

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
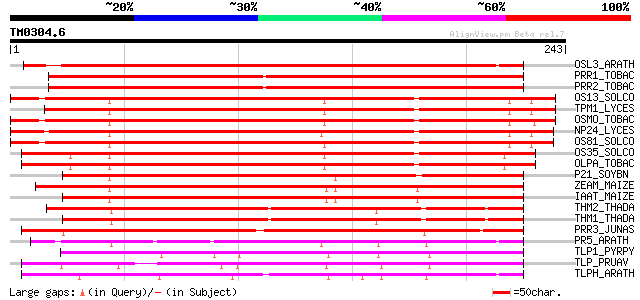
Query= TM0304.6
(243 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
OSL3_ARATH (P50700) Osmotin-like protein OSM34 precursor 361 e-100
PRR1_TOBAC (P13046) Pathogenesis-related protein R major form pr... 348 9e-96
PRR2_TOBAC (P07052) Pathogenesis-related protein R minor form pr... 344 1e-94
OS13_SOLCO (P50701) Osmotin-like protein OSML13 precursor (PA13) 343 3e-94
TPM1_LYCES (Q01591) Osmotin-like protein TPM-1 precursor (PR P23... 339 3e-93
OSMO_TOBAC (P14170) Osmotin precursor 338 9e-93
NP24_LYCES (P12670) NP24 protein precursor (Pathogenesis-related... 333 2e-91
OS81_SOLCO (P50702) Osmotin-like protein OSML81 precursor (PA81) 330 2e-90
OS35_SOLCO (P50703) Osmotin-like protein OSML15 precursor (PA15) 323 2e-88
OLPA_TOBAC (P25871) Osmotin-like protein precursor (Pathogenesis... 320 2e-87
P21_SOYBN (P25096) P21 protein 307 2e-83
ZEAM_MAIZE (P33679) Zeamatin precursor 278 6e-75
IAAT_MAIZE (P13867) Alpha-amylase/trypsin inhibitor (Antifungal ... 271 1e-72
THM2_THADA (P02884) Thaumatin II precursor 265 6e-71
THM1_THADA (P02883) Thaumatin I 251 9e-67
PRR3_JUNAS (P81295) Pathogenesis-related protein precursor (Poll... 248 9e-66
PR5_ARATH (P28493) Pathogenesis-related protein 5 precursor (PR-5) 200 2e-51
TLP1_PYRPY (O80327) Thaumatin-like protein 1 precursor 184 2e-46
TLP_PRUAV (P50694) Thaumatin-like protein precursor 177 2e-44
TLPH_ARATH (P50699) Thaumatin-like protein precursor 170 3e-42
>OSL3_ARATH (P50700) Osmotin-like protein OSM34 precursor
Length = 244
Score = 361 bits (927), Expect = e-100
Identities = 162/219 (73%), Positives = 183/219 (82%), Gaps = 7/219 (3%)
Query: 7 SSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMAR 66
S+LLL+ TA+ AA FEI+N C YTVWAAASPGGGRRLD GQ+W L V AGT MAR
Sbjct: 12 SALLLISTAT------AATFEILNQCSYTVWAAASPGGGRRLDAGQSWRLDVAAGTKMAR 65
Query: 67 IWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGF 126
IWGRT CNFD +GRGRCQTGDC+GGL C GWG PPNTLAE+ALNQ+ N DFYDISLVDGF
Sbjct: 66 IWGRTNCNFDSSGRGRCQTGDCSGGLQCTGWGQPPNTLAEYALNQFNNLDFYDISLVDGF 125
Query: 127 NIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPT 186
NIPM+F P + CH+I CTADINGQCPN LRAPGGCNNPCTV++TN++CCTNGQG+C T
Sbjct: 126 NIPMEFSPTSSNCHRILCTADINGQCPNVLRAPGGCNNPCTVFQTNQYCCTNGQGSCSDT 185
Query: 187 NFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
+SRFFK RC D+YSYPQDDPTSTFTC +NY+VVFCP
Sbjct: 186 EYSRFFKQRCPDAYSYPQDDPTSTFTC-TNTNYRVVFCP 223
>PRR1_TOBAC (P13046) Pathogenesis-related protein R major form
precursor (Thaumatin-like protein E22)
Length = 226
Score = 348 bits (892), Expect = 9e-96
Identities = 153/208 (73%), Positives = 173/208 (82%), Gaps = 1/208 (0%)
Query: 18 VTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDG 77
