

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
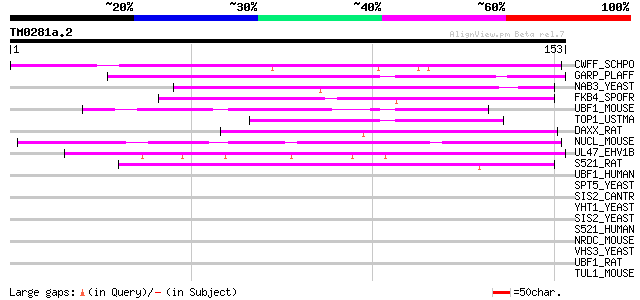
Query= TM0281a.2
(153 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CWFF_SCHPO (P78794) Cell cycle control protein cwf15 78 7e-15
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 49 3e-06
NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3 49 6e-06
FKB4_SPOFR (Q26486) 46 kDa FK506-binding nuclear protein (EC 5.2... 46 4e-05
UBF1_MOUSE (P25976) Nucleolar transcription factor 1 (Upstream b... 44 1e-04
TOP1_USTMA (P41511) DNA topoisomerase I (EC 5.99.1.2) 43 2e-04
DAXX_RAT (Q8VIB2) Death domain-associated protein 6 (Daxx) 42 4e-04
NUCL_MOUSE (P09405) Nucleolin (Protein C23) 42 5e-04
UL47_EHV1B (P28929) 97 kDa alpha trans-inducing protein (Tegumen... 42 7e-04
S521_RAT (Q9QY02) Putative splicing factor YT521 (RA301-binding ... 42 7e-04
UBF1_HUMAN (P17480) Nucleolar transcription factor 1 (Upstream b... 41 0.001
SPT5_YEAST (P27692) Transcription initiation protein SPT5 41 0.001
SIS2_CANTR (Q12600) SIS2 protein (Halotolerance protein HAL3) 41 0.001
YHT1_YEAST (P38835) Hypothetical 95.1 kDa protein in ACT5-YCK1 i... 40 0.002
SIS2_YEAST (P36024) SIS2 protein (Halotolerance protein HAL3) 40 0.002
S521_HUMAN (Q96MU7) Putative splicing factor YT521 40 0.002
NRDC_MOUSE (Q8BHG1) Nardilysin precursor (EC 3.4.24.61) (N-argin... 40 0.002
VHS3_YEAST (Q08438) Protein VHS3 (Viable in a HAL3 SIT4 backgrou... 40 0.002
UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream bin... 40 0.002
TUL1_MOUSE (Q9Z273) Tubby related protein 1 (Tubby-like protein 1) 40 0.002
>CWFF_SCHPO (P78794) Cell cycle control protein cwf15
Length = 265
Score = 78.2 bits (191), Expect = 7e-15
Identities = 60/192 (31%), Positives = 94/192 (48%), Gaps = 46/192 (23%)
Query: 1 MTTAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLR 60
MTTA RP + PA+G +E TRI SSR + +H LK R++ Q T++E+++++LR
Sbjct: 1 MTTAHRPQFDPARGHSEMAPTRIT------SSRALPAHLKLKYRQESQGTEEEVRKQDLR 54
Query: 61 DELEERERRHF-------SSKNKSYSDDRDHGKSSHLFLEGTKRDVE---DHIVARSVDA 110
+ L E HF SS+ S + G +S + DVE ++ ++++A
Sbjct: 55 EALLRAEAAHFATQEHGASSEEVSQNSKLIEGFTSPSTDDKPNNDVEVDYQELLRQTLEA 114
Query: 111 D----DSD--------------------------VEVKSDEESDDDDDDEDDTEALLAEL 140
D DSD V+ + E SD++ D ED+T+ LL EL
Sbjct: 115 DEDASDSDDSVDSSNKNSEVSIKRRKTESNSQESVDSSNSESSDEESDSEDETQQLLREL 174
Query: 141 EQIKKERAEEKL 152
E IK+ER E++
Sbjct: 175 ENIKQERKREQM 186
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 49.3 bits (116), Expect = 3e-06
Identities = 30/126 (23%), Positives = 60/126 (46%), Gaps = 7/126 (5%)
Query: 28 QKYSSRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKS 87
+K S+++ + + + +DE + +E EE E + + ++ D +
Sbjct: 551 KKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEE 610
Query: 88 SHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKER 147
