

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
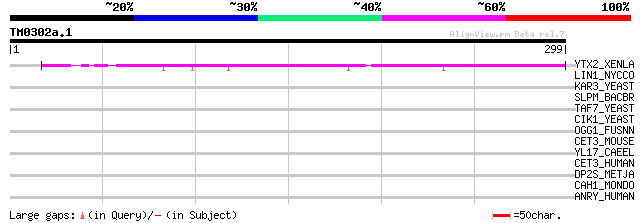
Query= TM0302a.1
(299 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 57 4e-08
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 40 0.007
KAR3_YEAST (P17119) Kinesin-like protein KAR3 (Nuclear fusion pr... 33 1.1
SLPM_BACBR (P06546) Middle cell wall protein precursor (MWP) 32 1.9
TAF7_YEAST (Q05021) Transcription initiation factor TFIID subuni... 32 2.5
CIK1_YEAST (Q01649) Spindle pole body associated protein 32 2.5
OGG1_FUSNN (Q8R689) Probable N-glycosylase/DNA lyase [Includes: ... 31 4.2
CET3_MOUSE (O35648) Centrin 3 31 4.2
YL17_CAEEL (Q11102) Hypothetical protein C02F12.7 in chromosome X 30 5.5
CET3_HUMAN (O15182) Centrin 3 30 7.2
DP2S_METJA (Q58113) DNA polymerase II small subunit (EC 2.7.7.7)... 30 9.4
CAH1_MONDO (Q8HY33) Carbonic anhydrase I (EC 4.2.1.1) (Carbonate... 30 9.4
ANRY_HUMAN (Q6UB99) Ankyrin repeat domain protein 11 (Ankyrin re... 30 9.4
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 57.4 bits (137), Expect = 4e-08
Identities = 70/314 (22%), Positives = 135/314 (42%), Gaps = 45/314 (14%)
Query: 18 NCWLQNHAFGECV-STWSSFDIQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDKICKDPV 76
N L++ F + V TW +GW A F+++ L NQW L +C++
Sbjct: 253 NSLLEDEGFAKSVRDTW-----RGWRA--FQDEFATL----NQWWDVGKVHLKLLCQEYT 301
Query: 77 TRINA---LDIKEGDGNLSSQEQK---QRDDFL-VEYWKVAKFNDSL-------LFQKSR 122
++ +I+ +G + EQ+ D L EY + + ++ F +SR
Sbjct: 302 KSVSGQRNAEIEALNGEVLDLEQRLSGSEDQALQCEYLERKEALRNMEQRQARGAFVRSR 361
Query: 123 DRWVKDGDSNTKYFHNSVNWRRRTNAIRGL-AVDGEWVEDPKVVKSKMIEFFEAQFTSNP 181
+ + D D +++F+ + I L A DG +EDP+ ++ + F++ F+ +P
Sbjct: 362 MQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNLFSPDP 421
Query: 182 ----EVGVFLEGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPDVLKV----- 232
+G P +S+ L L+E+ A+ ++S G D L +
Sbjct: 422 ISPDACEELWDGLPV--VSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFFQF 479
Query: 233 -------DILRVLKDFHRNGVWPKGGNSSFITLIPNISNLMSLNDYISISVIGCMYKIVS 285
D RVL + + G P + ++L+P +L + ++ +S++ YKIV+
Sbjct: 480 FWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIVA 539
Query: 286 KVLTTRLSKVMGHI 299
K ++ RL V+ +
Sbjct: 540 KAISLRLKSVLAEV 553
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 40.0 bits (92), Expect = 0.007
Identities = 53/274 (19%), Positives = 110/274 (39%), Gaps = 39/274 (14%)
Query: 38 IQGWGAYTFKEKLKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQK 97
+Q + T +E++ L L Q EKE + + +T+I A + + + Q K
