

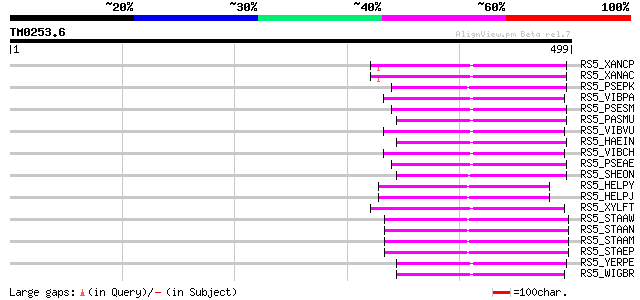
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0253.6
(499 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RS5_XANCP (P66587) 30S ribosomal protein S5 112 2e-24
RS5_XANAC (P66586) 30S ribosomal protein S5 112 2e-24
RS5_PSEPK (Q88QL8) 30S ribosomal protein S5 112 2e-24
RS5_VIBPA (Q87SZ6) 30S ribosomal protein S5 112 2e-24
RS5_PSESM (Q889V4) 30S ribosomal protein S5 112 2e-24
RS5_PASMU (Q9CL47) 30S ribosomal protein S5 112 3e-24
RS5_VIBVU (Q8DE57) 30S ribosomal protein S5 110 8e-24
RS5_HAEIN (P44374) 30S ribosomal protein S5 110 8e-24
RS5_VIBCH (Q9KP01) 30S ribosomal protein S5 108 3e-23
RS5_PSEAE (Q9HWF2) 30S ribosomal protein S5 108 3e-23
RS5_SHEON (P59124) 30S ribosomal protein S5 107 9e-23
RS5_HELPY (P66572) 30S ribosomal protein S5 107 9e-23
RS5_HELPJ (P66573) 30S ribosomal protein S5 107 9e-23
RS5_XYLFT (Q87E65) 30S ribosomal protein S5 105 2e-22
RS5_STAAW (P66580) 30S ribosomal protein S5 105 2e-22
RS5_STAAN (P66579) 30S ribosomal protein S5 105 2e-22
RS5_STAAM (P66578) 30S ribosomal protein S5 105 2e-22
RS5_STAEP (Q8CRH7) 30S ribosomal protein S5 105 4e-22
RS5_YERPE (Q8ZJ95) 30S ribosomal protein S5 103 1e-21
RS5_WIGBR (Q8D1Z5) 30S ribosomal protein S5 103 1e-21
>RS5_XANCP (P66587) 30S ribosomal protein S5
Length = 180
Score = 112 bits (281), Expect = 2e-24
Identities = 67/177 (37%), Positives = 98/177 (54%), Gaps = 4/177 (2%)
Query: 322 LAEER---NELTEKKRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVG 378
+AEER ++ R G KL+ VNR K KGG+ +TA+ G+ G VG
Sbjct: 1 MAEERAPRGRDRDRNREEKVDDGMIEKLVAVNRVSKTVKGGRQFTFTALTVVGDGLGKVG 60
Query: 379 FAKAKGPAVPVALQKAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMK 438
F K VPVA+QK+ E+ +NL V+ + T+ HA+++ +VY+ PA TG+
Sbjct: 61 FGYGKAREVPVAIQKSMEQARKNLATVDLNNG-TLWHAVKSGHGAARVYMQPASEGTGVI 119
Query: 439 AGRIVQTILHLAGFKNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
AG ++ +L G KNV +K VGSRNP N V+A K L+ +++P V K G+ V E
Sbjct: 120 AGGAMRAVLEAVGVKNVLAKAVGSRNPINLVRATLKGLSEVQSPARVAAKRGKKVEE 176
>RS5_XANAC (P66586) 30S ribosomal protein S5
Length = 180
Score = 112 bits (281), Expect = 2e-24
Identities = 67/177 (37%), Positives = 98/177 (54%), Gaps = 4/177 (2%)
Query: 322 LAEER---NELTEKKRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVG 378
+AEER ++ R G KL+ VNR K KGG+ +TA+ G+ G VG
Sbjct: 1 MAEERAPRGRDRDRNREEKVDDGMIEKLVAVNRVSKTVKGGRQFTFTALTVVGDGLGKVG 60
Query: 379 FAKAKGPAVPVALQKAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMK 438
F K VPVA+QK+ E+ +NL V+ + T+ HA+++ +VY+ PA TG+
Sbjct: 61 FGYGKAREVPVAIQKSMEQARKNLATVDLNNG-TLWHAVKSGHGAARVYMQPASEGTGVI 119
Query: 439 AGRIVQTILHLAGFKNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
AG ++ +L G KNV +K VGSRNP N V+A K L+ +++P V K G+ V E
