HUMAN (P35580) Myosin heavy chain, nonmuscle type B (Cellular
myosin heavy chain, type B) (Nonmuscle myosin heavy
chain-B) (NMMHC-B)
Length = 1976
Score = 38.9 bits (89), Expect = 0.027
Identities = 74/323 (22%), Positives = 125/323 (37%), Gaps = 49/323 (15%)
Query: 152 AKSKVLSN-RQQQKEAEEYE-EYEEQEEEKE--FTSSTTTPHSRCSRGVSTEELGEDEDR 207
A+ +V+ R+ Q + ++Y+ E EE ++ F S + S +L E+
Sbjct: 1639 ARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQEELAS 1698
Query: 208 HEALQNNTKTEKVV----VTNNVSSRGNVTNVPPPMKNHSAAAA-------SLAEKLNRR 256
E + + + E+ +TN+ S + + + ++ A S E LN R
Sbjct: 1699 SERARRHAEQERDELADEITNSASGKSALLDEKRRLEARIAQLEEELEEEQSNMELLNDR 1758
Query: 257 VVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTT 316
EL E SAA +A QL E KA+ Q+ G
Sbjct: 1759 FRKTTLQVDTLNAELAAER--SAAQKSDNARQQL---ERQNKELKAKLQELE-----GAV 1808
Query: 317 ASKQRQSVDRRDQKAFMMS--CEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSG 374
SK + ++ + K + E+E +E A A + R +E +L +Q +E+ +
Sbjct: 1809 KSKFKATISALEAKIGQLEEQLEQEAKERAAANKLVRRTEK-KLKEIFMQVEDERRHADQ 1867
Query: 375 SLVEMESMKANHPVLKRSNSYNEERRSKLGVEEVKLKKEMEE---------------ENR 419
+ME A LKR EE ++ KL++E+++ +NR
Sbjct: 1868 YKEQMEKANARMKQLKRQLEEAEEEATRANASRRKLQRELDDATEANEGLSREVSTLKNR 1927
Query: 420 ERKGGVKEKCIPIKKSSKESSRR 442
R+GG PI SS S RR
Sbjct: 1928 LRRGG------PISFSSSRSGRR 1944
Score = 34.3 bits (77), Expect = 0.66
Identities = 64/299 (21%), Positives = 122/299 (40%), Gaps = 21/299 (7%)
Query: 145 ETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEE--KEFTSSTTTPHSRCS--RGVSTEE 200
ET N S++ RQ ++E +E +E+EEE K S+ + + ++
Sbjct: 1327 ETRQKLNLSSRI---RQLEEEKNSLQEQQEEEEEARKNLEKQVLALQSQLADTKKKVDDD 1383
Query: 201 LGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNH-SAAAASLAEKLN--RRV 257
LG E EA + K + + + + + + KN L L+ R+V
Sbjct: 1384 LGTIESLEEAKKKLLKDAEAL-SQRLEEKALAYDKLEKTKNRLQQELDDLTVDLDHQRQV 1442
Query: 258 VVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTA 317
++ KQ+ +L E + +A RY+ + E KA + ++
Sbjct: 1443 ASNLEKKQKKFDQLLAEEKSISA--RYAEERDRAEAEAREKETKALSLARALEEALEAKE 1500
Query: 318 SKQRQSVDRRDQKAFMMSCEEE-----EEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYF 372
+RQ+ R +MS +++ E E + + E R L L++ +
Sbjct: 1501 EFERQNKQLRADMEDLMSSKDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATED 1560
Query: 373 SGSLVE--MESMKANHPV-LKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEK 428
+ +E M++MKA L+ + NEE++ L + +L+ E+E+E ++R V K
Sbjct: 1561 AKLRLEVNMQAMKAQFERDLQTRDEQNEEKKRLLIKQVRELEAELEDERKQRALAVASK 1619
>MPT2_HUMAN (O60237) Protein phosphatase 1 regulatory subunit 12B
(Myosin phosphatase targeting subunit 2) (Myosin
phosphatase target subunit 2)
Length = 982
Score = 38.5 bits (88), Expect = 0.035