V +T AA F+IVN C YTVWAAASPGGGRRLD GQ+W++ VN GT ARIWGRT CNFDG
Sbjct: 20 VAVTHAATFDIVNKCTYTVWAAASPGGGRRLDSGQSWSINVNPGTVQARIWGRTNCNFDG 79
Query: 78 AGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFPLNG 137
+GRG C+TGDC G L CQG+G PNTLAEFALNQ NQDF DISLVDGFNIPM+F P NG
Sbjct: 80 SGRGNCETGDCNGMLECQGYGKAPNTLAEFALNQ-PNQDFVDISLVDGFNIPMEFSPTNG 138
Query: 138 GCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPTNFSRFFKDRCH 197
GC + CTA IN QCP +L+ GGCNNPCTV KTNE+CCTNG G+CGPT+ SRFFK+RC
Sbjct: 139 GCRNLRCTAPINEQCPAQLKTQGGCNNPCTVIKTNEYCCTNGPGSCGPTDLSRFFKERCP 198
Query: 198 DSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
D+YSYPQDDPTS FTCP+G+NY+VVFCP
Sbjct: 199 DAYSYPQDDPTSLFTCPSGTNYRVVFCP 226
>PRR2_TOBAC (P07052) Pathogenesis-related protein R minor form
precursor (PR-R) (PROB12) (Thaumatin-like protein E2)
Length = 226
Score = 344 bits (882), Expect = 1e-94
Identities = 152/208 (73%), Positives = 171/208 (82%), Gaps = 1/208 (0%)
Query: 18 VTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDG 77
V +T AA F+IVN C YTVWAAASPGGGR+L+ GQ+W++ VN GT ARIWGRT CNFDG
Sbjct: 20 VAVTHAATFDIVNQCTYTVWAAASPGGGRQLNSGQSWSINVNPGTVQARIWGRTNCNFDG 79
Query: 78 AGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFPLNG 137
+GRG C+TGDC G L CQG+G PPNTLAEFALNQ NQDF DISLVDGFNIPM+F P NG
Sbjct: 80 SGRGNCETGDCNGMLECQGYGKPPNTLAEFALNQ-PNQDFVDISLVDGFNIPMEFSPTNG 138
Query: 138 GCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPTNFSRFFKDRCH 197
GC + CTA IN QCP +L+ GGCNNPCTV KTNEFCCTNG G+CGPT+ SRFFK RC
Sbjct: 139 GCRNLRCTAPINEQCPAQLKTQGGCNNPCTVIKTNEFCCTNGPGSCGPTDLSRFFKARCP 198
Query: 198 DSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
D+YSYPQDDP S FTCP G+NY+VVFCP
Sbjct: 199 DAYSYPQDDPPSLFTCPPGTNYRVVFCP 226
>OS13_SOLCO (P50701) Osmotin-like protein OSML13 precursor (PA13)
Length = 246
Score = 343 bits (879), Expect = 3e-94
Identities = 164/246 (66%), Positives = 186/246 (74%), Gaps = 11/246 (4%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVN 59
M YL S + +L + VT T AA E+ NNCPYTVWAA++P GGGRRLDRGQTW +
Sbjct: 1 MAYLRSSFVFFLL--AFVTYTYAATIEVRNNCPYTVWAASTPIGGGRRLDRGQTWVINAP 58
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
GT MARIWGRT CNFDGAGRG CQTGDC G L C GWG PPNTLAE+AL+Q+ N DF+D
Sbjct: 59 RGTKMARIWGRTNCNFDGAGRGSCQTGDCGGVLQCTGWGKPPNTLAEYALDQFSNLDFWD 118
Query: 120 ISLVDGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCC 176
ISLVDGFNIPM F P N G CH I CTA+ING+CP LR PGGCNNPCT + ++CC
Sbjct: 119 ISLVDGFNIPMTFAPTNPSGGKCHAIHCTANINGECPGSLRVPGGCNNPCTTFGGQQYCC 178
Query: 177 TNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTL 233
T QG CGPT+ SRFFK RC D+YSYPQDDPTSTFTCP+GS NY+VVFCP G SP+ L
Sbjct: 179 T--QGPCGPTDLSRFFKQRCPDAYSYPQDDPTSTFTCPSGSTNYRVVFCPNGVTSPNFPL 236
Query: 234 MMPGND 239
MP +D
Sbjct: 237 EMPASD 242
