

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
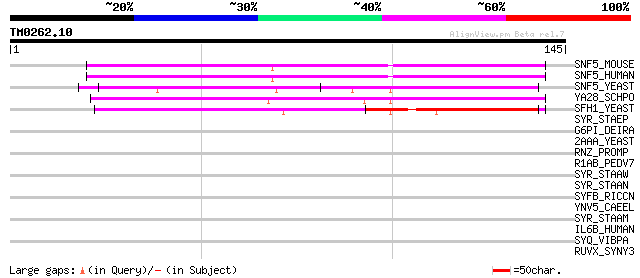
Query= TM0262.10
(145 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SNF5_MOUSE (Q9Z0H3) SWI/SNF related, matrix associated, actin de... 75 5e-14
SNF5_HUMAN (Q12824) SWI/SNF related, matrix associated, actin de... 75 5e-14
SNF5_YEAST (P18480) Transcription regulatory protein SNF5 (SWI/S... 64 9e-11
YA28_SCHPO (Q09699) Hypothetical protein C2F7.08c in chromosome I 62 6e-10
SFH1_YEAST (Q06168) Chromatin structure remodeling complex prote... 52 3e-07
SYR_STAEP (Q8CTN9) Arginyl-tRNA synthetase (EC 6.1.1.19) (Argini... 31 1.0
G6PI_DEIRA (Q9RTL8) Glucose-6-phosphate isomerase (EC 5.3.1.9) (... 30 1.8
2AAA_YEAST (P31383) Protein phosphatase PP2A regulatory subunit ... 29 3.0
RNZ_PROMP (Q7V0B9) Ribonuclease Z (EC 3.1.26.11) (RNase Z) (tRNA... 29 4.0
R1AB_PEDV7 (Q91AV2) Replicase polyprotein 1ab (pp1ab) (ORF1ab po... 29 4.0
SYR_STAAW (Q8NXT8) Arginyl-tRNA synthetase (EC 6.1.1.19) (Argini... 28 5.2
SYR_STAAN (Q99W05) Arginyl-tRNA synthetase (EC 6.1.1.19) (Argini... 28 5.2
SYFB_RICCN (Q92I38) Phenylalanyl-tRNA synthetase beta chain (EC ... 28 5.2
YNV5_CAEEL (P34568) Hypothetical protein T16H12.5 in chromosome III 28 6.8
SYR_STAAM (Q932F6) Arginyl-tRNA synthetase (EC 6.1.1.19) (Argini... 28 6.8
IL6B_HUMAN (P40189) Interleukin-6 receptor beta chain precursor ... 28 6.8
SYQ_VIBPA (Q87RG4) Glutaminyl-tRNA synthetase (EC 6.1.1.18) (Glu... 28 8.9
RUVX_SYNY3 (P74662) Putative Holliday junction resolvase (EC 3.1... 28 8.9
>SNF5_MOUSE (Q9Z0H3) SWI/SNF related, matrix associated, actin
dependent regulator of chromatin subfamily B member 1
(Integrase interactor 1 protein) (mSNF5)
Length = 385
Score = 75.1 bits (183), Expect = 5e-14
Identities = 43/121 (35%), Positives = 72/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +F+ DL L P FV IA +I
Sbjct: 184 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFSEILCDDLDLNPLTFVPAIASAIRQ 243
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 244 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 302
Query: 140 I 140
+
Sbjct: 303 L 303
Score = 36.2 bits (82), Expect = 0.025
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 258 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 317
Query: 80 QLA 82
QL+
Sbjct: 318 QLS 320
>SNF5_HUMAN (Q12824) SWI/SNF related, matrix associated, actin
dependent regulator of chromatin subfamily B member 1
(Integrase interactor 1 protein) (hSNF5) (BAF47)
Length = 385
Score = 75.1 bits (183), Expect = 5e-14
