

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0299.3
(762 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
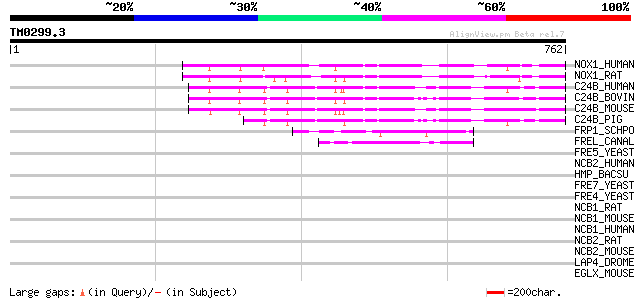
Score E
Sequences producing significant alignments: (bits) Value
NOX1_HUMAN (Q9Y5S8) NADPH oxidase homolog 1 (NOX-1) (NOH-1) (NAD... 243 1e-63
NOX1_RAT (Q9WV87) NADPH oxidase homolog 1 (NOX-1) (NOH-1) (NADH/... 243 2e-63
C24B_HUMAN (P04839) Cytochrome B-245 heavy chain (P22 phagocyte ... 239 3e-62
C24B_BOVIN (O46522) Cytochrome B-245 heavy chain (P22 phagocyte ... 235 3e-61
C24B_MOUSE (Q61093) Cytochrome B-245 heavy chain (P22 phagocyte ... 228 3e-59
C24B_PIG (P52649) Cytochrome B-245 heavy chain (P22 phagocyte B-... 194 7e-49
FRP1_SCHPO (Q04800) Ferric reductase transmembrane component (EC... 68 1e-10
FREL_CANAL (P78588) Probable ferric reductase transmembrane comp... 52 4e-06
FRE5_YEAST (Q08908) Ferric reductase transmembrane component 5 p... 42 0.006
NCB2_HUMAN (P80303) Nucleobindin 2 precursor (DNA-binding protei... 39 0.051
HMP_BACSU (P49852) Flavohemoprotein (Hemoglobin-like protein) (F... 39 0.051
FRE7_YEAST (Q12333) Ferric reductase transmembrane component 7 (... 39 0.051
FRE4_YEAST (P53746) Ferric reductase transmembrane component 4 p... 37 0.19
NCB1_RAT (Q63083) Nucleobindin 1 precursor (CALNUC) (Bone 63 kDa... 36 0.33
NCB1_MOUSE (Q02819) Nucleobindin 1 precursor (CALNUC) 36 0.33
NCB1_HUMAN (Q02818) Nucleobindin 1 precursor (CALNUC) 36 0.33
NCB2_RAT (Q9JI85) Nucleobindin 2 precursor (DNA-binding protein ... 36 0.43
NCB2_MOUSE (P81117) Nucleobindin 2 precursor (DNA-binding protei... 36 0.43
LAP4_DROME (Q7KRY7) LAP4 protein (Scribble protein) (Smell-impai... 35 0.74
EGLX_MOUSE (Q8BG58) Putative HIF-prolyl hydroxylase PH-4 (EC 1.1... 35 0.74
>NOX1_HUMAN (Q9Y5S8) NADPH oxidase homolog 1 (NOX-1) (NOH-1)
(NADH/NADPH mitogenic oxidase subunit P65-MOX)
(Mitogenic oxidase 1) (MOX1)
Length = 564
Score = 243 bits (620), Expect = 1e-63
Identities = 171/586 (29%), Positives = 254/586 (43%), Gaps = 121/586 (20%)
Query: 238 FIQDNWKRIWVMALWLSICVALFTWKFFQYKNRAVF----DVMGYCVVIAKGGAETLKFN 293
++ ++W + + +WL + V LF F +Y+ + ++G + A+ A L FN
Sbjct: 4 WVVNHWFSVLFLVVWLGLNVFLFVDAFLKYEKADKYYYTRKILGSTLACARASALCLNFN 63
Query: 294 MALILFPICRNTITWLRSKTKL---GMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHL--- 347
LIL P+CRN +++LR + D N+ FHK++A+ I + +H +AHL
Sbjct: 64 STLILLPVCRNLLSFLRGTCSFCSRTLRKQLDHNLTFHKLVAYMICLHTAIHIIAHLFNF 123
Query: 348 --------------TCDFPRLLHTTAAEYEPLKPFFGKNKPDNYWWFLKGTEGWTGVLMV 393
L H L P +N Y F G TGV+M
Sbjct: 124 DCYSRSRQATDGSLASILSSLSHDEKKGGSWLNPIQSRNTTVEYVTFTS-IAGLTGVIMT 182
Query: 394 VLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHG-------- 445
+ + + A + RR+ F FWY+HHL I + L IHG
Sbjct: 183 IALILMVTSATEFIRRSY------------FEVFWYTHHLFIFYILGLGIHGIGGIVRGQ 230
Query: 446 -------------------------HFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFR 480
H + + +W ++ P+ILY ER++R +R
Sbjct: 231 TEESMNESHPRKCAESFEMWDDRDSHCRRPKFEGHPPESWKWILAPVILYICERILRFYR 290
Query: 481 SCFKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPED 540
S K V I V + V L+M+K +GF GQYI+VNC IS EWHPF++TSAPE+
Sbjct: 291 SQQKVV-ITKVVMHPSKVLELQMNK-RGFSMEVGQYIFVNCPSISLLEWHPFTLTSAPEE 348
Query: 541 DYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGS 600