E + D E+ DA++ D E DEE DD+D+DED+ + E E+ ++E
Sbjct: 611 DEDDAEEDEDDAEED----EDDAEEDDDEEDDDEEDDDEDEDEDEED---EEEEEEEEEE 663
Query: 148 AEEKLR 153
+E+K++
Sbjct: 664 SEKKIK 669
Score = 36.6 bits (83), Expect = 0.022
Identities = 21/105 (20%), Positives = 46/105 (43%)
Query: 48 QDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARS 107
+D ++E + +E EE E + + D+ + + E + ED
Sbjct: 574 EDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDD 633
Query: 108 VDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKL 152
+ DD + + DE+ D++D++E++ E +E + + R K+
Sbjct: 634 DEEDDDEEDDDEDEDEDEEDEEEEEEEEEESEKKIKRNLRKNAKI 678
Score = 35.0 bits (79), Expect = 0.064
Identities = 25/119 (21%), Positives = 55/119 (46%), Gaps = 10/119 (8%)
Query: 43 PRRDGQDT------QDELKRRNLRDELEER--ERRHFSSKNKSYSDDRDHGKSSHLFLEG 94
PRRD + EL+++ D+ E++ E + ++K +D + + E
Sbjct: 522 PRRDNHKKKMAKIEEAELQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEE 581
Query: 95 TKRDVEDHIVARSVDADDSDVEVKSDEESDDDD--DDEDDTEALLAELEQIKKERAEEK 151
+ + E+ + ++ + E + ++E D+DD +DEDD E + E+ E +++
Sbjct: 582 EEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDE 640
Score = 33.9 bits (76), Expect = 0.14
Identities = 30/123 (24%), Positives = 55/123 (44%), Gaps = 20/123 (16%)
Query: 45 RDGQDTQDELKRRNLRDE---LEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVED 101
++G+ + E K ++E EE + + SK K +D GK + K V+
Sbjct: 385 KEGEHKEGEHKEEEHKEEEHKKEEHKSKEHKSKGKKDKGKKDKGKHK----KAKKEKVKK 440
Query: 102 HIVARSVDADDSD-VEVKSDEESDDDDDDEDDTEA---------LLAELEQI---KKERA 148
H+V ++ +D D VE+ + E+ + ++ E+ L+ E EQ+ K +
Sbjct: 441 HVVKNVIEDEDKDGVEIINLEDKEACEEQHITVESRPLSQPQCKLIDEPEQLTLMDKSKV 500
Query: 149 EEK 151
EEK
Sbjct: 501 EEK 503
Score = 28.5 bits (62), Expect = 6.0
Identities = 19/83 (22%), Positives = 42/83 (49%), Gaps = 5/83 (6%)
Query: 72 SSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADD---SDVEVKSDEESDDDDD 128
SS +++ +D +D+ K + E K D + + +++D + + E+K E+ +
Sbjct: 226 SSVHETSNDTKDNDKEN--ISEDKKEDHQQEEMLKTLDKKERKQKEKEMKEQEKIEKKKK 283
Query: 129 DEDDTEALLAELEQIKKERAEEK 151
+++ E E E+ K+E+ E K
Sbjct: 284 KQEEKEKKKQEKERKKQEKKERK 306
>NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3
Length = 802
Score = 48.5 bits (114), Expect = 6e-06
Identities = 30/106 (28%), Positives = 49/106 (45%), Gaps = 6/106 (5%)
Query: 46 DGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDH-GKSSHLFLEGTKRDVEDHIV 104
DG + + R E E E+ N +D++ G+ + + + + E+H
Sbjct: 37 DGIEFDAPEEEREAEREEENEEQHELEDVNDEEEEDKEEKGEENGEVINTEEEEEEEHQQ 96
Query: 105 ARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEE 150
D DD D E + +EE DDDDDD+DD + E+ ++E EE
Sbjct: 97 KGGNDDDDDDNEEEEEEEEDDDDDDDDDDDD-----EEEEEEEEEE 137
Score = 31.6 bits (70), Expect = 0.71
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 16/92 (17%)