Identities = 43/121 (35%), Positives = 72/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +F+ DL L P FV IA +I
Sbjct: 184 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFSEILCDDLDLNPLTFVPAIASAIRQ 243
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 244 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 302
Query: 140 I 140
+
Sbjct: 303 L 303
Score = 36.2 bits (82), Expect = 0.025
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 258 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 317
Query: 80 QLA 82
QL+
Sbjct: 318 QLS 320
>SNF5_YEAST (P18480) Transcription regulatory protein SNF5 (SWI/SNF
complex component SNF5) (Transcription factor TYE4)
Length = 905
Score = 64.3 bits (155), Expect = 9e-11
Identities = 41/131 (31%), Positives = 68/131 (51%), Gaps = 11/131 (8%)
Query: 19 TAENLVPIRLDIEIDAKRY--KDAFTWNPSDPDHEVGVFARRTVKDLKLPPP----FVTQ 72
T+E LVPIRL+ + D R+ +D WN +D ++ F ++D + +
Sbjct: 453 TSEQLVPIRLEFDQDRDRFFLRDTLLWNKNDKLIKIEDFVDDMLRDYRFEDATREQHIDT 512
Query: 73 IAQSIESQLAEFRSYE----GQDMYAGEKI-IPIKLDLCVNHMLVKDQFLWDLNNFESDL 127
I QSI+ Q+ EF+ QD G+ + I IKLD+ V + DQF W+++N ++
Sbjct: 513 ICQSIQEQIQEFQGNPYIELNQDRLGGDDLRIRIKLDIVVGQNQLIDQFEWEISNSDNCP 572
Query: 128 EELARIFCKDM 138
EE A C+++
Sbjct: 573 EEFAESMCQEL 583
Score = 41.2 bits (95), Expect = 8e-04
Identities = 22/58 (37%), Positives = 32/58 (54%)
Query: 24 VPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIESQL 81
+ I+LDI + + D F W S+ D+ FA ++L+LP FVT IA SI Q+
Sbjct: 544 IRIKLDIVVGQNQLIDQFEWEISNSDNCPEEFAESMCQELELPGEFVTAIAHSIREQV 601
>YA28_SCHPO (Q09699) Hypothetical protein C2F7.08c in chromosome I
Length = 632
Score = 61.6 bits (148), Expect = 6e-10
Identities = 45/156 (28%), Positives = 71/156 (44%), Gaps = 37/156 (23%)
Query: 22 NLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLP---PPFVTQIAQSIE 78
+LVPIRL+I+ D + +D+FTWN D + FA + D +P V I++SI+
Sbjct: 140 DLVPIRLEIDADRYKLRDSFTWNLYDKCISLDQFAEQICIDYDIPLHNVHIVQNISKSIQ 199
Query: 79 SQLAEFRSYEGQD---------------MYAGEKI-------------------IPIKLD 104
+Q+ ++ + Q +YA E I IKLD
Sbjct: 200 AQINDYEPRKAQSNLSFVSDVSSSTSETVYAHEPSDSLAKASKQQIPTVQNDLRILIKLD 259
Query: 105 LCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMGI 140
+ + + + DQF W+L ES EE A + C D+G+
Sbjct: 260 ITIGRLNLIDQFEWNLFAPESSAEEFATVMCLDLGL 295
Score = 39.3 bits (90), Expect = 0.003
Identities = 26/76 (34%), Positives = 34/76 (44%), Gaps = 2/76 (2%)
Query: 14 KFRMPTAEN--LVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVT 71
K ++PT +N + I+LDI I D F WN P+ FA DL L F T
Sbjct: 242 KQQIPTVQNDLRILIKLDITIGRLNLIDQFEWNLFAPESSAEEFATVMCLDLGLSGEFCT 301