D+ S+HIR GDWT L E+ P IPR+ +DGP+G+
Sbjct: 349 DFFSIHIRAAGDWTENLIRAFEQQYSP---------------------IPRIEVDGPFGT 387
Query: 601 PAQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQQDLENGVLESRVKTNKKKHFATK 660
++D YEV +LVG GIG TP SILK + + + TK
Sbjct: 388 ASEDVFQYEVAVLVGAGIGVTPFASILKSIWYKFQCAD----------------HNLKTK 431
Query: 661 QAYFYWVTREQGSFEWFKGLM----NEIAENDKEGVIELHNYCTGVYKEGDDGSALITML 716
+ YFYW+ RE G+F WF L+ E+ E K G + + TG + G A +
Sbjct: 432 KIYFYWICRETGAFSWFNNLLTSLEQEMEELGKVGFLNYRLFLTG-WDSNIVGHAALNFD 490
Query: 717 QSLYQSKYGLDKVSETRVKTHFGRPNWRNVYEHVSLKHPDKRVGKY 762
++ D V+ + KT FGRP W N + ++ HP VG +
Sbjct: 491 KA-------TDIVTGLKQKTSFGRPMWDNEFSTIATSHPKSVVGVF 529
>NOX1_RAT (Q9WV87) NADPH oxidase homolog 1 (NOX-1) (NOH-1)
(NADH/NADPH mitogenic oxidase subunit P65-MOX)
(Mitogenic oxidase 1) (MOX1)
Length = 563
Score = 243 bits (619), Expect = 2e-63
Identities = 173/585 (29%), Positives = 264/585 (44%), Gaps = 120/585 (20%)
Query: 238 FIQDNWKRIWVMALWLSICVALFTWKFFQYKNRAVF----DVMGYCVVIAKGGAETLKFN 293
++ ++W + + WL + + LF + F Y+ + +++G + +A+ A L FN
Sbjct: 4 WLVNHWLSVLFLVSWLGLNIFLFVYVFLNYEKSDKYYYTREILGTALALARASALCLNFN 63
Query: 294 MALILFPICRNTITWLRSKTKL---GMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHLTCD 350
+IL P+CRN +++LR + P D N+ FHK++A+ I I +H +AHL +
Sbjct: 64 SMVILIPVCRNLLSFLRGTCSFCNHTLRKPLDHNLTFHKLVAYMICIFTAIHIIAHLF-N 122
Query: 351 FPRLLHTTAAE----YEPLKPFFGKNKPDNY------------WWFLKGTEGWTGVLMVV 394
F R + A L F K D++ + G TGV+ V
Sbjct: 123 FERYSRSQQAMDGSLASVLSSLFHPEKEDSWLNPIQSPNVTVMYAAFTSIAGLTGVVATV 182
Query: 395 LMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHG--------- 445
+ + A + RRN +L FWY+HHL II + L IHG
Sbjct: 183 ALVLMVTSAMEFIRRNYFEL------------FWYTHHLFIIYIICLGIHGLGGIVRGQT 230
Query: 446 ----------HFLYLSKKWYKNT--------------TWMYLAVPMILYESERLIRAFRS 481
+ Y +W K +W ++ P+ Y ER++R +RS
Sbjct: 231 EESMSESHPRNCSYSFHEWDKYERSCRSPHFVGQPPESWKWILAPIAFYIFERILRFYRS 290
Query: 482 CFKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDD 541
K V I V + V L+M K +GF GQYI+VNC IS EWHPF++TSAPE++
Sbjct: 291 RQKVV-ITKVVMHPCKVLELQMRK-RGFTMGIGQYIFVNCPSISFLEWHPFTLTSAPEEE 348
Query: 542 YLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSP 601
+ S+HIR GDWT L E+ P +PR+ +DGP+G+
Sbjct: 349 FFSIHIRAAGDWTENLIRTFEQQHSP---------------------MPRIEVDGPFGTV 387
Query: 602 AQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQQDLENGVLESRVKTNKKKHFATKQ 661
++D YEV +LVG GIG TP S LK + ++ NK K T++
Sbjct: 388 SEDVFQYEVAVLVGAGIGVTPFASFLKSIWYKFQRAH-------------NKLK---TQK 431
Query: 662 AYFYWVTREQGSFEWFKGLMNEI-AENDKEGVIELHNY---CTGVYKEGDDGSALITMLQ 717
YFYW+ RE G+F WF L+N + E D+ G + NY TG + G A + +
Sbjct: 432 IYFYWICRETGAFAWFNNLLNSLEQEMDELGKPDFLNYRLFLTG-WDSNIAGHAALNFDR 490
Query: 718 SLYQSKYGLDKVSETRVKTHFGRPNWRNVYEHVSLKHPDKRVGKY 762
+ D ++ + KT FGRP W N + ++ HP VG +
Sbjct: 491 A-------TDVLTGLKQKTSFGRPMWDNEFSRIATAHPKSVVGVF 528
>C24B_HUMAN (P04839) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generat
Length = 569
Score = 239 bits (609), Expect = 3e-62
Identities = 169/568 (29%), Positives = 269/568 (46%), Gaps = 95/568 (16%)
Query: 246 IWVMALWLSICVALFTWKFFQYKNRAVF----DVMGYCVVIAKGGAETLKFNMALILFPI 301
I+V+ +WL + V LF W + Y F ++G + +A+ A L FN LIL P+
Sbjct: 11 IFVILVWLGLNVFLFVWYYRVYDIPPKFFYTRKLLGSALALARAPAACLNFNCMLILLPV 70
