

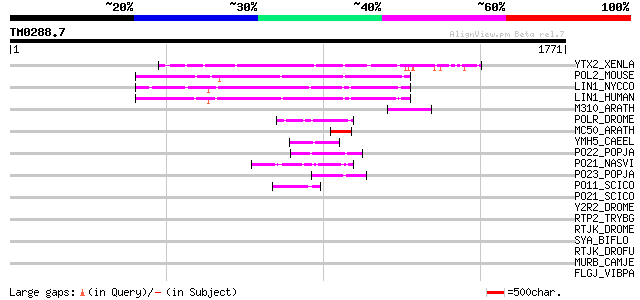
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0288.7
(1771 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 169 5e-41
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 155 7e-37
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 155 9e-37
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 149 9e-35
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 125 1e-27
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 80 4e-14
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 79 9e-14
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 72 1e-11
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 69 1e-10
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 61 3e-08
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 57 3e-07
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 49 2e-04
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 44 0.004
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 39 0.13
RTP2_TRYBG (P15594) Retrotransposable element SLACS 132 kDa prot... 34 4.1
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 34 4.1
SYA_BIFLO (Q8G5W9) Alanyl-tRNA synthetase (EC 6.1.1.7) (Alanine-... 33 5.4
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 33 7.0
MURB_CAMJE (Q9PM01) UDP-N-acetylenolpyruvoylglucosamine reductas... 33 7.0
FLGJ_VIBPA (Q9X9J3) Peptidoglycan hydrolase flgJ (EC 3.2.1.-) (M... 33 7.0
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein (ORF
2)
Length = 1308
Score = 169 bits (429), Expect = 5e-41
Identities = 267/1146 (23%), Positives = 469/1146 (40%), Gaps = 173/1146 (15%)
Query: 474 REVHIQILHSSQNYIHTSIFANNSVSDWDCTFVYGDPSSQRRRFLWPQITALRFRDNTPW 533
R +H+++ S + Y +++A + +R RF + D+
Sbjct: 91 RLLHLRVRESGRTYNLMNVYAPTT-------------GPERARFFESLSAYMETIDSDEA 137
Query: 534 CLLG-DFNEILFAHEKE--GMRPQPPSVLQRFREFVHNSGLMDVDLKGSRFT---WHSNP 587
++G DFN L A ++ R SVL RE + + L+DV + + T +
Sbjct: 138 LIIGGDFNYTLDARDRNVPKKRDSSESVL---RELIAHFSLVDVWREQNPETVAFTYVRV 194
Query: 588 RDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPILLDLS-PPTWSKTPF-KYE 645
RDG V++ +IDR+ + + +++ P SDH+ + L +S P+ K + +
Sbjct: 195 RDGHVSQSRIDRIYISSHLMSRAQSSTIRLAP--FSDHNCVSLRMSIAPSLPKAAYWHFN 252
Query: 646 VFWDEHEQCSEVVAEGWL--KPLHNE----DCWKD-----LENRIKSSKRTIWSWQRKTF 694
E E ++ V + W + +E + W D L+ + +++ +
Sbjct: 253 NSLLEDEGFAKSVRDTWRGWRAFQDEFATLNQWWDVGKVHLKLLCQEYTKSVSGQRNAEI 312
Query: 695 KVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDTLWRQEELFWGQRSRVKWA*YG 754
+ E+ L+Q L +D+ E K + + +++ RSR++
Sbjct: 313 EALNGEVLDLEQRLSGSEDQALQC----EYLERKEALRNMEQRQARGAFVRSRMQLLCDM 368
Query: 755 DKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQSEGVNQ--IE 812
D+ S+FF+A K+ NR +I + GT E + S Y+ +F + ++ E
Sbjct: 369 DRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNLFSPDPISPDACE 428
Query: 813 DCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDS 872
+ G P V+ RL P + E+++A+ L+ K+PG DGL FF+ W T+
Sbjct: 429 ELWDGLP-VVSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFFQFFWDTLGPD 487
Query: 873 VCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLK 932
+ E F L +++L+PK + +RP+S + K++++ + RLK
Sbjct: 488 FHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIVAKAISLRLK 547
Query: 933 DDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEW 992
L +I P+QS V GR I DN+ + ++ H +R + + +D K +DR++
Sbjct: 548 SVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTGLSL---AFLSLDQEKAFDRVDH 604
Query: 993 TFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVL 1052
+L TL AY F P + + T+ A +K+N L++ L GRG+RQG PLS L+ L
Sbjct: 605 QYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSL 664
Query: 1053 ATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSIIHILNLY 1112