Query: 64 EERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEES 123
EE E K + DD D+ + E + D +D D DD D + + +EE
Sbjct: 87 EEEEEEEHQQKGGNDDDDDDNEEE-----EEEEEDDDD-------DDDDDDDDEEEEEEE 134
Query: 124 DDDDDDE----DDTEALLAELEQIKKERAEEK 151
+++ +D D+ A E E+ KK++ ++K
Sbjct: 135 EEEGNDNSSVGSDSAAEDGEDEEDKKDKTKDK 166
>FKB4_SPOFR (Q26486) 46 kDa FK506-binding nuclear protein (EC
5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase)
Length = 412
Score = 45.8 bits (107), Expect = 4e-05
Identities = 30/113 (26%), Positives = 57/113 (49%), Gaps = 7/113 (6%)
Query: 42 KPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVED 101
K +R ++ D + + + + + ++++ S DD D + FL+G D ++
Sbjct: 120 KNKRKLENANDATANKKAKPDKKAGKNSAPAAESDSDDDDEDQLQK---FLDGEDIDTDE 176
Query: 102 HIVA----RSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEE 150
+ + S + DDSD E ++E D++DDDEDD E A ++ K+ AE+
Sbjct: 177 NDESFKMNTSAEGDDSDEEDDDEDEEDEEDDDEDDEEEEEAPKKKKKQPAAEQ 229
Score = 37.0 bits (84), Expect = 0.017
Identities = 26/123 (21%), Positives = 58/123 (47%), Gaps = 12/123 (9%)
Query: 41 LKPRRDGQDTQDELKRRNLRDELEER--ERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRD 98
L+ + ++ ++E + L +R E + ++ NK D+ GK+S E D
Sbjct: 98 LEDEEEAEEEEEEEEAPPLVPAKNKRKLENANDATANKKAKPDKKAGKNSAPAAESDSDD 157
Query: 99 VEDHIVARSVDADDSDVEVKSD----------EESDDDDDDEDDTEALLAELEQIKKERA 148
++ + + +D +D D + + ++SD++DDDED+ + + + ++E A
Sbjct: 158 DDEDQLQKFLDGEDIDTDENDESFKMNTSAEGDDSDEEDDDEDEEDEEDDDEDDEEEEEA 217
Query: 149 EEK 151
+K
Sbjct: 218 PKK 220
>UBF1_MOUSE (P25976) Nucleolar transcription factor 1 (Upstream
binding factor 1) (UBF-1)
Length = 765
Score = 44.3 bits (103), Expect = 1e-04
Identities = 32/112 (28%), Positives = 52/112 (45%), Gaps = 24/112 (21%)
Query: 21 TRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSD 80
T++ GP+ K +S TTL+ + + ++ DE + D+ EE E S D
Sbjct: 657 TKLRGPNPK------SSRTTLQSKSESEEDDDEEEE----DDEEEEEEEEDDENGDSSED 706
Query: 81 DRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDD 132
D +SS ED S D D++D + +++ DDDD+DED+
Sbjct: 707 GGDSSESSS----------EDE----SEDGDENDDDDDDEDDEDDDDEDEDN 744
>TOP1_USTMA (P41511) DNA topoisomerase I (EC 5.99.1.2)
Length = 1019
Score = 43.1 bits (100), Expect = 2e-04
Identities = 26/70 (37%), Positives = 33/70 (47%), Gaps = 4/70 (5%)
Query: 67 ERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDD 126
++R SS N DD D + L K + D D DD D E D+E DDD
Sbjct: 74 QKRSGSSNNDDNDDDSDSDSDAPLTALVKKSNGSDD----DEDDDDDDDEGDDDDEEDDD 129
Query: 127 DDDEDDTEAL 136
DDD+DD + L
Sbjct: 130 DDDDDDDKPL 139
>DAXX_RAT (Q8VIB2) Death domain-associated protein 6 (Daxx)
Length = 731
Score = 42.4 bits (98), Expect = 4e-04
Identities = 26/95 (27%), Positives = 52/95 (54%), Gaps = 2/95 (2%)
Query: 59 LRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTK--RDVEDHIVARSVDADDSDVE 116
