

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
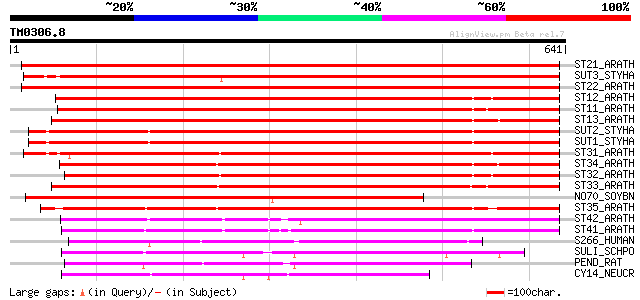
Query= TM0306.8
(641 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ST21_ARATH (O04722) Sulfate transporter 2.1 (AST68) 838 0.0
SUT3_STYHA (P53393) Low affinity sulphate transporter 3 806 0.0
ST22_ARATH (P92946) Sulfate transporter 2.2 (AST56) (AtH14) 774 0.0
ST12_ARATH (Q9MAX3) Sulfate transporter 1.2 658 0.0
ST11_ARATH (Q9SAY1) Sulfate transporter 1.1 (High-affinity sulfa... 649 0.0
ST13_ARATH (Q9FEP7) Sulfate transporter 1.3 648 0.0
SUT2_STYHA (P53392) High affinity sulphate transporter 2 631 e-180
SUT1_STYHA (P53391) High affinity sulphate transporter 1 629 e-180
ST31_ARATH (Q9SV13) Sulfate transporter 3.1 (AST12) (AtST1) 565 e-160
ST34_ARATH (Q9LW86) Probable sulfate transporter 3.4 557 e-158
ST32_ARATH (O04289) Sulfate transporter 3.2 (AST77) 540 e-153
ST33_ARATH (Q9SXS2) Probable sulfate transporter 3.3 (AST91) 539 e-152
NO70_SOYBN (Q02920) Early nodulin 70 522 e-147
ST35_ARATH (Q94LW6) Probable sulfate transporter 3.5 473 e-133
ST42_ARATH (Q8GYH8) Probable sulfate transporter 4.2 347 7e-95
ST41_ARATH (Q9FY46) Sulfate transporter 4.1, chloroplast precurs... 342 2e-93
S266_HUMAN (Q9BXS9) Solute carrier family 26 member 6 (Pendrin-l... 214 7e-55
SULI_SCHPO (Q9URY8) Probable sulfate permease C869.05c 203 1e-51
PEND_RAT (Q9R154) Pendrin (Sodium-independent chloride/iodide tr... 194 5e-49
CY14_NEUCR (P23622) Sulfate permease II 194 5e-49
>ST21_ARATH (O04722) Sulfate transporter 2.1 (AST68)
Length = 677
Score = 838 bits (2165), Expect = 0.0
Identities = 411/624 (65%), Positives = 504/624 (79%), Gaps = 2/624 (0%)
Query: 14 QEVRSQWVLNAPEPPTAWNMVTDSVKKTISQFPRKLSYLKDQPC-NTLMSFLQGIFPILS 72
Q RS+W+L+ PEPP+ W+ + VK + +K L+ QP ++S LQ IFPI
Sbjct: 46 QPDRSKWLLDCPEPPSPWHELKRQVKGSFLTKAKKFKSLQKQPFPKQILSVLQAIFPIFG 105
Query: 73 WGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSR 132
W RNY F+ D++AGLT+ASLCIPQSIGYATLA LDPQYGLYTSVVPPLIYA+MGTSR
Sbjct: 106 WCRNYKLTMFKNDLMAGLTLASLCIPQSIGYATLAKLDPQYGLYTSVVPPLIYALMGTSR 165
Query: 133 EVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGFLVDF 192
E+A+GPVAVVSLL+ SM+QKL+DP DP+ Y KLV TT FAGIFQ +FGLFRLGFLVDF
Sbjct: 166 EIAIGPVAVVSLLISSMLQKLIDPETDPLGYKKLVLTTTFFAGIFQASFGLFRLGFLVDF 225
Query: 193 LSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNPWQPRNFILG 252
LSHAAIVGF+ GAAIVIGLQQ KGLLGIT+FTT TDI+SV++AVW + W P FILG
Sbjct: 226 LSHAAIVGFMGGAAIVIGLQQLKGLLGITNFTTNTDIVSVLRAVWRSCQQQWSPHTFILG 285
Query: 253 SSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVKIVKHVKGGLN 312
SFL FIL TRF+GKK KKLFWL +IAPL+++++STL+VFLT+AD+ GVK V+H+KGGLN
Sbjct: 286 CSFLSFILITRFIGKKYKKLFWLPAIAPLIAVVVSTLMVFLTKADEHGVKTVRHIKGGLN 345
Query: 313 PSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGYQLDGNKEMMSIGFSN 372
P S+ +LDFN PH+G+ AKIGL++A+VALTE+IAVGRSFA IKGY+LDGNKEM++IGF N
Sbjct: 346 PMSIQDLDFNTPHLGQIAKIGLIIAIVALTEAIAVGRSFAGIKGYRLDGNKEMVAIGFMN 405
Query: 373 IIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQFFTKLLYYTPTAIIAS 432
++GS TSCY ATGSFSRTAVN+AAGCET +SNIVMA+TV ++L+ T+LLYYTP AI+AS
Sbjct: 406 VLGSFTSCYAATGSFSRTAVNFAAGCETAMSNIVMAVTVFVALECLTRLLYYTPIAILAS 465
Query: 433 VILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGLLAAVMISFLKIILIS 492
+ILSALPGLI+I EA IWKVDK DFLA GAFFGVLFASVEIGLL AV+ISF KIILIS
Sbjct: 466 IILSALPGLININEAIHIWKVDKFDFLALIGAFFGVLFASVEIGLLVAVVISFAKIILIS 525
Query: 493 IRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFANANFVRERIMKWVTQ 552
IRP E LG++PGT F D QYPM V+ PGV++ RVKSALLCFANA+ + ERIM WV +
Sbjct: 526 IRPGIETLGRMPGTDTFTDTNQYPMTVKTPGVLIFRVKSALLCFANASSIEERIMGWVDE 585
Query: 553 EESKDD-KGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLISNGKQLAIANPRWQVI 611
EE +++ K N+ I V+L+ S+L+++DTSGI +L E+ LI G +L I NP+WQVI
Sbjct: 586 EEEEENTKSNAKRKILFVVLDMSSLINVDTSGITALLELHNKLIKTGVELVIVNPKWQVI 645
Query: 612 HKLKVSNFVSKIGGRIYLTVEEAI 635
HKL + FV +IGG++YLT+ EA+
Sbjct: 646 HKLNQAKFVDRIGGKVYLTIGEAL 669
>SUT3_STYHA (P53393) Low affinity sulphate transporter 3
Length = 644
Score = 806 bits (2082), Expect = 0.0
Identities = 401/626 (64%), Positives = 500/626 (79%), Gaps = 12/626 (1%)
Query: 17 RSQWVLNAPEPPTAWNMVTDSVKKTISQFPRKLSYLKDQPCNTLMSFLQGIFPILSWGRN 76
RSQWVLN+P PP +K ++F S K+ +SFL +FPILSW R
Sbjct: 12 RSQWVLNSPNPPPLTKKFLGPLKD--NKFFTSSSSKKE---TRAVSFLASLFPILSWIRT 66
Query: 77 YTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSREVAV 136
Y+A KF+ D+L+GLT+ASL IPQSIGYA LA LDPQYGLYTSV+PP+IYA+MG+SRE+A+
Sbjct: 67 YSATKFKDDLLSGLTLASLSIPQSIGYANLAKLDPQYGLYTSVIPPVIYALMGSSREIAI 126
Query: 137 GPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGFLVDFLSHA 196
GPVAVVS+LL S+V K++DP P Y LVF TLFAGIFQTAFG+ RLGFLVDFLSHA
Sbjct: 127 GPVAVVSMLLSSLVPKVIDPDAHPNDYRNLVFTVTLFAGIFQTAFGVLRLGFLVDFLSHA 186
Query: 197 AIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNP------WQPRNFI 250
A+VGF+AGAAIVIGLQQ KGLLG+THFTTKTD ++V+K+V+ +LH W P NF+
Sbjct: 187 ALVGFMAGAAIVIGLQQLKGLLGLTHFTTKTDAVAVLKSVYTSLHQQITSSENWSPLNFV 246
Query: 251 LGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVKIVKHVKGG 310
+G SFLIF+L RF+G++ KK FWL +IAPL+S+ILSTLIVFL++ DK GV I+KHV+GG
Sbjct: 247 IGCSFLIFLLAARFIGRRNKKFFWLPAIAPLLSVILSTLIVFLSKGDKHGVNIIKHVQGG 306
Query: 311 LNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGYQLDGNKEMMSIGF 370
LNPSS+H+L N PHVG+AAKIGL+ A++ALTE+IAVGRSFA+IKGY LDGNKEM+++G
Sbjct: 307 LNPSSVHKLQLNGPHVGQAAKIGLISAIIALTEAIAVGRSFANIKGYHLDGNKEMLAMGC 366
Query: 371 SNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQFFTKLLYYTPTAII 430
NI GSLTSCYV+TGSFSRTAVN++AGC+T +SNIVMA+TVL+ L+ FT+LLYYTP AI+
Sbjct: 367 MNIAGSLTSCYVSTGSFSRTAVNFSAGCKTAVSNIVMAVTVLLCLELFTRLLYYTPMAIL 426
Query: 431 ASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGLLAAVMISFLKIIL 490
AS+ILSALPGLIDI EA IWKVDK DFLAC GAFFGVLF S+EIGLL A+ ISF KI+L
Sbjct: 427 ASIILSALPGLIDIGEAYHIWKVDKFDFLACLGAFFGVLFVSIEIGLLIALSISFAKILL 486