Identities = 59/252 (23%), Positives = 103/252 (40%), Gaps = 49/252 (19%)
Query: 127 ERFQQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTT 186
++ Q + S K ++ E++ +S N+++ EE + +E EEE + +SS++
Sbjct: 302 QKKQNVLRSEKETRNKLIESDLNSKIQSGFFKNKEKMLYEEETPKSQEMEEENKESSSSS 361
Query: 187 TPHSRCSRGVSTEELGEDE--DRHEALQNNTKTEKVVVTNNVSSRGNVT-NVPPPMKNHS 243
S EE GEDE + + + K E V +N S+ ++T +P P +N
Sbjct: 362 ----------SEEEEGEDEASESETEKEADKKPEAFVNHSNSESKSSITEQIPAPAQNTF 411
Query: 244 AAAASLAEKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIACGSSGPEFKAR 303
+A+ S +RF L + S S L L GS +
Sbjct: 412 SAS----------------SARRFSSGLFNKPEEPKDESPSSWRLGLRKTGSHNMLSEVA 455
Query: 304 TQQEP---RVSSVGTTASKQRQS--VDRRDQKAFMMSCEEEEEEEAVAMMMNRVSENPRL 358
+EP R SS+ ++S R S +D +D +E E + ++ + R
Sbjct: 456 NSREPIRDRGSSIYRSSSSPRISALLDNKD--------KERENKSYISSLAPR------- 500
Query: 359 MLGNLQNAEEKE 370
L + + EEKE
Sbjct: 501 KLNSTSDIEEKE 512
>INCE_MOUSE (Q9WU62) Inner centromere protein
Length = 880
Score = 38.1 bits (87), Expect = 0.046
Identities = 38/153 (24%), Positives = 62/153 (39%), Gaps = 8/153 (5%)
Query: 274 ESATSAAPSRYSALLQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFM 333
E A R + + A E +AR +QE + Q++ +RR Q+
Sbjct: 583 EKTEKAKEERLAEKAKKKATAKKMEEVEARRKQEEEARRLRWL---QQEEEERRHQEMLQ 639
Query: 334 MSCEEEEEE----EAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVL 389
EEE+E EA + R E R Q E+E E E + L
Sbjct: 640 RKKEEEQERRKAAEARRLAEQREQERRREQERREQERREQERREQERKEQERREQEQERL 699
Query: 390 KRSNSYNEERRSKLGVEEVKLKKEMEEENRERK 422
R+ +ER L +++ +L+KE+EE+ R+ +
Sbjct: 700 -RAKREMQEREKALRLQKERLQKELEEKKRKEE 731
Score = 32.0 bits (71), Expect = 3.3
Identities = 62/324 (19%), Positives = 114/324 (35%), Gaps = 31/324 (9%)
Query: 130 QQLNVSNKNGIHEPPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPH 189
Q+ + +G EPP++ + ++ K+A + E+ E+E+ TP
Sbjct: 405 QEAELDQTDGHREPPQS----------VRRKRSYKQAISEPDEEQLEDEELQPCQNKTPS 454
Query: 190 SRCSRGVSTEELG---EDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAA 246
C L +++ L T + V + R V P
Sbjct: 455 PPCPANKVVRPLRTFLHTVQKNQMLMTPTLASRSSVMKSFIKRNTPLRVDPKCSFVEKER 514
Query: 247 ASLAEKLNRRVVVPVQSKQRFGGELGT--ESATSAAPSRYSALLQLIACGSSGPEFKART 304
L E L R+ + +Q+ + E R +LQ E K +
Sbjct: 515 QRL-ESLRRKEEAEQRRRQKVEEDKRRRLEEVKLKREERLRKVLQARERVEQMKEEKKK- 572
Query: 305 QQEPRVSSVG--TTASKQRQSVDRRDQKAFMMSCEEEE-----EEEAVAMMMNRVSENPR 357
Q E + + + T +K+ + ++ +KA EE E EEEA + + E R
Sbjct: 573 QIEQKFAQIDEKTEKAKEERLAEKAKKKATAKKMEEVEARRKQEEEARRLRWLQQEEEER 632
Query: 358 LMLGNLQNAEEKEYFSGSLVEM-------ESMKANHPVLKRSNSYNEERRSKLGVEEVKL 410
LQ +E+E E E + + +ERR + E+ +
Sbjct: 633 RHQEMLQRKKEEEQERRKAAEARRLAEQREQERRREQERREQERREQERREQERKEQERR 692