++D+ EE ER+ ++ + S D K+S EG ED ++ D+ D E
Sbjct: 380 MQDKSEEGERQKRRARLLATSQSSDLPKASSDSGEGPSGVASQEDPTTPKAETEDEEDDE 439
Query: 117 VKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
DEE ++++++E+ TE +LEQ+++++ +E+
Sbjct: 440 ESDDEEEEEEEEEEEATEDEDEDLEQLQEDQDDEE 474
>NUCL_MOUSE (P09405) Nucleolin (Protein C23)
Length = 706
Score = 42.0 bits (97), Expect = 5e-04
Identities = 37/150 (24%), Positives = 67/150 (44%), Gaps = 17/150 (11%)
Query: 3 TAARPTWAPAKGGNEQGGTRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLRDE 62
T A+ P K G Q + P +K ++ T K ++G++ + E DE
Sbjct: 98 TPAKVIPTPGKKGAAQAKALVPTPGKKGAA------TPAKGAKNGKNAKKEDS-----DE 146
Query: 63 LEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEE 122
E+ E S +++ DD + + ++G K S D +D + E D+E
Sbjct: 147 DEDEEDEDDSDEDE---DDEEEDEFEPPIVKGVKPAKAAPAAPASEDEEDDEDE---DDE 200
Query: 123 SDDDDDDEDDTEALLAELEQIKKERAEEKL 152
DDD+++EDD+E + E+ K ++ K+
Sbjct: 201 EDDDEEEEDDSEEEVMEITTAKGKKTPAKV 230
Score = 35.0 bits (79), Expect = 0.064
Identities = 18/45 (40%), Positives = 28/45 (62%), Gaps = 2/45 (4%)
Query: 109 DADDSDVEVKSDEESDDDDDD--EDDTEALLAELEQIKKERAEEK 151
+ +D D E + DEE DD+DDD E++ E + A + KKE ++K
Sbjct: 245 EEEDEDDEDEDDEEEDDEDDDEEEEEEEPVKAAPGKRKKEMTKQK 289
Score = 31.6 bits (70), Expect = 0.71
Identities = 15/48 (31%), Positives = 29/48 (60%), Gaps = 1/48 (2%)
Query: 105 ARSV-DADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEK 151
A+SV + +D + E + DE+ DD+++D++D + E E +K + K
Sbjct: 235 AKSVAEEEDDEEEDEDDEDEDDEEEDDEDDDEEEEEEEPVKAAPGKRK 282
>UL47_EHV1B (P28929) 97 kDa alpha trans-inducing protein (Tegument
protein) (GP10)
Length = 871
Score = 41.6 bits (96), Expect = 7e-04
Identities = 35/154 (22%), Positives = 72/154 (46%), Gaps = 16/154 (10%)
Query: 16 NEQGGTRIFGPSQKYSSRDI-ASHTTLKPRRD-----GQDTQDELKRRN--LRDELEERE 67
N Q T +G ++ S D+ A +PR D G+++ E + + L+E +
Sbjct: 74 NAQQSTSFWGYLRRVFSDDVPAQPQAPRPRADFAPPAGEESSSEEEEEEGPAQAPLDEED 133
Query: 68 RRHFSSKNK--SYSDDRDHGKSSHLFLE----GTKRDVEDH--IVARSVDADDSDVEVKS 119
+ ++ + SD+ D + L + + DH +VA + +S+ ++ +
Sbjct: 134 QLMYADQYSVGDSSDENDEEEDPRLGSDYPTSAESSEYHDHGEMVAGAGAESESETDIDA 193
Query: 120 DEESDDDDDDEDDTEALLAELEQIKKERAEEKLR 153
+EE +DD+DDEDD E + E ++ + ++ +R
Sbjct: 194 EEEEEDDEDDEDDMEVIRDESYRLPRTWLDKSIR 227
>S521_RAT (Q9QY02) Putative splicing factor YT521 (RA301-binding
protein)
Length = 738
Score = 41.6 bits (96), Expect = 7e-04
Identities = 31/124 (25%), Positives = 53/124 (42%), Gaps = 4/124 (3%)
Query: 31 SSRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHL 90
S R S T R G + R + + E + S S + G ++
Sbjct: 140 SERRAKSPTPDGSERIGLEVDRRASRSSQSSKEEGNSEEYGSDHETGSSASSEQGNNTEN 199
Query: 91 FLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDD----DEDDTEALLAELEQIKKE 146
EG + DVE+ DD +V+ ++EE D+++D DE++ E E EQ +++
Sbjct: 200 EEEGGEEDVEEDEEVDEDGDDDEEVDEDAEEEEDEEEDEEEEDEEEEEEEEEEYEQDERD 259