Query: 491 ISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFANANFVRERIMKWV 550
+IRP E LG++P T +CDV QYPMAV PG++VIR+ S LCFANA FVRERI+KWV
Sbjct: 487 QAIRPGVEVLGRIPTTEAYCDVAQYPMAVTTPGILVIRISSGSLCFANAGFVRERILKWV 546
Query: 551 TQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLISNGKQLAIANPRWQV 610
EE + + + +Q +I++ ++L ++DTSGI +LEE+ K L+S G +LA+ NPRW+V
Sbjct: 547 EDEEQDNIEEAAKGRVQAIIIDMTDLTNVDTSGILALEELHKKLLSRGVELAMVNPRWEV 606
Query: 611 IHKLKVSNFVSKIG-GRIYLTVEEAI 635
IHKLKV+NFV KIG R++LTV EA+
Sbjct: 607 IHKLKVANFVDKIGKERVFLTVAEAV 632
>ST22_ARATH (P92946) Sulfate transporter 2.2 (AST56) (AtH14)
Length = 658
Score = 774 bits (1999), Expect = 0.0
Identities = 383/626 (61%), Positives = 496/626 (79%), Gaps = 4/626 (0%)
Query: 14 QEVRSQWVLNAPEPPTAWNMVTDSVKKTI-SQFPRKLSYLKDQPCNTLMSFLQGIFPILS 72
+E S+W++N PEPP+ W + ++ + ++ K + K+ N + S L+ FPILS
Sbjct: 19 EEPMSRWLINTPEPPSMWQELIGYIRTNVLAKKKHKRNKTKNSSSNLVYSCLKSAFPILS 78
Query: 73 WGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSR 132
WGR Y F+KD++AGLT+ASLCIPQSIGYA LA LDP+YGLYTSVVPPLIY+ MGTSR
Sbjct: 79 WGRQYKLNLFKKDLMAGLTLASLCIPQSIGYANLAGLDPEYGLYTSVVPPLIYSTMGTSR 138
Query: 133 EVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGFLVDF 192
E+A+GPVAVVSLLL SMV+ L DP DP+AY K+VF T FAG FQ FGLFRLGFLVDF
Sbjct: 139 ELAIGPVAVVSLLLSSMVRDLQDPVTDPIAYRKIVFTVTFFAGAFQAIFGLFRLGFLVDF 198
Query: 193 LSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNPWQPRNFILG 252
LSHAA+VGF+AGAAIVIGLQQ KGL G+THFT KTD++SV+ +V+ +LH+PWQP NF++G
Sbjct: 199 LSHAALVGFMAGAAIVIGLQQLKGLFGLTHFTNKTDVVSVLSSVFHSLHHPWQPLNFVIG 258
Query: 253 SSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVKIVKHVKGGLN 312
SSFLIFIL RF+GK+ KLFW+ ++APL+S++L+TLIV+L+ A+ GVKIVKH+K G N
Sbjct: 259 SSFLIFILLARFIGKRNNKLFWIPAMAPLISVVLATLIVYLSNAESRGVKIVKHIKPGFN 318
Query: 313 PSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGYQLDGNKEMMSIGFSN 372
S+++L F +PH+G+ AKIGL+ A++ALTE+IAVGRSFA+IKGY+LDGNKEMM++GF N
Sbjct: 319 QLSVNQLQFKSPHLGQIAKIGLISAIIALTEAIAVGRSFATIKGYRLDGNKEMMAMGFMN 378
Query: 373 IIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQFFTKLLYYTPTAIIAS 432
I GSL+SCYVATGSFSRTAVN++AGCET++SNIVMAITV+ISL+ T+ LY+TPTAI+AS
Sbjct: 379 IAGSLSSCYVATGSFSRTAVNFSAGCETVVSNIVMAITVMISLEVLTRFLYFTPTAILAS 438
Query: 433 VILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGLLAAVMISFLKIILIS 492
+ILSALPGLID+ A IWK+DKLDFL AFFGVLFASVEIGLL AV ISF +I+L S
Sbjct: 439 IILSALPGLIDVSGALHIWKLDKLDFLVLIAAFFGVLFASVEIGLLLAVGISFARIMLSS 498
Query: 493 IRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFANANFVRERIMKWVTQ 552
IRP EALG+L T +F D+ QYPMA + G++ +R+ S LLCFANANF+R+RI+ V +
Sbjct: 499 IRPSIEALGRLSKTDIFGDINQYPMANKTAGLLTLRISSPLLCFANANFIRDRILNSVQE 558
Query: 553 EESKDDKGN--STNTIQLVILETSNLVDIDTSGIASLEEMQKVLISNGKQLAIANPRWQV 610
E ++++ N +Q+VIL+ S ++ +DTSG+ +LEE+ + L SN +L IA+PRW+V
Sbjct: 559 IEGEENEQEVLKENGLQVVILDMSCVMGVDTSGVFALEELHQELASNDIRLVIASPRWRV 618
Query: 611 IHKLKVSNFVSKI-GGRIYLTVEEAI 635
+HKLK + KI IY+TV EA+
Sbjct: 619 LHKLKRAKLDEKIKTENIYMTVGEAV 644
>ST12_ARATH (Q9MAX3) Sulfate transporter 1.2
Length = 653
Score = 658 bits (1698), Expect = 0.0
Identities = 330/585 (56%), Positives = 442/585 (75%), Gaps = 4/585 (0%)
Query: 53 KDQP-CNTLMSFLQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDP 111
KDQP M LQ +FP+ WGRNYT KFR D+++GLTIASLCIPQ IGYA LA+LDP
Sbjct: 60 KDQPKSKQFMLGLQSVFPVFDWGRNYTFKKFRGDLISGLTIASLCIPQDIGYAKLANLDP 119
Query: 112 QYGLYTSVVPPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTT 171
+YGLY+S VPPL+YA MG+SR++A+GPVAVVSLLL ++++ +DP P Y +L F T
Sbjct: 120 KYGLYSSFVPPLVYACMGSSRDIAIGPVAVVSLLLGTLLRAEIDPNTSPDEYLRLAFTAT 179
Query: 172 LFAGIFQTAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIIS 231
FAGI + A G FRLGFL+DFLSHAA+VGF+ GAAI I LQQ KG LGI FT KTDIIS
Sbjct: 180 FFAGITEAALGFFRLGFLIDFLSHAAVVGFMGGAAITIALQQLKGFLGIKKFTKKTDIIS 239
Query: 232 VMKAVWEALHNPWQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIV 291
V+++V++A H+ W + ++G+SFL F+LT++ +GKK KKLFW+ +IAPL+S+I+ST V
Sbjct: 240 VLESVFKAAHHGWNWQTILIGASFLTFLLTSKIIGKKSKKLFWVPAIAPLISVIVSTFFV 299
Query: 292 FLTRADKSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSF 351
++TRADK GV+IVKH+ G+NPSS H + F ++ + +IG+V +VALTE++A+GR+F
Sbjct: 300 YITRADKQGVQIVKHLDQGINPSSFHLIYFTGDNLAKGIRIGVVAGMVALTEAVAIGRTF 359
Query: 352 ASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITV 411
A++K YQ+DGNKEM+++G N++GS++SCYVATGSFSR+AVN+ AGC+T +SNI+M+I V
Sbjct: 360 AAMKDYQIDGNKEMVALGMMNVVGSMSSCYVATGSFSRSAVNFMAGCQTAVSNIIMSIVV 419
Query: 412 LISLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFA 471
L++L F T L YTP AI+A++I++A+ LIDI A I+KVDKLDF+AC GAFFGV+F
Sbjct: 420 LLTLLFLTPLFKYTPNAILAAIIINAVIPLIDIQAAILIFKVDKLDFIACIGAFFGVIFV 479
Query: 472 SVEIGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKS 531
SVEIGLL AV ISF KI+L RP T LG +P TS++ ++ QYP A +PGV+ IRV S
Sbjct: 480 SVEIGLLIAVSISFAKILLQVTRPRTAVLGNIPRTSVYRNIQQYPEATMVPGVLTIRVDS 539
Query: 532 ALLCFANANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQ 591
A+ F+N+N+VRERI +W+ +EE K K S IQ +I+E S + DIDTSGI +LE++
Sbjct: 540 AIY-FSNSNYVRERIQRWLHEEEEK-VKAASLPRIQFLIIEMSPVTDIDTSGIHALEDLY 597
Query: 592 KVLISNGKQLAIANPRWQVIHKLKVSNFVSKIG-GRIYLTVEEAI 635
K L QL +ANP VI KL +S+F +G IYLTV +A+
Sbjct: 598 KSLQKRDIQLILANPGPLVIGKLHLSHFADMLGQDNIYLTVADAV 642
>ST11_ARATH (Q9SAY1) Sulfate transporter 1.1 (High-affinity sulfate
transporter 1) (Hst1At) (AST101)
Length = 649
Score = 649 bits (1673), Expect = 0.0
Identities = 324/581 (55%), Positives = 436/581 (74%), Gaps = 3/581 (0%)
Query: 56 PCNTLMSFLQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGL 115
P + +Q +FPI+ W R YT KFR D++AGLTIASLCIPQ IGYA LA++DP+YGL
Sbjct: 59 PAKKALLGIQAVFPIIGWAREYTLRKFRGDLIAGLTIASLCIPQDIGYAKLANVDPKYGL 118
Query: 116 YTSVVPPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAG 175