Sbjct: 301 LQAFLKKTEREEVNNLMGHLKQLEKEEHSNPKPSRRKEITKIRAELNEIENKRIIQQINK 360
Query: 98 QRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVDGE 157
+ F + K+ D L +R + VK S+ + ++ +
Sbjct: 361 SKSWFFEKINKI----DKPLANLTRKKRVKSLISSIRNGNDEIT---------------- 400
Query: 158 WVEDPKVVKSKMIEFFEAQFTSN----PEVGVFLEGTPFRSISDEDNLALTQTFNLEEIR 213
DP ++ + E+++ ++ E+ +LE +S ++ L + + EI
Sbjct: 401 --TDPSEIQKILNEYYKKLYSHKYENLKEIDQYLEACHLPRLSQKEVEMLNRPISSSEIA 458
Query: 214 TAVWECEGDRSLGPDVLKVD------------ILRVLKDFHRNGVWPKGGNSSFITLIPN 261
+ + +S GPD + +L + ++ + G+ P + ITLIP
Sbjct: 459 STIQNLPKKKSPGPDGFTSEFYQTFKEELVPILLNLFQNIEKEGILPNTFYEANITLIPK 518
Query: 262 I-SNLMSLNDYISISVIGCMYKIVSKVLTTRLSK 294
+ +Y IS++ KI++K+LT R+ +
Sbjct: 519 PGKDPTRKENYRPISLMNIDAKILNKILTNRIQQ 552
>KAR3_YEAST (P17119) Kinesin-like protein KAR3 (Nuclear fusion
protein)
Length = 729
Score = 32.7 bits (73), Expect = 1.1
Identities = 23/93 (24%), Positives = 48/93 (50%), Gaps = 19/93 (20%)
Query: 46 FKEKLKRLK-LKLNQWEKENVGDLDKI--CKDPVTRINALDIKEGDGNLSSQEQKQRDDF 102
+K K+++LK +K+ Q+E E LDKI ++ +T +N ++E ++ + +++++
Sbjct: 201 YKTKIEKLKFMKIKQFENERASLLDKIEEVRNKIT-MNPSTLQEMLNDVEQKHMLEKEEW 259
Query: 103 LVEYWKVAKFNDSLLFQKSRDRWVKDGDSNTKY 135
L EY + +W KD + N K+
Sbjct: 260 LTEY---------------QSQWKKDIELNNKH 277
>SLPM_BACBR (P06546) Middle cell wall protein precursor (MWP)
Length = 1053
Score = 32.0 bits (71), Expect = 1.9
Identities = 47/194 (24%), Positives = 80/194 (41%), Gaps = 27/194 (13%)
Query: 73 KDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEYWKVAKFNDSLLFQKSRDRWVKDGDSN 132
K+ + A +I + DG S++ K +DD L + +K D+LL + + D DG
Sbjct: 604 KEITVSLEAKNIYDFDGKNFSRDNKNQDD-LEDILVPSKDKDTLLLEVTLDA---DGKPG 659
Query: 133 TKYFHNSVNWRRRTNAIRGLAVDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVF------ 186
F V + + G A D ED +V + A F ++
Sbjct: 660 KVEFLKPVKVEQES----GKAWDDLADEDDDMVGDYEVTDKTAVFNMTGKLEESSKRKEL 715
Query: 187 --LEGTPFRSISDEDNLALTQTFNLEEIRTAVWECEGDRSLGPDVLKVDILRVLKDFHRN 244
+ F+ ++DE++L++ T N ++ A++ EGD G D ++ DF R
Sbjct: 716 KNAKTAKFKDVADENDLSVIYTVNDKDEVEAIFVVEGDGLTG-DAHYGQVI----DFGRK 770
Query: 245 G------VWPKGGN 252
G VW K G+
Sbjct: 771 GGKDTIRVWEKDGD 784
>TAF7_YEAST (Q05021) Transcription initiation factor TFIID subunit 7
(TBP-associated factor 7) (TBP-associated factor 67 kDa)
(TAFII-67)
Length = 590
Score = 31.6 bits (70), Expect = 2.5
Identities = 29/103 (28%), Positives = 47/103 (45%), Gaps = 9/103 (8%)
Query: 47 KEKLKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEY 106