Query: 411 KKEMEEENRERKGGVKEKCIPIKK 434
++E E +R+ +EK + ++K
Sbjct: 693 EQEQERLRAKREMQEREKALRLQK 716
>CENB_HUMAN (P07199) Major centromere autoantigen B (Centromere
protein B) (CENP-B)
Length = 599
Score = 38.1 bits (87), Expect = 0.046
Identities = 21/65 (32%), Positives = 31/65 (47%)
Query: 145 ETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGED 204
E +G + + ++ + + E EE EE EE+EEE E G EELGE+
Sbjct: 386 EAGFGGGPNATITTSLKSEGEEEEEEEEEEEEEEGEGEEEEEEGEEEEEEGGEGEELGEE 445
Query: 205 EDRHE 209
E+ E
Sbjct: 446 EEVEE 450
>CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2L1
(EC 2.7.1.37) (Galactosyltransferase associated protein
kinase p58/GTA) (Cell division cycle 2-like 1) (CLK-1)
(CDK11) (p58 CLK-1)
Length = 795
Score = 38.1 bits (87), Expect = 0.046
Identities = 19/43 (44%), Positives = 27/43 (62%)
Query: 165 EAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELGEDEDR 207
E E EE EE+EEE+E S++ + + VS EE+ EDE+R
Sbjct: 321 EESEEEEEEEEEEEEETGSNSEEASEQSAEEVSEEEMSEDEER 363
>YWIE_CAEEL (Q23525) Hypothetical protein ZK546.14 in chromosome II
Length = 472
Score = 37.7 bits (86), Expect = 0.060
Identities = 56/288 (19%), Positives = 114/288 (39%), Gaps = 14/288 (4%)
Query: 165 EAEEYEEYEEQEEEKEFTSSTTTPHSRCSRGVSTEELG--EDEDRHEALQNNTKTEKVVV 222
EAE+ + +E+EE + T + S G +E+ ED++ +E+ + + + +
Sbjct: 182 EAEDSDSTDEEEETPSKPNKTVAQSTLKSNGKIDKEIQKLEDDEDNESPEIRRQIALLRL 241
Query: 223 TNNVSSRGNVTNVPPPMKNHSAAAASLAE--KLNRRVVVPVQSKQRFGGELGTESATSAA 280
+ P K SA A +AE +L RR ++R + G E+A
Sbjct: 242 QKKLKEMKVERKGKGPAKVTSAMAEKMAEEKRLKRRESKLKLKQRRAEEKKGKEAAAQVK 301
Query: 281 PSRYSAL------LQLIACGSSGPEFKARTQQEPRVSSVGTTASKQRQ-SVDRRDQKAFM 333
+ ++ G S K +++ + TA K R + RD K+ +
Sbjct: 302 KETVESTENGNEPVEKQKSGISFNNLKFEIKEDKQRGKKQRTAKKDRALKLTGRDYKSLI 361
Query: 334 MSCEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSN 393
+ EE + +A + +M L+ + + +G V+ +++ L + N
Sbjct: 362 --AKVEETKATIAKVREADPHKATIMEDELKWEKTLKRAAGGKVK-DNLGMLKKALVKKN 418
Query: 394 SYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEKCIPIKKSSKESSR 441
+ R+ K E K + E + + +RK ++++ +KK R
Sbjct: 419 KMKDRRKQKWENRENKTEGEKQTKQDKRKKNLQKRIDDVKKRKMNKLR 466
Score = 32.0 bits (71), Expect = 3.3
Identities = 63/299 (21%), Positives = 114/299 (38%), Gaps = 30/299 (10%)
Query: 153 KSKVLSNRQQQKEAEEYEEYEEQ----EEEKEFTSSTTTPHSRCSRGVSTEELGEDEDRH 208
K KV+ Q++++ + + EE +EEKE T + +E ED +
Sbjct: 115 KEKVVKKDQKKQDKKADSDSEEDDSSDDEEKEETDEPVAKKQKKEESSDDDEDSEDGEEP 174
Query: 209 EALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSAAAASLAEKLNRRVVVPVQSKQRFG 268
E + E S + P N + A ++L K N ++ +Q +
Sbjct: 175 EGNNGAVEAED-------SDSTDEEEETPSKPNKTVAQSTL--KSNGKIDKEIQKLE--- 222