A + +L + ++G + +ADD +L A+ V + +Y
Sbjct: 665 AIEPFLCLLRK-----RLTGLVLKEPDMRVVLSAYADDVILVAQDLV-DLERAQECQEVY 718
Query: 1113 NRASGQRINVAK-SGLIFGRSASNRVK---RDISNLLHMAIWDSP-GQYLGL-PAIWGRN 1166
AS RIN +K SGL+ G + + RDIS W+S +YLG+ +
Sbjct: 719 AAASSARINWSKSSGLLEGSLKVDFLPPAFRDIS-------WESKIIKYLGVYLSAEEYP 771
Query: 1167 KCHSLAWIEERVKEKLEGWK--ETLLNQAGKEVLIKAIIQAIPLYAMAMVHFPKTFCNSL 1224
+ +EE V +L WK +L+ G+ ++I ++ + Y + + + F +
Sbjct: 772 VSQNFIELEECVLTRLGKWKGFAKVLSMRGRALVINQLVASQIWYRLICLSPTQEFIAKI 831
Query: 1225 NSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMG-------FREFRMQNL-------- 1269
+ +F W G+ HW S +L EGG G FR+Q +
Sbjct: 832 QRRLLDFLWIGK------HWVSAGVSSLPLKEGGQGVVCIRSQVHTFRLQQIQRYLYADP 885
Query: 1270 ----AILAKQAWRVLTN-----------PEAL-----------------W--VCVLKSVY 1295
LA +R + N PE W V VL+
Sbjct: 886 SPQWCTLASSFYRQVRNMGYDRQLFIIEPEGFLRNLSTLPAYYQDTLKTWSMVSVLRQGA 945
Query: 1296 FPTSDFLNASLGRNPSWMWRSL--LAGRDFISRYIRWAVGGGLLIEVWGDNWLSSGEALI 1353
D LN L NPS+ R L ++ R + + VG L++ +W+ S +A++
Sbjct: 946 TEGEDILNEPLLYNPSFKTRMLESISIRRRLCQAQLTRVGD--LLDFEKSDWVDS-QAVM 1002
Query: 1354 Q---------PHNAPDMKVSELISPHGE--------------GWNSALVRSLFQPELALK 1390
Q PH ++ + ISP WNS+ + P+
Sbjct: 1003 QRMGFLTTRVPHRLL-KEIKDTISPDSHTFIDGVLHAGEPRPPWNSSPPDIIIAPKTRQS 1061
Query: 1391 VLQTPVSKLNQTDVLFWPHTPKGDYTVSSGFEIARSNSIKPQALSSRSHNIPELLWRTI- 1449
P L+Q + +P T D T + + +++ AL SR + +WR +
Sbjct: 1062 PQAPPSPNLSQLE--NFPLTRFHDITRKLLYSL-MLHTVHFLALISRY----DTIWRRVL 1114
Query: 1450 -----------WKSQLPQKVKVFLWKASHNSLAVKYNLWRKRILPSGQCPVCCEAQETVE 1498
+ S +P+ WK H +L+ L R P+ CP C + E+V
Sbjct: 1115 NEGERPQWRAFYSSLVPRPTGDLSWKVLHGALSTGEYLARFTDSPAA-CPFCGKG-ESVF 1172
Query: 1499 HMFFVC 1504
H +F C
Sbjct: 1173 HAYFTC 1178
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 155 bits (393), Expect = 7e-37
Identities = 187/907 (20%), Positives = 378/907 (41%), Gaps = 60/907 (6%)
Query: 403 ILSWNCRGVAATPTGRELRTLCSKNKPTIVFLMETRSKKENLEVIRRSLGFDYLFTVNPR 462
++S N G+ + L K PT L ET ++++ +R G+ +F N
Sbjct: 36 LISLNINGLNSPIKRHRLTDWLHKQDPTFCCLQETHLREKDRHYLRVK-GWKTIFQANGL 94
Query: 463 GLSGGLALFWKREVHIQILHSSQNYIHTSIFANNSVSDWDCTF--VYGDPSSQRRRFLWP 520
G+A+ ++ Q ++ I + + + +Y P+++ F+
Sbjct: 95 KKQAGVAILISDKIDFQPKVIKKDKEGHFILIKGKILQEELSILNIYA-PNARAATFIRD 153
Query: 521 QITALRFRDNTPWCLLGDFNEILFAHEKEGMRPQPPSVLQRFREFVHNSGLMDVDL---- 576
+ L+ ++GDFN L + ++ + + + E + L D+
Sbjct: 154 TLVKLKAYIAPHTIIVGDFNTPLSSKDRSWKQKLNRDTV-KLTEVMKQMDLTDIYRTFYP 212
Query: 577 KGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPILLDLSPPT 636
K +T+ S P F KID ++ + + N + N + P I SDH + L +
Sbjct: 213 KTKGYTFFSAPHGTF---SKIDHIIGHKTGLNRYKNIEIV--PCILSDHHGLRLIFNNNI 267
Query: 637 WSKTPFKYEVFWDEHEQC--SEVVAEGWLKPL-------HNE-----DCWKDLENRIKSS 682
+ P W + +V EG K + NE + W ++ ++
Sbjct: 268 NNGKP---TFTWKLNNTLLNDTLVKEGIKKEIKDFLEFNENEATTYPNLWDTMKAFLRGK 324
Query: 683 KRTIWSWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDTLWRQEELFW 742
+ + ++K + + L+ K+ E+ ++ EI+ + + +
Sbjct: 325 LIALSASKKKRETAHTSSLTTHLKALEKKEANSPKRSRRQEIIKLRGEINQVETRRTIQR 384
Query: 743 GQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEI 802
++R + +K K T R++ I+++ + +G E+ + S Y+ +
Sbjct: 385 INQTRSWFFEKINKIDKPLARLTKGHRDKILINKIRNEKGDITTDPEEIQNTIRSFYKRL 444
Query: 803 FQS--EGVNQIEDCLRGFP-RKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNG 859
+ + E +++++ L + K+ + + L P S EI +N L K+PGPDG +
Sbjct: 445 YSTKLENLDEMDKFLDRYQVPKLNQDQVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSA 504
Query: 860 LFFKNHWATIQDSVCYAIKEFFST*NL*AEIN----ETIVALVPKVDS-PESVVQFRPIS 914
F++ T ++ + + + F + + E + L+PK P + FRPIS
Sbjct: 505 EFYQ----TFKEDLIPILHKLFHKIEVEGTLPNSFYEATITLIPKPQKDPTKIENFRPIS 560