Y+S VPPLIYA MG+SR++A+GPVAVVSLL+ ++ Q ++DP +P Y +LVF T FAG
Sbjct: 119 YSSFVPPLIYAGMGSSRDIAIGPVAVVSLLVGTLCQAVIDPKKNPEDYLRLVFTATFFAG 178
Query: 176 IFQTAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKA 235
IFQ G RLGFL+DFLSHAA+VGF+ GAAI I LQQ KG LGI FT KTDI+SVM +
Sbjct: 179 IFQAGLGFLRLGFLIDFLSHAAVVGFMGGAAITIALQQLKGFLGIKTFTKKTDIVSVMHS 238
Query: 236 VWEALHNPWQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTR 295
V++ + W + ++G+SFL F+L T+F+GK+ +KLFW+ +IAPL+S+I+ST VF+ R
Sbjct: 239 VFKNAEHGWNWQTIVIGASFLTFLLVTKFIGKRNRKLFWVPAIAPLISVIISTFFVFIFR 298
Query: 296 ADKSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIK 355
ADK GV+IVKH+ G+NP S+H++ F+ + E +IG + +VALTE++A+ R+FA++K
Sbjct: 299 ADKQGVQIVKHIDQGINPISVHKIFFSGKYFTEGIRIGGIAGMVALTEAVAIARTFAAMK 358
Query: 356 GYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISL 415
YQ+DGNKEM+++G N++GS+TSCY+ATGSFSR+AVN+ AG ET +SNIVMAI V ++L
Sbjct: 359 DYQIDGNKEMIALGTMNVVGSMTSCYIATGSFSRSAVNFMAGVETAVSNIVMAIVVALTL 418
Query: 416 QFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEI 475
+F T L YTP AI+A++I+SA+ GLIDI A IW++DKLDFLAC GAF GV+F SVEI
Sbjct: 419 EFITPLFKYTPNAILAAIIISAVLGLIDIDAAILIWRIDKLDFLACMGAFLGVIFISVEI 478
Query: 476 GLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLC 535
GLL AV+ISF KI+L RP T LGKLP ++++ + QYP A QIPG+++IRV SA+
Sbjct: 479 GLLIAVVISFAKILLQVTRPRTTVLGKLPNSNVYRNTLQYPDAAQIPGILIIRVDSAIY- 537
Query: 536 FANANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLI 595
F+N+N+VRER +WV +EE ++ K I+ VI+E S + DIDTSGI S+EE+ K L
Sbjct: 538 FSNSNYVRERASRWV-REEQENAKEYGMPAIRFVIIEMSPVTDIDTSGIHSIEELLKSLE 596
Query: 596 SNGKQLAIANPRWQVIHKLKVSNFVSKIGGR-IYLTVEEAI 635
QL +ANP VI KL S FV +IG + I+LTV +A+
Sbjct: 597 KQEIQLILANPGPVVIEKLYASKFVEEIGEKNIFLTVGDAV 637
>ST13_ARATH (Q9FEP7) Sulfate transporter 1.3
Length = 656
Score = 648 bits (1671), Expect = 0.0
Identities = 326/589 (55%), Positives = 443/589 (74%), Gaps = 4/589 (0%)
Query: 49 LSYLKDQP-CNTLMSFLQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLA 107
L + KDQ LM +Q +FP++ WGR Y FR D++AGLTIASLCIPQ IGYA LA
Sbjct: 59 LRHFKDQSKSKKLMLGIQSVFPVIEWGRKYNLKLFRGDLIAGLTIASLCIPQDIGYAKLA 118
Query: 108 HLDPQYGLYTSVVPPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLV 167
LDP+YGLY+S VPPL+YA MG+S+++A+GPVAVVSLLL ++++ +DP +P Y +L
Sbjct: 119 SLDPKYGLYSSFVPPLVYACMGSSKDIAIGPVAVVSLLLGTLLRAEIDPNTNPNEYLRLA 178
Query: 168 FLTTLFAGIFQTAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKT 227
F +T FAG+ Q A G FRLGFL+DFLSHAA+VGF+ GAAI I LQQ KG LGI FT KT
Sbjct: 179 FTSTFFAGVTQAALGFFRLGFLIDFLSHAAVVGFMGGAAITIALQQLKGFLGINKFTKKT 238
Query: 228 DIISVMKAVWEALHNPWQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILS 287
DII+V+ +V + H+ W + ++ +SFLIF+L ++F+GK+ KKLFW+ +IAPLVS+I+S
Sbjct: 239 DIIAVLSSVISSAHHGWNWQTILISASFLIFLLISKFIGKRNKKLFWIPAIAPLVSVIIS 298
Query: 288 TLIVFLTRADKSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAV 347
T V++TRADK GV+IVKH+ GLNPSSL + F+ ++ + +IG+V +VALTE++A+
Sbjct: 299 TFFVYITRADKKGVQIVKHLDKGLNPSSLRLIYFSGDYLLKGFRIGVVSGMVALTEAVAI 358
Query: 348 GRSFASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVM 407
GR+FA++K YQ+DGNKEM+++G N+IGS+TSCYV+TGSFSR+AVN+ AGC+T +SNI+M
Sbjct: 359 GRTFAAMKDYQIDGNKEMVALGAMNVIGSMTSCYVSTGSFSRSAVNFMAGCQTAVSNIIM 418
Query: 408 AITVLISLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFG 467
+I VL++L F T L YTP AI+A++I++A+ L+D+ I+K+DKLDF+AC GAFFG
Sbjct: 419 SIVVLLTLLFLTPLFKYTPNAILAAIIINAVIPLVDVNATILIFKIDKLDFVACMGAFFG 478
Query: 468 VLFASVEIGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVI 527
V+F SVEIGLL AV ISF KI+L RP T LGK+PGTS++ ++ QYP A +IPGV+ I
Sbjct: 479 VIFVSVEIGLLIAVGISFAKILLQVTRPRTAILGKIPGTSVYRNINQYPEATRIPGVLTI 538
Query: 528 RVKSALLCFANANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASL 587
RV SA+ F+N+N+VRERI +W+T EE + IQ +I+E S + DIDTSGI +L
Sbjct: 539 RVDSAIY-FSNSNYVRERIQRWLTDEEEMVEAARLPR-IQFLIIEMSPVTDIDTSGIHAL 596
Query: 588 EEMQKVLISNGKQLAIANPRWQVIHKLKVSNFVSKIG-GRIYLTVEEAI 635
E++ K L QL +ANP VI+KL VS+F IG +I+LTV EA+
Sbjct: 597 EDLYKSLQKRDIQLVLANPGPPVINKLHVSHFADLIGHDKIFLTVAEAV 645
>SUT2_STYHA (P53392) High affinity sulphate transporter 2
Length = 662
Score = 631 bits (1628), Expect = e-180
Identities = 327/617 (52%), Positives = 441/617 (70%), Gaps = 8/617 (1%)
Query: 22 LNAPEPPTAWNMVTDSVKKTISQFPRK-LSYLKDQP-CNTLMSFLQGIFPILSWGRNYTA 79
+ AP T + + S +T FP K KDQ + LQ IFPIL WGR+Y
Sbjct: 41 VGAPPKQTLFQEIKHSFNETF--FPDKPFGNFKDQSGSRKFVLGLQYIFPILEWGRHYDL 98
Query: 80 AKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSREVAVGPV 139
KFR D +AGLTIASLCIPQ + YA LA+LDP YGLY+S V PL+YA MGTSR++A+GPV
Sbjct: 99 KKFRGDFIAGLTIASLCIPQDLAYAKLANLDPWYGLYSSFVAPLVYAFMGTSRDIAIGPV 158
Query: 140 AVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGFLVDFLSHAAIV 199
AVVSLLL +++ + Y +L F T FAG+ Q G+ RLGFL+DFLSHAAIV
Sbjct: 159 AVVSLLLGTLLSNEISNTKSH-DYLRLAFTATFFAGVTQMLLGVCRLGFLIDFLSHAAIV 217
Query: 200 GFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNPWQPRNFILGSSFLIFI 259
GF+AGAAI IGLQQ KGLLGI FT +DI+SVM +VW +H+ W ++G SFLIF+
Sbjct: 218 GFMAGAAITIGLQQLKGLLGIKDFTKNSDIVSVMHSVWSNVHHGWNWETILIGLSFLIFL 277
Query: 260 LTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVKIVKHVKGGLNPSSLHEL 319
L T+++ KK KKLFW+++I+P++ +I+ST V++TRADK GV IVKH+K G+NPSS +E+
Sbjct: 278 LITKYIAKKNKKLFWVSAISPMICVIVSTFFVYITRADKRGVTIVKHIKSGVNPSSANEI 337
Query: 320 DFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGYQLDGNKEMMSIGFSNIIGSLTS 379
F+ ++G ++G+V +VALTE++A+GR+FA++K Y +DGNKEM+++G NI+GSLTS
Sbjct: 338 FFHGKYLGAGVRVGVVAGLVALTEAMAIGRTFAAMKDYSIDGNKEMVAMGTMNIVGSLTS 397
Query: 380 CYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQFFTKLLYYTPTAIIASVILSALP 439
CYV TGSFSR+AVNY AGC+T +SNIVMAI VL++L T L YTP A++AS+I++A+
Sbjct: 398 CYVTTGSFSRSAVNYMAGCKTAVSNIVMAIVVLLTLLVITPLFKYTPNAVLASIIIAAVV 457
Query: 440 GLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGLLAAVMISFLKIILISIRPDTEA 499