Query: 269 GELGTESATSAAPSRYSALLQLIACGSSGPEFKARTQQE--PRVSSVGTTASKQRQSVDR 326
E S R ALL+L E K + + +V+S + + + R
Sbjct: 223 ---DDEDNESPEIRRQIALLRL---QKKLKEMKVERKGKGPAKVTSAMAEKMAEEKRLKR 276
Query: 327 RDQKAFMMS--CEEEEEEEAVAMMMNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKA 384
R+ K + EE++ +EA A + E+ GN ++K S + ++ E +K
Sbjct: 277 RESKLKLKQRRAEEKKGKEAAAQVKKETVESTE--NGNEPVEKQKSGISFNNLKFE-IKE 333
Query: 385 NHPVLKRSNSYNEERRSKLGVEEVKLKKEMEEENRERKGGVKEKCIPIKKSSKESSRRW 443
+ K+ + ++R KL + K EE + V+E P K + E +W
Sbjct: 334 DKQRGKKQRTAKKDRALKLTGRDYKSLIAKVEETKATIAKVRE-ADPHKATIMEDELKW 391
>NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130
kDa protein) (140 kDa nucleolar phosphoprotein)
(Nopp140) (Nucleolar and coiled-body phosphoprotein 1)
Length = 699
Score = 37.7 bits (86), Expect = 0.060
Identities = 60/335 (17%), Positives = 122/335 (35%), Gaps = 41/335 (12%)
Query: 143 PPETNYGYNAKSKVLSNRQQQKEAEEYEEYEEQEEEKE-------FTSSTTTPHSRCSRG 195
PP+ + G K + +Q + +E+ + + E+E S TT +
Sbjct: 299 PPKKSLGTQPPKKAVEKQQPVESSEDSSDESDSSSEEEKKPPTKAVVSKATTKPPPAKKA 358
Query: 196 VSTEELGEDEDRHEALQNNTKTEKVVVTNNVSSRGNVTNVPPPMKNHSA----------- 244
+ D D E + ++ T N S++ VT P +K +A
Sbjct: 359 AESSSDSSDSDSSE--DDEAPSKPAGTTKNSSNKPAVTTKSPAVKPAAAPKQPVGGGQKL 416
Query: 245 ------------AAASLAEKLNRRVVVPVQSKQRFGGELGTESATSAAPSRYSALLQLIA 292
++S E+ +++V + K L + + SR S+ +
Sbjct: 417 LTRKADSSSSEEESSSSEEEKTKKMVATTKPKATAKAALSLPAKQAPQGSRDSSSDSDSS 476
Query: 293 CGSSGPE--FKARTQQEPRVSSVGTTASKQRQSVDRRDQKAFMMSCEE--EEEEEAVAMM 348
E K+ +++P+ + G SK + + + + S ++ EEEEE +
Sbjct: 477 SSEEEEEKTSKSAVKKKPQKVAGGAAPSKPASAKKGKAESSNSSSSDDSSEEEEEKLKGK 536
Query: 349 MNRVSENPRLMLGNLQNAEEKEYFSGSLVEMESMKANHPVLKRSNSYNEERRSKLGVE-- 406
+ + P+ + A+ + S E E K V+ +S S + ++++ E
Sbjct: 537 GSPRPQAPKANGTSALTAQNGKAAKNSEEEEEEKKKAAVVVSKSGSLKKRKQNEAAKEAE 596
Query: 407 ---EVKLKKEMEEENRERKGGVKEKCIPIKKSSKE 438
K+K + +RK G K P ++ +E
Sbjct: 597 TPQAKKIKLQTPNTFPKRKKGEKRASSPFRRVREE 631
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.310 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,092,500
Number of Sequences: 164201
Number of extensions: 2192120
Number of successful extensions: 12813
Number of sequences better than 10.0: 294
Number of HSP's better than 10.0 without gapping: 62
Number of HSP's successfully gapped in prelim test: 241
Number of HSP's that attempted gapping in prelim test: 10945
Number of HSP's gapped (non-prelim): 1093
length of query: 443
length of database: 59,974,054
effective HSP length: 113
effective length of query: 330
effective length of database: 41,419,341
effective search space: 13668382530
effective search space used: 13668382530
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0307.3