Query: 915 CCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMK 974
N K++++I+ R+++ + +I P+Q F+ G NI S H + + + K
Sbjct: 561 LMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYINKLKD--K 618
Query: 975 DYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLT 1034
++M I +D K +D+++ F+ K L G + + + +KVNG +
Sbjct: 619 NHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMIKAIYSKPVANIKVNGEKLEAIP 678
Query: 1035 PGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLF 1094
G RQG PLSPYLF + +VL+ + Q + EI G + ++ L ADD +++
Sbjct: 679 LKSGTRQGCPLSPYLFNIVLEVLARAIRQQK---EIKGIQIGKEEVKISLL--ADDMIVY 733
Query: 1095 AEATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPG 1154
+++++N + G +IN KS + F + + + +++I +I +
Sbjct: 734 ISDPKNSTRELLNLINSFGEVVGYKINSNKS-MAFLYTKNKQAEKEIRETTPFSIVTNNI 792
Query: 1155 QYLGLPAIWGRNKCH--SLAWIEERVKEKLEGWKETLLNQAGKEVLIKAII--QAIPLYA 1210
+YLG+ + + +++ +KE L WK+ + G+ ++K I +AI +
Sbjct: 793 KYLGVTLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLPCSWIGRINIVKMAILPKAIYRFN 852
Query: 1211 MAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGFREFRMQNLA 1270
+ P F N L + F W + K R +K T GG+ + ++ A
Sbjct: 853 AIPIKIPTQFFNELEGAICKFVWNNK-KPRIAKSLLKDKRT----SGGITMPDLKLYYRA 907
Query: 1271 ILAKQAW 1277
I+ K AW
Sbjct: 908 IVIKTAW 914
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 155 bits (392), Expect = 9e-37
Identities = 196/914 (21%), Positives = 376/914 (40%), Gaps = 70/914 (7%)
Query: 401 MMILSWNCRGVAATPTGRELRTLCSKNKPTIVFLME---TRSKKENLEVIRRSLGFDYLF 457
+ I S N G+ L K KP I + E T K L+V G+ +F
Sbjct: 7 LSIFSINVNGLNCPLKRHRLADWIQKLKPDICCIQESHLTLKDKYRLKV----KGWSSIF 62
Query: 458 TVNPRGLSGGLALFWKREVHIQILHSSQNYIHTSIFA--NNSVSDWDCTFVYGDPSSQRR 515
N + G+A+ + + + ++ IF N + +Y P+
Sbjct: 63 QANGKQKKAGIAILFADAIGFKPTKIRKDKDGHFIFVKGNTQYDEISIINIYA-PNHNAP 121
Query: 516 RFLWPQITALRFRDNTPWCLLGDFNEILFAHEKEGMRPQPPSVLQRFREFVHNSGLMDVD 575
+F+ +T + ++ ++GDFN L ++ + +L NS + +D
Sbjct: 122 QFIRETLTDMSNLISSTSIVVGDFNTPLAVLDRSSKKKLSKEILDL------NSTIQHLD 175
Query: 576 LKGSRFTWHSNPRD------GFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPIL 629
L T+H N + T KID +L + S + F + P I SDH I
Sbjct: 176 LTDIYRTFHPNKTEYTFFSSAHGTYSKIDHILGHKSNLSKFKKIEII--PCIFSDHHGIK 233
Query: 630 LDLS--------PPTWSKTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDC-WKDLENRIK 680
++L+ TW + W E E+ +L+ +N+D +++L + K
Sbjct: 234 VELNNNRNLHTHTKTWKLNNLMLKDTWVIDEIKKEITK--FLEQNNNQDTNYQNLWDTAK 291
Query: 681 SSKRTIWSWQRKTFKVAEKE-----IGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDTLW 735
+ R + + K E+E +G LKQ + + P E+ I+ E++ +
Sbjct: 292 AVLRGKFIALQAFLKKTEREEVNNLMGHLKQLEKEEHSNPKPSRR-KEITKIRAELNEIE 350
Query: 736 RQEELFWGQRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAF 795
+ + +S+ + +K K T K+R ++ I + + +E+
Sbjct: 351 NKRIIQQINKSKSWFFEKINKIDKPLANLTRKKRVKSLISSIRNGNDEITTDPSEIQKIL 410
Query: 796 VSHYEEIFQS--EGVNQIEDCLRG--FPRKVTAELNNRLIMPASDAEIAEAVNLLGGLKA 851
+Y++++ E + +I+ L PR E+ L P S +EIA + L K+
Sbjct: 411 NEYYKKLYSHKYENLKEIDQYLEACHLPRLSQKEVE-MLNRPISSSEIASTIQNLPKKKS 469
Query: 852 PGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDS-PESVVQF 910
PGPDG F++ + + + L E + L+PK P +
Sbjct: 470 PGPDGFTSEFYQTFKEELVPILLNLFQNIEKEGILPNTFYEANITLIPKPGKDPTRKENY 529
Query: 911 RPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQR-G 969
RPIS N K++++I+ R++ + +I +Q F+ G NI +++ +++Q
Sbjct: 530 RPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNI---RKSINVIQHIN 586
Query: 970 ESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFL 1029
+ + KD+M + +D K +D ++ F+ +TL G + + + T + +NG
Sbjct: 587 KLKNKDHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNGVK 646
Query: 1030 SSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFAD 1089
