

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0269b.6
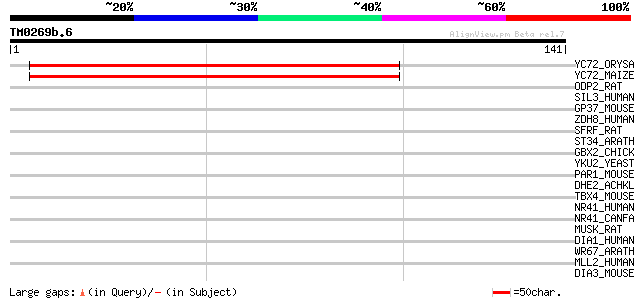
(141 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YC72_ORYSA (Q36996) Hypothetical 14.9 kDa protein ycf72 (ORF137) 174 7e-44
YC72_MAIZE (Q37082) Hypothetical 14.9 kDa protein ycf72 (ORF137) 172 3e-43
ODP2_RAT (P08461) Dihydrolipoyllysine-residue acetyltransferase ... 33 0.15
SIL3_HUMAN (O60292) Signal-induced proliferation-associated 1 li... 31 0.75
GP37_MOUSE (Q9QY42) Probable G protein-coupled receptor 37 precu... 31 0.98
ZDH8_HUMAN (Q9ULC8) Zinc finger DHHC domain containing protein 8... 30 1.3
SFRF_RAT (Q63627) Splicing factor, arginine/serine-rich 15 (CTD-... 30 1.7
ST34_ARATH (Q9LW86) Probable sulfate transporter 3.4 30 2.2
GBX2_CHICK (O42230) Homeobox protein GBX-2 (Gastrulation and bra... 30 2.2
YKU2_YEAST (P36042) Hypothetical 21.2 kDa protein in TOR2-MNN4 i... 28 4.9
PAR1_MOUSE (P30558) Proteinase activated receptor 1 precursor (P... 28 4.9
DHE2_ACHKL (P41755) NAD-specific glutamate dehydrogenase (EC 1.4... 28 4.9
TBX4_MOUSE (P70325) T-box transcription factor TBX4 (T-box prote... 28 6.4
NR41_HUMAN (P22736) Orphan nuclear receptor NR4A1 (Orphan nuclea... 28 6.4
NR41_CANFA (P51666) Orphan nuclear receptor NR4A1 (Orphan nuclea... 28 6.4
MUSK_RAT (Q62838) Muscle, skeletal receptor tyrosine protein kin... 28 6.4
DIA1_HUMAN (O60610) Diaphanous protein homolog 1 (Diaphanous-rel... 28 6.4
WR67_ARATH (Q93WV7) Probable WRKY transcription factor 67 (WRKY ... 28 8.3
MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia p... 28 8.3
DIA3_MOUSE (Q9Z207) Diaphanous protein homolog 3 (Diaphanous-rel... 28 8.3
>YC72_ORYSA (Q36996) Hypothetical 14.9 kDa protein ycf72 (ORF137)
Length = 137
Score = 174 bits (440), Expect = 7e-44
Identities = 79/94 (84%), Positives = 82/94 (87%)
Query: 6 FTPPPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRTSP 65
F PPPWGWSTGFITTPLTTGRLPSQH DPALPKLFWFTPT PTCPTVA+QF D KRTSP
Sbjct: 4 FPSPPPWGWSTGFITTPLTTGRLPSQHLDPALPKLFWFTPTLPTCPTVAKQFWDTKRTSP 63
Query: 66 EGNFNVADFPSFAISFATAPAALANFPPFPNVIT 99
+GN VAD PSFAISFATAPAALAN PP P VI+
Sbjct: 64 DGNLKVADLPSFAISFATAPAALANCPPLPRVIS 97
>YC72_MAIZE (Q37082) Hypothetical 14.9 kDa protein ycf72 (ORF137)
Length = 137
Score = 172 bits (435), Expect = 3e-43
Identities = 78/94 (82%), Positives = 82/94 (86%)
Query: 6 FTPPPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRTSP 65
F PPPWGWSTGFITTPLTTGRLPSQH DPALPKLFWFTPT PTCPTVA+QF D KRTSP
Sbjct: 4 FPSPPPWGWSTGFITTPLTTGRLPSQHLDPALPKLFWFTPTLPTCPTVAKQFWDTKRTSP 63
Query: 66 EGNFNVADFPSFAISFATAPAALANFPPFPNVIT 99
+GN VA+ PSFAISFATAPAALAN PP P VI+
Sbjct: 64 DGNLKVANLPSFAISFATAPAALANCPPLPRVIS 97
>ODP2_RAT (P08461) Dihydrolipoyllysine-residue acetyltransferase
component of pyruvate dehydrogenase complex (EC