>TPM1_LYCES (Q01591) Osmotin-like protein TPM-1 precursor (PR P23)
(Fragment)
Length = 238
Score = 339 bits (870), Expect = 3e-93
Identities = 159/231 (68%), Positives = 179/231 (76%), Gaps = 9/231 (3%)
Query: 16 SLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVNAGTAMARIWGRTGCN 74
+ VT T AA FE+ NNCPYTVWAA++P GGGRRLDRGQTW + GT MARIWGRT CN
Sbjct: 6 AFVTYTYAATFEVRNNCPYTVWAASTPIGGGRRLDRGQTWVINAPRGTKMARIWGRTNCN 65
Query: 75 FDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFP 134
FDG GRG CQTGDC G L C GWG PPNTLAE+AL+Q+ N DF+DISLVDGFNIPM F P
Sbjct: 66 FDGDGRGSCQTGDCGGVLQCTGWGKPPNTLAEYALDQFSNLDFWDISLVDGFNIPMTFAP 125
Query: 135 LN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPTNFSRF 191
N G CH I CTA+ING+CP LR PGGCNNPCT + ++CCT QG CGPT+ SRF
Sbjct: 126 TNPSGGKCHAIHCTANINGECPGSLRVPGGCNNPCTTFGGQQYCCT--QGPCGPTDLSRF 183
Query: 192 FKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTLMMPGND 239
FK RC D+YSYPQDDPTSTFTCP+GS NY+VVFCP G SP+ L MP +D
Sbjct: 184 FKQRCPDAYSYPQDDPTSTFTCPSGSTNYRVVFCPNGVTSPNFPLEMPSSD 234
>OSMO_TOBAC (P14170) Osmotin precursor
Length = 246
Score = 338 bits (866), Expect = 9e-93
Identities = 161/246 (65%), Positives = 185/246 (74%), Gaps = 11/246 (4%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVN 59
MG L S + +L +LVT T AA E+ NNCPYTVWAA++P GGGRRLDRGQTW +
Sbjct: 1 MGNLRSSFVFFLL--ALVTYTYAATIEVRNNCPYTVWAASTPIGGGRRLDRGQTWVINAP 58
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
GT MAR+WGRT CNF+ AGRG CQTGDC G L C GWG PPNTLAE+AL+Q+ DF+D
Sbjct: 59 RGTKMARVWGRTNCNFNAAGRGTCQTGDCGGVLQCTGWGKPPNTLAEYALDQFSGLDFWD 118
Query: 120 ISLVDGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCC 176
ISLVDGFNIPM F P N G CH I CTA+ING+CP ELR PGGCNNPCT + ++CC
Sbjct: 119 ISLVDGFNIPMTFAPTNPSGGKCHAIHCTANINGECPRELRVPGGCNNPCTTFGGQQYCC 178
Query: 177 TNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLGS--PHVTL 233
T QG CGPT FS+FFK RC D+YSYPQDDPTSTFTCP GS NY+V+FCP G P+ L
Sbjct: 179 T--QGPCGPTFFSKFFKQRCPDAYSYPQDDPTSTFTCPGGSTNYRVIFCPNGQAHPNFPL 236
Query: 234 MMPGND 239
MPG+D
Sbjct: 237 EMPGSD 242
>NP24_LYCES (P12670) NP24 protein precursor (Pathogenesis-related
protein PR P23) (Salt-induced protein)
Length = 247
Score = 333 bits (855), Expect = 2e-91
Identities = 160/245 (65%), Positives = 182/245 (73%), Gaps = 11/245 (4%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVN 59
MGYLT S +L L VT T AA E+ NNCPYTVWAA++P GGGRRL+RGQTW +
Sbjct: 1 MGYLTSSFVLFFLLC--VTYTYAATIEVRNNCPYTVWAASTPIGGGRRLNRGQTWVINAP 58
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
GT MARIWGRTGCNF+ AGRG CQTGDC G L C GWG PPNTLAE+AL+Q+ N DF+D
Sbjct: 59 RGTKMARIWGRTGCNFNAAGRGTCQTGDCGGVLQCTGWGKPPNTLAEYALDQFSNLDFWD 118
Query: 120 ISLVDGFNIPMDFFPL---NGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCC 176
ISLVDGFNIPM F P G CH I CTA+ING+CP L+ PGGCNNPCT + ++CC