+ G RQG PLSP LF + +VL+ + + + I G L+ FAD
Sbjct: 647 LKSFPLRSGTRQGCPLSPLLFNIVMEVLAIAI---REEKAIKGIHIGSEEIKLS--LFAD 701
Query: 1090 DSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAI 1149
D +++ E T T ++ ++ Y+ SG +IN KS + F + +N+ ++ + + + +
Sbjct: 702 DMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKS-VAFIYTNNNQAEKTVKDSIPFTV 760
Query: 1150 WDSPGQYLGLPAIWGRNKCH--SLAWIEERVKEKLEGWKETLLNQAGKEVLIKAII--QA 1205
+YLG+ + + + + + E + WK + G+ ++K I +A
Sbjct: 761 VPKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSWLGRINIVKMSILPKA 820
Query: 1206 IPLYAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDE--GGMGFRE 1263
I + + P ++ L ++ +F W Q K + K LS GG+ +
Sbjct: 821 IYNFNAIPIKAPLSYFKDLEKIILHFIW-NQKKPQ------IAKTLLSNKNKAGGITLPD 873
Query: 1264 FRMQNLAILAKQAW 1277
R+ +I+ K AW
Sbjct: 874 LRLYYKSIVIKTAW 887
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 149 bits (375), Expect = 9e-35
Identities = 191/908 (21%), Positives = 374/908 (41%), Gaps = 62/908 (6%)
Query: 403 ILSWNCRGVAATPTGRELRTLCSKNKPTIVFLMETRSKKENLEVIRRSLGFDYLFTVNPR 462
IL+ N G+ A L P++ + ET + ++ G+ ++ N +
Sbjct: 9 ILTLNVNGLNAPIKRHRLANWIKSQDPSVCCIQETHLTCRDTHRLKIK-GWRNIYQANGK 67
Query: 463 GLSGGLALFWKREVHIQILHSSQNYIHTSIFANNSVSDWDCTF--VYGDPSSQRRRFLWP 520
G+A+ + + ++ I S+ + T +Y P++ RF+
Sbjct: 68 QKKAGVAILVSDKTDFKPTKIKRDKEGHYIMVKGSIQQEELTILNIYA-PNTGAPRFIKQ 126
Query: 521 QITALRFRDNTPWCLLGDFNEILFAHEKEGMRPQPPSVLQRFREFVHNSGLMDV----DL 576
++ L+ ++ ++GDFN L ++ R + +Q +H + L+D+
Sbjct: 127 VLSDLQRDLDSHTIIMGDFNTPLSTLDRS-TRQKINKDIQELNSALHQADLIDIYRTLHP 185
Query: 577 KGSRFTWHSNPRDGFVTREKIDRVLTNWSWRNIFPNASAMAYPQISSDHSPILLDLS--- 633
K + +T+ S P T K D +L + + + + SDHS I L+L
Sbjct: 186 KSTEYTFFSAPHH---TYSKTDHILGSKTLLSKCKRTEIIT--NCLSDHSAIKLELRIKK 240
Query: 634 -----PPTWSKTPFKYEVFWDEHEQCSEVVAEGWLKPLHNEDC-----WKDLENRIKSSK 683
TW +W +E +E+ + + + N+D W + +
Sbjct: 241 LTQNHSTTWKLNNLLLNDYWVHNEMKAEI--KKFFETNENKDTTYQNLWDTAKAVCRGKF 298
Query: 684 RTIWSWQRKTFKVAEKEIGRLKQELQSKQDRPNHLVDWNEVKNIKLEIDTLWRQEELFWG 743
+ + +RK + + +EL+ ++ + E+ I+ E+ + Q+ L
Sbjct: 299 IALNAHKRKQERSKIDTLISQLKELEKQEQTNSKASRRQEIIKIRAELKEIETQKTLQKI 358
Query: 744 QRSRVKWA*YGDKNSKFFHASTLKRRNRNRIDRVCDAQGTWKEGQTEVMHAFVSHYEEIF 803
SR + +K + K+R +N+ID + + +G TE+ +Y+ ++
Sbjct: 359 NESRSWFFEKINKIDRPLARLIKKKREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLY 418
Query: 804 QS--EGVNQIEDCLRGF--PRKVTAELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNG 859
+ E + +++ L + PR E+ + L P + +EI +N L K+PGP+G
Sbjct: 419 ANKLENLEEMDKFLDTYTLPRLNQEEVES-LNRPITSSEIEAIINSLPNKKSPGPEGFTA 477
Query: 860 LFFKNHWATIQDSVCYAIKEFFST*N---L*AEINETIVALVPKVDSPESVVQ-FRPISC 915
F++ + ++ V + +K F S L E + L+PK + + FRPIS
Sbjct: 478 EFYQRYK---EELVPFLLKLFQSIEKEGILPNSFYEASIILIPKPGRDTTKKENFRPISL 534
Query: 916 CNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKD 975
N K++++I+ +++ + LI +Q F+ NI S + R ++ +
Sbjct: 535 MNIDAKILNKILANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKSINIIQHINR--TKDTN 592
Query: 976 YMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTP 1035
+M I +D K +D+++ F+ K L G + + + T + +NG
Sbjct: 593 HMIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPL 652
Query: 1036 GRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFA 1095
G RQG PLSP L + +VL+ + Q + EI G + L+ FADD +++
Sbjct: 653 KTGTRQGCPLSPLLPNIVLEVLARAIRQEK---EIKGIQLGKEEVKLS--LFADDMIVYL 707
Query: 1096 EATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNRVKRDISNLLHMAIWDSPGQ 1155
E + A +++ +++ +++ SG +INV KS F + + + + I + L I +
Sbjct: 708 ENPIVSAQNLLKLISNFSKVSGYKINVQKS-QAFLYTNNRQTESQIMSELPFTIASKRIK 766
Query: 1156 YLGLPAIWGRNKCHSLAW--IEERVKEKLEGWKETLLNQAGKEVLIKAII--QAIPLYAM 1211
YLG+ + + +KE WK + G+ ++K I + I +