Query: 302 CRNTITWLRSKT---KLGMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHLT----CDFPRL 354
CRN +++LR + + D N+ FHK++A+ IA+ +H +AHL C R+
Sbjct: 71 CRNLLSFLRGSSACCSTRVRRQLDRNLTFHKMVAWMIALHSAIHTIAHLFNVEWCVNARV 130
Query: 355 LHTTAAEYEPLKPFFGKNKPDNYWWF----LKGTEGWTGVLMVVLMAIPFILAQPWARRN 410
+ Y G + ++Y F +K EG + + +L I ++
Sbjct: 131 --NNSDPYSVALSELGDRQNESYLNFARKRIKNPEGGLYLAVTLLAGITGVVITLCLILI 188
Query: 411 RLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHG---------------HFLYLSKK-- 453
K +++ + F FWY+HHL +I ++ L IHG H + + ++
Sbjct: 189 ITSSTKTIRR-SYFEVFWYTHHLFVIFFIGLAIHGAERIVRGQTAESLAVHNITVCEQKI 247
Query: 454 --WYK-------------NTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNV 498
W K TW ++ PM LY ERL+R +RS K V I V +
Sbjct: 248 SEWGKIKECPIPQFAGNPPMTWKWIVGPMFLYLCERLVRFWRSQQKVV-ITKVVTHPFKT 306
Query: 499 FALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQ 558
L+M K +GFK GQYI+V C +S EWHPF++TSAPE+D+ S+HIR +GDWT L
Sbjct: 307 IELQMKK-KGFKMEVGQYIFVKCPKVSKLEWHPFTLTSAPEEDFFSIHIRIVGDWTEGL- 364
Query: 559 AILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGI 618
C D K +P++ +DGP+G+ ++D +YEV++LVG GI
Sbjct: 365 ------------FNACGCDKQEFQDAWK--LPKIAVDGPFGTASEDVFSYEVVMLVGAGI 410
Query: 619 GATPLISILKGMLNNIKQQQDLENGVLESRVKTNKKKHFATKQAYFYWVTREQGSFEWFK 678
G TP SILK + N + K+ YFYW+ R+ +FEWF
Sbjct: 411 GVTPFASILKSVWYKY----------------CNNATNLKLKKIYFYWLCRDTHAFEWFA 454
Query: 679 GLM----NEIAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRV 734
L+ +++ E + G + + Y TG D+ A + + D ++ +
Sbjct: 455 DLLQLLESQMQERNNAGFLSYNIYLTG----WDESQANHFAVHHDEEK----DVITGLKQ 506
Query: 735 KTHFGRPNWRNVYEHVSLKHPDKRVGKY 762
KT +GRPNW N ++ ++ +HP+ R+G +
Sbjct: 507 KTLYGRPNWDNEFKTIASQHPNTRIGVF 534
>C24B_BOVIN (O46522) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generat
Length = 570
Score = 235 bits (600), Expect = 3e-61
Identities = 176/567 (31%), Positives = 265/567 (46%), Gaps = 93/567 (16%)
Query: 246 IWVMALWLSICVALFTWKFFQYKNRAVF----DVMGYCVVIAKGGAETLKFNMALILFPI 301
I+V+ +WL + V LF W + Y F ++G + +A+ A L FN LIL P+
Sbjct: 12 IFVILVWLGMNVFLFVWYYRVYDIPDKFFYTRKLLGSALALARAPAACLNFNCMLILLPV 71
Query: 302 CRNTITWLRSKT---KLGMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHLT----CDFPRL 354
CRN +++LR + + D N+ FHK++A+ IA+ +H +AHL C R+
Sbjct: 72 CRNLLSFLRGSSACCSTRIRRQLDRNLTFHKMVAWMIALHTAIHTIAHLFNVEWCVNARV 131
Query: 355 LHTTAAEYEPLKPFFGKNKPDNYWWF----LKGTEGWTGVLMVVLMAIPFILAQPWARRN 410
+ Y G + Y F +K EG V + L I ++
Sbjct: 132 --NNSDPYSIALSDIGDKPNETYLNFVRQRIKNPEGGLYVAVTRLAGITGVVITLCLILI 189
Query: 411 RLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHGH------------FLYLSKKWYKNT 458
K +++ + F FWY+HHL +I ++ L IHG + + Y+N
Sbjct: 190 ITSSTKTIRR-SYFEVFWYTHHLFVIFFIGLAIHGAQRIVRGQTAESLLKHQPRNCYQNI 248
Query: 459 --------------------TWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNV 498
TW ++ PM LY ERL+R +RS K V I V +
Sbjct: 249 SQWGKIENCPIPEFSGNPPMTWKWIVGPMFLYLCERLVRFWRSQQKVV-ITKVVTHPFKT 307
Query: 499 FALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQ 558
L+M K +GFK GQYI+V C +S EWHPF++TSAPE+D+ S+HIR +GDWT
Sbjct: 308 IELQMKK-KGFKMEVGQYIFVKCPVVSKLEWHPFTLTSAPEEDFFSIHIRIVGDWTEG-- 364
Query: 559 AILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGI 618
L KAC D Q+ D K +P++ +DGP+G+ ++D +YEV++LVG GI