Query: 72 QIAQSIESQLAEFRSY 87
+A SI Q + Y
Sbjct: 302 AVAHSIREQCQMYIKY 317
>SFH1_YEAST (Q06168) Chromatin structure remodeling complex protein
SFH1 (SNF5 homolog 1)
Length = 426
Score = 52.4 bits (124), Expect = 3e-07
Identities = 36/119 (30%), Positives = 55/119 (45%), Gaps = 2/119 (1%)
Query: 23 LVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFV-TQIAQSIESQL 81
++PI LDIE KD F WN +D FA KDL + + TQIA I+ QL
Sbjct: 203 MIPITLDIEHMGHTIKDQFLWNYNDDSISPEEFASIYCKDLDMTSATLQTQIANIIKEQL 262
Query: 82 AEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMGI 140
+ + ++ + +I I L + +D F W+LN+ E A +D+G+
Sbjct: 263 KDLENIAATEIMSDLHVI-INLTCNLQDRFFEDNFQWNLNDKSLTPERFATSIVQDLGL 320
Score = 42.0 bits (97), Expect = 5e-04
Identities = 24/48 (50%), Positives = 31/48 (64%), Gaps = 5/48 (10%)
Query: 94 AGEKI-IPIKLDLCVNHM--LVKDQFLWDLNNFESDLEELARIFCKDM 138
+GE I IPI LD + HM +KDQFLW+ N+ EE A I+CKD+
Sbjct: 198 SGEAIMIPITLD--IEHMGHTIKDQFLWNYNDDSISPEEFASIYCKDL 243
Score = 35.0 bits (79), Expect = 0.056
Identities = 30/116 (25%), Positives = 46/116 (38%), Gaps = 21/116 (18%)
Query: 24 VPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIESQLAE 83
V I L + + ++D F WN +D FA V+DL L F+ I+QS+
Sbjct: 279 VIINLTCNLQDRFFEDNFQWNLNDKSLTPERFATSIVQDLGLTREFIPLISQSLH----- 333
Query: 84 FRSYEGQDMYAGEKIIPIKLDLCVNHML----VKDQFLWDLNNFESDLEELARIFC 135
E I+ IK D H++ D L+ D++EL +C
Sbjct: 334 ------------ETILKIKKDWVDGHLIQDHVPNDAAFGYLSGIRLDIDELGSNWC 377
>SYR_STAEP (Q8CTN9) Arginyl-tRNA synthetase (EC 6.1.1.19)
(Arginine--tRNA ligase) (ArgRS)
Length = 553
Score = 30.8 bits (68), Expect = 1.0
Identities = 21/49 (42%), Positives = 28/49 (56%), Gaps = 4/49 (8%)
Query: 70 VTQIAQSIESQLAEF---RSYE-GQDMYAGEKIIPIKLDLCVNHMLVKD 114
+T +A+SIE++ E SYE D Y G+ II I DL V H +KD
Sbjct: 174 ITNLARSIEARYFEALGDTSYEMPADGYNGKDIIEIGKDLAVKHPEIKD 222
>G6PI_DEIRA (Q9RTL8) Glucose-6-phosphate isomerase (EC 5.3.1.9)
(GPI) (Phosphoglucose isomerase) (PGI) (Phosphohexose
isomerase) (PHI)
Length = 541
Score = 30.0 bits (66), Expect = 1.8
Identities = 24/104 (23%), Positives = 44/104 (42%), Gaps = 11/104 (10%)
Query: 42 TWNPSDPDHEV---GVFARRTVKDLKLPPPFVTQIAQSIESQLAEFRSYEGQ---DMYAG 95
T NP P H++ VFA+ + L +A + ++A R +EG
Sbjct: 407 TLNPLPPHHDLLMANVFAQ--TEALAFGKTLEQVLADGVAPEVAPHRVFEGNRPTSTILA 464
Query: 96 EKIIPIKLDLCV---NHMLVKDQFLWDLNNFESDLEELARIFCK 136
+++ P L + H + +WD+N+F+ EL ++ K
Sbjct: 465 DRLTPRTLGALIALYEHKVFVQGAVWDINSFDQWGVELGKVLAK 508
>2AAA_YEAST (P31383) Protein phosphatase PP2A regulatory subunit A
(PR65)
Length = 635
Score = 29.3 bits (64), Expect = 3.0