Query: 147 RAEE 150
+ EE
Sbjct: 260 QKEE 263
>UBF1_HUMAN (P17480) Nucleolar transcription factor 1 (Upstream
binding factor 1) (UBF-1) (Autoantigen NOR-90)
Length = 764
Score = 40.8 bits (94), Expect = 0.001
Identities = 31/114 (27%), Positives = 54/114 (47%), Gaps = 28/114 (24%)
Query: 21 TRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSD 80
T++ GP+ K +S TTL+ + + ++ +E + DE EE + SS+
Sbjct: 657 TKLRGPNPK------SSRTTLQSKSESEEDDEEDEDDEDEDEEEEDDENGDSSE------ 704
Query: 81 DRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
D G SS E + D +++D D + DE+ DDD+DD++D +
Sbjct: 705 --DGGDSS----ESSSED----------ESEDGDENEEDDEDEDDDEDDDEDED 742
Score = 32.3 bits (72), Expect = 0.41
Identities = 23/84 (27%), Positives = 37/84 (43%), Gaps = 3/84 (3%)
Query: 68 RRHFSSKNKSYSDDRD-HGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDD 126
+ + S+K KS + R + KSS L+ ED D +D D E + DE D
Sbjct: 646 KEYISNKRKSMTKLRGPNPKSSRTTLQSKSESEEDD--EEDEDDEDEDEEEEDDENGDSS 703
Query: 127 DDDEDDTEALLAELEQIKKERAEE 150
+D D +E+ + + E E+
Sbjct: 704 EDGGDSSESSSEDESEDGDENEED 727
>SPT5_YEAST (P27692) Transcription initiation protein SPT5
Length = 1063
Score = 40.8 bits (94), Expect = 0.001
Identities = 37/133 (27%), Positives = 60/133 (44%), Gaps = 15/133 (11%)
Query: 32 SRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDR------DHG 85
S + A T+ P DG++ + + + + D E SS N +DD D
Sbjct: 51 STERAESTSNIPPLDGEEKEAKSEPQQPEDNAETAATEQVSSSNGPATDDAQATLNTDSS 110
Query: 86 KSSHLFL--EGT---KRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAEL 140
+++ + EG+ KR E+ ++ D D D DE+ D+DDDD+DD E +
Sbjct: 111 EANEIVKKEEGSDERKRPREED--TKNSDGDTKDEGDNKDEDDDEDDDDDDDDED--DDD 166
Query: 141 EQIKKERAEEKLR 153
E K R +E+ R
Sbjct: 167 EAPTKRRRQERNR 179
>SIS2_CANTR (Q12600) SIS2 protein (Halotolerance protein HAL3)
Length = 531
Score = 40.8 bits (94), Expect = 0.001
Identities = 18/57 (31%), Positives = 32/57 (55%), Gaps = 5/57 (8%)
Query: 78 YSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
Y +D D ++ + +K ++++ + D DD D + D++ DDDDDD+DD E
Sbjct: 461 YPEDEDEDEA-----DDSKDNIDESAIIDDDDDDDDDDDDDDDDDDDDDDDDDDDEE 512
Score = 34.3 bits (77), Expect = 0.11
Identities = 15/38 (39%), Positives = 21/38 (54%)
Query: 109 DADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKE 146
D DD D + D++ DDDDDDE+D + + K E
Sbjct: 490 DDDDDDDDDDDDDDDDDDDDDEEDPPQQQSTTDNSKDE 527
Score = 33.1 bits (74), Expect = 0.24
Identities = 15/43 (34%), Positives = 22/43 (50%)
Query: 92 LEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
L G D ++ S D D + D++ DDDDDD+DD +
Sbjct: 458 LGGYPEDEDEDEADDSKDNIDESAIIDDDDDDDDDDDDDDDDD 500
>YHT1_YEAST (P38835) Hypothetical 95.1 kDa protein in ACT5-YCK1
intergenic region
Length = 840
Score = 40.4 bits (93), Expect = 0.002
Identities = 22/55 (40%), Positives = 28/55 (50%), Gaps = 5/55 (9%)