Sbjct: 365 --LFKAC--GCDKQE-------FQDAWK--LPKIAVDGPFGTASEDVFSYEVVMLVGAGI 411
Query: 619 GATPLISILKGMLNNIKQQQDLENGVLESRVKTNKKKHFATKQAYFYWVTREQGSFEWFK 678
G TP SILK + NK + K+ YFYW+ R+ +FEWF
Sbjct: 412 GVTPFASILKSVWYKY----------------CNKAPNLRLKKIYFYWLCRDTHAFEWFA 455
Query: 679 GLMN--EIAENDKEGVIEL-HNYCTGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVK 735
L+ E +K L +N C G D S + K D ++ + K
Sbjct: 456 DLLQLLETQMQEKNNTDFLSYNICL----TGWDESQASHFAMHHDEEK---DVITGLKQK 508
Query: 736 THFGRPNWRNVYEHVSLKHPDKRVGKY 762
T +GRPNW N ++ + +HP+ R+G +
Sbjct: 509 TLYGRPNWDNEFKTIGSQHPNTRIGVF 535
>C24B_MOUSE (Q61093) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generat
Length = 570
Score = 228 bits (582), Expect = 3e-59
Identities = 168/564 (29%), Positives = 263/564 (45%), Gaps = 87/564 (15%)
Query: 246 IWVMALWLSICVALFTWKFFQYKNRAVFD----VMGYCVVIAKGGAETLKFNMALILFPI 301
I+V+ +WL + V LF + Y + ++ ++G + +A+ A L FN LIL P+
Sbjct: 12 IFVILVWLGLNVFLFINYYKVYDDGPKYNYTRKLLGSALALARAPAACLNFNCMLILLPV 71
Query: 302 CRNTITWLRSKT---KLGMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHLT----CDFPRL 354
CRN +++LR + + D N+ FHK++A+ IA+ +H +AHL C R+
Sbjct: 72 CRNLLSFLRGSSACCSTRIRRQLDRNLTFHKMVAWMIALHTAIHTIAHLFNVEWCVNARV 131
Query: 355 LHTTAAEYEPLKPFFGKNKPDNYWWF----LKGTEGWTGVLMVVLMAIPFILAQPWARRN 410
+ Y G N+ + Y F +K EG V + L I I+
Sbjct: 132 --GISDRYSIALSDIGDNENEEYLNFAREKIKNPEGGLYVAVTRLAGITGIVITLCLILI 189
Query: 411 RLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHG---------------HFLYLS---- 451
K +++ + F FWY+HHL +I ++ L IHG H L +
Sbjct: 190 ITSSTKTIRR-SYFEVFWYTHHLFVIFFIGLAIHGAERIVRGQTAESLEEHNLDICADKI 248
Query: 452 KKWYK-------------NTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNV 498
++W K TW ++ PM LY ERL+R +RS K V I V +
Sbjct: 249 EEWGKIKECPVPKFAGNPPMTWKWIVGPMFLYLCERLVRFWRSQQKVV-ITKVVTHPFKT 307
Query: 499 FALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQ 558
L+M K +GFK GQYI+V C +S EWHPF++TSAPE+D+ S+HIR +GDWT L
Sbjct: 308 IELQMKK-KGFKMEVGQYIFVKCPKVSKLEWHPFTLTSAPEEDFFSIHIRIVGDWTEGL- 365
Query: 559 AILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGI 618
C D K +P++ +DGP+G+ ++D +YEV++LVG GI
Sbjct: 366 ------------FNACGCDKQEFQDAWK--LPKIAVDGPFGTASEDVFSYEVVMLVGAGI 411
Query: 619 GATPLISILKGMLNNIKQQQDLENGVLESRVKTNKKKHFATKQAYFYWVTREQGSFEWFK 678
G TP SILK + + K+ YFYW+ R+ +FEWF
Sbjct: 412 GVTPFASILKSVWYKY----------------CDNATSLKLKKIYFYWLCRDTHAFEWFA 455
Query: 679 GLMNEIAENDKEGVIELHNYCTGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVKTHF 738
L+ ++ E + + +Y G D S + K D ++ + KT +
Sbjct: 456 DLL-QLLETQMQERNNANFLSYNIYLTGWDESQANHFAVHHDEEK---DVITGLKQKTLY 511
Query: 739 GRPNWRNVYEHVSLKHPDKRVGKY 762
GRPNW N ++ ++ +HP+ +G +
Sbjct: 512 GRPNWDNEFKTIASEHPNTTIGVF 535
>C24B_PIG (P52649) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generatin
Length = 484
Score = 194 bits (493), Expect = 7e-49
Identities = 144/485 (29%), Positives = 225/485 (45%), Gaps = 88/485 (18%)
Query: 322 DDNVNFHKVIAFGIAIGIGLHAVAHLT----CDFPRLLHTTAAEYEPLKPFFGKNKPDNY 377
D N+ FHK++A+ IA+ +H +AHL C R+ + Y G + Y
Sbjct: 9 DRNLTFHKMVAWMIALHATIHTIAHLFNVEWCVNARV--NNSDPYSIALSDIGDKPNETY 66
Query: 378 WWF----LKGTEGWTGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHL 433
F +K EG V + L I ++ K +++ + F FWY+HHL
Sbjct: 67 LNFVRQRIKNPEGGLYVAVTRLAGITGVVITLCLILIITSSTKTIRR-SYFEVFWYTHHL 125
