

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0278.2
(507 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
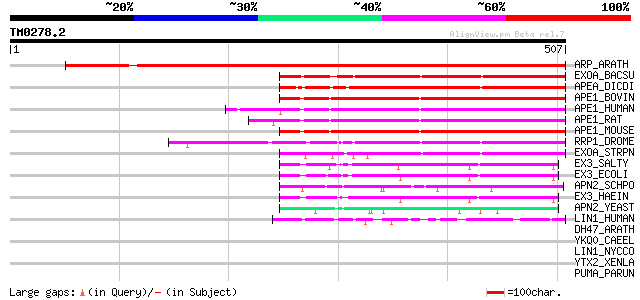
Score E
Sequences producing significant alignments: (bits) Value
ARP_ARATH (P45951) Apurinic endonuclease-redox protein (DNA-(apu... 584 e-166
EXOA_BACSU (P37454) Exodeoxyribonuclease (EC 3.1.11.2) 263 6e-70
APEA_DICDI (P51173) DNA-(apurinic or apyrimidinic site) lyase (E... 245 2e-64
APE1_BOVIN (P23196) DNA-(apurinic or apyrimidinic site) lyase (E... 242 1e-63
APE1_HUMAN (P27695) DNA-(apurinic or apyrimidinic site) lyase (E... 242 2e-63
APE1_RAT (P43138) DNA-(apurinic or apyrimidinic site) lyase (EC ... 241 3e-63
APE1_MOUSE (P28352) DNA-(apurinic or apyrimidinic site) lyase (E... 240 5e-63
RRP1_DROME (P27864) Recombination repair protein 1 (DNA-(apurini... 224 4e-58
EXOA_STRPN (P21998) Exodeoxyribonuclease (EC 3.1.11.2) 197 7e-50
EX3_SALTY (Q9Z612) Exodeoxyribonuclease III (EC 3.1.11.2) (Exonu... 100 2e-20
EX3_ECOLI (P09030) Exodeoxyribonuclease III (EC 3.1.11.2) (Exonu... 99 3e-20
APN2_SCHPO (P87175) DNA-(apurinic or apyrimidinic site) lyase 2 ... 92 2e-18
EX3_HAEIN (P44318) Exodeoxyribonuclease III (EC 3.1.11.2) (Exonu... 92 4e-18
APN2_YEAST (P38207) DNA-(apurinic or apyrimidinic site) lyase 2 ... 66 2e-10
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 46 2e-04
DH47_ARATH (P31168) Dehydrin COR47 (Cold-induced COR47 protein) 43 0.002
YKQ0_CAEEL (P34305) Putative ATP-dependent RNA helicase C06E1.10... 43 0.002
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 42 0.003
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 39 0.041
PUMA_PARUN (O61308) 227 kDa spindle- and centromere-associated p... 39 0.041
>ARP_ARATH (P45951) Apurinic endonuclease-redox protein
(DNA-(apurinic or apyrimidinic site) lyase) (EC
4.2.99.18)
Length = 536
Score = 584 bits (1506), Expect = e-166
Identities = 288/458 (62%), Positives = 358/458 (77%), Gaps = 9/458 (1%)
Query: 52 EIERLRNDPSIVDTMTVQELRKTLKSIRVPAKGRKEDLLSTLKSFMDNNMGEQHPQIEEE 111
E+ L++D ++ MTVQELR TL+ + VP KGRK++L+STL+ MD+N+ +Q
Sbjct: 86 EMGTLQDDRKEIEAMTVQELRSTLRKLGVPVKGRKQELISTLRLHMDSNLPDQKETSS-- 143
Query: 112 HGLLISSENTSVEVKTKKVD-EDHVDDINDNPEVFEHSRGKRRLKQSGSESETVKVTTKK 170
S+ + SV +K K + E+ +D N E ++ G++R+KQS ++ KV+ K
Sbjct: 144 -----STRSDSVTIKRKISNREEPTEDECTNSEAYDIEHGEKRVKQSTEKNLKAKVSAKA 198
Query: 171 KLLVESDEVSDFKPS-RAKRRVSSDIVRVVSQSDEISTTTIPTEPWTVLAHKKPQKDWIA 229
+ + K ++K SS I + +++EI ++ +EPWTVLAHKKPQKDW A
Sbjct: 199 IAKEQKSLMRTGKQQIQSKEETSSTISSELLKTEEIISSPSQSEPWTVLAHKKPQKDWKA 258
Query: 230 YNPKTMRPQPLTRDTKFVKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQE 289
YNPKTMRP PL TK VK+++WNVNGLR LLK E FSAL+LAQRE+FD+LCLQETKLQ
Sbjct: 259 YNPKTMRPPPLPEGTKCVKVMTWNVNGLRGLLKFESFSALQLAQRENFDILCLQETKLQV 318
Query: 290 KDINEIKRQLIDGYENSFWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTA 349
KD+ EIK+ LIDGY++SFW+CSVSKLGYSGTAIISRIKPLSVRYG G+S HDTEGR+VTA
Sbjct: 319 KDVEEIKKTLIDGYDHSFWSCSVSKLGYSGTAIISRIKPLSVRYGTGLSGHDTEGRIVTA 378
Query: 350 EFDTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEID 409
EFD+FYLI Y+PNSGDGLKRLSYR+ +WD +LSN++KELEK+KPVVLTGDLNCAHEEID