Sbjct: 119 ISLVDGFNIPMTFAPTKPSGGKCHAIHCTANINGECPRALKVPGGCNNPCTTFGGQQYCC 178
Query: 177 TNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTL 233
T QG CGPT S+FFK RC D+YSYPQDDPTSTFTCP GS NY+VVFCP G P+ L
Sbjct: 179 T--QGPCGPTELSKFFKKRCPDAYSYPQDDPTSTFTCPGGSTNYRVVFCPNGVADPNFPL 236
Query: 234 MMPGN 238
MP +
Sbjct: 237 EMPAS 241
>OS81_SOLCO (P50702) Osmotin-like protein OSML81 precursor (PA81)
Length = 247
Score = 330 bits (846), Expect = 2e-90
Identities = 157/245 (64%), Positives = 183/245 (74%), Gaps = 11/245 (4%)
Query: 1 MGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVN 59
MGYL S + +L + VT T AA E+ NNCPYTVWAA++P GGGRRL++GQTW +
Sbjct: 1 MGYLRSSFIFSLL--AFVTYTYAATIEVRNNCPYTVWAASTPIGGGRRLNKGQTWVINAP 58
Query: 60 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 119
GT MARIWGRTGCNF+ AGRG CQTGDC G L C GWG PPNTLAE+AL+Q+ N DF+D
Sbjct: 59 RGTKMARIWGRTGCNFNAAGRGSCQTGDCGGVLQCTGWGKPPNTLAEYALDQFSNLDFWD 118
Query: 120 ISLVDGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCC 176
ISLVDGFNIPM F P G CH I CTA+ING+CP L+ PGGCNNPCT + ++CC
Sbjct: 119 ISLVDGFNIPMTFAPTKPSAGKCHAIHCTANINGECPRALKVPGGCNNPCTTFGGQQYCC 178
Query: 177 TNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGS-NYKVVFCPLG--SPHVTL 233
T QG CGPT S+FFK RC D+YSYPQDDPTSTFTCP+GS NY+VVFCP G P+ L
Sbjct: 179 T--QGPCGPTELSKFFKKRCPDAYSYPQDDPTSTFTCPSGSTNYRVVFCPNGVADPNFPL 236
Query: 234 MMPGN 238
MP +
Sbjct: 237 EMPAS 241
>OS35_SOLCO (P50703) Osmotin-like protein OSML15 precursor (PA15)
Length = 250
Score = 323 bits (829), Expect = 2e-88
Identities = 150/231 (64%), Positives = 176/231 (75%), Gaps = 8/231 (3%)
Query: 6 LSSLLLMLTASLVTLTRAAN-FEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVNAGTA 63
L++ L+ + VT T A+ FE+ NNCPYTVWAAA+P GGGRRL+RGQ+W GT
Sbjct: 4 LTTCLVFFLLAFVTYTNASGVFEVHNNCPYTVWAAATPIGGGRRLERGQSWWFWAPPGTK 63
Query: 64 MARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLV 123
MARIWGRT CNFDGAGRG CQTGDC G L C+GWG PPNTLAE+ALNQ+ N DF+DIS++
Sbjct: 64 MARIWGRTNCNFDGAGRGWCQTGDCGGVLECKGWGKPPNTLAEYALNQFSNLDFWDISVI 123
Query: 124 DGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQ 180
DGFNIPM F P N G CH I C A+ING+CP LR PGGCNNPCT + ++CCT Q
Sbjct: 124 DGFNIPMSFGPTNPGPGKCHPIQCVANINGECPGSLRVPGGCNNPCTTFGGQQYCCT--Q 181
Query: 181 GTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPA-GSNYKVVFCPLGSPH 230
G CGPT+ SRFFK RC D+YSYPQDDPTSTFTC + ++YKV+FCP GS H
Sbjct: 182 GPCGPTDLSRFFKQRCPDAYSYPQDDPTSTFTCQSWTTDYKVMFCPYGSTH 232
>OLPA_TOBAC (P25871) Osmotin-like protein precursor
(Pathogenesis-related protein PR-5d)
Length = 251
Score = 320 bits (820), Expect = 2e-87
Identities = 149/231 (64%), Positives = 175/231 (75%), Gaps = 8/231 (3%)
Query: 6 LSSLLLMLTASLVTLTRAAN-FEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVNAGTA 63