Sbjct: 767 YLGIQLTRDVKDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRINIVKMAILPKVIYRFNA 826
Query: 1212 AMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSKDE--GGMGFREFRMQNL 1269
+ P TF L F W + H K TLS+ GG+ +F++
Sbjct: 827 IPIKLPMTFFTELEKTTLKFIW----NQKRAH---IAKSTLSQKNKAGGITLPDFKLYYK 879
Query: 1270 AILAKQAW 1277
A + K AW
Sbjct: 880 ATVTKTAW 887
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 125 bits (314), Expect = 1e-27
Identities = 57/143 (39%), Positives = 86/143 (59%), Gaps = 1/143 (0%)
Query: 1205 AIPLYAMAMVHFPKTFCNSLNSLVANFWWKGQGKDRGIHWRSWEKMTLSK-DEGGMGFRE 1263
A+P+YAM+ K C L S + FWW R I W +W+K+ SK D+GG+GFR+
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 1264 FRMQNLAILAKQAWRVLTNPEALWVCVLKSVYFPTSDFLNASLGRNPSWMWRSLLAGRDF 1323
N A+LAKQ++R++ P L +L+S YFP S + S+G PS+ WRS++ GR+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 1324 ISRYIRWAVGGGLLIEVWGDNWL 1346
+SR + +G G+ +VW D W+
Sbjct: 122 LSRGLLRTIGDGIHTKVWLDRWI 144
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 80.5 bits (197), Expect = 4e-14
Identities = 71/246 (28%), Positives = 105/246 (41%), Gaps = 16/246 (6%)
Query: 851 APGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEINETIVALVPKVDSPESVVQF 910
+PGPDG+ + + I + I NL I +PK + + F
Sbjct: 359 SPGPDGITPKSAREVPSGIMLRIMNLI---LWCGNLPHSIRLARTVFIPKTVTAKRPQDF 415
Query: 911 RPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGE 970
RPIS + +++ ++ I+ TRL +N P Q F+ DN T+ +L+
Sbjct: 416 RPISVPSVLVRQLNAILATRLNSSINW--DPRQRGFLPTDGCADNATIVDL---VLRHSH 470
Query: 971 SRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLS 1030
+ +D+SK +D L + TL AYG + V +G L +G+ S
Sbjct: 471 KHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQNTYEGGGTSLNGDGWSS 530
Query: 1031 STLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADD 1090
P RG++QGDPLSP LF L D L + T G G T + FADD
Sbjct: 531 EEFVPARGVKQGDPLSPILFNLVMDRL--LRTLPSEIGAKVGNAITNAAA------FADD 582
Query: 1091 SLLFAE 1096
+LFAE
Sbjct: 583 LVLFAE 588
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 79.3 bits (194), Expect = 9e-14
Identities = 36/67 (53%), Positives = 47/67 (69%)
Query: 1025 VNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTH 1084
+NG +TP RGLRQGDPLSPYLF+L T+VLS + +AQ G + G R + +SP + H
Sbjct: 14 INGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRINH 73
Query: 1085 LFFADDS 1091
L FADD+
Sbjct: 74 LLFADDT 80
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 72.4 bits (176), Expect = 1e-11
Identities = 42/160 (26%), Positives = 80/160 (49%), Gaps = 4/160 (2%)
Query: 894 IVALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQ 953
++ +PK +P S +RPIS + +++ RI+ +R++ + + L+SP+Q F+ R
Sbjct: 669 VIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCP 728
Query: 954 DNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVL 1013
++ S +H + + E K + D +K +D++ L K L +G + +
Sbjct: 729 SSLVRSISLYHSILKNE---KSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFK 785
Query: 1014 TLVKGATYKLKVNGFLSSTLTP-GRGLRQGDPLSPYLFVL 1052
+ T+ +K+N F+SS P G+ QG P LF+L
Sbjct: 786 EFLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFIL 825
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 68.9 bits (167), Expect = 1e-10
Identities = 61/230 (26%), Positives = 107/230 (46%), Gaps = 13/230 (5%)
Query: 897 LVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNI 956
L+PK E+ +RPI+ + + +++ RI+ RL+ + L + A + G L+ +
Sbjct: 63 LIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVELHPAQKGYARIDGTLVNSLL 122
Query: 957 TVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLV 1016
+ ++ R E R K Y + +D+ K +D + + + + L G + + +
Sbjct: 123 LDT----YISSRREQR-KTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYITGSL 177