Sbjct: 379 EFDSFYLINTYVPNSGDGLKRLSYRIEEWDRTLSNHIKELEKSKPVVLTGDLNCAHEEID 438
Query: 410 IYNPAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVGYTYWGYRHGGRKTNRGW 469
I+NPAGNK+SAGFT EER+SF AN L +GFVDTFR+QHP VVGYTYWGYRHGGRKTN+GW
Sbjct: 439 IFNPAGNKRSAGFTIEERQSFGANLLDKGFVDTFRKQHPGVVGYTYWGYRHGGRKTNKGW 498
Query: 470 RLDYFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
RLDYFLVS+SIA VHDSYILPD+ GSDHCP+GLI+KL
Sbjct: 499 RLDYFLVSQSIAANVHDSYILPDINGSDHCPIGLILKL 536
>EXOA_BACSU (P37454) Exodeoxyribonuclease (EC 3.1.11.2)
Length = 252
Score = 263 bits (673), Expect = 6e-70
Identities = 129/262 (49%), Positives = 187/262 (71%), Gaps = 11/262 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+KL+SWNVNGLRA+++ F L + ED D++CLQETK+Q+ ++ L +
Sbjct: 1 MKLISWNVNGLRAVMRKMDF--LSYLKEEDADIICLQETKIQDGQVD-----LQPEDYHV 53
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W +V K GYSGTA+ S+ +PL V YG+G+ +HD EGR++T EF+ +++ Y PNS
Sbjct: 54 YWNYAVKK-GYSGTAVFSKQEPLQVIYGIGVEEHDQEGRVITLEFENVFVMTVYTPNSRR 112
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL+R+ YR+ QW+ +L +Y+ EL++ KPV+L GDLN AH+EID+ NP N+ +AGF+D+E
Sbjct: 113 GLERIDYRM-QWEEALLSYILELDQKKPVILCGDLNVAHQEIDLKNPKANRNNAGFSDQE 171
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R++F FL GFVD+FR +PD+ G Y++W YR G R N GWR+DYF+VSES+ +++
Sbjct: 172 REAFT-RFLEAGFVDSFRHVYPDLEGAYSWWSYRAGARDRNIGWRIDYFVVSESLKEQIE 230
Query: 486 DSYILPDVMGSDHCPVGLIIKL 507
D+ I DVMGSDHCPV LII +
Sbjct: 231 DASISADVMGSDHCPVELIINI 252
>APEA_DICDI (P51173) DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (Class II
apurinic/apyrimidinic(AP)-endonuclease)
Length = 361
Score = 245 bits (625), Expect = 2e-64
Identities = 125/264 (47%), Positives = 180/264 (67%), Gaps = 11/264 (4%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+K++SWNV G +++L +GF+ E ++E+ DVLCLQETK+ +I K Q+ GYE
Sbjct: 105 MKIISWNVAGFKSVLS-KGFT--ECVEKENPDVLCLQETKINPSNIK--KDQMPKGYEYH 159
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
F + G+ GT ++++ KP ++ +G+GI+ HD EGR++T E+ FY++ YIPN+G
Sbjct: 160 F--IEADQKGHHGTGVLTKKKPNAITFGIGIAKHDNEGRVITLEYVQFYIVNTYIPNAGT 217
Query: 367 -GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDE 425
GL+RL YR+ +WD YL++L TKP++ GDLN AH EID+ NP NKKSAGFT E
Sbjct: 218 RGLQRLDYRIKEWDVDFQAYLEKLNATKPIIWCGDLNVAHTEIDLKNPKTNKKSAGFTIE 277
Query: 426 ERKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKV 484
ER SF +NFL +G+VD++R +P G YT+W Y GGR N GWRLDYF+VS+ + D +
Sbjct: 278 ERTSF-SNFLEKGYVDSYRHFNPGKEGSYTFWSYLGGGRSKNVGWRLDYFVVSKRLMDSI 336
Query: 485 HDS-YILPDVMGSDHCPVGLIIKL 507
S + VMGSDHCP+G+++ L
Sbjct: 337 KISPFHRTSVMGSDHCPIGVVVDL 360
>APE1_BOVIN (P23196) DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN)
Length = 317
Score = 242 bits (618), Expect = 1e-63
Identities = 121/262 (46%), Positives = 166/262 (63%), Gaps = 6/262 (2%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+K+ SWNV+GLRA +K +G L+ + E D+LCLQETK E + ++ Q + G +
Sbjct: 61 LKICSWNVDGLRAWIKKKG---LDWVKEEAPDILCLQETKCSENKL-PVELQELSGLSHQ 116
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W+ K GYSG ++SR PL V YG+G +HD EGR++ AE+D F L+ Y+PN+G
Sbjct: 117 YWSAPSDKEGYSGVGLLSRQCPLKVSYGIGEEEHDQEGRVIVAEYDAFVLVTAYVPNAGR 176