L++ L+ + VT T A+ FE+ NNCPYTVWAAA+P GGGRRL+RGQ+W GT
Sbjct: 4 LTTFLVFFLLAFVTYTYASGVFEVHNNCPYTVWAAATPVGGGRRLERGQSWWFWAPPGTK 63
Query: 64 MARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLV 123
MARIWGRT CNFDGAGRG CQTGDC G L C+GWG PPNTLAE+ALNQ+ N DF+DIS++
Sbjct: 64 MARIWGRTNCNFDGAGRGWCQTGDCGGVLECKGWGKPPNTLAEYALNQFSNLDFWDISVI 123
Query: 124 DGFNIPMDFFPLN---GGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQ 180
DGFNIPM F P G CH I CTA+ING+CP LR PGGCNNPCT + ++CCT Q
Sbjct: 124 DGFNIPMSFGPTKPGPGKCHGIQCTANINGECPGSLRVPGGCNNPCTTFGGQQYCCT--Q 181
Query: 181 GTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPA-GSNYKVVFCPLGSPH 230
G CGPT SR+FK RC D+YSYPQDDPTSTFTC + ++YKV+FCP GS H
Sbjct: 182 GPCGPTELSRWFKQRCPDAYSYPQDDPTSTFTCTSWTTDYKVMFCPYGSAH 232
>P21_SOYBN (P25096) P21 protein
Length = 202
Score = 307 bits (786), Expect = 2e-83
Identities = 135/204 (66%), Positives = 167/204 (81%), Gaps = 4/204 (1%)
Query: 24 ANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDGAGRGR 82
A FEI N C YTVWAA+ P GGG +L+ GQ+W++ V AGT AR+W RTGCNFDG+GRG
Sbjct: 1 ARFEITNRCTYTVWAASVPVGGGVQLNPGQSWSVDVPAGTKGARVWARTGCNFDGSGRGG 60
Query: 83 CQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFPLNGGCHK- 141
CQTGDC G L+C+ +G PPNTLAE+ LN + N DF+DISLVDGFN+PMDF P + GC +
Sbjct: 61 CQTGDCGGVLDCKAYGAPPNTLAEYGLNGFNNLDFFDISLVDGFNVPMDFSPTSNGCTRG 120
Query: 142 ISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTCGPTNFSRFFKDRCHDSYS 201
ISCTADINGQCP+EL+ GGCNNPCTV+KT+++CC + G+CGPT++SRFFK RC D+YS
Sbjct: 121 ISCTADINGQCPSELKTQGGCNNPCTVFKTDQYCCNS--GSCGPTDYSRFFKQRCPDAYS 178
Query: 202 YPQDDPTSTFTCPAGSNYKVVFCP 225
YP+DDP STFTC G++Y+VVFCP
Sbjct: 179 YPKDDPPSTFTCNGGTDYRVVFCP 202
>ZEAM_MAIZE (P33679) Zeamatin precursor
Length = 227
Score = 278 bits (712), Expect = 6e-75
Identities = 127/218 (58%), Positives = 157/218 (71%), Gaps = 4/218 (1%)
Query: 12 MLTASLVTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVNAGTAMARIWGR 70
+ A L AA F +VN CP+TVWAA+ P GGGR+L+RG++W + AGT ARIW R
Sbjct: 10 IFVALLAVAGEAAVFTVVNQCPFTVWAASVPVGGGRQLNRGESWRITAPAGTTAARIWAR 69
Query: 71 TGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPM 130
TGC FD +GRG C+TGDC G L C G+G PNTLAE+AL Q+ N DF+DISL+DGFN+PM
Sbjct: 70 TGCKFDASGRGSCRTGDCGGVLQCTGYGRAPNTLAEYALKQFNNLDFFDISLIDGFNVPM 129
Query: 131 DFFPLNG-GCHK-ISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCT-NGQGTCGPTN 187
F P G GC + C D+N +CP ELR G CNN C V+K +E+CC + C PTN
Sbjct: 130 SFLPDGGSGCSRGPRCAVDVNARCPAELRQDGVCNNACPVFKKDEYCCVGSAANDCHPTN 189
Query: 188 FSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
+SR+FK +C D+YSYP+DD TSTFTCPAG+NYKVVFCP
Sbjct: 190 YSRYFKGQCPDAYSYPKDDATSTFTCPAGTNYKVVFCP 227
>IAAT_MAIZE (P13867) Alpha-amylase/trypsin inhibitor (Antifungal
protein)
Length = 206
Score = 271 bits (692), Expect = 1e-72