Query: 80 DDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
D DHG+ +G D ED D DD D E D++ DDDDDD+DD +
Sbjct: 786 DAEDHGED-----DGDGDDGEDDDDDDDDDDDDDDDEDDDDDDDDDDDDDDDDDD 835
Score = 31.6 bits (70), Expect = 0.71
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 11/41 (26%)
Query: 94 GTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
GT +D EDH DD D ++ +DDDDD+DD +
Sbjct: 782 GTTKDAEDH------GEDDGD-----GDDGEDDDDDDDDDD 811
Score = 29.3 bits (64), Expect = 3.5
Identities = 17/57 (29%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Query: 82 RDHGKSSHLFLEGTKRDVEDHI----VARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
R +SS ++L G + E +RS+ + D E+ DE + + D+DE+D E
Sbjct: 577 RPKPQSSSIYLSGLAPNGESATDLSQSSRSLCLTNRDAEINDDESATETDEDENDGE 633
>SIS2_YEAST (P36024) SIS2 protein (Halotolerance protein HAL3)
Length = 562
Score = 40.4 bits (93), Expect = 0.002
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 5/64 (7%)
Query: 71 FSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDE 130
+ N+ DD D + + + D ED + D DD D + D++ DDDDDD+
Sbjct: 491 YPKNNEEEDDDEDEEED-----DDEEEDTEDKNENNNDDDDDDDDDDDDDDDDDDDDDDD 545
Query: 131 DDTE 134
D+ E
Sbjct: 546 DEDE 549
Score = 36.6 bits (83), Expect = 0.022
Identities = 20/61 (32%), Positives = 30/61 (48%)
Query: 74 KNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDT 133
KN DD + + E T+ E++ D DD D + D++ DDDD+DED+
Sbjct: 493 KNNEEEDDDEDEEEDDDEEEDTEDKNENNNDDDDDDDDDDDDDDDDDDDDDDDDEDEDEA 552
Query: 134 E 134
E
Sbjct: 553 E 553
Score = 32.0 bits (71), Expect = 0.54
Identities = 21/95 (22%), Positives = 39/95 (40%), Gaps = 28/95 (29%)
Query: 43 PRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDH 102
P+ + ++ DE +E ++ E KN++ +DD D
Sbjct: 492 PKNNEEEDDDE------DEEEDDDEEEDTEDKNENNNDDDDDDD---------------- 529
Query: 103 IVARSVDADDSDVEVKSDEESDDDDDDEDDTEALL 137
D DD D + D++ DD+D+DE +T ++
Sbjct: 530 ------DDDDDDDDDDDDDDDDDEDEDEAETPGII 558
Score = 29.3 bits (64), Expect = 3.5
Identities = 12/37 (32%), Positives = 23/37 (61%)
Query: 98 DVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
++ + IV + ++ E DE+ ++DDD+E+DTE
Sbjct: 479 EIVNKIVMKLGGYPKNNEEEDDDEDEEEDDDEEEDTE 515
>S521_HUMAN (Q96MU7) Putative splicing factor YT521
Length = 727
Score = 40.4 bits (93), Expect = 0.002
Identities = 31/116 (26%), Positives = 54/116 (45%), Gaps = 8/116 (6%)
Query: 44 RRDGQDTQDELKRRNLRDELEERERRHFSSK-----NKSYSDDRDHGKSSHLFLEGTKRD 98
RR T D +R L E++ R R S ++ Y D + G S +G +
Sbjct: 142 RRAKSPTPDGSERIGL--EVDRRASRSSQSSKEEVNSEEYGSDHETGSSGSSDEQGNNTE 199
Query: 99 VEDHIVARSVDADDS-DVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKLR 153
E+ V V+ D+ + + + DEE D+D ++E++ E E E+ ++E E+ R
Sbjct: 200 NEEEGVEEDVEEDEEVEEDAEEDEEVDEDGEEEEEEEEEEEEEEEEEEEEYEQDER 255
Score = 40.0 bits (92), Expect = 0.002