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL RL YR +WD + +LK L KP+VL GDLN AHEEID+ NP GNKK+AGFT +E
Sbjct: 177 GLVRLEYR-QRWDEAFRKFLKGLASRKPLVLCGDLNVAHEEIDLRNPKGNKKNAGFTPQE 235
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVV-GYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R+ F + D+FR +P+ YT+W Y R N GWRLDYFL+S+S+ +
Sbjct: 236 RQGFGELLQAVPLTDSFRHLYPNTAYAYTFWTYMMNARSKNVGWRLDYFLLSQSVLPALC 295
Query: 486 DSYILPDVMGSDHCPVGLIIKL 507
DS I +GSDHCP+ L + L
Sbjct: 296 DSKIRSKALGSDHCPITLYLAL 317
>APE1_HUMAN (P27695) DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN)
(REF-1 protein)
Length = 317
Score = 242 bits (617), Expect = 2e-63
Identities = 133/316 (42%), Positives = 181/316 (57%), Gaps = 13/316 (4%)
Query: 198 VVSQSDEISTTTIPTEPWTVLAHKKPQKDWIAYNPKTMRPQPLTRDTKF-----VKLLSW 252
V DE+ T P + A KK K+ P P + + +K+ SW
Sbjct: 9 VAEDGDELRTE--PEAKKSKTAAKKNDKEAAGEGPALYEDPPDQKTSPSGKPATLKICSW 66
Query: 253 NVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENSFWTCSV 312
NV+GLRA +K +G L+ + E D+LCLQETK E + + Q + G + +W+
Sbjct: 67 NVDGLRAWIKKKG---LDWVKEEAPDILCLQETKCSENKL-PAELQELPGLSHQYWSAPS 122
Query: 313 SKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGDGLKRLS 372
K GYSG ++SR PL V YG+G +HD EGR++ AEFD+F L+ Y+PN+G GL RL
Sbjct: 123 DKEGYSGVGLLSRQCPLKVSYGIGDEEHDQEGRVIVAEFDSFVLVTAYVPNAGRGLVRLE 182
Query: 373 YRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEERKSFAA 432
YR +WD + +LK L KP+VL GDLN AHEEID+ NP GNKK+AGFT +ER+ F
Sbjct: 183 YR-QRWDEAFRKFLKGLASRKPLVLCGDLNVAHEEIDLRNPKGNKKNAGFTPQERQGFGE 241
Query: 433 NFLSRGFVDTFRRQHPDV-VGYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVHDSYILP 491
+ D+FR +P+ YT+W Y R N GWRLDYFL+S S+ + DS I
Sbjct: 242 LLQAVPLADSFRHLYPNTPYAYTFWTYMMNARSKNVGWRLDYFLLSHSLLPALCDSKIRS 301
Query: 492 DVMGSDHCPVGLIIKL 507
+GSDHCP+ L + L
Sbjct: 302 KALGSDHCPITLYLAL 317
>APE1_RAT (P43138) DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN)
Length = 316
Score = 241 bits (615), Expect = 3e-63
Identities = 127/295 (43%), Positives = 176/295 (59%), Gaps = 11/295 (3%)
Query: 219 AHKKPQKDWIAYNPKTMRPQP-----LTRDTKFVKLLSWNVNGLRALLKLEGFSALELAQ 273
A KK +K+ P P + + +K+ SWNV+GLRA +K +G L+ +
Sbjct: 27 AAKKTEKEAAGEGPVLYEDPPDQKTSASGKSATLKICSWNVDGLRAWIKKKG---LDWVK 83
Query: 274 REDFDVLCLQETKLQEKDINEIKRQLIDGYENSFWTCSVSKLGYSGTAIISRIKPLSVRY 333
E D+LCLQETK E + + Q + G + +W+ K GYSG ++SR PL V Y
Sbjct: 84 EEAPDILCLQETKCSENKL-PAELQELPGLTHQYWSAPSDKEGYSGVGLLSRQCPLKVSY 142
Query: 334 GLGISDHDTEGRLVTAEFDTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKTK 393
G+G +HD EGR++ AEF++F L+ Y+PN+G GL RL YR +WD + +LK+L K
Sbjct: 143 GIGEEEHDQEGRVIVAEFESFILVTAYVPNAGRGLVRLEYR-QRWDEAFRKFLKDLASRK 201
Query: 394 PVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVV-G 452
P+VL GDLN AHEEID+ NP GNKK+AGFT +ER+ F + D+FR +P+
Sbjct: 202 PLVLCGDLNVAHEEIDLRNPKGNKKNAGFTPQERQGFGEMLQAVPLADSFRHLYPNTAYA 261
Query: 453 YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
YT+W Y R N GWRLDYFL+S S+ + DS I +GSDHCP+ L + L
Sbjct: 262 YTFWTYMMNARSKNVGWRLDYFLLSHSLLPALCDSKIRSKALGSDHCPITLYLAL 316
>APE1_MOUSE (P28352) DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (AP endonuclease 1) (APEX nuclease) (APEN)