Sbjct: 120 AGGAMRAVLEAVGVKNVLAKAVGSRNPINLVRATLKGLSEVQSPARVAAKRGKKVEE 176
>RS5_PSEPK (Q88QL8) 30S ribosomal protein S5
Length = 166
Score = 112 bits (281), Expect = 2e-24
Identities = 60/156 (38%), Positives = 93/156 (59%), Gaps = 1/156 (0%)
Query: 340 KGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCF 399
+G+ KL+ VNR K KGG++ +TA+ G+ G VGF + K VP A+QKA E
Sbjct: 10 EGYIEKLVQVNRVAKTVKGGRIFTFTALTVVGDGKGRVGFGRGKSREVPAAIQKAMEAAR 69
Query: 400 QNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKV 459
+N+ V+ + T+ +A + + +KVY+ PA TG+ AG ++ +L +AG +NV +K
Sbjct: 70 RNMIQVDL-KGTTLQYATKAAHGASKVYMQPASEGTGIIAGGAMRAVLEVAGVQNVLAKC 128
Query: 460 VGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
GS NP N V A FK L A+++P + K G++V E
Sbjct: 129 YGSTNPVNVVYATFKGLKAMQSPESIAAKRGKSVEE 164
>RS5_VIBPA (Q87SZ6) 30S ribosomal protein S5
Length = 167
Score = 112 bits (280), Expect = 2e-24
Identities = 63/161 (39%), Positives = 91/161 (56%), Gaps = 1/161 (0%)
Query: 333 KRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQ 392
K + + KLI VNR K KGG+++ +TA+ G+ NG VGF K VP A+Q
Sbjct: 3 KEQQVQANDLQEKLIAVNRVSKTVKGGRIMSFTALTVVGDGNGRVGFGYGKAREVPAAIQ 62
Query: 393 KAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGF 452
KA EK +N+ + +E T+ H ++ +KVY+ PA TG+ AG ++ +L +AG
Sbjct: 63 KAMEKARRNMTTIALNEG-TLHHPVKGRHSGSKVYMQPAAEGTGVIAGGAMRAVLEVAGV 121
Query: 453 KNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTV 493
NV SK GS NP N V+A AL ++++P V K G TV
Sbjct: 122 HNVLSKAYGSTNPINIVRATIDALGSMKSPEMVAAKRGLTV 162
>RS5_PSESM (Q889V4) 30S ribosomal protein S5
Length = 166
Score = 112 bits (280), Expect = 2e-24
Identities = 59/156 (37%), Positives = 93/156 (58%), Gaps = 1/156 (0%)
Query: 340 KGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCF 399
+G+ KL+ VNR K KGG++ +TA+ G+ G VGF + K VP A+QKA E
Sbjct: 10 EGYIEKLVQVNRVAKTVKGGRIFTFTALTVVGDGKGRVGFGRGKSREVPAAIQKAMEAAR 69
Query: 400 QNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKV 459
+N+ V+ + T+ +A++++ +KVY+ PA TG+ AG ++ +L +AG +NV +K
Sbjct: 70 RNMIQVDLNGT-TLQYAMKSAHGASKVYMQPASEGTGIIAGGAMRAVLEVAGVQNVLAKC 128
Query: 460 VGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
GS NP N V A FK L +++P + K G+ V E
Sbjct: 129 YGSTNPVNVVHATFKGLKGMQSPESIAAKRGKRVEE 164
>RS5_PASMU (Q9CL47) 30S ribosomal protein S5
Length = 166
Score = 112 bits (279), Expect = 3e-24
Identities = 63/151 (41%), Positives = 91/151 (59%), Gaps = 1/151 (0%)
Query: 345 KLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHF 404
KLI VNR K KGG+++ +TA+ G+ NG VGF K VP A+QKA EK +N+
Sbjct: 14 KLIAVNRVSKTVKGGRIMSFTALTVVGDGNGRVGFGYGKAREVPAAIQKAMEKARRNMIN 73
Query: 405 VERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGSRN 464
V +E T+ H I+ + ++V++ PA TG+ AG ++ +L +AG +NV SK GS N