2.3.1.12) (E2) (Dihydrolipoamide acetyltransferase
component of pyruvate dehydrogenase complex) (PDC-E2)
(70 kDa mitochondrial autoantigen of
Length = 555
Score = 33.5 bits (75), Expect = 0.15
Identities = 32/97 (32%), Positives = 38/97 (38%), Gaps = 11/97 (11%)
Query: 4 PPFTPPPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLD---I 60
PP PPP P+ P+ PA PK F AE+ +D +
Sbjct: 232 PPPVPPPVAA------VPPIPQPLAPTPSAAPAGPKGRVFVSPLAK-KLAAEKGIDLTQV 284
Query: 61 KRTSPEGNFNVADFPSFAISFATAPAALANFPPFPNV 97
K T PEG D SF + APAA A PP P V
Sbjct: 285 KGTGPEGRIIKKDIDSF-VPTKAAPAAAAAAPPGPRV 320
>SIL3_HUMAN (O60292) Signal-induced proliferation-associated 1 like
protein 3 (Fragment)
Length = 1368
Score = 31.2 bits (69), Expect = 0.75
Identities = 24/99 (24%), Positives = 35/99 (35%), Gaps = 22/99 (22%)
Query: 2 GGPPFTPPPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFP------TCPTVAE 55
G PP+ PW WS P+ H +LP W TPT P + P
Sbjct: 659 GRPPYRSNAPWQWSG------------PASH--NSLPASKWATPTTPGHAQSLSRPLKQT 704
Query: 56 QFLDIKRTSPEGNFNVADFPSFAISFATAPAALANFPPF 94
+ + + P + FP + +PA PP+
Sbjct: 705 PIVPFRESQPLHSKRPVSFPE--TPYTVSPAGADRVPPY 741
>GP37_MOUSE (Q9QY42) Probable G protein-coupled receptor 37
precursor
Length = 600
Score = 30.8 bits (68), Expect = 0.98
Identities = 17/45 (37%), Positives = 21/45 (45%), Gaps = 10/45 (22%)
Query: 3 GPPFTPPPPWGWSTGFITTPLTTGRLPSQHF---DPALPKLFWFT 44
GPP PP W W + G+ PS H DP P+LF+ T
Sbjct: 108 GPPTKPPSAWRWKS-------AQGKEPSGHLRRRDPTDPQLFFPT 145
>ZDH8_HUMAN (Q9ULC8) Zinc finger DHHC domain containing protein 8
(Zinc finger protein 378)
Length = 673
Score = 30.4 bits (67), Expect = 1.3
Identities = 18/54 (33%), Positives = 22/54 (40%), Gaps = 3/54 (5%)
Query: 3 GPPFTPPPPWGWSTGFITTPL---TTGRLPSQHFDPALPKLFWFTPTFPTCPTV 53
GPP P G + + TP L Q P P ++ F P FPT P V
Sbjct: 222 GPPLPPKIEAGTFSSDLQTPRPGSAESALSVQRTSPPTPAMYKFRPAFPTGPKV 275
>SFRF_RAT (Q63627) Splicing factor, arginine/serine-rich 15
(CTD-binding SR-like protein RA4) (Fragment)
Length = 1048
Score = 30.0 bits (66), Expect = 1.7
Identities = 16/37 (43%), Positives = 16/37 (43%)
Query: 2 GGPPFTPPPPWGWSTGFITTPLTTGRLPSQHFDPALP 38
G PP PPPP W GF L G LP P P
Sbjct: 628 GVPPPPPPPPPFWGPGFNPMHLPPGFLPPGPPPPITP 664
>ST34_ARATH (Q9LW86) Probable sulfate transporter 3.4
Length = 653
Score = 29.6 bits (65), Expect = 2.2
Identities = 16/52 (30%), Positives = 21/52 (39%), Gaps = 10/52 (19%)
Query: 12 WGWST-----GFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFL 58
W W T GF++ LTT +H PKLFW + P + L
Sbjct: 250 WSWETIVMGIGFLSILLTT-----RHISMRKPKLFWISAASPLASVIISTLL 296
>GBX2_CHICK (O42230) Homeobox protein GBX-2 (Gastrulation and
brain-specific homeobox protein 2)
Length = 340
Score = 29.6 bits (65), Expect = 2.2
Identities = 23/69 (33%), Positives = 27/69 (38%), Gaps = 9/69 (13%)
Query: 5 PFTPPPPWGWSTGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRTS 64
P PPPP G L G + ALP F P P P A +F +
Sbjct: 72 PPAPPPPLPALPGAFCPGLAQGMALTSTLMAALPGSF---PASPPRPEAARKF------A 122
Query: 65 PEGNFNVAD 73
P GNF+ AD
Sbjct: 123 PPGNFDKAD 131
>YKU2_YEAST (P36042) Hypothetical 21.2 kDa protein in TOR2-MNN4
intergenic region
Length = 193
Score = 28.5 bits (62), Expect = 4.9
Identities = 23/95 (24%), Positives = 41/95 (42%), Gaps = 7/95 (7%)
Query: 15 STGFITTPLTTGRLPSQHFDPALPKLFWFTPTFPTCPTVAEQFLDIKRTSPEGNFNVADF 74
S+ F+ P ++ L F P P +F P+ + L + P +F+
Sbjct: 100 SSSFLLLPASSSLLLPSSFSPLPP-----ASSFLLLPSFSSLLLPSFSSLPLPSFSFLLL 154
Query: 75 PSFAISFATAPAALANFPPFPNVITHFFINRTFYK 109
PS SF P++ + PP + + FF+ T++K
Sbjct: 155 PSS--SFPLLPSSSSPLPPSSSSPSAFFLQVTWHK 187
>PAR1_MOUSE (P30558) Proteinase activated receptor 1 precursor
(PAR-1) (Thrombin receptor)
Length = 430
Score = 28.5 bits (62), Expect = 4.9
Identities = 13/29 (44%), Positives = 20/29 (68%), Gaps = 2/29 (6%)
Query: 4 PPFTPPPPW--GWSTGFITTPLTTGRLPS 30
PP TPPPP+ ++G++T+P T +PS
Sbjct: 84 PPHTPPPPFISEDASGYLTSPWLTLFMPS 112
>DHE2_ACHKL (P41755) NAD-specific glutamate dehydrogenase (EC
1.4.1.2) (NAD-GDH)
Length = 1063
Score = 28.5 bits (62), Expect = 4.9
Identities = 15/35 (42%), Positives = 19/35 (53%), Gaps = 3/35 (8%)
Query: 85 PAALANFPPFPNVITHFFINRTFYKRHKTKRDNPS 119
P A +FPP+ THF++N T Y K DN S
Sbjct: 245 PLAQKHFPPY---FTHFYLNVTIYAVVKNILDNIS 276
>TBX4_MOUSE (P70325) T-box transcription factor TBX4 (T-box protein
4)
Length = 552
Score = 28.1 bits (61), Expect = 6.4
Identities = 22/87 (25%), Positives = 33/87 (37%), Gaps = 7/87 (8%)
Query: 8 PPPPWGWSTGFITTPLTTGRLPSQHFDP--ALPKLFWFTPTFPTCPTVAEQFLDIKRTSP 65
PPPP + +P T+ + + P P F T P PT+A Q P
Sbjct: 418 PPPPLSCNMWTSVSPYTSYSVQTMETVPYQPFPAHFTATTVMPRLPTIAAQ-----SAQP 472
Query: 66 EGNFNVADFPSFAISFATAPAALANFP 92
GN + + + + S A+FP
Sbjct: 473 PGNAHFSVYNQLSQSQVRERGPSASFP 499
>NR41_HUMAN (P22736) Orphan nuclear receptor NR4A1 (Orphan nuclear
receptor HMR) (Early response protein NAK1) (TR3 orphan
receptor)
Length = 598
Score = 28.1 bits (61), Expect = 6.4
Identities = 20/66 (30%), Positives = 27/66 (40%), Gaps = 9/66 (13%)
Query: 4 PPFTPPP--PWGWSTGFITTPLT-------TGRLPSQHFDPALPKLFWFTPTFPTCPTVA 54
P F PP PW S G + T T +LP P P F F+P P++A
Sbjct: 144 PSFQPPQLSPWDGSFGHFSPSQTYEGLRAWTEQLPKASGPPQPPAFFSFSPPTGPSPSLA 203
Query: 55 EQFLDI 60
+ L +
Sbjct: 204 QSPLKL 209
>NR41_CANFA (P51666) Orphan nuclear receptor NR4A1 (Orphan nuclear
receptor HMR) (Orphan nuclear receptor NGFI-B)
Length = 598
Score = 28.1 bits (61), Expect = 6.4
Identities = 24/88 (27%), Positives = 36/88 (40%), Gaps = 11/88 (12%)
Query: 4 PPFTPPP--PWGWSTGFITTPLT-------TGRLPSQHFDPALPKLFWFTPTFPTCPTVA 54
P F PP PW S G + T T +LP P P F F+P P++A
Sbjct: 144 PSFQPPQLSPWDGSFGPFSPSQTYEGLRAWTEQLPKASGHPQPPAFFSFSPPTGPSPSLA 203