Length = 316
Score = 240 bits (613), Expect = 5e-63
Identities = 121/262 (46%), Positives = 166/262 (63%), Gaps = 6/262 (2%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENS 306
+K+ SWNV+GLRA +K +G L+ + E D+LCLQETK E + + Q + G +
Sbjct: 60 LKICSWNVDGLRAWIKKKG---LDWVKEEAPDILCLQETKCSENKL-PAELQELPGLTHQ 115
Query: 307 FWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGD 366
+W+ K GYSG ++SR PL V YG+G +HD EGR++ AEF++F L+ Y+PN+G
Sbjct: 116 YWSAPSDKEGYSGVGLLSRQCPLKVSYGIGEEEHDQEGRVIVAEFESFVLVTAYVPNAGR 175
Query: 367 GLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEE 426
GL RL YR +WD + +LK+L KP+VL GDLN AHEEID+ NP GNKK+AGFT +E
Sbjct: 176 GLVRLEYR-QRWDEAFRKFLKDLASRKPLVLCGDLNVAHEEIDLRNPKGNKKNAGFTPQE 234
Query: 427 RKSFAANFLSRGFVDTFRRQHPDVV-GYTYWGYRHGGRKTNRGWRLDYFLVSESIADKVH 485
R+ F + D+FR +P+ YT+W Y R N GWRLDYFL+S S+ +
Sbjct: 235 RQGFGELLQAVPLADSFRHLYPNTAYAYTFWTYMMNARSKNVGWRLDYFLLSHSLLPALC 294
Query: 486 DSYILPDVMGSDHCPVGLIIKL 507
DS I +GSDHCP+ L + L
Sbjct: 295 DSKIRSKALGSDHCPITLYLAL 316
>RRP1_DROME (P27864) Recombination repair protein 1 (DNA-(apurinic
or apyrimidinic site) lyase) (EC 4.2.99.18)
Length = 679
Score = 224 bits (571), Expect = 4e-58
Identities = 138/375 (36%), Positives = 200/375 (52%), Gaps = 25/375 (6%)
Query: 146 EHSRGKRRLKQSGSES-----------ETVKVTTKKKLLVESDEVSDFKPSRAKRRVSSD 194
E + GK++ K + E+ ET +KK V++++V D + + + + +
Sbjct: 317 EPAPGKKQKKSADKENGVVEEEAKPSTETKPAKGRKKAPVKAEDVEDIEEAAEESKPARG 376
Query: 195 IVRVVSQSDEISTTTIPTEPWTVLAHKKPQKDWIAYNPKTMRPQPLTRDTKF-VKLLSWN 253
+ ++++E T A K K + + L D +F +K+ SWN
Sbjct: 377 RKKAAAKAEEPDVDEESGSKTTKKAKKAETKTTVTLDKDAFA---LPADKEFNLKICSWN 433
Query: 254 VNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYENSFWTCSVS 313
V GLRA LK +G ++L E+ D+ CLQETK + E +L GY + +W C
Sbjct: 434 VAGLRAWLKKDGLQLIDL---EEPDIFCLQETKCANDQLPEEVTRL-PGY-HPYWLCMPG 488
Query: 314 KLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFYLICGYIPNSGDGLKRLSY 373
GY+G AI S+I P+ V YG+G + D GR++TAE++ FYLI Y+PNSG L L
Sbjct: 489 --GYAGVAIYSKIMPIHVEYGIGNEEFDDVGRMITAEYEKFYLINVYVPNSGRKLVNLEP 546
Query: 374 RVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAGNKKSAGFTDEERKSFAAN 433
R+ +W+ Y+K+L+ KPVV+ GD+N +H ID+ NP N K+AGFT EER
Sbjct: 547 RM-RWEKLFQAYVKKLDALKPVVICGDMNVSHMPIDLENPKNNTKNAGFTQEERDKM-TE 604
Query: 434 FLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVHDSYILPD 492
L GFVDTFR +PD G YT+W Y R N GWRLDY LVSE KV + I
Sbjct: 605 LLGLGFVDTFRHLYPDRKGAYTFWTYMANARARNVGWRLDYCLVSERFVPKVVEHEIRSQ 664
Query: 493 VMGSDHCPVGLIIKL 507
+GSDHCP+ + +
Sbjct: 665 CLGSDHCPITIFFNI 679
>EXOA_STRPN (P21998) Exodeoxyribonuclease (EC 3.1.11.2)
Length = 275
Score = 197 bits (500), Expect = 7e-50
Identities = 122/278 (43%), Positives = 159/278 (56%), Gaps = 20/278 (7%)
Query: 247 VKLLSWNVNGLRALLKLEGFSA------LELAQREDFDVLCLQETKLQEKDIN----EIK 296
+KL+SWN++ L A L + A L+ E+ D++ +QETKL K EI
Sbjct: 1 MKLISWNIDSLNAALTSDSARAKLSQEVLQTLVAENADIIAIQETKLSAKGPTKKHVEIL 60
Query: 297 RQLIDGYENSFWTCSV--SKLGYSGTAIISR--IKPLSVRYGLGI-SDHDTEGRLVTAEF 351
+L GYEN+ W S ++ GY+GT + + + P +G S D EGR++T EF
Sbjct: 61 EELFPGYENT-WRSSQEPARKGYAGTMFLYKKELTPTISFPEIGAPSTMDLEGRIITLEF 119