Query: 1017 KGATYKLKVN-GFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRF 1075
+T ++V G + + RG++QGDPLSP+LF VL +L Q I G
Sbjct: 178 SDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLF---NAVLDELLCSLQSTPGIGG--- 231
Query: 1076 TPHSPPLTHLFFADDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKS 1125
T + L FADD LL + V T++ + N + R G +N KS
Sbjct: 232 TIGEEKIPVLAFADDLLLLEDNDVLLPTTLATVANFF-RLRGMSLNAKKS 280
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 60.8 bits (146), Expect = 3e-08
Identities = 83/340 (24%), Positives = 141/340 (41%), Gaps = 43/340 (12%)
Query: 773 RIDRVCDAQGTWKEGQTEVMHAFVSHYEEIFQS--EGVNQIEDCLR-GFPR------KVT 823
R + ++ +WK+ ++ H + + + EG +R G P ++
Sbjct: 250 RRQTIKESNNSWKKNMSKAAHIVLDGDTDACPAGLEGTEASGAIMRAGCPTTRHLRSRMQ 309
Query: 824 AELNNRLIMPASDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST 883
E+ N L P S+ EI E A GPDG+ T +S+ IK F+
Sbjct: 310 GEIKN-LWRPISNDEIKEVEACKR--TAAGPDGMT--------TTAWNSIDECIKSLFNM 358
Query: 884 *NL*AEIN----ETIVALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLKDDLNLLI 939
+ ++ L+PK FRP+S + L+ RI+ R+ + + L+
Sbjct: 359 IMYHGQCPRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLL 416
Query: 940 SPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKD---YMAIKVDMSKTYDRLEWTFLE 996
Q AF+ + +N ++ E+RMK Y+AI +D+ K +D +E +
Sbjct: 417 DTRQRAFIVADGVAENTSLLSAMIK-----EARMKIKGLYIAI-LDVKKAFDSVEHRSIL 470
Query: 997 KTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDV 1056
L ++ + + + +L+V + P RG+RQGDPLSP LF D
Sbjct: 471 DALRRKKLPLEMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCVMDA 530
Query: 1057 LSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAE 1096
+ +R E +GF + + L FADD +L AE
Sbjct: 531 V------LRRLPENTGFLM--GAEKIGALVFADDLVLLAE 562
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 57.4 bits (137), Expect = 3e-07
Identities = 44/173 (25%), Positives = 74/173 (42%), Gaps = 7/173 (4%)
Query: 964 HLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKL 1023
H ++ + K Y + +D+ K +D + + + + A+G +++ + A +
Sbjct: 11 HYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMSTITDAYTNI 70
Query: 1024 KVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLT 1083
V G ++ + G++QGDPLSP LF + D L L Q G TP + +
Sbjct: 71 VVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQ-----PGASMTP-ACKIA 124
Query: 1084 HLFFADDSLLFAEATVKEATSIIHILNLYNRASGQRINVAKSGLIFGRSASNR 1136
L FADD LL + + S+ Y R G +N K I + S R
Sbjct: 125 SLAFADDLLLLEDRDIDVPNSLATTC-AYFRTRGMTLNPEKCASISAATVSGR 176
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 48.5 bits (114), Expect = 2e-04
Identities = 44/160 (27%), Positives = 74/160 (45%), Gaps = 12/160 (7%)
Query: 838 EIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEIN-ETIVA 896
E++++V K+PGPDG+ G + W I + + K+ + ++V
Sbjct: 409 EVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIASLVI 468
Query: 897 LVPKVDSPES-VVQFRPISCCNYILKVISRIMVTRLKDDL-NLLISPNQSAFVGGRLIQD 954
L+ +D S +RPI + + KV+ IMV RL L ++ +SP Q AF G+ +D
Sbjct: 469 LLKLLDRIRSDPGSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKSTED 528
Query: 955 NITVSQEAFHLLQRGE--SRMKDYMAIKVDMSKTYDRLEW 992
A+ +QR S MK + + +D +D L W
Sbjct: 529 -------AWRCVQRHVECSEMKYVIGLNIDFQGAFDNLGW 561
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 43.9 bits (102), Expect = 0.004
Identities = 79/344 (22%), Positives = 134/344 (37%), Gaps = 43/344 (12%)
Query: 791 VMHAFVSHYEEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVNLLGGLK 850
V H F +++++F + G + + R P A L P S EI A K
Sbjct: 121 VRHMF-DYWKDVFATGGGSAATNINRAPP----APHMETLWDPVSLIEIKSA--RASNEK 173
Query: 851 APGPDGLNGLFFKNHWATIQDS---VCYAIKEFFST*NL*AEINETIVALVPKVDSPESV 907
GPDG+ W + D + Y I F+ + + I + PK++