Identities = 23/101 (22%), Positives = 50/101 (48%), Gaps = 2/101 (1%)
Query: 52 DELKRRNLRDELEERERRHFSSKNKSYSDDR--DHGKSSHLFLEGTKRDVEDHIVARSVD 109
D R+ + EE + S +++ S + G ++ EG + DVE+
Sbjct: 160 DRRASRSSQSSKEEVNSEEYGSDHETGSSGSSDEQGNNTENEEEGVEEDVEEDEEVEEDA 219
Query: 110 ADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEE 150
+D +V+ +EE ++++++E++ E E EQ ++++ EE
Sbjct: 220 EEDEEVDEDGEEEEEEEEEEEEEEEEEEEEYEQDERDQKEE 260
>NRDC_MOUSE (Q8BHG1) Nardilysin precursor (EC 3.4.24.61) (N-arginine
dibasic convertase) (NRD convertase) (NRD-C)
Length = 1161
Score = 40.4 bits (93), Expect = 0.002
Identities = 36/138 (26%), Positives = 61/138 (44%), Gaps = 13/138 (9%)
Query: 26 PSQKYSSRDIASHTTLKP--RRDGQDTQDELKRRNLRDELEERERRHFSSKN-----KSY 78
P+ R+ A T P + +GQD + + L + E E R FS+ KS
Sbjct: 45 PTLTMPGRNKAKSTCSCPDLQPNGQDLGESGRLARLGADESEEEGRSFSNVGDPEIIKSP 104
Query: 79 SDDRDHG-----KSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDT 133
SD + + L +VE + D ++ + E + +E+ DDDDDD+DD
Sbjct: 105 SDPKQYRYIKLQNGLQALLISDLSNVEGK-TGNATDEEEEEEEEEEEEDDDDDDDDDDDD 163
Query: 134 EALLAELEQIKKERAEEK 151
E AE++ +E +++
Sbjct: 164 EDSGAEIQDDDEEGFDDE 181
Score = 38.5 bits (88), Expect = 0.006
Identities = 30/100 (30%), Positives = 41/100 (41%), Gaps = 19/100 (19%)
Query: 53 ELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADD 112
E K N DE EE E + DD D + S ++ DD
Sbjct: 131 EGKTGNATDEEEEEEEEEEEEDDDDDDDDDDDDEDSGAEIQ-----------------DD 173
Query: 113 SDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERAEEKL 152
+ +EE DDDDDDE D + L E E+ + E EE++
Sbjct: 174 DEEGFDDEEEFDDDDDDEHDDDDL--ENEENELEELEERV 211
>VHS3_YEAST (Q08438) Protein VHS3 (Viable in a HAL3 SIT4 background
protein 3)
Length = 674
Score = 40.0 bits (92), Expect = 0.002
Identities = 26/91 (28%), Positives = 41/91 (44%), Gaps = 10/91 (10%)
Query: 61 DELEERERRHFSSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSD 120
DE ++ ++ K++ DD D D +D D DD D + D
Sbjct: 588 DEEDDNKKNDTGGKDEDNDDDDDDDDD----------DDDDDDDDDDDDDDDDDDDDDDD 637
Query: 121 EESDDDDDDEDDTEALLAELEQIKKERAEEK 151
++ DDDDDD+DD E E E + ++ E+K
Sbjct: 638 DDDDDDDDDDDDDEDDEDEDEDDEGKKKEDK 668
Score = 37.4 bits (85), Expect = 0.013
Identities = 20/62 (32%), Positives = 29/62 (46%)
Query: 73 SKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDD 132
S K +D D+ + T ED+ D DD D + D++ DDDDDD+DD
Sbjct: 576 SAGKEEEEDEDNDEEDDNKKNDTGGKDEDNDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 635
Query: 133 TE 134
+
Sbjct: 636 DD 637
Score = 37.0 bits (84), Expect = 0.017
Identities = 18/56 (32%), Positives = 28/56 (49%)
Query: 79 SDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTE 134
+D+ D K + + D +D D DD D + D++ DDDDDD+DD +
Sbjct: 587 NDEEDDNKKNDTGGKDEDNDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 642
Score = 36.2 bits (82), Expect = 0.029
Identities = 19/63 (30%), Positives = 29/63 (45%)