Query: 352 DTFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIY 411
D F++ Y PN+GDGLKRL R WD + YL EL+K KPV+ TGD N AH EID+
Sbjct: 120 DAFFVTQVYTPNAGDGLKRLEERQV-WDAKYAEYLAELDKEKPVLATGDYNVAHNEIDLA 178
Query: 412 NPAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDV-VGYTYWGYRHGGRK-TNRGW 469
NPA N++S GFTDEER F N L+ GF DTFR H DV YT+W R K N GW
Sbjct: 179 NPASNRRSPGFTDEERAGF-TNLLATGFTDTFRHVHGDVPERYTWWAQRSKTSKINNTGW 237
Query: 470 RLDYFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
R+DY+L S IADKV S ++ DH P+ L I L
Sbjct: 238 RIDYWLTSNRIADKVTKSDMIDSGARQDHTPIVLEIDL 275
>EX3_SALTY (Q9Z612) Exodeoxyribonuclease III (EC 3.1.11.2)
(Exonuclease III) (EXO III) (AP endonuclease VI)
Length = 268
Score = 99.8 bits (247), Expect = 2e-20
Identities = 84/275 (30%), Positives = 142/275 (51%), Gaps = 33/275 (12%)
Query: 247 VKLLSWNVNGLRALL-KLEGFSALELAQREDFDVLCLQETKLQEK--DINEIKRQLIDGY 303
+K +S+N+NGLRA +LE + ++ DV+ LQETK+ ++ + E+ + GY
Sbjct: 1 MKFVSFNINGLRARPHQLEA-----IVEKHQPDVIGLQETKVHDEMFPLEEVAKL---GY 52
Query: 304 ENSFWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDT----FYLICG 359
N F+ + G+ G A++++ P+SVR G + + R++ AE + +I G
Sbjct: 53 -NVFYH---GQKGHYGVALLTKATPISVRRGFPDDGEEAQRRIIMAEIPSPLGNITVING 108
Query: 360 YIPNSGDGLKRLSYRV-TQWDPSLSNYLK-ELEKTKPVVLTGDLNCAHEEIDIYNPAGNK 417
Y P L + Q+ +L NYL+ EL+ PV++ GD+N + ++DI N+
Sbjct: 109 YFPQGESRDHPLKFPAKAQFYQNLQNYLETELKCDNPVLIMGDMNISPTDLDIGIGEENR 168
Query: 418 K------SAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWR 470
K F EER+ + + L G VDTFR+ +P + ++++ YR G NRG R
Sbjct: 169 KRWLRTGKCSFLPEERE-WMSRLLKWGLVDTFRQANPQTMDKFSWFDYRSKGFVDNRGLR 227
Query: 471 LDYFLVSESIADKVHDSYILPDVMG----SDHCPV 501
+D L S +A++ ++ I D+ SDH PV
Sbjct: 228 IDLLLASAPLAERCAETGIDYDIRSMEKPSDHAPV 262
>EX3_ECOLI (P09030) Exodeoxyribonuclease III (EC 3.1.11.2)
(Exonuclease III) (EXO III) (AP endonuclease VI)
Length = 268
Score = 99.0 bits (245), Expect = 3e-20
Identities = 82/273 (30%), Positives = 141/273 (51%), Gaps = 29/273 (10%)
Query: 247 VKLLSWNVNGLRALL-KLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGYEN 305
+K +S+N+NGLRA +LE + ++ DV+ LQETK+ + D+ ++ GY N
Sbjct: 1 MKFVSFNINGLRARPHQLEA-----IVEKHQPDVIGLQETKVHD-DMFPLEEVAKLGY-N 53
Query: 306 SFWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFY----LICGYI 361
F+ + G+ G A++++ P++VR G D + + R++ AE + +I GY
Sbjct: 54 VFYH---GQKGHYGVALLTKETPIAVRRGFPGDDEEAQRRIIMAEIPSLLGNVTVINGYF 110
Query: 362 PNSGDGLKRLSYRV-TQWDPSLSNYLK-ELEKTKPVVLTGDLNCAHEEIDIYNPAGNKK- 418
P + + Q+ +L NYL+ EL++ PV++ GD+N + ++DI N+K
Sbjct: 111 PQGESRDHPIKFPAKAQFYQNLQNYLETELKRDNPVLIMGDMNISPTDLDIGIGEENRKR 170
Query: 419 -----SAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLD 472
F EER+ + +S G VDTFR +P ++++ YR G NRG R+D
Sbjct: 171 WLRTGKCSFLPEERE-WMDRLMSWGLVDTFRHANPQTADRFSWFDYRSKGFDDNRGLRID 229
Query: 473 YFLVSESIADKVHDSYILPDVMG----SDHCPV 501
L S+ +A+ ++ I ++ SDH PV
Sbjct: 230 LLLASQPLAECCVETGIDYEIRSMEKPSDHAPV 262
>APN2_SCHPO (P87175) DNA-(apurinic or apyrimidinic site) lyase 2 (EC
4.2.99.18) (AP endonuclease 2) (Apurinic-apyrimidinic
endonuclease 2)
Length = 523
Score = 92.4 bits (228), Expect = 2e-18
Identities = 90/308 (29%), Positives = 143/308 (46%), Gaps = 53/308 (17%)