L++I +WK+DK DF+AC GAFFGV+F SVEIGLL AV ISF KI+L RP T
Sbjct: 458 NLVNIEAMVLLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRTAV 517
Query: 500 LGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFANANFVRERIMKWVTQEESKDDK 559
LGKLPGTS++ ++ QYP A QIPG+++IRV SA+ F+N+N+++ERI++W+ E ++ +
Sbjct: 518 LGKLPGTSVYRNIQQYPKAEQIPGMLIIRVDSAIY-FSNSNYIKERILRWLIDEGAQRTE 576
Query: 560 GNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLISNGKQLAIANPRWQVIHKLKVSNF 619
+ IQ +I+E S + DIDTSGI + EE+ K L QL +ANP VI KL SN
Sbjct: 577 -SELPEIQHLIVEMSPVTDIDTSGIHAFEELYKTLQKREVQLMLANPGPVVIEKLHASNL 635
Query: 620 VSKIG-GRIYLTVEEAI 635
IG +I+LTV +A+
Sbjct: 636 AELIGEDKIFLTVADAV 652
>SUT1_STYHA (P53391) High affinity sulphate transporter 1
Length = 667
Score = 629 bits (1622), Expect = e-180
Identities = 330/619 (53%), Positives = 443/619 (71%), Gaps = 10/619 (1%)
Query: 22 LNAPEPPTAWNMVTDSVKKTISQFPRK-LSYLKDQP-CNTLMSFLQGIFPILSWGRNYTA 79
+ P T + + S +T FP K KDQ L LQ IFPIL WGR+Y
Sbjct: 44 VGTPPKQTLFQEIKHSFNETF--FPDKPFGKFKDQSGFRKLELGLQYIFPILEWGRHYDL 101
Query: 80 AKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSREVAVGPV 139
KFR D +AGLTIASLCIPQ + YA LA+LDP YGLY+S V PL+YA MGTSR++A+GPV
Sbjct: 102 KKFRGDFIAGLTIASLCIPQDLAYAKLANLDPWYGLYSSFVAPLVYAFMGTSRDIAIGPV 161
Query: 140 AVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGFLVDFLSHAAIV 199
AVVSLLL +++ + Y +L F T FAG+ Q G+ RLGFL+DFLSHAAIV
Sbjct: 162 AVVSLLLGTLLSNEISNTKSH-DYLRLAFTATFFAGVTQMLLGVCRLGFLIDFLSHAAIV 220
Query: 200 GFVAGAAIVIGLQQFKGLLGITH--FTTKTDIISVMKAVWEALHNPWQPRNFILGSSFLI 257
GF+AGAAI IGLQQ KGLLGI++ FT KTDIISVM++VW +H+ W ++G SFLI
Sbjct: 221 GFMAGAAITIGLQQLKGLLGISNNNFTKKTDIISVMRSVWTHVHHGWNWETILIGLSFLI 280
Query: 258 FILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVKIVKHVKGGLNPSSLH 317
F+L T+++ KK KKLFW+++I+P++S+I+ST V++TRADK GV IVKH+K G+NPSS +
Sbjct: 281 FLLITKYIAKKNKKLFWVSAISPMISVIVSTFFVYITRADKRGVSIVKHIKSGVNPSSAN 340
Query: 318 ELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGYQLDGNKEMMSIGFSNIIGSL 377
E+ F+ ++G ++G+V +VALTE+IA+GR+FA++K Y LDGNKEM+++G NI+GSL
Sbjct: 341 EIFFHGKYLGAGVRVGVVAGLVALTEAIAIGRTFAAMKDYALDGNKEMVAMGTMNIVGSL 400
Query: 378 TSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQFFTKLLYYTPTAIIASVILSA 437
+SCYV TGSFSR+AVNY AGC+T +SNIVM+I VL++L T L YTP A++AS+I++A
Sbjct: 401 SSCYVTTGSFSRSAVNYMAGCKTAVSNIVMSIVVLLTLLVITPLFKYTPNAVLASIIIAA 460
Query: 438 LPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGLLAAVMISFLKIILISIRPDT 497
+ L++I +WK+DK DF+AC GAFFGV+F SVEIGLL AV ISF KI+L RP T
Sbjct: 461 VVNLVNIEAMVLLWKIDKFDFVACMGAFFGVIFKSVEIGLLIAVAISFAKILLQVTRPRT 520
Query: 498 EALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFANANFVRERIMKWVTQEESKD 557
LGKLPGTS++ ++ QYP A QIPG+++IRV SA+ F+N+N+++ERI++W+ E ++
Sbjct: 521 AVLGKLPGTSVYRNIQQYPKAAQIPGMLIIRVDSAIY-FSNSNYIKERILRWLIDEGAQR 579
Query: 558 DKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLISNGKQLAIANPRWQVIHKLKVS 617
+ + IQ +I E S + DIDTSGI + EE+ K L QL +ANP VI KL S
Sbjct: 580 TE-SELPEIQHLITEMSPVPDIDTSGIHAFEELYKTLQKREVQLILANPGPVVIEKLHAS 638
Query: 618 NFVSKIG-GRIYLTVEEAI 635
IG +I+LTV +A+
Sbjct: 639 KLTELIGEDKIFLTVADAV 657
>ST31_ARATH (Q9SV13) Sulfate transporter 3.1 (AST12) (AtST1)
Length = 658
Score = 565 bits (1455), Expect = e-160
Identities = 289/625 (46%), Positives = 420/625 (66%), Gaps = 13/625 (2%)
Query: 17 RSQWVLNAPEPPTAWNMVTDSVKKTISQFPRK-LSYLKDQPCNTLMSFLQGI---FPILS 72
R + AP+P + SVK+T+ FP K+Q N F+ G+ PI
Sbjct: 17 RRHHTVEAPQPQPFLKSLQYSVKETL--FPDDPFRQFKNQ--NASRKFVLGLKYFLPIFE 72
Query: 73 WGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSR 132
W Y F+ D++AG+TIASL IPQ I YA LA+L P GLY+S VPPL+YAV+G+SR
Sbjct: 73 WAPRYNLKFFKSDLIAGITIASLAIPQGISYAKLANLPPILGLYSSFVPPLVYAVLGSSR 132
Query: 133 EVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGFLVDF 192
++AVG VAV SLL +M+ K VD DP Y L F T FAG+ + + G+FRLGF+VDF
Sbjct: 133 DLAVGTVAVASLLTGAMLSKEVDAEKDPKLYLHLAFTATFFAGVLEASLGIFRLGFIVDF 192
Query: 193 LSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNPWQPRNFILG 252
LSHA IVGF+ GAA V+ LQQ KG+ G+ HFT TD+ISVM++V+ H W+ + +LG
Sbjct: 193 LSHATIVGFMGGAATVVSLQQLKGIFGLKHFTDSTDVISVMRSVFSQTHE-WRWESGVLG 251
Query: 253 SSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVKIVKHVKGGLN 312
FL F+L+TR+ KK K FW+A++APL S+IL +L+V+ T A++ GV+++ +K GLN
Sbjct: 252 CGFLFFLLSTRYFSIKKPKFFWVAAMAPLTSVILGSLLVYFTHAERHGVQVIGDLKKGLN 311
Query: 313 PSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGYQLDGNKEMMSIGFSN 372
P S +L F +P++ A K GL+ ++AL E +AVGRSFA K Y +DGNKEM++ G N
Sbjct: 312 PLSGSDLIFTSPYMSTAVKTGLITGIIALAEGVAVGRSFAMFKNYNIDGNKEMIAFGMMN 371
Query: 373 IIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQFFTKLLYYTPTAIIAS 432
I+GS TSCY+ TG FSR+AVNY AGC+T +SNIVMAI V+ +L F T L +YTP ++++
Sbjct: 372 IVGSFTSCYLTTGPFSRSAVNYNAGCKTAMSNIVMAIAVMFTLLFLTPLFHYTPLVVLSA 431
Query: 433 VILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGLLAAVMISFLKIILIS 492
+I+SA+ GLID A +WKVDK DFL C A+ GV+F SVEIGL+ AV IS +++L
Sbjct: 432 IIISAMLGLIDYQAAIHLWKVDKFDFLVCMSAYVGVVFGSVEIGLVVAVAISIARLLLFV 491
Query: 493 IRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFANANFVRERIMKWVTQ 552
RP T G +P + ++ + QYP + +PG++++ + A + FANA+++RERI++W+ +
Sbjct: 492 SRPKTAVKGNIPNSMIYRNTEQYPSSRTVPGILILEI-DAPIYFANASYLRERIIRWIDE 550
Query: 553 EESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLISNGKQLAIANPRWQVIH 612
EE + K + +++Q +IL+ S + +IDTSGI+ + E++KV+ +L ++NP+ +V+
Sbjct: 551 EEER-VKQSGESSLQYIILDMSAVGNIDTSGISMMVEIKKVIDRRALKLVLSNPKGEVVK 609
Query: 613 KLKVSNFVSKIGGR--IYLTVEEAI 635
KL S F+ G+ ++LTV EA+
Sbjct: 610 KLTRSKFIGDHLGKEWMFLTVGEAV 634
>ST34_ARATH (Q9LW86) Probable sulfate transporter 3.4
Length = 653
Score = 557 bits (1435), Expect = e-158
Identities = 277/579 (47%), Positives = 399/579 (68%), Gaps = 4/579 (0%)
Query: 58 NTLMSFLQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYT 117