Identities = 121/206 (58%), Positives = 152/206 (73%), Gaps = 4/206 (1%)
Query: 24 ANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDGAGRGR 82
A F +VN CP+TVWAA+ P GGGR+L+RG++W + AGT ARIW RTGC FD +GRG
Sbjct: 1 AVFTVVNQCPFTVWAASVPVGGGRQLNRGESWRITAPAGTTAARIWARTGCQFDASGRGS 60
Query: 83 CQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFPLNG-GCHK 141
C+TGDC G + C G+G PNTLAE+AL Q+ N DF+DIS++DGFN+P F P G GC +
Sbjct: 61 CRTGDCGGVVQCTGYGRAPNTLAEYALKQFNNLDFFDISILDGFNVPYSFLPDGGSGCSR 120
Query: 142 -ISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCT-NGQGTCGPTNFSRFFKDRCHDS 199
C D+N +CP ELR G CNN C V+K +E+CC + C PTN+SR+FK +C D+
Sbjct: 121 GPRCAVDVNARCPAELRQDGVCNNACPVFKKDEYCCVGSAANNCHPTNYSRYFKGQCPDA 180
Query: 200 YSYPQDDPTSTFTCPAGSNYKVVFCP 225
YSYP+DD TSTFTCPAG+NYKVVFCP
Sbjct: 181 YSYPKDDATSTFTCPAGTNYKVVFCP 206
>THM2_THADA (P02884) Thaumatin II precursor
Length = 235
Score = 265 bits (678), Expect = 6e-71
Identities = 126/216 (58%), Positives = 156/216 (71%), Gaps = 11/216 (5%)
Query: 17 LVTLTRAANFEIVNNCPYTVWAAASPG------GGRRLDRGQTWNLGVNAGTAMARIWGR 70
L+TL+RAA FEIVN C YTVWAAAS G GGR+L+ G++W + V GT +IW R
Sbjct: 16 LLTLSRAATFEIVNRCSYTVWAAASKGDAALDAGGRQLNSGESWTINVEPGTKGGKIWAR 75
Query: 71 TGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPM 130
T C FD +GRG C+TGDC G L C+ +G PP TLAEF+LNQYG +D+ DIS + GFN+PM
Sbjct: 76 TDCYFDDSGRGICRTGDCGGLLQCKRFGRPPTTLAEFSLNQYG-KDYIDISNIKGFNVPM 134
Query: 131 DFFPLNGGCHKISCTADINGQCPNELRAP-GGCNNPCTVYKTNEFCCTNGQGTCGPTNFS 189
DF P GC + C ADI GQCP +L+AP GGCN+ CTV++T+E+CCT G+ CGPT +S
Sbjct: 135 DFSPTTRGCRGVRCAADIVGQCPAKLKAPGGGCNDACTVFQTSEYCCTTGK--CGPTEYS 192
Query: 190 RFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
RFFK C D++SY D PT T TCP SNY+V FCP
Sbjct: 193 RFFKRLCPDAFSYVLDKPT-TVTCPGSSNYRVTFCP 227
>THM1_THADA (P02883) Thaumatin I
Length = 207
Score = 251 bits (642), Expect = 9e-67
Identities = 119/209 (56%), Positives = 148/209 (69%), Gaps = 11/209 (5%)
Query: 24 ANFEIVNNCPYTVWAAASPG------GGRRLDRGQTWNLGVNAGTAMARIWGRTGCNFDG 77
A FEIVN C YTVWAAAS G GGR+L+ G++W + V GT +IW RT C FD
Sbjct: 1 ATFEIVNRCSYTVWAAASKGDAALDAGGRQLNSGESWTINVEPGTNGGKIWARTDCYFDD 60
Query: 78 AGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFFPLNG 137
+G G C+TGDC G L C+ +G PP TLAEF+LNQYG +D+ DIS + GFN+PM+F P
Sbjct: 61 SGSGICKTGDCGGLLRCKRFGRPPTTLAEFSLNQYG-KDYIDISNIKGFNVPMNFSPTTR 119
Query: 138 GCHKISCTADINGQCPNELRAP-GGCNNPCTVYKTNEFCCTNGQGTCGPTNFSRFFKDRC 196
GC + C ADI GQCP +L+AP GGCN+ CTV++T+E+CCT G+ CGPT +SRFFK C
Sbjct: 120 GCRGVRCAADIVGQCPAKLKAPGGGCNDACTVFQTSEYCCTTGK--CGPTEYSRFFKRLC 177
Query: 197 HDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
D++SY D PT T TCP SNY+V FCP
Sbjct: 178 PDAFSYVLDKPT-TVTCPGSSNYRVTFCP 205
>PRR3_JUNAS (P81295) Pathogenesis-related protein precursor (Pollen
allergen Jun a 3)
Length = 225