Query: 247 VKLLSWNVNGLRALLKLEGF----SALELAQREDFDVLCLQETKLQEKDINEIKRQLIDG 302
+++LSWNVNG++ + S E+ Q DV+C+QE K+Q KD + +++G
Sbjct: 1 MRILSWNVNGIQNPFNYFPWNKKNSYKEIFQELQADVICVQELKMQ-KDSFPQQYAVVEG 59
Query: 303 YENSFWTCSVSKLGYSGTAI-ISRIKPLSVRYGLGIS----------------DH----- 340
++ S++T + GYSG + + + V+ GI+ +H
Sbjct: 60 FD-SYFTFPKIRKGYSGVGFYVKKDVAIPVKAEEGITGILPVRGQKYSYSEAPEHEKIGF 118
Query: 341 -------------DTEGRLVTAEFDTFYLICGYIP-NSGDGLKRLSYRVTQWDPSLSNYL 386
D+EGR + +F F LI Y P NSG+ RL YR + +L +
Sbjct: 119 FPKDIDRKTANWIDSEGRCILLDFQMFILIGVYCPVNSGEN--RLEYRRAFYK-ALRERI 175
Query: 387 KEL--EKTKPVVLTGDLNCAHEEIDIYNPAGN-KKSAGFTDEERKSFAANFLSRG----F 439
+ L E + ++L GD+N ID + ++S + E + + + L
Sbjct: 176 ERLIKEGNRKIILVGDVNILCNPIDTADQKDIIRESLIPSIMESRQWIRDLLLPSRLGLL 235
Query: 440 VDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRLDYFLVSESIADKVHDSYILPDVMGSDH 498
+D R QHP G +T W R R TN G R+DY L + + V D+ I+ +VMGSDH
Sbjct: 236 LDIGRIQHPTRKGMFTCWNTRLNTRPTNYGTRIDYTLATPDLLPWVQDADIMAEVMGSDH 295
Query: 499 CPVGLIIK 506
CPV L +K
Sbjct: 296 CPVYLDLK 303
>EX3_HAEIN (P44318) Exodeoxyribonuclease III (EC 3.1.11.2)
(Exonuclease III) (EXO III)
Length = 267
Score = 91.7 bits (226), Expect = 4e-18
Identities = 77/274 (28%), Positives = 135/274 (49%), Gaps = 31/274 (11%)
Query: 247 VKLLSWNVNGLRALL-KLEGFSALELAQREDFDVLCLQETKLQEKDIN-EIKRQLIDGYE 304
+K +S+N+NGLRA +LE + ++ DV+ LQE K+ ++ EI L GY
Sbjct: 1 MKFISFNINGLRARPHQLEA-----IIEKYQPDVIGLQEIKVADEAFPYEITENL--GYH 53
Query: 305 NSFWTCSVSKLGYSGTAIISRIKPLSVRYGLGISDHDTEGRLVTAEFDTFY----LICGY 360
+ G+ G A++++ +P +R G + D + R++ A+ +T + +I GY
Sbjct: 54 ----VFHHGQKGHYGVALLTKQEPKVIRRGFPTDNEDAQKRIIMADLETEFGLLTVINGY 109
Query: 361 IPNSGDGLKRLSYRVTQ-WDPSLSNYL-KELEKTKPVVLTGDLNCAHEEIDIYNPAGNKK 418
P + + + L YL KE +K+ P+++ GD+N + ++DI N+K
Sbjct: 110 FPQGESRAHETKFPAKEKFYADLQQYLEKEHDKSNPILIMGDMNISPSDLDIGIGDENRK 169
Query: 419 ------SAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVG-YTYWGYRHGGRKTNRGWRL 471
F EER ++ G D+FR+ +P ++++ YR G NRG R+
Sbjct: 170 RWLRTGKCSFLPEER-AWYQRLYDYGLEDSFRKLNPTANDKFSWFDYRSKGFDDNRGLRI 228
Query: 472 DYFLVSESIADKVHDSYILPDVMG----SDHCPV 501
D+ LVS+ +A++ D I D+ SDH P+
Sbjct: 229 DHILVSQKLAERCVDVGIALDIRAMEKPSDHAPI 262
>APN2_YEAST (P38207) DNA-(apurinic or apyrimidinic site) lyase 2 (EC
4.2.99.18) (AP endonuclease 2) (Apurinic-apyrimidinic
endonuclease 2)
Length = 520
Score = 66.2 bits (160), Expect = 2e-10
Identities = 86/344 (25%), Positives = 135/344 (39%), Gaps = 92/344 (26%)
Query: 247 VKLLSWNVNGLRALLKLEGFSALELAQREDFD-----VLCLQETKLQEKDINEIKRQLID 301
++ L++NVNG+R + FS + + R FD ++ QE K ++ I++ R +D
Sbjct: 17 IRFLTFNVNGIRTFFHYQPFSQMNQSLRSVFDFFRADIITFQELKTEKLSISKWGR--VD 74
Query: 302 GYENSFWTCSVSKLGYSGTAIISRIK----PL---------------------------- 329
G+ SF + ++ GYSG RI PL
Sbjct: 75 GFY-SFISIPQTRKGYSGVGCWIRIPEKNHPLYHALQVVKAEEGITGYLTIKNGKHSAIS 133
Query: 330 ---SVRYGLGISDH-------------DTEGRLVTAEFDT-FYLICGYIPNSGDGLKRLS 372
V G+G D D+EGR V E +I Y P + + +
Sbjct: 134 YRNDVNQGIGGYDSLDPDLDEKSALELDSEGRCVMVELACGIVIISVYCPANSNSSEEGE 193
Query: 373 YRVTQWDPSLSNYLKELEKT-KPVVLTGDLNCAHEEID-----------IYNPAGNKK-S 419
++ L ++ L+K K +VL GD+N + ID I +P G K