N ++ LQ +FPI +WG Y R D+++GLTIASL IPQ I YA LA+L P GLY+
Sbjct: 65 NRVILGLQSLFPIFTWGSQYDLKLLRSDVISGLTIASLAIPQGISYAKLANLPPIVGLYS 124
Query: 118 SVVPPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIF 177
S VPPLIYAV+G+SR +AVGPV++ SL++ SM+ + V P D + Y KL F +T FAG+F
Sbjct: 125 SFVPPLIYAVLGSSRHLAVGPVSIASLVMGSMLSESVSPTQDSILYLKLAFTSTFFAGVF 184
Query: 178 QTAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVW 237
Q + GL RLGF++DFLS A ++GF AGAA+++ LQQ KGLLGI HFT K I+ VM +V+
Sbjct: 185 QASLGLLRLGFMIDFLSKATLIGFTAGAAVIVSLQQLKGLLGIVHFTGKMQIVPVMSSVF 244
Query: 238 EALHNPWQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRAD 297
+ W ++G FL +LTTR + +K KLFW+++ +PL S+I+STL+V+L R+
Sbjct: 245 NH-RSEWSWETIVMGIGFLSILLTTRHISMRKPKLFWISAASPLASVIISTLLVYLIRSK 303
Query: 298 KSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGY 357
+ + H+ GLNP SL+ L F+ H+ A K G++ +++LTE IAVGR+FAS+K Y
Sbjct: 304 THAISFIGHLPKGLNPPSLNMLYFSGAHLALAIKTGIITGILSLTEGIAVGRTFASLKNY 363
Query: 358 QLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQF 417
Q++GNKEMM+IGF N+ GS TSCYV TGSFSR+AVNY AG +T +SNIVMA VL++L F
Sbjct: 364 QVNGNKEMMAIGFMNMAGSCTSCYVTTGSFSRSAVNYNAGAKTAVSNIVMASAVLVTLLF 423
Query: 418 FTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGL 477
L YYTP I+A++IL+A+ GLID A K+WKVDK DF C +FFGVLF SV +GL
Sbjct: 424 LMPLFYYTPNVILAAIILTAVIGLIDYQAAYKLWKVDKFDFFTCLCSFFGVLFVSVPLGL 483
Query: 478 LAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFA 537
AV +S +KI+L RP+T G +PGT ++ + +Y A +IPG +++ ++S + FA
Sbjct: 484 AIAVAVSVIKILLHVTRPNTSEFGNIPGTQIYQSLGRYREASRIPGFLILAIESPIY-FA 542
Query: 538 NANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLISN 597
N+ ++++RI++W +EE++ + N T T++ +IL+ + + IDTSG+ ++ E+++ L
Sbjct: 543 NSTYLQDRILRWAREEENRIKENNGT-TLKCIILDMTAVSAIDTSGLEAVFELRRRLEKQ 601
Query: 598 GKQLAIANPRWQVIHKLKVSNFVSKIG-GRIYLTVEEAI 635
QL + NP V+ KL S + +G +YLTV EA+
Sbjct: 602 SLQLVLVNPVGTVMEKLHKSKIIEALGLSGLYLTVGEAV 640
>ST32_ARATH (O04289) Sulfate transporter 3.2 (AST77)
Length = 646
Score = 540 bits (1391), Expect = e-153
Identities = 263/573 (45%), Positives = 396/573 (68%), Gaps = 4/573 (0%)
Query: 64 LQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPL 123
L+ +FPIL W R Y+ + D+++G+TIASL IPQ I YA LA+L P GLY+S+VPPL
Sbjct: 55 LRHVFPILEWARGYSLEYLKSDVISGITIASLAIPQGISYAQLANLPPILGLYSSLVPPL 114
Query: 124 IYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGL 183
+YA+MG+SR++AVG VAV SLL +M+ K V+ V+P Y L F T FAG+ QT GL
Sbjct: 115 VYAIMGSSRDLAVGTVAVASLLTAAMLGKEVNAVVNPKLYLHLAFTATFFAGLMQTCLGL 174
Query: 184 FRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNP 243
RLGF+V+ LSHAAIVGF+ GAA V+ LQQ KGLLG+ HFT TDI++V+++++ H
Sbjct: 175 LRLGFVVEILSHAAIVGFMGGAATVVCLQQLKGLLGLHHFTHSTDIVTVLRSIFSQSHM- 233
Query: 244 WQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVKI 303
W+ + +LG FLIF+LTT+++ KK+ KLFW+++++PLVS+I T+ ++ G++
Sbjct: 234 WRWESGVLGCCFLIFLLTTKYISKKRPKLFWISAMSPLVSVIFGTIFLYFLHDQFHGIQF 293
Query: 304 VKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGYQLDGNK 363
+ +K G+NP S+ L F P+V A K+G++ V+AL E IAVGRSFA K Y +DGNK
Sbjct: 294 IGELKKGINPPSITHLVFTPPYVMLALKVGIITGVIALAEGIAVGRSFAMYKNYNIDGNK 353
Query: 364 EMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQFFTKLLY 423
EM++ G NI+GS +SCY+ TG FSR+AVNY AGC+T +SN+VMA+ V ++L F T L +
Sbjct: 354 EMIAFGMMNILGSFSSCYLTTGPFSRSAVNYNAGCKTALSNVVMAVAVAVTLLFLTPLFF 413
Query: 424 YTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGLLAAVMI 483
YTP +++S+I++A+ GL+D A +WK+DK DF C A+ GV+F ++EIGL+ +V I
Sbjct: 414 YTPLVVLSSIIIAAMLGLVDYEAAIHLWKLDKFDFFVCLSAYLGVVFGTIEIGLILSVGI 473
Query: 484 SFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFANANFVR 543
S ++++L RP +G + + ++ ++ YP A+ ++++ + + FAN+ ++R
Sbjct: 474 SVMRLVLFVGRPKIYVMGNIQNSEIYRNIEHYPQAITRSSLLILHIDGPIY-FANSTYLR 532
Query: 544 ERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLISNGKQLAI 603
+RI +W+ +EE K + + ++Q ++L+ S + +IDTSGI+ LEE+ K+L +L I
Sbjct: 533 DRIGRWIDEEEDK-LRTSGDISLQYIVLDMSAVGNIDTSGISMLEELNKILGRRELKLVI 591
Query: 604 ANPRWQVIHKLKVSNFVSKIG-GRIYLTVEEAI 635
ANP +V+ KL S F+ IG RIYLTV EA+
Sbjct: 592 ANPGAEVMKKLSKSTFIESIGKERIYLTVAEAV 624
>ST33_ARATH (Q9SXS2) Probable sulfate transporter 3.3 (AST91)
Length = 631
Score = 539 bits (1388), Expect = e-152
Identities = 269/590 (45%), Positives = 408/590 (68%), Gaps = 6/590 (1%)
Query: 49 LSYLKDQPCNT-LMSFLQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLA 107
L + QP T L+ Q IFPIL W Y+ + + D+++GLTIASL IPQ I YA LA
Sbjct: 32 LRQFRGQPNRTKLIRAAQYIFPILQWCPEYSFSLLKSDVVSGLTIASLAIPQGISYAKLA 91
Query: 108 HLDPQYGLYTSVVPPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLV 167
+L P GLY+S VPPL+YAV+G+SR++AVGPV++ SL+L SM+++ V P DPV + +L
Sbjct: 92 NLPPIVGLYSSFVPPLVYAVLGSSRDLAVGPVSIASLILGSMLRQQVSPVDDPVLFLQLA 151
Query: 168 FLTTLFAGIFQTAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKT 227
F +T FAG+FQ + G+ RLGF++DFLS A ++GF+ GAAI++ LQQ KGLLGITHFT
Sbjct: 152 FSSTFFAGLFQASLGILRLGFIIDFLSKATLIGFMGGAAIIVSLQQLKGLLGITHFTKHM 211
Query: 228 DIISVMKAVWEALHNPWQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILS 287
++ V+ +V++ N W + ++G FL+F+L+TR L KK KLFW+++ APL+S+I+S
Sbjct: 212 SVVPVLSSVFQHT-NEWSWQTIVMGVCFLLFLLSTRHLSMKKPKLFWVSAGAPLLSVIVS 270
Query: 288 TLIVFLTRADKSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAV 347
TL+VF+ RA++ G+ ++ + GLNP S + L F+ H+ AK GLV +V+LTE IAV
Sbjct: 271 TLLVFVFRAERHGISVIGKLPEGLNPPSWNMLQFHGSHLALVAKTGLVTGIVSLTEGIAV 330
Query: 348 GRSFASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVM 407
GR+FA++K Y +DGNKEM++IG N++GS TSCYV TG+FSR+AVN AG +T +SNIVM
Sbjct: 331 GRTFAALKNYHVDGNKEMIAIGLMNVVGSATSCYVTTGAFSRSAVNNNAGAKTAVSNIVM 390
Query: 408 AITVLISLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFG 467
++TV+++L F L YTP ++ ++I++A+ GLID+P AC IWK+DK DFL AFFG
Sbjct: 391 SVTVMVTLLFLMPLFEYTPNVVLGAIIVTAVIGLIDLPAACHIWKIDKFDFLVMLCAFFG 450