Sbjct: 74 VALNEG-TLQHPIKGAHTGSRVFMQPASEGTGIIAGGAMRAVLEVAGVRNVLSKAYGSTN 132
Query: 465 PHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
P N V+A AL +++P V K G+TV E
Sbjct: 133 PINVVRATIDALANMKSPEMVAAKRGKTVDE 163
>RS5_VIBVU (Q8DE57) 30S ribosomal protein S5
Length = 167
Score = 110 bits (275), Expect = 8e-24
Identities = 63/161 (39%), Positives = 90/161 (55%), Gaps = 1/161 (0%)
Query: 333 KRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQ 392
K + + KLI VNR K KGG+++ +TA+ G+ NG VGF K VP A+Q
Sbjct: 3 KEQQVQANDLQEKLIAVNRVSKTVKGGRIMSFTALTVVGDGNGRVGFGYGKAREVPAAIQ 62
Query: 393 KAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGF 452
KA EK +N+ + +E T+ H ++ +KVY+ PA TG+ AG ++ +L +AG
Sbjct: 63 KAMEKARRNMVTIALNEG-TLHHPVKGRHSGSKVYMQPAAEGTGVIAGGAMRAVLEVAGV 121
Query: 453 KNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTV 493
NV SK GS NP N V+A AL +++P V K G TV
Sbjct: 122 HNVLSKAYGSTNPINIVRATIDALVDVKSPEMVAAKRGLTV 162
>RS5_HAEIN (P44374) 30S ribosomal protein S5
Length = 166
Score = 110 bits (275), Expect = 8e-24
Identities = 62/151 (41%), Positives = 90/151 (59%), Gaps = 1/151 (0%)
Query: 345 KLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHF 404
KLI VNR K KGG+++ +TA+ G+ NG VGF K VP A+QKA EK +N+
Sbjct: 14 KLIAVNRVSKTVKGGRIMSFTALTVVGDGNGRVGFGYGKAREVPAAIQKAMEKARRNMIN 73
Query: 405 VERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGSRN 464
V +E T+ H ++ ++V++ PA TG+ AG ++ +L +AG +NV SK GS N
Sbjct: 74 VALNEG-TLQHPVKGVHTGSRVFMQPASEGTGIIAGGAMRAVLEVAGVRNVLSKAYGSTN 132
Query: 465 PHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
P N V+A AL +++P V K G+TV E
Sbjct: 133 PINVVRATIDALANMKSPEMVAAKRGKTVDE 163
>RS5_VIBCH (Q9KP01) 30S ribosomal protein S5
Length = 167
Score = 108 bits (270), Expect = 3e-23
Identities = 62/161 (38%), Positives = 90/161 (55%), Gaps = 1/161 (0%)
Query: 333 KRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQ 392
K + + KLI VNR K KGG+++ +TA+ G+ NG VGF K VP A+Q
Sbjct: 3 KEQQVQANDLQEKLIAVNRVSKTVKGGRIMSFTALTVVGDGNGRVGFGYGKAREVPAAIQ 62
Query: 393 KAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGF 452
KA EK +++ + +E T+ H ++ +KVY+ PA TG+ AG ++ +L +AG
Sbjct: 63 KAMEKARRSMVTIALNEG-TLHHPVKGRHSGSKVYMQPAAEGTGVIAGGAMRAVLEVAGV 121
Query: 453 KNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTV 493
NV SK GS NP N V+A AL +++P V K G TV
Sbjct: 122 HNVLSKAYGSTNPINIVRATIDALVDVKSPEMVAAKRGLTV 162
>RS5_PSEAE (Q9HWF2) 30S ribosomal protein S5
Length = 166
Score = 108 bits (270), Expect = 3e-23
Identities = 59/156 (37%), Positives = 91/156 (57%), Gaps = 1/156 (0%)
Query: 340 KGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCF 399
+G+ KL+ VNR K KGG++ +TA+ G+ G VGF + K VP A+QKA E
Sbjct: 10 EGYIEKLVQVNRVAKTVKGGRIFAFTALTVVGDGKGRVGFGRGKAREVPAAIQKAMEAAR 69