Query: 72 SSKNKSYSDDRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDED 131
+ K + +D D + G K + D D DD D + D++ DDDDDD+D
Sbjct: 577 AGKEEEEDEDNDEEDDNKKNDTGGKDEDNDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 636
Query: 132 DTE 134
D +
Sbjct: 637 DDD 639
Score = 28.1 bits (61), Expect = 7.8
Identities = 10/21 (47%), Positives = 15/21 (70%)
Query: 114 DVEVKSDEESDDDDDDEDDTE 134
DV +EE D+D+D+EDD +
Sbjct: 574 DVSAGKEEEEDEDNDEEDDNK 594
>UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream
binding factor 1) (UBF-1)
Length = 764
Score = 40.0 bits (92), Expect = 0.002
Identities = 31/115 (26%), Positives = 54/115 (46%), Gaps = 26/115 (22%)
Query: 21 TRIFGPSQKYSSRDIASHTTLKPRRDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSD 80
T++ GP+ K +S TTL+ + + ++ DE D+ ++ E +N S+
Sbjct: 657 TKLRGPNPK------SSRTTLQSKSESEEDDDE------EDDDDDDEEEEEDDENGDSSE 704
Query: 81 DRDHGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEA 135
D G SS E D +++ DD D DE+ D+DDD+++D E+
Sbjct: 705 DG--GDSSESSSEDESEDGDEN-------EDDDD-----DEDDDEDDDEDEDNES 745
Score = 31.2 bits (69), Expect = 0.92
Identities = 23/84 (27%), Positives = 38/84 (44%), Gaps = 4/84 (4%)
Query: 68 RRHFSSKNKSYSDDRD-HGKSSHLFLEGTKRDVEDHIVARSVDADDSDVEVKSDEESDDD 126
+ + S+K K+ + R + KSS L+ ED D DD D E + D+E+ D
Sbjct: 646 KEYISNKRKNMTKLRGPNPKSSRTTLQSKSESEEDD---DEEDDDDDDEEEEEDDENGDS 702
Query: 127 DDDEDDTEALLAELEQIKKERAEE 150
+D D+ +E E + E+
Sbjct: 703 SEDGGDSSESSSEDESEDGDENED 726
>TUL1_MOUSE (Q9Z273) Tubby related protein 1 (Tubby-like protein 1)
Length = 543
Score = 40.0 bits (92), Expect = 0.002
Identities = 38/119 (31%), Positives = 58/119 (47%), Gaps = 17/119 (14%)
Query: 41 LKPR-RDGQDTQDELKRRNLRDELEERERRHFSSKNKSYSDDRDHGKSSHL------FL- 92
L+P+ R Q + K+ D LE + R+ + + K D + FL
Sbjct: 29 LRPKQRPAQGQKLRKKKPETPDSLESKPRKAGAGRRKHEEPPADSAEPRAAQTVYAKFLR 88
Query: 93 --EGTKRDV-EDHIVARSVDADDSDVEVKSDEESDDDDDDEDDTEALLAELEQIKKERA 148
E KRD E+ +VAR A D E S+E+SDDDD+D+D+ E E ++ KKE++
Sbjct: 89 DPEAKKRDPRENFLVAR---APDLGGEENSEEDSDDDDNDDDEEE---EEKKEGKKEKS 141
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.306 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,384,649
Number of Sequences: 164201
Number of extensions: 866808
Number of successful extensions: 12934
Number of sequences better than 10.0: 783
Number of HSP's better than 10.0 without gapping: 400
Number of HSP's successfully gapped in prelim test: 399
Number of HSP's that attempted gapping in prelim test: 6448
Number of HSP's gapped (non-prelim): 2810
length of query: 153
length of database: 59,974,054
effective HSP length: 101
effective length of query: 52
effective length of database: 43,389,753
effective search space: 2256267156
effective search space used: 2256267156
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0281a.2