KEK LKLKLN K+N + KI K P R+ + I G+ S+ DD L+E
Sbjct: 59 KEKKNSLKLKLNL--KKNEEPVKKIHKAPKLRLKPIRI-PGEA-YDSEASDIEDDPLIES 114
Query: 107 WKVAKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNWRRRTNAI 149
+ + + + +VK+ + Y S+ W+ +A+
Sbjct: 115 GVILRILPDIQLE-----FVKNSLESGDYSGISIKWKNERHAV 152
>CIK1_YEAST (Q01649) Spindle pole body associated protein
Length = 594
Score = 31.6 bits (70), Expect = 2.5
Identities = 19/65 (29%), Positives = 38/65 (58%), Gaps = 5/65 (7%)
Query: 47 KEKLKRLKLKLNQWEKENVGDLDKICKDPVTRIN-ALDIKEGDGNLS----SQEQKQRDD 101
+E+ K+++ + N W+K+ V +L+K+ ++ TR N A I+E G + E++ D+
Sbjct: 333 QEEKKQVEGEANNWKKKYVNELEKVQQELYTRQNLATSIEEIKGYTRCFAYANERQMPDE 392
Query: 102 FLVEY 106
F + Y
Sbjct: 393 FHINY 397
>OGG1_FUSNN (Q8R689) Probable N-glycosylase/DNA lyase [Includes:
8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic
or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]
Length = 217
Score = 30.8 bits (68), Expect = 4.2
Identities = 28/101 (27%), Positives = 47/101 (45%), Gaps = 9/101 (8%)
Query: 46 FKEKLKRLKLKLNQWEKENVGDLDKICKDPVT--RINALDIKEGDGNLSSQE--QKQRDD 101
FKE+LK K N WE D+ + + AL+ + NL + + +
Sbjct: 21 FKERLKEFK---NIWENGTNKDIHLELSFCILTPQSKALNAWQAITNLKKDDLIYNGKAE 77
Query: 102 FLVEYWKVAKF--NDSLLFQKSRDRWVKDGDSNTKYFHNSV 140
LVE+ + +F N S + R++ +KDG+ TK F N++
Sbjct: 78 ELVEFLNIVRFKNNKSKYLVELREKMMKDGEIITKDFFNTL 118
>CET3_MOUSE (O35648) Centrin 3
Length = 167
Score = 30.8 bits (68), Expect = 4.2
Identities = 28/92 (30%), Positives = 43/92 (46%), Gaps = 19/92 (20%)
Query: 152 LAVDGEWVEDPKVVKSKMIEFFEAQFTSNPEVGVFLEGTPFRSISDEDNLALTQTFNLEE 211
LA+ GE V D K + K E E Q + + +D+D Q + E
Sbjct: 3 LALRGELVVD-KTKRKKRRELSEEQKQEIKDAFELFD-------TDKD-----QAIDYHE 49
Query: 212 IRTAVWECEGDRSLGPDVLKVDILRVLKDFHR 243
++ A+ R+LG DV K D+L++LKD+ R
Sbjct: 50 LKVAM------RALGFDVKKADVLKILKDYDR 75
>YL17_CAEEL (Q11102) Hypothetical protein C02F12.7 in chromosome X
Length = 1130
Score = 30.4 bits (67), Expect = 5.5
Identities = 19/77 (24%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query: 46 FKEKLKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVE 105
F+ ++K + +++E+E L+K+ K+ I L+ + D L + EQK +++E
Sbjct: 188 FEARMKSFQALQDKFEREKEQALEKLRKEHQKEIQVLEQRFSDTQLLNLEQK----YIIE 243
Query: 106 YWKVAKFNDSLLFQKSR 122
++ + SL +K R
Sbjct: 244 IQRLEEERKSLRTEKER 260
>CET3_HUMAN (O15182) Centrin 3
Length = 167
Score = 30.0 bits (66), Expect = 7.2
Identities = 19/62 (30%), Positives = 30/62 (47%), Gaps = 17/62 (27%)
Query: 193 RSISDEDNLALTQTFNL-----------EEIRTAVWECEGDRSLGPDVLKVDILRVLKDF 241