Query: 468 VLFASVEIGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVI 527
V+F SV+ GL AV +S KI++ RP +G +PGT ++ D++ Y A +IPG +V+
Sbjct: 451 VIFLSVQNGLAIAVGLSLFKILMQVTRPKMVIMGNIPGTDIYRDLHHYKEAQRIPGFLVL 510
Query: 528 RVKSALLCFANANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASL 587
++S + FAN+N++ ER +W+ +E +++ +++Q +ILE S + +DT+G++
Sbjct: 511 SIESP-VNFANSNYLTERTSRWI-EECEEEEAQEKHSSLQFLILEMSAVSGVDTNGVSFF 568
Query: 588 EEMQKVLISNGKQLAIANPRWQVIHKLKVSNFVSKI--GGRIYLTVEEAI 635
+E++K +L NP +V+ KL+ ++ + ++LTV EA+
Sbjct: 569 KELKKTTAKKDIELVFVNPLSEVVEKLQRADEQKEFMRPEFLFLTVAEAV 618
>NO70_SOYBN (Q02920) Early nodulin 70
Length = 485
Score = 522 bits (1344), Expect = e-147
Identities = 271/470 (57%), Positives = 351/470 (74%), Gaps = 11/470 (2%)
Query: 19 QWVLNAPEPPTAWNMVTDSVKKTISQFPRK--LSYLKDQPCNT-LMSFLQGIFPILSWGR 75
QWVLNAPEPP+ V D+VK+T+ P SYL++QP + + LQ +FPIL+ +
Sbjct: 4 QWVLNAPEPPSMLRQVVDNVKETLLPHPNPNTFSYLRNQPFSKRAFALLQNLFPILASLQ 63
Query: 76 NYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSREVA 135
NY A K + D++AGLT+A IPQ +G ATLA L P+YGLYT +VPPLIYA++ +SRE+
Sbjct: 64 NYNAQKLKCDLMAGLTLAIFAIPQCMGNATLARLSPEYGLYTGIVPPLIYAMLASSREIV 123
Query: 136 VGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGFLVDFLSH 195
+GP +V SLLL SM+Q L P D Y +LVF T FAGIFQ AFGLFR GFLV+ LS
Sbjct: 124 IGPGSVDSLLLSSMIQTLKVPIHDSSTYIQLVFTVTFFAGIFQVAFGLFRFGFLVEHLSQ 183
Query: 196 AAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEALHNP--WQPRNFILGS 253
A IVGF+A AA+ IGLQQ KGL GI +F KTD+ SV+K++W + N W P N I+G
Sbjct: 184 ATIVGFLAAAAVGIGLQQLKGLFGIDNFNNKTDLFSVVKSLWTSFKNQSAWHPYNLIIGF 243
Query: 254 SFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADKSGVK-----IVKHVK 308
SFL FIL TRFLGK+ KKL WL+ +APL+S+I S+ I + ++ VK ++ +K
Sbjct: 244 SFLCFILFTRFLGKRNKKLMWLSHVAPLLSVIGSSAIAYKINFNELQVKDYKVAVLGPIK 303
Query: 309 GG-LNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGYQLDGNKEMMS 367
GG LNPSSLH+L F++ VG +IGL +A+++LT SIAVGRSFAS+KG+ +D N+E++S
Sbjct: 304 GGSLNPSSLHQLTFDSQVVGHLIRIGLTIAIISLTGSIAVGRSFASLKGHSIDPNREVVS 363
Query: 368 IGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQFFTKLLYYTPT 427
+G NI+GSLTSCY+A+GS SRTAVNY AG ET++S IVMA+TVL+SL+F T LLY+TP
Sbjct: 364 LGIMNIVGSLTSCYIASGSLSRTAVNYNAGSETMVSIIVMALTVLMSLKFLTGLLYFTPK 423
Query: 428 AIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGL 477
AI+A++ILSA+PGLID+ +A +IWKVDK+DFLAC GAF GVLFASVEIGL
Sbjct: 424 AILAAIILSAVPGLIDLNKAREIWKVDKMDFLACTGAFLGVLFASVEIGL 473
>ST35_ARATH (Q94LW6) Probable sulfate transporter 3.5
Length = 634
Score = 473 bits (1217), Expect = e-133
Identities = 244/602 (40%), Positives = 383/602 (63%), Gaps = 23/602 (3%)
Query: 36 DSVKKTISQFPRKLSYLKDQPCNTLMSFLQGIFPILSWGRNYTAAKFRKDILAGLTIASL 95
D K ISQ P +L K L+ PI W Y K + D+LAG+TI SL
Sbjct: 40 DDPFKPISQEPNRLLKTK--------KLLEYFVPIFEWLPKYDMQKLKYDVLAGITITSL 91
Query: 96 CIPQSIGYATLAHLDPQYGLYTSVVPPLIYAVMGTSREVAVGPVAVVSLLLFSMV-QKLV 154
+PQ I YA LA + P GLY+S VPP +YAV G+S +AVG VA SLL+ ++++
Sbjct: 92 AVPQGISYAKLASIPPIIGLYSSFVPPFVYAVFGSSNNLAVGTVAACSLLIAETFGEEMI 151
Query: 155 DPAVDPVAYTKLVFLTTLFAGIFQTAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQF 214
+P Y L+F TL G+FQ A G RLG LVDFLSH+ I GF+ G AI+I LQQ
Sbjct: 152 KN--EPELYLHLIFTATLITGLFQFAMGFLRLGILVDFLSHSTITGFMGGTAIIILLQQL 209
Query: 215 KGLLGITHFTTKTDIISVMKAVWEALHNPWQPRNFILGSSFLIFILTTRFLGKKKKKLFW 274
KG+ G+ HFT KTD++SV+ ++ + W+ ++ + G FL+F+ +TR++ ++ KLFW
Sbjct: 210 KGIFGLVHFTHKTDVVSVLHSILDN-RAEWKWQSTLAGVCFLVFLQSTRYIKQRYPKLFW 268
Query: 275 LASIAPLVSIILSTLIVFLTRADKSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGL 334
++++ P+V +++ ++ +L + G+ V +K GLNP S+ L+F++ ++G K G+
Sbjct: 269 VSAMGPMVVVVVGCVVAYLVKGTAHGIATVGPLKKGLNPPSIQLLNFDSKYLGMVFKAGI 328
Query: 335 VVAVVALTESIAVGRSFASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNY 394
V ++AL E IA+GRSFA +K Q DGNKEM++ G N+IGS TSCY+ TG FS+TAVNY
Sbjct: 329 VTGLIALAEGIAIGRSFAVMKNEQTDGNKEMIAFGLMNVIGSFTSCYLTTGPFSKTAVNY 388
Query: 395 AAGCETLISNIVMAITVLISLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVD 454
AG +T +SN+VM + +++ L F L YTP ++++I+SA+ GLI+ E ++KVD
Sbjct: 389 NAGTKTPMSNVVMGVCMMLVLLFLAPLFSYTPLVGLSAIIMSAMLGLINYEEMYHLFKVD 448
Query: 455 KLDFLACAGAFFGVLFASVEIGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQ 514
K DFL C AFFGV F S++ GL+ +V S ++ +L RP T LG++P + +F D+ Q
Sbjct: 449 KFDFLVCMSAFFGVSFLSMDYGLIISVGFSIVRALLYVARPSTCKLGRIPNSVMFRDIEQ 508
Query: 515 YPMAVQIPGVVVIRVKSALLCFANANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETS 574
YP + ++ G +++++ S + FAN+ +VRERI++W+ E I+ ++L+ S
Sbjct: 509 YPASEEMLGYIILQLGSPVF-FANSTYVRERILRWIRDE---------PEAIEFLLLDLS 558
Query: 575 NLVDIDTSGIASLEEMQKVLISNGKQLAIANPRWQVIHKLKVSNFVSKIGGR-IYLTVEE 633
+ ID +G+ +L E+Q++L S ++ I NPR++V+ K+ +S+FV KIG ++L++++
Sbjct: 559 GVSTIDMTGMETLLEIQRILGSKNIKMVIINPRFEVLEKMMLSHFVEKIGKEYMFLSIDD 618
Query: 634 AI 635
A+
Sbjct: 619 AV 620
>ST42_ARATH (Q8GYH8) Probable sulfate transporter 4.2
Length = 677
Score = 347 bits (889), Expect = 7e-95
Identities = 204/583 (34%), Positives = 330/583 (55%), Gaps = 20/583 (3%)
Query: 59 TLMSFLQGIFPILSWGRNYTAAK-FRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYT 117
T ++ IFP W R Y + F+ D++AG+T+ + +PQ++ YA LA L P YGLY+
Sbjct: 58 TFFDWIDAIFPCFLWIRTYRWHQYFKLDLMAGITVGIMLVPQAMSYARLAGLQPIYGLYS 117
Query: 118 SVVPPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIF 177
S VP +YAV G+SR++AVGPVA+VSLL+ + + +VDP+ + YT+L L L GIF
Sbjct: 118 SFVPVFVYAVFGSSRQLAVGPVALVSLLVSNALSGIVDPSEE--LYTELAILLALMVGIF 175
Query: 178 QTAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVW 237
++ G RLG+L+ F+SH+ I GF +A+VIGL Q K LG + + + + +
Sbjct: 176 ESIMGFLRLGWLIRFISHSVISGFTTASAVVIGLSQLKYFLGYSVSRSSKIMPVIDSIIA 235
Query: 238 EALHNPWQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRAD 297
A W P F+LG + L+ +L + +GK KK+L ++ + PL + L T+I +