Query: 434 LIIVYVLLVIHGHFLYLSKKWYKNT--------------------------------TWM 461
+I ++ L IHG + ++ K+ TW
Sbjct: 126 FVIFFIGLAIHGAERIVRRQTPKSLLVHDPKACAQNISQWGKIKDCPIPEFAGNPPMTWK 185
Query: 462 YLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNC 521
++ PM LY ERL+R +RS K V I V + L+M K +GF+ GQYI+V
Sbjct: 186 WIVGPMFLYLCERLVRFWRSQQKVV-ITKVVTHPFKTIELQMKK-KGFRMEVGQYIFVKR 243
Query: 522 SDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTL 581
+S EWHPF++TSAPE+D+ S+HIR +GDWT L KAC D Q+
Sbjct: 244 PAVSKLEWHPFTLTSAPEEDFFSIHIRIVGDWTEG----LFKAC--GCDKQE-------F 290
Query: 582 PDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQQDLE 641
D K +P++ +DGP+G+ ++D +Y+V++LVG GIG TP SILK +
Sbjct: 291 QDAWK--LPKIAVDGPFGTASEDVFSYQVVMLVGAGIGVTPFASILKSVWYKY------- 341
Query: 642 NGVLESRVKTNKKKHFATKQAYFYWVTREQGSFEWFKGLM----NEIAENDKEGVIELHN 697
N + K+ YFYW+ R+ +FEWF L+ ++ E + G + +
Sbjct: 342 ---------CNNATNLRLKKIYFYWLCRDTHAFEWFADLLQLLETQMQERNNAGFLSYNI 392
Query: 698 YCTGVYKEGDDGSALITMLQSLYQSKYGLDKVSETRVKTHFGRPNWRNVYEHVSLKHPDK 757
Y TG D+ A + + D ++ + KT +GRPNW N ++ ++ +HP
Sbjct: 393 YLTG----WDESQANHFAVHHDEEK----DVITGLKQKTLYGRPNWDNEFKTIASQHPTT 444
Query: 758 RVGKY 762
R+G +
Sbjct: 445 RIGVF 449
>FRP1_SCHPO (Q04800) Ferric reductase transmembrane component (EC
1.16.1.7) (Ferric-chelate reductase)
Length = 564
Score = 67.8 bits (164), Expect = 1e-10
Identities = 61/276 (22%), Positives = 128/276 (46%), Gaps = 57/276 (20%)
Query: 389 GVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLIIVYVLLVIHGHFL 448
G +++ LM I + + P+ RR + F+ HH+ I +++ + H
Sbjct: 195 GYVILGLMVIMIVSSLPFFRRRF------------YEWFFVLHHMCSIGFLITIWLHH-- 240
Query: 449 YLSKKWYKNTTWMYLAVPMILYESE-RLIRAF--RSCFKYVRIQSVSWYTGNVFALKMSK 505
+ +M + V + +++ R++R+F RS F V ++ ++ +K +
Sbjct: 241 ------RRCVVYMKVCVAVYVFDRGCRMLRSFLNRSKFDVVLVED------DLIYMKGPR 288
Query: 506 PQ----GFKYMSGQYIYVNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQL-QAI 560
P+ G + +G ++Y+N +S ++ HPF+I S P DD++ + + +T +L + +
Sbjct: 289 PKKSFFGLPWGAGNHMYINIPSLSYWQIHPFTIASVPSDDFIELFVAVRAGFTKRLAKKV 348
Query: 561 LEKACQPSSD-------------------DQKCILQANTLPDTSKPRIPRLWIDGPYGSP 601
K+ SD ++ + Q + + ++S ++ L +DGPYG
Sbjct: 349 SSKSLSDVSDINISDEKIEKNGDVGIEVMERHSLSQEDLVFESSAAKVSVL-MDGPYGPV 407
Query: 602 AQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQ 637
+ YK+Y L L G+G + ++ I+ L+ IK+Q
Sbjct: 408 SNPYKDYSYLFLFAGGVGVSYILPII---LDTIKKQ 440
>FREL_CANAL (P78588) Probable ferric reductase transmembrane
component (EC 1.16.1.7) (Ferric-chelate reductase)
Length = 669
Score = 52.4 bits (124), Expect = 4e-06
Identities = 46/215 (21%), Positives = 95/215 (43%), Gaps = 28/215 (13%)
Query: 424 FNAFWYSHHLLIIVYVLLVIHGHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCF 483
+ F+ H +L++ +V+ G + +L + Y + W +AV + +R++R R F
Sbjct: 338 YEVFFLIHIVLVVFFVV----GGYYHLESQGYGDFMWAAIAV----WAFDRVVRLGRIFF 389
Query: 484 KYVRIQSVSWYTGNVFALKMSKPQGFKYMSGQYIYVNCSDISPF-EWHPFSITSAPEDDY 542
R +VS + +++ KP+ +K ++G + +++ + F + HPF+ T+ +D
Sbjct: 390 FGARKATVSIKGDDTLKIEVPKPKYWKSVAGGHAFIHFLKPTLFLQSHPFTFTTTESNDK 449
Query: 543 LSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPA 602
+ ++ + TS + L L NT R+ ++GPYG P+
Sbjct: 450 IVLYAKIKNGITSNIAKYLSP------------LPGNTATI-------RVLVEGPYGEPS 490
Query: 603 QDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQ 637