Identities = 19/70 (27%), Positives = 33/70 (47%), Gaps = 8/70 (11%)
Query: 68 PFVTQIAQSIES--------QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWD 119
PF+T++AQ E QL +F Y G YA + +++ LV+++ +
Sbjct: 79 PFLTEVAQDDEDEVFAVLAEQLGKFVPYIGGPQYATILLPVLEILASAEETLVREKAVDS 138
Query: 120 LNNFESDLEE 129
LNN +L +
Sbjct: 139 LNNVAQELSQ 148
>RNZ_PROMP (Q7V0B9) Ribonuclease Z (EC 3.1.26.11) (RNase Z) (tRNA 3
endonuclease)
Length = 312
Score = 28.9 bits (63), Expect = 4.0
Identities = 17/53 (32%), Positives = 29/53 (54%), Gaps = 2/53 (3%)
Query: 40 AFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIESQLAEFRSYEGQD 91
A+ + S+ D + GVF + +D +PP P +++ QL + RS+ GQD
Sbjct: 148 AYGYRVSEKD-KPGVFDIKKAEDSNIPPGPIYSELQAGKTVQLKDGRSFNGQD 199
>R1AB_PEDV7 (Q91AV2) Replicase polyprotein 1ab (pp1ab) (ORF1ab
polyprotein) [Includes: Replicase polyprotein 1a (pp1a)
(ORF1a)] [Contains: p9; p87; p195 (EC 3.4.24.-)
(Papain-like proteinases 1/2) (PL1-PRO/PL2-PRO); Peptide
HD2; Unknown protein 1; 3C-like
Length = 6781
Score = 28.9 bits (63), Expect = 4.0
Identities = 15/51 (29%), Positives = 24/51 (46%), Gaps = 2/51 (3%)
Query: 60 VKDLKLPPPFVTQIAQSIESQLAEFRSYEGQDMYAGE--KIIPIKLDLCVN 108
VKD+ P V + +E + F SY+ D Y G+ ++ + D VN
Sbjct: 1269 VKDVNWTAPLVPAVDSVVEPVVKPFYSYKNVDFYQGDFSDLVKLPCDFVVN 1319
>SYR_STAAW (Q8NXT8) Arginyl-tRNA synthetase (EC 6.1.1.19)
(Arginine--tRNA ligase) (ArgRS)
Length = 553
Score = 28.5 bits (62), Expect = 5.2
Identities = 19/49 (38%), Positives = 27/49 (54%), Gaps = 4/49 (8%)
Query: 70 VTQIAQSIESQLAEF---RSYE-GQDMYAGEKIIPIKLDLCVNHMLVKD 114
+T +A+SIE++ E SY +D Y G+ II I DL H +KD
Sbjct: 174 ITNLARSIETRFFEALGDNSYSMPEDGYNGKDIIEIGKDLAEKHPEIKD 222
>SYR_STAAN (Q99W05) Arginyl-tRNA synthetase (EC 6.1.1.19)
(Arginine--tRNA ligase) (ArgRS)
Length = 553
Score = 28.5 bits (62), Expect = 5.2
Identities = 19/49 (38%), Positives = 27/49 (54%), Gaps = 4/49 (8%)
Query: 70 VTQIAQSIESQLAEF---RSYE-GQDMYAGEKIIPIKLDLCVNHMLVKD 114
+T +A+SIE++ E SY +D Y G+ II I DL H +KD
Sbjct: 174 ITNLARSIETRFFEALGDNSYSMPEDGYNGKDIIEIGKDLAEKHPEIKD 222
>SYFB_RICCN (Q92I38) Phenylalanyl-tRNA synthetase beta chain (EC
6.1.1.20) (Phenylalanine--tRNA ligase beta chain)
(PheRS)
Length = 818
Score = 28.5 bits (62), Expect = 5.2
Identities = 15/59 (25%), Positives = 28/59 (47%), Gaps = 3/59 (5%)
Query: 69 FVTQIAQSIESQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDL 127
++ + + + F Y G + G+K I +K++L + + D DLN+F DL
Sbjct: 749 YINNFNKKLIKSVILFDIYSGDKLPEGKKSIAVKIELQADDRTLSDT---DLNSFSKDL 804
>YNV5_CAEEL (P34568) Hypothetical protein T16H12.5 in chromosome III
Length = 451
Score = 28.1 bits (61), Expect = 6.8