Query: 400 QNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKV 459
+N+ V+ + T+ + +++ +KVY+ PA TG+ AG ++ +L +AG +NV +K
Sbjct: 70 RNMIQVDLNGT-TLQYPTKSAHGASKVYMQPASEGTGIIAGGAMRAVLEVAGVQNVLAKC 128
Query: 460 VGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
GS NP N V A FK L ++ P V K G++V E
Sbjct: 129 YGSTNPVNVVYATFKGLKNMQAPEAVAAKRGKSVEE 164
>RS5_SHEON (P59124) 30S ribosomal protein S5
Length = 167
Score = 107 bits (266), Expect = 9e-23
Identities = 61/151 (40%), Positives = 87/151 (57%), Gaps = 1/151 (0%)
Query: 345 KLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHF 404
KLI VNR KV KGG++ +TA+ G+ NG VG+ K VP A+QKA EK +N+
Sbjct: 15 KLIAVNRVSKVVKGGRIFSFTALTVVGDGNGKVGYGYGKAREVPAAIQKAMEKARRNMVT 74
Query: 405 VERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGSRN 464
VE + T+ H ++ ++VY+ PA TG+ AG ++ +L +AG NV SK GS N
Sbjct: 75 VELN-AGTLHHPVKGRHTGSRVYMQPASQGTGIIAGGAMRAVLEVAGVHNVLSKAYGSTN 133
Query: 465 PHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
P N V+A AL +++P + K G V E
Sbjct: 134 PINIVRATVDALVHMKSPSQIAAKRGLNVDE 164
>RS5_HELPY (P66572) 30S ribosomal protein S5
Length = 153
Score = 107 bits (266), Expect = 9e-23
Identities = 59/153 (38%), Positives = 92/153 (59%), Gaps = 2/153 (1%)
Query: 329 LTEKK-RRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAV 387
+TE+K + R+ F ++N+ R KV KGG+ ++ A++ GN NG+VGF K V
Sbjct: 1 MTERKGMEEINREEFQEVVVNIGRVTKVVKGGRRFRFNALVVVGNKNGLVGFGLGKAKEV 60
Query: 388 PVALQKAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTIL 447
P A++KA + F+NL V + TIAH I+ + +++ L PA TG+ AG + I+
Sbjct: 61 PDAIKKAVDDAFKNLIHVTI-KGTTIAHDIEHKYNASRILLKPASEGTGVIAGGSTRPIV 119
Query: 448 HLAGFKNVKSKVVGSRNPHNTVKAVFKALNAIE 480
LAG K++ +K +GS NP+N V+A F AL I+
Sbjct: 120 ELAGIKDILTKSLGSNNPYNVVRATFDALAKIK 152
>RS5_HELPJ (P66573) 30S ribosomal protein S5
Length = 153
Score = 107 bits (266), Expect = 9e-23
Identities = 59/153 (38%), Positives = 92/153 (59%), Gaps = 2/153 (1%)
Query: 329 LTEKK-RRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAV 387
+TE+K + R+ F ++N+ R KV KGG+ ++ A++ GN NG+VGF K V
Sbjct: 1 MTERKGMEEINREEFQEVVVNIGRVTKVVKGGRRFRFNALVVVGNKNGLVGFGLGKAKEV 60
Query: 388 PVALQKAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTIL 447
P A++KA + F+NL V + TIAH I+ + +++ L PA TG+ AG + I+
Sbjct: 61 PDAIKKAVDDAFKNLIHVTI-KGTTIAHDIEHKYNASRILLKPASEGTGVIAGGSTRPIV 119
Query: 448 HLAGFKNVKSKVVGSRNPHNTVKAVFKALNAIE 480
LAG K++ +K +GS NP+N V+A F AL I+
Sbjct: 120 ELAGIKDILTKSLGSNNPYNVVRATFDALAKIK 152
>RS5_XYLFT (Q87E65) 30S ribosomal protein S5
Length = 179
Score = 105 bits (263), Expect = 2e-22
Identities = 61/173 (35%), Positives = 94/173 (54%), Gaps = 2/173 (1%)
Query: 322 LAEERNELT-EKKRRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFA 380
+AEER++ + ++ R G KL+ VNR K KGG+ +TA+ GN G VGF