+N + ++ V G G + S + K Q
Sbjct: 491 SAGRNCKNVVFVAGGNGIPGIYSECVDLAKKSKNQ 525
>FRE5_YEAST (Q08908) Ferric reductase transmembrane component 5
precursor (EC 1.16.1.7) (Ferric-chelate reductase 5)
Length = 694
Score = 42.0 bits (97), Expect = 0.006
Identities = 93/460 (20%), Positives = 173/460 (37%), Gaps = 105/460 (22%)
Query: 296 LILFPICRNTITWLRSKTKLGMVVPFDDNVNFHKVIAFGIAIGIGLHAVAHLTCDFPRLL 355
++LF +T+TWL + + + +HK + + + +HA+ +
Sbjct: 285 IVLFGGKNSTMTWLTG-------IRYTAFITYHKWLGRFMLVDCTIHAIGY--------- 328
Query: 356 HTTAAEYEPLKPFFGKNKPDNYWWFLKGTEGWT-GVLMVVLMAIPFILAQPWARRNRLKL 414
T A E NYW ++K ++ WT G ++++ I
Sbjct: 329 -TYHAYIE------------NYWKYVKYSDLWTSGRHAMIIVGI---------------- 359
Query: 415 PKPLKKLTGFNAFWYSHH---LLIIVYVLLVIHGHFLYLSKKWYKNTTWMYLAVPMILYE 471
L F+ F++ H L +I +++L I G F K YK ++ + +
Sbjct: 360 ------LVFFSFFFFRRHYYELFVITHIILAI-GFFHACWKHCYKLGWGEWIMACALFWI 412
Query: 472 SERLIRAFRSCFKYVRIQSVSWYTGNVFALKMSK-PQGFKYMSGQYIYVNCSDISPF-EW 529
++R++R + + + ++ +++SK + +K GQYIY+ F +
Sbjct: 413 ADRILRLIKIAIFGMPWAKLKLCGESMIEVRISKSSKWWKAEPGQYIYLYFLRPKIFWQS 472
Query: 530 HPFSI-TSAPEDDYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPR 588
HPF++ S ED L V I T +LQ L L++ +
Sbjct: 473 HPFTVMDSLVEDGELVVVITVKNGLTKKLQEYL--------------LESEGYTEM---- 514
Query: 589 IPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISILKGMLNNIKQQQDLENGVLESR 648
R+ +GPYG + + +E LL + G G +S+ IK + +++
Sbjct: 515 --RVLAEGPYGQSTRTHL-FESLLFIAGGAGVPGPLSMA------IKAGRQVKSNDSHQM 565
Query: 649 VKTNKKKHFATKQAYFYWVTREQGSFEWFKGLMNEIAENDKEGVIELHNYCTGVYKEGDD 708
+K F W R E ++ + + KE I+ Y TG K D+
Sbjct: 566 IK-------------FVWSVRNLDLLEVYRKEIMVL----KELNIDTKIYFTGERK--DE 606
Query: 709 GSALITMLQSLYQSKYGLDKVSETRVKTHFGRPNWRNVYE 748
+ + ++ L + T FGRPN + E
Sbjct: 607 SNTEEGAIANMSTEGRLLTTSKSAEMITDFGRPNIDEIIE 646
>NCB2_HUMAN (P80303) Nucleobindin 2 precursor (DNA-binding protein
NEFA)
Length = 420
Score = 38.9 bits (89), Expect = 0.051
Identities = 23/75 (30%), Positives = 44/75 (58%), Gaps = 11/75 (14%)
Query: 109 TKDELREIWEQ---ITNQSFDSRVQTFFDLVDKNADGRINEEQVKEIISLSASSNKLSKI 165
+KD+L+E+WE+ + FD + TFF L D N+DG ++E++++ + + +L K+
Sbjct: 225 SKDQLKEVWEETDGLDPNDFDPK--TFFKLHDVNSDGFLDEQELEALF-----TKELEKV 277
Query: 166 QERSGEYASLI-MEE 179
+ E ++ MEE
Sbjct: 278 YDPKNEEDDMVEMEE 292
>HMP_BACSU (P49852) Flavohemoprotein (Hemoglobin-like protein)
(Flavohemoglobin) (Nitric oxide dioxygenase) (EC
1.14.12.17) (NO oxygenase) (NOD)
Length = 399
Score = 38.9 bits (89), Expect = 0.051
Identities = 42/187 (22%), Positives = 75/187 (39%), Gaps = 39/187 (20%)
Query: 510 KYMSGQYIY--VNCSDISPFEWHPFSITSAPEDDYLSVHIRTLGDWTSQLQAILEKACQP 567
++ +GQYI V D +S++ P DY + ++ G +S L L++
Sbjct: 184 EFQAGQYISIKVRIPDSEYTHIRQYSLSDMPGKDYYRISVKKDGVVSSYLHDGLQEG--- 240
Query: 568 SSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGIGATPLISIL 627
+ I P G D+ + + L+L+ G+G TP+IS+L
Sbjct: 241 ----------------------DSIEISAPAGDFVLDHASQKDLVLISAGVGITPMISML 278
Query: 628 KGMLNNIKQQQDL--------ENGVLESRVKTNKKKHFATKQAYFYWVTREQ---GSFEW 676
K ++ ++Q L E L V+ KH A K A+ Y E+ G +
Sbjct: 279 KTSVSKQPERQILFIHAAKNSEYHALRHEVE-EAAKHSAVKTAFVYREPTEEDRAGDLHF 337
Query: 677 FKGLMNE 683
+G +++
Sbjct: 338 HEGQIDQ 344
>FRE7_YEAST (Q12333) Ferric reductase transmembrane component 7 (EC