Identities = 11/40 (27%), Positives = 23/40 (57%), Gaps = 1/40 (2%)
Query: 95 GEKIIPIKLDLCVNHM-LVKDQFLWDLNNFESDLEELARI 133
G+ ++P+ + C + +VK ++W +NNF EE+ +
Sbjct: 76 GDPLLPVAENWCHTQVKVVKFNYMWTINNFSFCREEMGEV 115
>SYR_STAAM (Q932F6) Arginyl-tRNA synthetase (EC 6.1.1.19)
(Arginine--tRNA ligase) (ArgRS)
Length = 553
Score = 28.1 bits (61), Expect = 6.8
Identities = 19/49 (38%), Positives = 26/49 (52%), Gaps = 4/49 (8%)
Query: 70 VTQIAQSIESQLAEF---RSYE-GQDMYAGEKIIPIKLDLCVNHMLVKD 114
+T +A+SIE+ E SY +D Y G+ II I DL H +KD
Sbjct: 174 ITNLARSIETHFFEALGDNSYSMPEDGYNGKDIIEIGKDLAEKHPEIKD 222
>IL6B_HUMAN (P40189) Interleukin-6 receptor beta chain precursor
(IL-6R-beta) (Interleukin 6 signal transducer) (Membrane
glycoprotein 130) (gp130) (Oncostatin M receptor)
(CDw130) (CD130 antigen)
Length = 918
Score = 28.1 bits (61), Expect = 6.8
Identities = 16/56 (28%), Positives = 31/56 (54%), Gaps = 3/56 (5%)
Query: 18 PTAENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQI 73
P+ ++++ ++ +I+ R KDA TW+ P+ + TV+DLK +V +I
Sbjct: 247 PSIKSVIILKYNIQY---RTKDASTWSQIPPEDTASTRSSFTVQDLKPFTEYVFRI 299
>SYQ_VIBPA (Q87RG4) Glutaminyl-tRNA synthetase (EC 6.1.1.18)
(Glutamine--tRNA ligase) (GlnRS)
Length = 556
Score = 27.7 bits (60), Expect = 8.9
Identities = 16/62 (25%), Positives = 31/62 (49%), Gaps = 3/62 (4%)
Query: 57 RRTVKDLKLPPPFVTQIAQSIESQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQF 116
R T+K+ P P+ + S+E LA F + G+ + K+D+ + M+++D
Sbjct: 134 RGTLKEPGKPSPYRDR---SVEENLALFEKMRAGEFEEGKACLRAKIDMGSSFMVMRDPV 190
Query: 117 LW 118
L+
Sbjct: 191 LY 192
>RUVX_SYNY3 (P74662) Putative Holliday junction resolvase (EC
3.1.-.-)
Length = 152
Score = 27.7 bits (60), Expect = 8.9
Identities = 21/67 (31%), Positives = 33/67 (48%), Gaps = 7/67 (10%)
Query: 51 EVGVFARRTVKDLKLPPPFVTQIAQSIESQ-----LAEFRSYEGQ--DMYAGEKIIPIKL 103
+V FARR + L LP ++ + S+E++ F SY+ D A E I+ L
Sbjct: 77 QVQKFARRVAEQLHLPLEYMDERLSSVEAENQLKARKRFSSYDKGLIDQQAAEIILQQWL 136
Query: 104 DLCVNHM 110
DL +H+
Sbjct: 137 DLRRSHL 143
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,039,948
Number of Sequences: 164201
Number of extensions: 710063
Number of successful extensions: 1408
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1388
Number of HSP's gapped (non-prelim): 27
length of query: 145
length of database: 59,974,054
effective HSP length: 100
effective length of query: 45
effective length of database: 43,553,954
effective search space: 1959927930
effective search space used: 1959927930
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0262.10