Sbjct: 1 MAEERSQRSRDRSREEKIDDGMIEKLVAVNRVSKTVKGGRQFTFTALTIVGNGEGSVGFG 60
Query: 381 KAKGPAVPVALQKAYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAG 440
K VPVA+QK+ E + + V + T+ H ++ + V++ PA TG+ AG
Sbjct: 61 YGKAREVPVAIQKSMEYARKRMSNVSLNNG-TLWHPVKANHGAASVFMKPASEGTGVIAG 119
Query: 441 RIVQTILHLAGFKNVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTV 493
++ +L G KNV +K +GSRNP N V+A K L +++P + K G+ V
Sbjct: 120 GAMRAVLEAVGVKNVLAKAIGSRNPINLVRATLKGLEDMQSPTHIALKRGKNV 172
>RS5_STAAW (P66580) 30S ribosomal protein S5
Length = 166
Score = 105 bits (263), Expect = 2e-22
Identities = 61/164 (37%), Positives = 90/164 (54%), Gaps = 1/164 (0%)
Query: 334 RRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQK 393
RR K F+ +++ +NR KV KGG+ ++TA++ G+ NG VGF K VP A++K
Sbjct: 3 RREEETKEFEERVVTINRVAKVVKGGRRFRFTALVVVGDKNGRVGFGTGKAQEVPEAIKK 62
Query: 394 AYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFK 453
A E ++L V R E T H I + V++ PA TG+ AG V+ +L LAG
Sbjct: 63 AVEAAKKDLVVVPR-VEGTTPHTITGRYGSGSVFMKPAAPGTGVIAGGPVRAVLELAGIT 121
Query: 454 NVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEKY 497
++ SK +GS P N V+A L ++ DV + G+TV E Y
Sbjct: 122 DILSKSLGSNTPINMVRATIDGLQNLKNAEDVAKLRGKTVEELY 165
>RS5_STAAN (P66579) 30S ribosomal protein S5
Length = 166
Score = 105 bits (263), Expect = 2e-22
Identities = 61/164 (37%), Positives = 90/164 (54%), Gaps = 1/164 (0%)
Query: 334 RRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQK 393
RR K F+ +++ +NR KV KGG+ ++TA++ G+ NG VGF K VP A++K
Sbjct: 3 RREEETKEFEERVVTINRVAKVVKGGRRFRFTALVVVGDKNGRVGFGTGKAQEVPEAIKK 62
Query: 394 AYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFK 453
A E ++L V R E T H I + V++ PA TG+ AG V+ +L LAG
Sbjct: 63 AVEAAKKDLVVVPR-VEGTTPHTITGRYGSGSVFMKPAAPGTGVIAGGPVRAVLELAGIT 121
Query: 454 NVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEKY 497
++ SK +GS P N V+A L ++ DV + G+TV E Y
Sbjct: 122 DILSKSLGSNTPINMVRATIDGLQNLKNAEDVAKLRGKTVEELY 165
>RS5_STAAM (P66578) 30S ribosomal protein S5
Length = 166
Score = 105 bits (263), Expect = 2e-22
Identities = 61/164 (37%), Positives = 90/164 (54%), Gaps = 1/164 (0%)
Query: 334 RRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQK 393
RR K F+ +++ +NR KV KGG+ ++TA++ G+ NG VGF K VP A++K
Sbjct: 3 RREEETKEFEERVVTINRVAKVVKGGRRFRFTALVVVGDKNGRVGFGTGKAQEVPEAIKK 62
Query: 394 AYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFK 453
A E ++L V R E T H I + V++ PA TG+ AG V+ +L LAG
Sbjct: 63 AVEAAKKDLVVVPR-VEGTTPHTITGRYGSGSVFMKPAAPGTGVIAGGPVRAVLELAGIT 121
Query: 454 NVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEKY 497
++ SK +GS P N V+A L ++ DV + G+TV E Y
Sbjct: 122 DILSKSLGSNTPINMVRATIDGLQNLKNAEDVAKLRGKTVEELY 165
>RS5_STAEP (Q8CRH7) 30S ribosomal protein S5
Length = 166