Sbjct: 194 MFRLRFLKVLLRRVRNLDKIGKKIVLMGDVNVCRDLIDSADTLEQFSIPITDPMGGTKLE 253
Query: 420 AGFTDEERK-------------------SFAANFLSRGF-VDTFR--RQHPDVVGYTYWG 457
A + D+ + S + RG +DT R + + YT W
Sbjct: 254 AQYRDKAIQFIINPDTPHRRIFNQILADSLLPDASKRGILIDTTRLIQTRNRLKMYTVWN 313
Query: 458 YRHGGRKTNRGWRLDYFLVSESIADKVHDSYILPDVMGSDHCPV 501
R +N G R+D+ LVS + + + ILPD++GSDHCPV
Sbjct: 314 MLKNLRPSNYGSRIDFILVSLKLERCIKAADILPDILGSDHCPV 357
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 46.2 bits (108), Expect = 2e-04
Identities = 59/275 (21%), Positives = 117/275 (42%), Gaps = 45/275 (16%)
Query: 241 TRDTKFVKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLI 300
T + +L+ NVNGL A +K + + +D V C+QET L +D + +K I
Sbjct: 1 TGSNSHITILTLNVNGLNAPIKRHRLA--NWIKSQDPSVCCIQETHLTCRDTHRLK---I 55
Query: 301 DGYENSFWTCSVSKLGYSGTAII----SRIKPLSVRYGLGISDHDTEGRLV----TAEFD 352
G+ N + K +G AI+ + KP ++ D EG + + + +
Sbjct: 56 KGWRNIYQANGKQK--KAGVAILVSDKTDFKPTKIK-------RDKEGHYIMVKGSIQQE 106
Query: 353 TFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYN 412
++ Y PN+ G R +V LS+ ++L+ ++ GD N +D
Sbjct: 107 ELTILNIYAPNT--GAPRFIKQV------LSDLQRDLD--SHTIIMGDFNTPLSTLD--- 153
Query: 413 PAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVGYTYWGYRHGGRKTNRGWRLD 472
+ + +++ + + +D +R HP YT++ H + + D
Sbjct: 154 ----RSTRQKINKDIQELNSALHQADLIDIYRTLHPKSTEYTFFSAPH-----HTYSKTD 204
Query: 473 YFLVSESIADKVHDSYILPDVMGSDHCPVGLIIKL 507
+ L S+++ K + I+ + + SDH + L +++
Sbjct: 205 HILGSKTLLSKCKRTEIITNCL-SDHSAIKLELRI 238
>DH47_ARATH (P31168) Dehydrin COR47 (Cold-induced COR47 protein)
Length = 265
Score = 43.1 bits (100), Expect = 0.002
Identities = 34/148 (22%), Positives = 63/148 (41%), Gaps = 9/148 (6%)
Query: 84 GRKEDLL-----STLKSFMDNNMGEQHPQIEEEHGLLISSENTSVEVKTKKVDEDHVDDI 138
G+KE+ + +TL+S D+ P++ EH + ++ T +E +K +ED
Sbjct: 39 GKKEEEVKPQETTTLESEFDHKAQISEPELAAEHEEVKENKITLLEELQEKTEEDE---- 94
Query: 139 NDNPEVFEHSRGKRRLKQSGSESETVKVTTKKKLLVESDEVSDFKPSRAKRRVSSDIVRV 198
+ P V E S S+ E + KKK +VE +E + K ++ +
Sbjct: 95 ENKPSVIEKLHRSNSSSSSSSDEEGEEKKEKKKKIVEGEEDKKGLVEKIKEKLPGHHDKT 154
Query: 199 VSQSDEISTTTIPTEPWTVLAHKKPQKD 226
+STT +V+ H P+++
Sbjct: 155 AEDDVPVSTTIPVPVSESVVEHDHPEEE 182
>YKQ0_CAEEL (P34305) Putative ATP-dependent RNA helicase C06E1.10 in
chromosome III (EC 3.6.1.-)
Length = 1148
Score = 42.7 bits (99), Expect = 0.002
Identities = 40/153 (26%), Positives = 70/153 (45%), Gaps = 14/153 (9%)
Query: 79 RVPAKGRKEDLLSTLKSFMDNNMGEQHPQIEEEHGLLISSENTSVEVKTKKVDEDHVDD- 137
+ AK ++D T D+ + + EEE I +VEVK + +D D VD+
Sbjct: 126 KTEAKRTQQDYYPT-----DDESSSEEEEEEEEGDNDIEDAGNTVEVKIEPIDLDDVDEA 180
Query: 138 INDNPEVFEHSRGKRRLKQSGSESE------TVKVTTKKKLLVESDEVSDFKPSRAKRRV 191
I+ NPE +R S++E T V +KK++VE + + + SRA+ +
Sbjct: 181 IDGNPETNLDQIVVKREDDEESDNEDILALPTTTVINRKKVIVERSK--EIQKSRAELPI 238
Query: 192 SSDIVRVVSQSDEISTTTIPTEPWTVLAHKKPQ 224
++ +R+V +E T + E + + PQ
Sbjct: 239 FAEEMRIVEAINENLVTVVCGETGSGKTTQIPQ 271
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 42.4 bits (98), Expect = 0.003
Identities = 57/263 (21%), Positives = 98/263 (36%), Gaps = 44/263 (16%)
Query: 244 TKFVKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINEIKRQLIDGY 303