Sbjct: 236 GADQFKWPP--FLLGCTILVILLVMKHVGKAKKELRFIRAAGPLTGLALGTIIAKVFHPP 293
Query: 298 KSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGL----VVAVVALTESIAVGRSFAS 353
+ +V + GL F+ P + AK+ L ++ VA+ ES+ + ++ A+
Sbjct: 294 S--ITLVGDIPQGLPK-------FSFPKSFDHAKLLLPTSALITGVAILESVGIAKALAA 344
Query: 354 IKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLI 413
Y+LD N E+ +G +NI GSL S Y TGSFSR+AVN + +T +S +V I +
Sbjct: 345 KNRYELDSNSELFGLGVANIFGSLFSAYPTTGSFSRSAVNSESEAKTGLSGLVTGIIIGC 404
Query: 414 SLQFFTKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASV 473
SL F T + + P +A++++SA+ GL+D A +W+VDK DF LF +
Sbjct: 405 SLLFLTPMFKFIPQCALAAIVISAVSGLVDYEGAIFLWRVDKRDFTLWTITSTTTLFFGI 464
Query: 474 EIGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSAL 533
EIG+L V S +I S P LG+LPGT+++ ++ QYP A G+V++R+ A
Sbjct: 465 EIGVLIGVGFSLAFVIHESANPHIAVLGRLPGTTVYRNMKQYPEAYTYNGIVIVRI-DAP 523
Query: 534 LCFANANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKV 593
+ FAN +++++R+ ++ + KG I VILE S + ID+S + +L+++ +
Sbjct: 524 IYFANISYIKDRLREYEVAIDKHTSKGPDMERIYFVILEMSPVTYIDSSAVEALKDLYEE 583
Query: 594 LISNGKQLAIANPRWQVIHKLKVSNFVSKIGGR-IYLTVEEAI 635
+ G QLAI+NP +V+ L + V IG ++ V +A+
Sbjct: 584 YKTRGIQLAISNPNKEVLLTLARAGIVELIGKEWFFVRVHDAV 626
>ST41_ARATH (Q9FY46) Sulfate transporter 4.1, chloroplast precursor
(AST82)
Length = 685
Score = 342 bits (877), Expect = 2e-93
Identities = 199/578 (34%), Positives = 327/578 (56%), Gaps = 12/578 (2%)
Query: 60 LMSFLQGIFPILSWGRNYTAAK-FRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTS 118
L+ ++ +FP W R Y ++ F+ D++AG+T+ + +PQ++ YA LA L P YGLY+S
Sbjct: 72 LVDWIDTLFPCFRWIRTYRWSEYFKLDLMAGITVGIMLVPQAMSYAKLAGLPPIYGLYSS 131
Query: 119 VVPPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQ 178
VP +YA+ G+SR++A+GPVA+VSLL+ + + + D + + +L L L GI +
Sbjct: 132 FVPVFVYAIFGSSRQLAIGPVALVSLLVSNALGGIAD--TNEELHIELAILLALLVGILE 189
Query: 179 TAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWE 238
GL RLG+L+ F+SH+ I GF + +AIVIGL Q K LG + + + V +
Sbjct: 190 CIMGLLRLGWLIRFISHSVISGFTSASAIVIGLSQIKYFLGYSIARSSKIVPIVESIIAG 249
Query: 239 ALHNPWQPRNFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVFLTRADK 298
A W P F++GS L+ + + +GK KK+L +L + APL I+L T I +
Sbjct: 250 ADKFQWPP--FVMGSLILVILQVMKHVGKAKKELQFLRAAAPLTGIVLGTTIAKVFHPPS 307
Query: 299 SGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIKGYQ 358
+ +V + GL P+ F+ H ++ VA+ ES+ + ++ A+ Y+
Sbjct: 308 --ISLVGEIPQGL-PTFSFPRSFD--HAKTLLPTSALITGVAILESVGIAKALAAKNRYE 362
Query: 359 LDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISLQFF 418
LD N E+ +G +NI+GSL S Y ATGSFSR+AVN + +T +S ++ I + SL F
Sbjct: 363 LDSNSELFGLGVANILGSLFSAYPATGSFSRSAVNNESEAKTGLSGLITGIIIGCSLLFL 422
Query: 419 TKLLYYTPTAIIASVILSALPGLIDIPEACKIWKVDKLDFLACAGAFFGVLFASVEIGLL 478
T + Y P +A++++SA+ GL+D EA +W+VDK DF LF +EIG+L
Sbjct: 423 TPMFKYIPQCALAAIVISAVSGLVDYDEAIFLWRVDKRDFSLWTITSTITLFFGIEIGVL 482
Query: 479 AAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALLCFAN 538
V S +I S P LG+LPGT+++ ++ QYP A G+V++R+ S + FAN
Sbjct: 483 VGVGFSLAFVIHESANPHIAVLGRLPGTTVYRNIKQYPEAYTYNGIVIVRIDSPIY-FAN 541
Query: 539 ANFVRERIMKWVTQEESKDDKGNSTNTIQLVILETSNLVDIDTSGIASLEEMQKVLISNG 598
+++++R+ ++ + ++G + I VILE S + ID+S + +L+E+ + +
Sbjct: 542 ISYIKDRLREYEVAVDKYTNRGLEVDRINFVILEMSPVTHIDSSAVEALKELYQEYKTRD 601
Query: 599 KQLAIANPRWQVIHKLKVSNFVSKIGGR-IYLTVEEAI 635
QLAI+NP V + S V +G ++ V +A+
Sbjct: 602 IQLAISNPNKDVHLTIARSGMVELVGKEWFFVRVHDAV 639
>S266_HUMAN (Q9BXS9) Solute carrier family 26 member 6 (Pendrin-like
protein 1) (Pendrin L1)
Length = 759
Score = 214 bits (544), Expect = 7e-55
Identities = 134/492 (27%), Positives = 259/492 (52%), Gaps = 20/492 (4%)
Query: 69 PILSWGRNYTAAKFRK-DILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPPLIYAV 127
P+L W Y + D+L+GL++A + +PQ + YA LA L P +GLY+S P IY +
Sbjct: 76 PVLVWLPRYPVRDWLLGDLLSGLSVAIMQLPQGLAYALLAGLPPVFGLYSSFYPVFIYFL 135
Query: 128 MGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDP---------VAYTKLVFLTTLFAGIFQ 178
GTSR ++VG AV+S+++ S+ + L A++ A ++ ++ G+FQ
Sbjct: 136 FGTSRHISVGTFAVMSVMVGSVTESLAPQALNDSMINETARDAARVQVASTLSVLVGLFQ 195
Query: 179 TAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWE 238
GL GF+V +LS + G+ AA+ + + Q K + G+ H ++ + +S++ V E
Sbjct: 196 VGLGLIHFGFVVTYLSEPLVRGYTTAAAVQVFVSQLKYVFGL-HLSSHSGPLSLIYTVLE 254
Query: 239 ALHNPWQPR--NFILGSSFLIFILTTRFLGKKKKKLFWLASIAPLVSIILSTLIVF-LTR 295
Q + + + + ++ + L K ++ + L+++I +T I + +
Sbjct: 255 VCWKLPQSKVGTVVTAAVAGVVLVVVKLLNDKLQQQLPMPIPGELLTLIGATGISYGMGL 314
Query: 296 ADKSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGRSFASIK 355
+ V +V ++ GL P + VG A +AVV +I++G+ FA
Sbjct: 315 KHRFEVDVVGNIPAGLVPPVAPNTQLFSKLVGSA----FTIAVVGFAIAISLGKIFALRH 370
Query: 356 GYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAITVLISL 415
GY++D N+E++++G SN+IG + C+ + S SR+ V + G + ++ + ++ +L+ +
Sbjct: 371 GYRVDSNQELVALGLSNLIGGIFQCFPVSCSMSRSLVQESTGGNSQVAGAISSLFILLII 430
Query: 416 QFFTKLLYYTPTAIIASVILSALPGLI-DIPEACKIWKVDKLDFLACAGAFFGVLFASVE 474
+L + P A++A++I+ L G++ + + +WK ++ D L F + +++
Sbjct: 431 VKLGELFHDLPKAVLAAIIIVNLKGMLRQLSDMRSLWKANRADLLIWLVTFTATILLNLD 490
Query: 475 IGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQYPMAVQIPGVVVIRVKSALL 534
+GL+ AV+ S L +++ + P LG++P T ++ DV +Y A ++ GV V R SA +
Sbjct: 491 LGLVVAVIFSLLLVVVRTQMPHYSVLGQVPDTDIYRDVAEYSEAKEVRGVKVFR-SSATV 549
Query: 535 CFANANFVRERI 546
FANA F + +
Sbjct: 550 YFANAEFYSDAL 561
>SULI_SCHPO (Q9URY8) Probable sulfate permease C869.05c
Length = 840
Score = 203 bits (517), Expect = 1e-51
Identities = 154/595 (25%), Positives = 279/595 (46%), Gaps = 73/595 (12%)
Query: 61 MSFLQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVV 120
+ +L+ +FPI+ W NY D++AG+T+ + +PQ + YA +A L +YGLY+S V
Sbjct: 100 LHYLKSLFPIIEWLPNYNPYWLINDLIAGITVGCVVVPQGMSYAKVATLPSEYGLYSSFV 159
Query: 121 PPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYT--KLVFLTTLFAGIFQ 178