Sbjct: 174 GAGPDGVT----PRSWNALDDRYKRLLYNIFVFYG--RVPSPIKGSRTVFTPKIEGGPDP 227
Query: 908 VQFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQ 967
FRP+S C+ IL+ ++I+ R Q+A+ L D + ++ +
Sbjct: 228 GVFRPLSICSVILREFNKILARRFVSCYT--YDERQTAY----LPIDGVCINVSMLTAII 281
Query: 968 RGESRMKDYMAIKV-DMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYKLKVN 1026
R++ + I + D+ K ++ + + L + G P + + +++
Sbjct: 282 AEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEAGCPPGVVDYIADMYNNVITEMQFE 341
Query: 1027 G--FLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEISGFRFTPHSPPLTH 1084
G L+S L G+ QGDPLS LF LA + L +G RF +
Sbjct: 342 GKCELASILA---GVYQGDPLSGPLFTLAYEK---ALRALNNEG-----RFDIADVRVNA 390
Query: 1085 LFFADDSLLFAEATVKEATSIIHILNLYNRA---SGQRINVAKS 1125
++DD LL A + + H L+ + G RIN KS
Sbjct: 391 SAYSDDGLLLAMTVI----GLQHNLDKFGETLAKIGLRINSRKS 430
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 38.9 bits (89), Expect = 0.13
Identities = 65/310 (20%), Positives = 118/310 (37%), Gaps = 41/310 (13%)
Query: 838 EIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL*AEIN-ETIVA 896
E+ V L ++PG DG+NG K W I + + AE +V+
Sbjct: 438 EVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVS 497
Query: 897 LVPKVDSPE-SVVQFRPISCCNYILKVISRIMVTRLKDDLNLLISPN----QSAFVGGRL 951
L+ D + +R I KV+ IMV R+++ + P Q F GR
Sbjct: 498 LLKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVRE-----VLPEGCRWQFGFRQGRC 552
Query: 952 IQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFLEKTLLAYGFHPIWTAR 1011
++D + + G S + + VD +D +EW+ L G + +
Sbjct: 553 VEDAWRHVKSSV-----GASAAQYVLGTFVDFKGAFDNVEWSAALSRLADLGCREMGLWQ 607
Query: 1012 VLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATDVLSFMLTQAQRDGEIS 1071
+ A + +G + +T RG QG P+++ + DVL L + Q ++S
Sbjct: 608 SFFSGRRAVIR-SSSGTVEVPVT--RGCPQGSISGPFIWDILMDVL---LQRLQPYCQLS 661
Query: 1072 GFRFTPHSPPLTHLFFADDSLLFAEATVK-----EATSIIHILNLYNRASGQRINVAKSG 1126
+ADD LL E + + ++ I+ + G ++ +K+
Sbjct: 662 A--------------YADDLLLLVEGNSRAVLEEKGAQLMSIVETWGAEVGDCLSTSKTV 707
Query: 1127 LIFGRSASNR 1136
++ + A R
Sbjct: 708 IMLLKGALRR 717
>RTP2_TRYBG (P15594) Retrotransposable element SLACS 132 kDa protein
(ORF2)
Length = 1182
Score = 33.9 bits (76), Expect = 4.1
Identities = 43/154 (27%), Positives = 67/154 (42%), Gaps = 26/154 (16%)
Query: 909 QFRPISCCNYILKVISRIMVTRLKDDLNLLISPNQSAFVGGRLIQDNITVSQEAFHLLQR 968
++RPI + K+ S I ++R+ S Q VGG I++ I ++ F +
Sbjct: 585 KYRPIGAESVWAKLASHIAISRVMKTAEKKFSGIQFG-VGGH-IEEAIAKIRKDF--ATK 640
Query: 969 GESRMKD----YMAI--KVDMSKTYDRLEWTFLEKTLLAYGFHPIWTARVLTLVKGATYK 1022
G M D Y AI + + Y W+ P+W R+++L+ G T +
Sbjct: 641 GSLAMLDGRNAYNAISRRAILEAVYGDSTWS------------PLW--RLVSLLLGTTGE 686
Query: 1023 LKV--NGFLSSTLTPGRGLRQGDPLSPYLFVLAT 1054
+ NG L T RG+RQG L P LF + T
Sbjct: 687 VGFYENGKLCHTWESTRGVRQGMVLGPLLFSIGT 720
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 33.9 bits (76), Expect = 4.1
Identities = 68/297 (22%), Positives = 114/297 (37%), Gaps = 44/297 (14%)
Query: 782 GTWKEGQTEVMHAFVSHYEEIFQSEGVNQIEDCLRGFPRKVTAELNN--RLIMPASDAEI 839
G W E F ++ E+ F C R+V L + ++ +P S +
Sbjct: 386 GGWCRTSLEKTEVFANNLEQRFTPYNYAPESLC-----RQVEEYLESPFQMSLPLSAVTL 440
Query: 840 AEAVNLLGGL---KAPGPDGLNGLFFK-------NHWATIQDSVC---YAIKEFFST*NL 886
E NL+ L KAPG D L+ + A I +SV Y K + S
Sbjct: 441 EEVKNLIAKLPLKKAPGEDLLDNRTIRLLPDQALQFLALIFNSVLDVGYFPKAWKSA--- 497
Query: 887 *AEINETIVALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRLKD--DLNLLISPNQS 944
+I+ + +P V +RP S + K++ R+++ RL D+ I Q
Sbjct: 498 ------SIIMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQF 551
Query: 945 AFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEW----TFLEKTLL 1000
F RL + + + +D+ + +DR+ W + K L