1.16.1.7) (Ferric-chelate reductase 7)
Length = 629
Score = 38.9 bits (89), Expect = 0.051
Identities = 70/309 (22%), Positives = 115/309 (36%), Gaps = 62/309 (20%)
Query: 382 KGTEGW-TGVLMVVLMAIPFILAQPWARRNRLKLPKPLKKLTGFNAFWYSHHLLII-VYV 439
K ++ W +GV ++ + + ++ + P ARR+ ++ F H +L + Y+
Sbjct: 233 KASDMWRSGVPPILFLNLLWLSSLPIARRHFYEI------------FLQLHWILAVGFYI 280
Query: 440 LLVIHGHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAFRSCFKYVRIQSVSWYTGNVF 499
L H + S + T ++ A + +R RS F I +VS G
Sbjct: 281 SLFYHVYPELNSHMYLVATIVVWFAQLFYRLAVKGYLRPGRS-FMASTIANVS-IVGEGC 338
Query: 500 ALKMSKPQGFKYMSGQYIYVNCSDISPFEWHPFSI-TSAPEDDYLSVHIRTLGDWTSQLQ 558
+ K Y GQ+I+V D HPFSI SA + + IR Q
Sbjct: 339 VELIVKDVEMAYSPGQHIFVRTIDKGIISNHPFSIFPSAKYPGGIKMLIRA--------Q 390
Query: 559 AILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDGPYGSPAQDYKNYEVLLLVGLGI 618
K S+DD K IL IDGPYG +D +++ + L+ G
Sbjct: 391 KGFSKRLYESNDDMKKIL-----------------IDGPYGGIERDIRSFTNVYLICSGS 433
Query: 619 GATPLISILKGMLNNIKQQQDLENGVLESRVKTNKKKHFATKQAYFYWVTREQGSFEWFK 678
G + + L+ I + +LE L+ WV R + W +
Sbjct: 434 GISTCLPFLQ-KYGPILHKTNLEVITLD-------------------WVVRHREDISWIR 473
Query: 679 GLMNEIAEN 687
M ++ N
Sbjct: 474 DEMCTLSNN 482
>FRE4_YEAST (P53746) Ferric reductase transmembrane component 4
precursor (EC 1.16.1.7) (Ferric-chelate reductase 4)
Length = 719
Score = 37.0 bits (84), Expect = 0.19
Identities = 41/199 (20%), Positives = 86/199 (42%), Gaps = 28/199 (14%)
Query: 421 LTGFNAFWYSHHLLI-IVYVLLVIHGHFLYLSKKWYKNTTWMYLAVPMILYESERLIRAF 479
+ F +Y + + IV+ L ++ + +++ + W+Y A+ ++ +R++R
Sbjct: 364 IAAFRRHYYETFMALHIVFAALFLYTCWEHVTN--FSGIEWIYAAIA--IWGVDRIVRIT 419
Query: 480 RSCFKYVRIQSVSWYTGNVFALKMSKPQGF-KYMSGQYIYVN-CSDISPFEWHPFSIT-S 536
R + ++ + + KP+ F K GQY++V+ + ++ HPF++ S
Sbjct: 420 RIALLGFPKADLQLVGSDLVRVTVKKPKKFWKAKPGQYVFVSFLRPLCFWQSHPFTVMDS 479
Query: 537 APEDDYLSVHIRTLGDWTSQLQAILEKACQPSSDDQKCILQANTLPDTSKPRIPRLWIDG 596
D L + ++ T ++ +E+ +S RL I+G
Sbjct: 480 CVNDRELVIVLKAKKGVTKLVRNFVERKGGKAS--------------------MRLAIEG 519
Query: 597 PYGSPAQDYKNYEVLLLVG 615
PYGS + ++ VLLL G
Sbjct: 520 PYGSKSTAHRFDNVLLLAG 538
>NCB1_RAT (Q63083) Nucleobindin 1 precursor (CALNUC) (Bone 63 kDa
calcium-binding protein)
Length = 459
Score = 36.2 bits (82), Expect = 0.33
Identities = 27/115 (23%), Positives = 59/115 (50%), Gaps = 10/115 (8%)
Query: 74 KTRFNECIGMSELKDFACKLFDALARCRGITSTSI--TKDELREIWEQITNQSFDS-RVQ 130
+ R+ E +G + K+ KL + R R ++ ++ +L+E+WE++ + +
Sbjct: 186 RRRYLESLGEEQRKEAERKLQEQQRRHREHPKVNVPGSQAQLKEVWEELDGLDPNRFNPK 245
Query: 131 TFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNL 185
TFF L D N+DG ++E++++ + + +L K+ + E M E++ + L
Sbjct: 246 TFFILHDINSDGVLDEQELEALF-----TKELEKVYDPKNEEDD--MREMEEERL 293
>NCB1_MOUSE (Q02819) Nucleobindin 1 precursor (CALNUC)
Length = 459
Score = 36.2 bits (82), Expect = 0.33
Identities = 27/115 (23%), Positives = 59/115 (50%), Gaps = 10/115 (8%)
Query: 74 KTRFNECIGMSELKDFACKLFDALARCRGITSTSI--TKDELREIWEQITNQSFDS-RVQ 130
+ R+ E +G + K+ KL + R R ++ ++ +L+E+WE++ + +
Sbjct: 186 RRRYLESLGEEQRKEAERKLQEQQRRHREHPKVNVPGSQAQLKEVWEELDGLDPNRFNPK 245
Query: 131 TFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNL 185
TFF L D N+DG ++E++++ + + +L K+ + E M E++ + L
Sbjct: 246 TFFILHDINSDGVLDEQELEALF-----TKELEKVYDPKNEEDD--MREMEEERL 293