+K + + S NVNGL LK + + Q+ D+ C+QE+ L KD +K + G+
Sbjct: 4 SKGLSIFSINVNGLNCPLKRHRLA--DWIQKLKPDICCIQESHLTLKDKYRLK---VKGW 58
Query: 304 ENSFWTCSVSKLGYSGTAIISR----IKPLSVRYGLGISDHDTEGRLV----TAEFDTFY 355
+ F K +G AI+ KP +R D +G + ++D
Sbjct: 59 SSIFQANGKQKK--AGIAILFADAIGFKPTKIR-------KDKDGHFIFVKGNTQYDEIS 109
Query: 356 LICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYNPAG 415
+I Y PN + + +SN + ++ GD N +D
Sbjct: 110 IINIYAPNHN-----APQFIRETLTDMSNLISSTS-----IVVGDFNTPLAVLD------ 153
Query: 416 NKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVGYTYWGYRHGGRKTNRGWRLDYFL 475
+ S +E + D +R HP+ YT++ HG ++D+ L
Sbjct: 154 -RSSKKKLSKEILDLNSTIQHLDLTDIYRTFHPNKTEYTFFSSAHGTYS-----KIDHIL 207
Query: 476 VSESIADKVHDSYILPDVMGSDH 498
+S K I+P + H
Sbjct: 208 GHKSNLSKFKKIEIIPCIFSDHH 230
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 38.5 bits (88), Expect = 0.041
Identities = 33/137 (24%), Positives = 58/137 (42%), Gaps = 17/137 (12%)
Query: 353 TFYLICGYIPNSGDGLKRLSYRVTQWDPSLSNYLKELEKTKPVVLTGDLNCAHEEIDIYN 412
T+ L+ Y P +G R SLS Y++ ++ + +++ GD N + D
Sbjct: 103 TYNLMNVYAPTTGPERARFF-------ESLSAYMETIDSDEALIIGGDFNYTLDARDRNV 155
Query: 413 PAGNKKSAGFTDEERKSFAANFLSRGFVDTFRRQHPDVVGYTYWGYRHGGRKTNRGWRLD 472
P K ++ + A+F VD +R Q+P+ V +TY R G + R+D
Sbjct: 156 P----KKRDSSESVLRELIAHF---SLVDVWREQNPETVAFTYVRVRDGHVSQS---RID 205
Query: 473 YFLVSESIADKVHDSYI 489
+S + + S I
Sbjct: 206 RIYISSHLMSRAQSSTI 222
>PUMA_PARUN (O61308) 227 kDa spindle- and centromere-associated
protein (PUMA1)
Length = 1955
Score = 38.5 bits (88), Expect = 0.041
Identities = 66/305 (21%), Positives = 122/305 (39%), Gaps = 49/305 (16%)
Query: 14 MLPFVEEKKDMKLNKGLEASPGSKKHVIKGNDANSYSIEIERLRNDPSIVDTMTVQELRK 73
M +EE K + EA+ +KH I N++ EIER+R+D ++++K
Sbjct: 944 MEKMLEEAKHRETVLREEATEWEEKHDIISNESLKLRNEIERIRSDAE-------EDIQK 996
Query: 74 TLKSIRVPAKGRK--EDLLSTLKSFMDNNMGEQHPQIEEEHGLLISSENTSVEVKTKKVD 131
K + + K E + TL+S Q+ + ++S NT++ +T K+
Sbjct: 997 WKKDVHMAQNELKNLERVCETLRS-----------QLTAAND-RVASLNTTINEQTSKIR 1044
Query: 132 EDHVDDINDNPEVFEHSRGKRRLKQSGSESETVKVTTKKKLLVESDEVSDFKPSRAKRRV 191
E +N + E R S E++ T + + E + + + ++K +
Sbjct: 1045 E-----LNSHEHRLEEDLADSRATSSAIENDLGNATGRLRSSEEHNAILQSENRKSKTEI 1099
Query: 192 SS-----DIVRVVSQSDE----------ISTTTIPTEPWTVLAHKKPQKDWIA--YNPKT 234
+ D + +S E + TTTI E + + + D + Y KT
Sbjct: 1100 EALKHQIDTIMNTKESCESEVERLKKKIVQTTTITKEQNEKIEKLRIEHDHLERDYREKT 1159
Query: 235 MRPQPLTRDTKFVKLLSWNVNGLRALLKLEGFSALELAQREDFDVLCLQETKLQEKDINE 294
L K +L VN RA +L+ FS + + + + + KL +K++
Sbjct: 1160 KEVDRLKEVEKTFEL---KVN--RARQELDEFSKKLIVTETERNAISGEAQKL-DKEVQL 1213
Query: 295 IKRQL 299
+K QL
Sbjct: 1214 VKEQL 1218
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,399,676
Number of Sequences: 164201
Number of extensions: 2711351
Number of successful extensions: 6335
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 6242
Number of HSP's gapped (non-prelim): 98
length of query: 507
length of database: 59,974,054
effective HSP length: 115
effective length of query: 392
effective length of database: 41,090,939
effective search space: 16107648088
effective search space used: 16107648088
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0278.2