Score = 105 bits (261), Expect = 4e-22
Identities = 60/164 (36%), Positives = 90/164 (54%), Gaps = 1/164 (0%)
Query: 334 RRPLYRKGFDVKLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQK 393
RR K F+ +++ +NR KV KGG+ ++TA++ G+ NG VGF K VP A++K
Sbjct: 3 RREEETKEFEERVVTINRVAKVVKGGRRFRFTALVVVGDKNGRVGFGTGKAQEVPEAIKK 62
Query: 394 AYEKCFQNLHFVERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFK 453
A E ++L V R E T H I + V++ PA TG+ AG V+ +L LAG
Sbjct: 63 AVEAAKKDLVVVPR-VEGTTPHTITGQYGSGSVFMKPAAPGTGVIAGGPVRAVLELAGIT 121
Query: 454 NVKSKVVGSRNPHNTVKAVFKALNAIETPRDVQEKFGRTVVEKY 497
++ SK +GS P N V+A L ++ DV + G++V E Y
Sbjct: 122 DILSKSLGSNTPINMVRATINGLQNLKNAEDVAKLRGKSVEELY 165
>RS5_YERPE (Q8ZJ95) 30S ribosomal protein S5
Length = 167
Score = 103 bits (256), Expect = 1e-21
Identities = 58/151 (38%), Positives = 87/151 (57%), Gaps = 1/151 (0%)
Query: 345 KLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHF 404
KLI VNR K KGG++ +TA+ G+ NG VGF K VP A+QKA EK + +
Sbjct: 14 KLIAVNRVSKTVKGGRIFSFTALTVVGDGNGRVGFGYGKAREVPAAIQKAMEKARRAMIN 73
Query: 405 VERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGSRN 464
V + T+ H ++ + ++V++ PA TG+ AG ++ +L +AG NV +K GS N
Sbjct: 74 VALNNG-TLQHPVKGAHTGSRVFMQPASEGTGIIAGGAMRAVLEVAGVHNVLAKAYGSTN 132
Query: 465 PHNTVKAVFKALNAIETPRDVQEKFGRTVVE 495
P N V+A AL +++P V K G++V E
Sbjct: 133 PINVVRATIAALEDMKSPEMVAAKRGKSVEE 163
>RS5_WIGBR (Q8D1Z5) 30S ribosomal protein S5
Length = 166
Score = 103 bits (256), Expect = 1e-21
Identities = 55/149 (36%), Positives = 86/149 (56%), Gaps = 1/149 (0%)
Query: 345 KLINVNRTCKVTKGGQVVKYTAMLACGNYNGVVGFAKAKGPAVPVALQKAYEKCFQNLHF 404
KLI+VNR K KGG++ +TA+ G+ G +GF K VP+A+QKA +K +N+
Sbjct: 14 KLISVNRVSKTVKGGRIFSFTALTVVGDCQGKIGFGYGKSKEVPLAIQKAMDKARKNMII 73
Query: 405 VERHEEHTIAHAIQTSFKKTKVYLWPAPTATGMKAGRIVQTILHLAGFKNVKSKVVGSRN 464
+ + T+ H I ++V++ PA TG+ AG ++ +L +AG NV +KV GS N
Sbjct: 74 IPIVCK-TLQHEINGYHTGSRVFMKPASEGTGIIAGGAMRAVLEVAGVHNVLAKVYGSTN 132
Query: 465 PHNTVKAVFKALNAIETPRDVQEKFGRTV 493
P N V+A AL + P + EK G+++
Sbjct: 133 PINVVRATVNALKKMRFPMHIAEKRGKSI 161
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 59,794,536
Number of Sequences: 164201
Number of extensions: 2717337
Number of successful extensions: 14103
Number of sequences better than 10.0: 416
Number of HSP's better than 10.0 without gapping: 134
Number of HSP's successfully gapped in prelim test: 290
Number of HSP's that attempted gapping in prelim test: 13268
Number of HSP's gapped (non-prelim): 855
length of query: 499
length of database: 59,974,054
effective HSP length: 114
effective length of query: 385
effective length of database: 41,255,140
effective search space: 15883228900
effective search space used: 15883228900
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0253.6