Query: 55 EQFLDIKRTSPEGNFNVADFPSFAISFA 82
+ +K + + + S++IS A
Sbjct: 204 QS--PLKLFPSQATCQLGERESYSISTA 229
>MUSK_RAT (Q62838) Muscle, skeletal receptor tyrosine protein kinase
precursor (EC 2.7.1.112) (Muscle-specific kinase
receptor) (MuSK)
Length = 868
Score = 28.1 bits (61), Expect = 6.4
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 13/64 (20%)
Query: 27 RLPSQHFDPA----LPKLFWFTPTFPTCPTVAEQFLDIKRTSPEGNFNVADFPSFAISFA 82
+LPS H DP LP L + T P++ TS + + ++ + P+ SFA
Sbjct: 436 KLPSMHQDPTACTRLPYLDYKKENITTFPSI---------TSSKPSVDIPNLPASTSSFA 486
Query: 83 TAPA 86
+PA
Sbjct: 487 VSPA 490
>DIA1_HUMAN (O60610) Diaphanous protein homolog 1
(Diaphanous-related formin 1) (DRF1)
Length = 1248
Score = 28.1 bits (61), Expect = 6.4
Identities = 19/48 (39%), Positives = 20/48 (41%), Gaps = 4/48 (8%)
Query: 2 GGPPFTPPPPWGWSTGFITTPLTTGRLPSQHFD---PALPKL-FWFTP 45
GGP PPPP+ G P G P F PA P L F TP
Sbjct: 701 GGPGIPPPPPFPGGPGIPPPPPGMGMPPPPPFGFGVPAAPVLPFGLTP 748
>WR67_ARATH (Q93WV7) Probable WRKY transcription factor 67 (WRKY
DNA-binding protein 67)
Length = 254
Score = 27.7 bits (60), Expect = 8.3
Identities = 20/73 (27%), Positives = 30/73 (40%), Gaps = 8/73 (10%)
Query: 33 FDPALPKLFWFTPTFPTCPTVAEQFLDIKRTSPEGNFNV-ADFPSFAISFATAPAALANF 91
FD +P+ P P T+ Q + ++ S E N D + + P + F
Sbjct: 184 FDQVVPE-----PIMPQLTTIDHQVITVEENSAEHIMNQECDINDYLVD--DDPFWASQF 236
Query: 92 PPFPNVITHFFIN 104
PPFP+ T F N
Sbjct: 237 PPFPSSDTMFLEN 249
>MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia
protein 2 (ALL1-related protein)
Length = 5262
Score = 27.7 bits (60), Expect = 8.3
Identities = 14/36 (38%), Positives = 17/36 (46%)
Query: 3 GPPFTPPPPWGWSTGFITTPLTTGRLPSQHFDPALP 38
GPP P PP STG + P+ PS +P P
Sbjct: 4010 GPPRLPAPPGALSTGPVLGPVHPTPPPSSPQEPKRP 4045
>DIA3_MOUSE (Q9Z207) Diaphanous protein homolog 3
(Diaphanous-related formin 3) (DRF3) (mDIA2) (p134mDIA2)
Length = 1171
Score = 27.7 bits (60), Expect = 8.3
Identities = 16/44 (36%), Positives = 22/44 (49%), Gaps = 4/44 (9%)
Query: 3 GPPFTPPPPWGWSTGFITTPLTT--GRLPSQHFDP--ALPKLFW 42
G P PPPP G+ G + PL G P + F P ++ +L W
Sbjct: 587 GGPVPPPPPLGFLGGQSSIPLNLPFGLKPKKEFKPEISMRRLNW 630
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.329 0.144 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,185,191
Number of Sequences: 164201
Number of extensions: 887941
Number of successful extensions: 2520
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 2508
Number of HSP's gapped (non-prelim): 25
length of query: 141
length of database: 59,974,054
effective HSP length: 99
effective length of query: 42
effective length of database: 43,718,155
effective search space: 1836162510
effective search space used: 1836162510
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0269b.6