Score = 248 bits (633), Expect = 9e-66
Identities = 115/226 (50%), Positives = 150/226 (65%), Gaps = 10/226 (4%)
Query: 6 LSSLLLMLTASLVTLTR-----AANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNA 60
+S L +L A+L F+I N C YTVWAA PGGG+RLD+GQTW + + A
Sbjct: 4 VSELAFLLAATLAISLHMQEAGVVKFDIKNQCGYTVWAAGLPGGGKRLDQGQTWTVNLAA 63
Query: 61 GTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDI 120
GTA AR WGRTGC FD +G+G CQTGDC G L+C G P TLAE+ +QD+YD+
Sbjct: 64 GTASARFWGRTGCTFDASGKGSCQTGDCGGQLSCTVSGAVPATLAEYT---QSDQDYYDV 120
Query: 121 SLVDGFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQ 180
SLVDGFNIP+ P N C +C ADIN CP+EL+ GGCN+ C V+KT+++CC N
Sbjct: 121 SLVDGFNIPLAINPTNAQCTAPACKADINAVCPSELKVDGGCNSACNVFKTDQYCCRNAY 180
Query: 181 -GTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
C TN+S+ FK++C +YSY +DD T+TF C +G++Y +VFCP
Sbjct: 181 VDNCPATNYSKIFKNQCPQAYSYAKDD-TATFACASGTDYSIVFCP 225
>PR5_ARATH (P28493) Pathogenesis-related protein 5 precursor (PR-5)
Length = 239
Score = 200 bits (509), Expect = 2e-51
Identities = 104/233 (44%), Positives = 136/233 (57%), Gaps = 22/233 (9%)
Query: 10 LLMLTASLVTLTRAANFEIVNNCPYTVWAAASPG-------GGRRLDRGQTWNLGVNAGT 62
L+ +T+ + + A +F + NNCP TVWA G GG L G + L AG
Sbjct: 12 LVFITSGIAVM--ATDFTLRNNCPTTVWAGTLAGQGPKLGDGGFELTPGASRQLTAPAGW 69
Query: 63 AMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISL 122
+ R W RTGCNFD +G GRC TGDC GGL C G GVPP TLAEF L G +DFYD+SL
Sbjct: 70 S-GRFWARTGCNFDASGNGRCVTGDC-GGLRCNGGGVPPVTLAEFTLVGDGGKDFYDVSL 127
Query: 123 VDGFNIPMDFFPL--NGGCHKISCTADINGQCPNELRAPG-----GCNNPCTVYKTNEFC 175
VDG+N+ + P +G C C +D+N CP+ L+ C + C + T+++C
Sbjct: 128 VDGYNVKLGIRPSGGSGDCKYAGCVSDLNAACPDMLKVMDQNNVVACKSACERFNTDQYC 187
Query: 176 CTNGQG---TCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
C TC PT++SR FK+ C D+YSY DD TSTFTC G+NY++ FCP
Sbjct: 188 CRGANDKPETCPPTDYSRIFKNACPDAYSYAYDDETSTFTC-TGANYEITFCP 239
>TLP1_PYRPY (O80327) Thaumatin-like protein 1 precursor
Length = 244
Score = 184 bits (467), Expect = 2e-46
Identities = 94/223 (42%), Positives = 123/223 (55%), Gaps = 20/223 (8%)
Query: 23 AANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTAMA-------RIWGRTGCNF 75
+A F N CP TVW GGG + L A T++ R WGR+ C+
Sbjct: 22 SAKFTFTNKCPNTVWPGTLTGGGGPQLLSTGFELASGASTSLTVQAPWSGRFWGRSHCSI 81
Query: 76 DGAGRGRCQTGDC-TGGLNCQGWGV-PPNTLAEFALNQYGNQDFYDISLVDGFNIPMDFF 133
D +G+ +C TGDC +G ++C G G PP +L E L G QDFYD+SLVDGFN+P+
Sbjct: 82 DSSGKFKCSTGDCGSGQISCNGAGASPPASLVELTLATNGGQDFYDVSLVDGFNLPIKLA 141
Query: 134 PLNGG--CHKISCTADINGQCPNELRAPG------GCNNPCTVYKTNEFCCTNGQGT--- 182
P G C+ SC A+IN CP EL G GC + C ++CCT GT
Sbjct: 142 PRGGSGDCNSTSCAANINTVCPAELSDKGSDGSVIGCKSACLALNQPQYCCTGAYGTPDT 201
Query: 183 CGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 225
C PT+FS+ FK++C +YSY DD +STFTC G NY++ FCP