>NCB1_HUMAN (Q02818) Nucleobindin 1 precursor (CALNUC)
Length = 461
Score = 36.2 bits (82), Expect = 0.33
Identities = 27/115 (23%), Positives = 59/115 (50%), Gaps = 10/115 (8%)
Query: 74 KTRFNECIGMSELKDFACKLFDALARCRGITSTSI--TKDELREIWEQITNQSFDS-RVQ 130
+ R+ E +G + K+ KL + R R ++ ++ +L+E+WE++ + +
Sbjct: 187 RRRYLESLGEEQRKEAERKLEEQQRRHREHPKVNVPGSQAQLKEVWEELDGLDPNRFNPK 246
Query: 131 TFFDLVDKNADGRINEEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNL 185
TFF L D N+DG ++E++++ + + +L K+ + E M E++ + L
Sbjct: 247 TFFILHDINSDGVLDEQELEALF-----TKELEKVYDPKNEEDD--MREMEEERL 294
>NCB2_RAT (Q9JI85) Nucleobindin 2 precursor (DNA-binding protein
NEFA)
Length = 420
Score = 35.8 bits (81), Expect = 0.43
Identities = 17/49 (34%), Positives = 32/49 (64%), Gaps = 5/49 (10%)
Query: 109 TKDELREIWEQ---ITNQSFDSRVQTFFDLVDKNADGRINEEQVKEIIS 154
+KD+L+E+WE+ + FD + TFF L D N DG ++E++++ + +
Sbjct: 225 SKDQLKEVWEETDGLDPNDFDPK--TFFKLHDVNNDGFLDEQELEALFT 271
>NCB2_MOUSE (P81117) Nucleobindin 2 precursor (DNA-binding protein
NEFA)
Length = 420
Score = 35.8 bits (81), Expect = 0.43
Identities = 17/49 (34%), Positives = 32/49 (64%), Gaps = 5/49 (10%)
Query: 109 TKDELREIWEQ---ITNQSFDSRVQTFFDLVDKNADGRINEEQVKEIIS 154
+KD+L+E+WE+ + FD + TFF L D N DG ++E++++ + +
Sbjct: 225 SKDQLKEVWEETDGLDPNDFDPK--TFFKLHDVNNDGFLDEQELEALFT 271
>LAP4_DROME (Q7KRY7) LAP4 protein (Scribble protein) (Smell-impaired
protein)
Length = 1851
Score = 35.0 bits (79), Expect = 0.74
Identities = 40/151 (26%), Positives = 65/151 (42%), Gaps = 20/151 (13%)
Query: 92 KLFDALARCRGITSTSITKDELREIWEQITNQSFDSRVQTFFDLVDKNADGRINEE--QV 149
+L D L C + +T++ L E+ I + + + VD+NA + E Q
Sbjct: 281 RLNDTLGNCENMQELILTENFLSELPASIGQMTKLNNLN-----VDRNALEYLPLEIGQC 335
Query: 150 KEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIELYSLEMLLLQA--------- 200
+ LS NKL K+ G L + ++ + L Y+ YSL L L+A
Sbjct: 336 ANLGVLSLRDNKLKKLPPELGNCTVLHVLDVSGNQLLYLP-YSLVNLQLKAVWLSENQSQ 394
Query: 201 ---SAQSTTDSKVVSQMLSQKFVPTKERNPI 228
+ Q TD++ Q+LS +P +E PI
Sbjct: 395 PLLTFQPDTDAETGEQVLSCYLLPQQEYQPI 425
>EGLX_MOUSE (Q8BG58) Putative HIF-prolyl hydroxylase PH-4 (EC
1.14.11.-) (Hypoxia-inducible factor prolyl
4-hydroxylase)
Length = 503
Score = 35.0 bits (79), Expect = 0.74
Identities = 30/106 (28%), Positives = 48/106 (44%), Gaps = 11/106 (10%)
Query: 86 LKDFACKLFDALARCRGITSTSITK-DELREIWEQITNQSFDSRVQTFFDLVDKNADGRI 144
L D C+L LA+ +G+ + I +E E + D F L+D+N DGR+
Sbjct: 152 LSDEECRLIIHLAQMKGLQRSQILPTEEYEEAMSAMQVSQLD-----LFQLLDQNHDGRL 206
Query: 145 NEEQVKEIISLSASSNKLSKIQERSGEYASLIMEELDPDNLGYIEL 190
Q++E+++ + N E E S I + DPD G + L
Sbjct: 207 ---QLREVLAQTRLGNGRWMTPENIQEMYSAI--KADPDGDGVLSL 247
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.137 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 91,184,708
Number of Sequences: 164201
Number of extensions: 3879586
Number of successful extensions: 9816
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 53
Number of HSP's that attempted gapping in prelim test: 9729
Number of HSP's gapped (non-prelim): 106
length of query: 762
length of database: 59,974,054
effective HSP length: 118
effective length of query: 644
effective length of database: 40,598,336
effective search space: 26145328384
effective search space used: 26145328384
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0299.3