Sbjct: 552 GF---RLQHGTPEQLHRVVNFALEAMENKEYAVGAFLDIQQAFDRV-WHPGLLYKAKRL- 606
Query: 1001 AYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLF-VLATDV 1056
F P V + ++ T+ + V+G+ SS G+ QG L P L+ V A+D+
Sbjct: 607 ---FPPQLYLVVKSFLEERTFHVSVDGYKSSIKPIAAGVPQGSVLGPTLYSVFASDM 660
>SYA_BIFLO (Q8G5W9) Alanyl-tRNA synthetase (EC 6.1.1.7) (Alanine--tRNA
ligase) (AlaRS)
Length = 893
Score = 33.5 bits (75), Expect = 5.4
Identities = 15/35 (42%), Positives = 23/35 (64%), Gaps = 1/35 (2%)
Query: 1227 LVANFWWKGQGKDRGIHWRSWEKMTLSKDEGGMGF 1261
++ NF + K+ IH+ +WE +T S+DEGG GF
Sbjct: 86 MLGNFSFGDYFKEEAIHY-AWELLTTSQDEGGYGF 119
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 33.1 bits (74), Expect = 7.0
Identities = 61/291 (20%), Positives = 113/291 (37%), Gaps = 37/291 (12%)
Query: 782 GTWKEGQTEVMHAFVSHYEEIF-------QSEGVNQIEDCLRGFPRKVTAELNNRLIMPA 834
G W E F +H E+ F +S + E F + A+ P
Sbjct: 386 GGWCRTSREKTEVFANHLEQRFKALAFAPESHSLMVAESLQTPFQMALPAD-------PV 438
Query: 835 SDAEIAEAVNLLGGLKAPGPDGLNGLFFKNHWATIQDSVCYAIKEFFST*NL----*AEI 890
+ E+ E V+ L KAPG D L+ + ++ Y + F S + A
Sbjct: 439 TLEEVKELVSKLKPKKAPGEDLLDNRTIR---LLPDQALLYLVLIFNSILRVGYFPKARP 495
Query: 891 NETIVALVPKVDSPESVVQFRPISCCNYILKVISRIMVTRL--KDDLNLLISPNQSAFVG 948
+I+ ++ P V +RP S + K++ R+++ R+ +++ I Q F
Sbjct: 496 TASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRILTSEEVTRAIPKFQFGFRL 555
Query: 949 GRLIQDNI-TVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEW----TFLEKTLLAYG 1003
+ + V A L++ E ++ D+ + +DR+ W + K+LL+
Sbjct: 556 QHGTPEQLHRVVNFALEALEKKEYAGSCFL----DIQQAFDRV-WHPGLLYKAKSLLS-- 608
Query: 1004 FHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLAT 1054
P + + +G + + +G SS G+ QG L P L+ + T
Sbjct: 609 --PQLFQLIKSFWEGRKFSVTADGCRSSVKFIEAGVPQGSVLGPTLYSIFT 657
>MURB_CAMJE (Q9PM01) UDP-N-acetylenolpyruvoylglucosamine reductase (EC
1.1.1.158) (UDP-N-acetylmuramate dehydrogenase)
Length = 258
Score = 33.1 bits (74), Expect = 7.0
Identities = 55/214 (25%), Positives = 83/214 (38%), Gaps = 50/214 (23%)
Query: 936 NLLISPNQSAFVGGRLIQDNITVSQEAFHLLQRGESRMKDYMAIKVDMSKTYDRLEWTFL 995
NLL+SP NI + + F+ +Q + R KD++ +++ KT + F
Sbjct: 38 NLLVSPKPK----------NIGILGDGFNFIQILD-RNKDFIHLRIGC-KTKSSKMYRFA 85
Query: 996 EKTLLAYGFHPIWTARVLTLVKGATYKLKVNGFLSSTLTPGRGLRQGDPLSPYLFVLATD 1055
++ L KG Y K+ G L L GL+ G+ +S L +AT
Sbjct: 86 KENNL----------------KGFEYLSKIPGTLGGLLKMNAGLK-GECISQNLIKIATS 128
Query: 1056 VLSFMLTQAQRDGEISGFRFTPHSPPLTHLFFADDSLLFAEATVKEATSIIHILNLYNRA 1115
+ D +RF P + TH F+A+ L F T+K+ L N
Sbjct: 129 QGEILRANINFD-----YRFCPLN---THFFWAEFKLNFGFDTLKDEA-------LKNAR 173
Query: 1116 SGQRINVAKSGLIFGRSASNRVKRDISNLLHMAI 1149
S Q SG FG S K D + L A+
Sbjct: 174 SNQ-----PSGASFG-SIFKNPKNDFAGRLIEAV 201
>FLGJ_VIBPA (Q9X9J3) Peptidoglycan hydrolase flgJ (EC 3.2.1.-)
(Muramidase flgJ)
Length = 308
Score = 33.1 bits (74), Expect = 7.0
Identities = 15/65 (23%), Positives = 29/65 (44%)
Query: 785 KEGQTEVMHAFVSHYEEIFQSEGVNQIEDCLRGFPRKVTAELNNRLIMPASDAEIAEAVN 844
K+G+ E +HA +E IF S + + + GF + N + D ++A ++
Sbjct: 29 KDGEQEALHAAARQFESIFTSMMLKSMREANEGFESNIMNSQNEKFYRQMLDEQMASELS 88
Query: 845 LLGGL 849
G +
Sbjct: 89 ANGSM 93
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.334 0.144 0.487
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 204,344,427
Number of Sequences: 164201
Number of extensions: 8544964
Number of successful extensions: 28605
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 28558
Number of HSP's gapped (non-prelim): 36
length of query: 1771
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1647
effective length of database: 39,613,130
effective search space: 65242825110
effective search space used: 65242825110
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0288.7