Sbjct: 202 CPPTDFSKVFKNQCPQAYSYAYDDKSSTFTCFGGPNYEITFCP 244
>TLP_PRUAV (P50694) Thaumatin-like protein precursor
Length = 245
Score = 177 bits (450), Expect = 2e-44
Identities = 99/253 (39%), Positives = 128/253 (50%), Gaps = 42/253 (16%)
Query: 6 LSSLLLMLTASLVTLT----RAANFEIVNNCPYTVWAAASPGGGR--------------- 46
+ +L+++L+ SL L+ AA NNCPY VW +
Sbjct: 2 MKTLVVVLSLSLTILSFGGAHAATISFKNNCPYMVWPGTLTSDQKPQLSTTGFELASQAS 61
Query: 47 -RLDRGQTWNLGVNAGTAMARIWGRTGCNFDGAGRGRCQTGDCTGG-LNCQGWG-VPPNT 103
+LD WN R W RTGC+ D +G+ C T DC G + C G G +PP T
Sbjct: 62 FQLDTPVPWN---------GRFWARTGCSTDASGKFVCATADCASGQVMCNGNGAIPPAT 112
Query: 104 LAEFALNQYGNQDFYDISLVDGFNIPMDFFPLNG--GCHKISCTADINGQCPNELRAPGG 161
LAEF + G QDFYD+SLVDGFN+PM P G C SC A++N CP+EL+ G
Sbjct: 113 LAEFNIPAGGGQDFYDVSLVDGFNLPMSVTPQGGTGDCKTASCPANVNAVCPSELQKKGS 172
Query: 162 ------CNNPCTVYKTNEFCCTNGQGT---CGPTNFSRFFKDRCHDSYSYPQDDPTSTFT 212
C + C + T ++CCT Q T C PTN+S F + C D+YSY DD TFT
Sbjct: 173 DGSVVACLSACVKFGTPQYCCTPPQNTPETCPPTNYSEIFHNACPDAYSYAYDDKRGTFT 232
Query: 213 CPAGSNYKVVFCP 225
C G NY + FCP
Sbjct: 233 CNGGPNYAITFCP 245
>TLPH_ARATH (P50699) Thaumatin-like protein precursor
Length = 243
Score = 170 bits (430), Expect = 3e-42
Identities = 95/242 (39%), Positives = 131/242 (53%), Gaps = 25/242 (10%)
Query: 6 LSSLLLMLTASLVTLTRAANFEIV--NNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTA 63
++S+ L L A L+ L+ A+ ++ N C + VW P G+ L G + L N +
Sbjct: 1 MASINLFLFAFLLLLSHASASTVIFYNKCKHPVWPGIQPSAGQNLLAGGGFKLPANKAHS 60
Query: 64 M-------ARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQG-WGVPPNTLAEFALNQYGNQ 115
+ R WGR GC FD +GRG C TGDC G L+C G G PP TLAE L
Sbjct: 61 LQLPPLWSGRFWGRHGCTFDRSGRGHCATGDCGGSLSCNGAGGEPPATLAEITLGP--EL 118
Query: 116 DFYDISLVDGFNIPMDFFPLNGG--CHKISCTADINGQCP--NELRAPGG-----CNNPC 166
DFYD+SLVDG+N+ M P+ G C C +D+N CP ++R+ G C + C
Sbjct: 119 DFYDVSLVDGYNLAMSIMPVKGSGQCSYAGCVSDLNQMCPVGLQVRSRNGKRVVACKSAC 178
Query: 167 TVYKTNEFCCTNGQG---TCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVF 223
+ + + ++CCT G +C PT +S+ FK C +YSY DDPTS TC + +NY V F
Sbjct: 179 SAFNSPQYCCTGLFGNPQSCKPTAYSKIFKVACPKAYSYAYDDPTSIATC-SKANYIVTF 237
Query: 224 CP 225
CP
Sbjct: 238 CP 239
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.140 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,893,923
Number of Sequences: 164201
Number of extensions: 1657045
Number of successful extensions: 3020
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 2814
Number of HSP's gapped (non-prelim): 104
length of query: 243
length of database: 59,974,054
effective HSP length: 107
effective length of query: 136
effective length of database: 42,404,547
effective search space: 5767018392
effective search space used: 5767018392
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0304.6