IY TS++V++GPVAV+SL+ ++ ++ YT ++ L AG
Sbjct: 160 GVAIYCFFATSKDVSIGPVAVMSLITAKVIANVM---AKDETYTAPQIATCLALLAGAIT 216
Query: 179 TAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWE 238
GL RLGF+++F+ A+ GF G+A+ I Q L+G + T V+ +
Sbjct: 217 CGIGLLRLGFIIEFIPVPAVAGFTTGSALNILSGQVPALMGYKNKVTAKATYMVIIQSLK 276
Query: 239 ALHNPWQPRNFILGSSFLIFILT--TRFLGKK----KKKLFWLASIAPLVSIILSTLIVF 292
L + F L S F++F ++LGK+ ++ F ++ V +I+ T I +
Sbjct: 277 HLPDTTVDAAFGLVSLFILFFTKYMCQYLGKRYPRWQQAFFLTNTLRSAVVVIVGTAISY 336
Query: 293 LTRADKSGVKIVKHVKGGLNPSSLHELDFNNPHVG----------EAAKIGLVVAVVALT 342
I KH + S + + HVG + A V +V L
Sbjct: 337 A---------ICKHHRSDPPISIIKTVPRGFQHVGVPLITKKLCRDLASELPVSVIVLLL 387
Query: 343 ESIAVGRSFASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLI 402
E I++ +SF + Y++ ++E++++G +N+IG + Y ATGSFSR+A+ AG +T I
Sbjct: 388 EHISIAKSFGRVNDYRIVPDQELIAMGVTNLIGIFFNAYPATGSFSRSAIKAKAGVKTPI 447
Query: 403 SNIVMAITVLISLQFFTKLLYYTPTAIIASVILSALPGLI-DIPEACKIWKVDKLDFLAC 461
+ I A V++SL T YY P AI+++VI+ A+ LI + + W++ L+
Sbjct: 448 AGIFTAAVVILSLYCLTDAFYYIPNAILSAVIIHAVTDLILPMKQTILFWRLQPLEACIF 507
Query: 462 AGAFFGVLFASVEIGLLAAVMISFLKIILISIRPDTEALGKL------------------ 503
+ +F+S+E G+ +V ++ ++L +P LGK+
Sbjct: 508 FISVIVSVFSSIENGIYVSVCLAAALLLLRIAKPHGSFLGKIQAANKYGSDNIANVRDIY 567
Query: 504 ------------------PGTSLF--CDVYQYPMAVQIPGVVVIRVKSALLCFANANFVR 543
PG +F + + YP A ++ ++ R+K + +V+
Sbjct: 568 VPLEMKEENPNLEIQSPPPGVFIFRLQESFTYPNASRVSTMISRRIKDLTRRGIDNIYVK 627
Query: 544 ERIMKWVTQEESKDDKGNSTN----TIQLVILETSNLVDIDTSGIASLEEMQKVL 594
+ W + K + + +Q +I + S + ++DT+ + SL +++K L
Sbjct: 628 DIDRPWNVPRQRKKKENSEIEDLRPLLQAIIFDFSAVNNLDTTAVQSLIDIRKEL 682
>PEND_RAT (Q9R154) Pendrin (Sodium-independent chloride/iodide
transporter)
Length = 780
Score = 194 bits (494), Expect = 5e-49
Identities = 136/498 (27%), Positives = 240/498 (47%), Gaps = 36/498 (7%)
Query: 64 LQGIFPILSWGRNYTAAKFR-KDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVVPP 122
L+ + PIL W Y ++ DI++G++ + Q + YA LA + QYGLY++ P
Sbjct: 65 LKALLPILDWLPKYRVKEWLLSDIISGVSTGLVGTLQGMAYALLAAVPVQYGLYSAFFPI 124
Query: 123 LIYAVMGTSREVAVGPVAVVSLLLFSMVQKL--------------------VDPAVDPVA 162
L Y V GTSR ++VGP VVSL++ S+V + +D A
Sbjct: 125 LTYFVFGTSRHISVGPFPVVSLMVGSVVLSMAPDDHFLVPSGNGSTLNTTTLDTGTRDAA 184
Query: 163 YTKLVFLTTLFAGIFQTAFGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITH 222
L TL GI Q FG ++GF+V +L+ + GF AA + + Q K +L ++
Sbjct: 185 RVLLASTLTLLVGIIQLVFGGLQIGFIVRYLADPLVGGFTTAAAFQVLVSQLKIVLNVST 244
Query: 223 FTTKTDIISVMKAVWEALHNPWQPR--NFILGSSFLIFILTTRFLGKK-KKKLFWLASIA 279
++S++ + E N +FI G +I + + L + K K+ I
Sbjct: 245 -KNYNGVLSIIYTLIEIFQNIGDTNIADFIAGLLTIIVCMAVKELNDRFKHKIPVPIPIE 303
Query: 280 PLVSIILSTLIVFLTRADKSGVKIVKHVKGGLNPSSLHELDFNNPHVG---EAAKIGLVV 336
+V+II + + IVK + G P L P VG + +
Sbjct: 304 VIVTIIATAISYGANLEANYNAGIVKSIPSGFLPPVL-------PSVGLFSDMLAASFSI 356
Query: 337 AVVALTESIAVGRSFASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAA 396
AVVA +++VG+ +A+ Y +DGN+E ++ G SN+ SC+VAT + SRTAV +
Sbjct: 357 AVVAYAIAVSVGKVYATKHDYIIDGNQEFIAFGISNVFSGFFSCFVATTALSRTAVQEST 416
Query: 397 GCETLISNIVMAITVLISLQFFTKLLYYTPTAIIASVILSALPGL-IDIPEACKIWKVDK 455
G +T ++ ++ A+ V++++ KLL +++A+V+++ L G+ + + + ++WK +K
Sbjct: 417 GGKTQVAGLISAVIVMVAIVALGKLLEPLQKSVLAAVVIANLKGMFMQVCDVPRLWKQNK 476
Query: 456 LDFLACAGAFFGVLFASVEIGLLAAVMISFLKIILISIRPDTEALGKLPGTSLFCDVYQY 515
D + + +++GLLA ++ L ++L P LG +P T ++ + Y
Sbjct: 477 TDAVIWVFTCIMSIILGLDLGLLAGLLFGLLTVVLRVQFPSWNGLGSVPSTDIYKSITHY 536
Query: 516 PMAVQIPGVVVIRVKSAL 533
+ GV ++R S +
Sbjct: 537 KNLEEPEGVKILRFSSPI 554
>CY14_NEUCR (P23622) Sulfate permease II
Length = 819
Score = 194 bits (494), Expect = 5e-49
Identities = 127/436 (29%), Positives = 223/436 (51%), Gaps = 15/436 (3%)
Query: 61 MSFLQGIFPILSWGRNYTAAKFRKDILAGLTIASLCIPQSIGYATLAHLDPQYGLYTSVV 120
+++L+ +FP ++W +Y D +AG+T+ + +PQ + YA LA+L P+YGLYTS V
Sbjct: 56 LNYLRELFPFVNWIFHYNLTWLLGDFIAGVTVGFVVVPQGMAYAKLANLAPEYGLYTSFV 115
Query: 121 PPLIYAVMGTSREVAVGPVAVVSLLLFSMVQKLVDPAVDPVAYTKLVFLTTLFAGIFQTA 180
++Y TS+++ +G VAV+S ++ +++ + D A + +G
Sbjct: 116 GFVLYWAFATSKDITIGAVAVMSTIVGNIIANVQKDHPDFDA-GDIARTLAFISGAMLLF 174
Query: 181 FGLFRLGFLVDFLSHAAIVGFVAGAAIVIGLQQFKGLLGITHFTTKTDIISVMKAVWEAL 240
GL R GF+V+F+ AI F+ G+AI I Q L+GI + ++ + V+ + L
Sbjct: 175 LGLIRFGFIVEFIPIVAISAFMTGSAISIAAGQVSTLMGIPNINSREETYKVIINTLKGL 234
Query: 241 HNPWQPRNFILGSSFLIFILT--TRFLGKK----KKKLFWLASIAPLVSIILSTLIVFLT 294
N L + F ++ + +GK+ ++ F+++++ + IIL L+ +L
Sbjct: 235 PNTHLDAAMGLTALFGLYFIRWFCTQMGKRYPRQQRAWFFVSTLRMVFIIILYILVSWLV 294
Query: 295 RAD-----KSGVKIVKHVKGGLNPSSLHELDFNNPHVGEAAKIGLVVAVVALTESIAVGR 349
K+ KI+ HV G LD N + + +V L E IA+ +
Sbjct: 295 NRHVKDPKKAHFKILGHVPSGFQHKGAPRLD--NEILSAISGDIPTTILVLLIEHIAISK 352
Query: 350 SFASIKGYQLDGNKEMMSIGFSNIIGSLTSCYVATGSFSRTAVNYAAGCETLISNIVMAI 409
SF + Y ++ ++E+++IGF+N++G Y ATGSFSRTA+ AG T ++ I A+
Sbjct: 353 SFGRVNNYIINPSQELVAIGFTNLLGPFLGGYPATGSFSRTAIKAKAGVRTPLAGIFTAV 412
Query: 410 TVLISLQFFTKLLYYTPTAIIASVILSALPGLIDIP-EACKIWKVDKLDFLACAGAFFGV 468
VL++L T + +Y P + +A++I+ A+ LI P E K W L+ + F
Sbjct: 413 LVLLALYALTSVFFYIPNSALAAMIIHAVGDLITPPREVYKFWLTSPLEVVIFFAGVFVS 472
Query: 469 LFASVEIGLLAAVMIS 484
+F S+E G+ V S
Sbjct: 473 IFTSIENGIYVTVAAS 488
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.139 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,037,060
Number of Sequences: 164201
Number of extensions: 2724167
Number of successful extensions: 9267
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 9078
Number of HSP's gapped (non-prelim): 70
length of query: 641
length of database: 59,974,054
effective HSP length: 117
effective length of query: 524
effective length of database: 40,762,537
effective search space: 21359569388
effective search space used: 21359569388
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0306.8