R +S+E + F L E++ A+ R+LG DV K D+L++LKD+
Sbjct: 20 RELSEEQKQEIKDAFELFDTDKDEAIDYHELKVAM------RALGFDVKKADVLKILKDY 73
Query: 242 HR 243
R
Sbjct: 74 DR 75
>DP2S_METJA (Q58113) DNA polymerase II small subunit (EC 2.7.7.7)
(Pol II)
Length = 594
Score = 29.6 bits (65), Expect = 9.4
Identities = 33/123 (26%), Positives = 61/123 (48%), Gaps = 15/123 (12%)
Query: 28 ECVSTWSSFD-IQGWGAYTFKEKLKRLKLKLNQW-----EKENVGDLDKICKDPVTRINA 81
E ++ + FD I + KEK K +K ++ + EKE + + K K+ + +
Sbjct: 65 EIINEYKDFDFIFYYTGEEEKEKPKEVKKEIKKETEEKIEKEKIEFVKKEEKEQFIKKSD 124
Query: 82 LDIKEGDGNLSSQEQKQRDDFLVE----YWKVAKFNDSLLFQKSRDRWV-KDGDSNTKYF 136
D++E L S+E+K ++DF E Y + K +S+ SR +W+ KD D+ + +
Sbjct: 125 EDVEEKLKQLISKEEK-KEDFDAERAKRYEHITKIKESV---NSRIKWIAKDIDAVIEIY 180
Query: 137 HNS 139
+S
Sbjct: 181 EDS 183
>CAH1_MONDO (Q8HY33) Carbonic anhydrase I (EC 4.2.1.1) (Carbonate
dehydratase I) (CA-I)
Length = 262
Score = 29.6 bits (65), Expect = 9.4
Identities = 25/93 (26%), Positives = 34/93 (35%), Gaps = 4/93 (4%)
Query: 54 KLKLNQWEKENVGDLDKICKDPV-TRINALDIKEGDGNLSSQEQKQRDDFLVEYWK---V 109
+L + W E + + P I A+ IK G N Q+ + K
Sbjct: 118 ELHIVHWNSEKYSSFSEAAEKPDGLAIIAVFIKAGQANPGLQKVIDALSSIKTKGKKAPF 177
Query: 110 AKFNDSLLFQKSRDRWVKDGDSNTKYFHNSVNW 142
A F+ SLL +S D W G H SV W
Sbjct: 178 ANFDPSLLIPQSSDYWSYHGSLTHPPLHESVTW 210
>ANRY_HUMAN (Q6UB99) Ankyrin repeat domain protein 11 (Ankyrin
repeat-containing cofactor-1)
Length = 2664
Score = 29.6 bits (65), Expect = 9.4
Identities = 29/114 (25%), Positives = 46/114 (39%), Gaps = 10/114 (8%)
Query: 47 KEKLKRLKLKLNQWEKENVGDLDKICKDPVTRINALDIKEGDGNLSSQEQKQRDDFLVEY 106
+E+ K+ K + ++ EK+N DKI K+ KE L ++ + D EY
Sbjct: 764 EERKKKSKDRPSKLEKKNDLKEDKISKE-----KEKIFKEDKEKLKKEKVYREDSAFDEY 818
Query: 107 WKVAKF--NDSLLFQKS---RDRWVKDGDSNTKYFHNSVNWRRRTNAIRGLAVD 155
+F N+ F S RDRW D ++ F +W R + D
Sbjct: 819 CNKNQFLENEDTKFSLSDDQRDRWFSDLSDSSFDFKGEDSWDSPVTDYRDMKSD 872
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.140 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,646,962
Number of Sequences: 164201
Number of extensions: 1496665
Number of successful extensions: 4289
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 4282
Number of HSP's gapped (non-prelim): 15
length of query: 299
length of database: 59,974,054
effective HSP length: 110
effective length of query: 189
effective length of database: 41,911,944
effective search space: 7921357416
effective search space used: 7921357416
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0302a.1


