

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252a.4
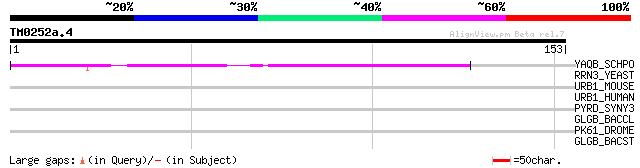
(153 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YAQB_SCHPO (Q10110) Hypothetical protein C18G6.11c in chromosome I 51 9e-07
RRN3_YEAST (P36070) RNA polymerase I specific transcription init... 38 0.010
URB1_MOUSE (Q7TMY8) E3 ubiquitin protein ligase URE-B1 (EC 6.3.2... 31 1.2
URB1_HUMAN (Q7Z6Z7) E3 ubiquitin protein ligase URE-B1 (EC 6.3.2... 31 1.2
PYRD_SYNY3 (P74782) Dihydroorotate dehydrogenase (EC 1.3.3.1) (D... 29 4.6
GLGB_BACCL (P30537) 1,4-alpha-glucan branching enzyme (EC 2.4.1.... 29 4.6
PK61_DROME (Q9W0V1) Serine/threonine-protein kinase Pk61C (EC 2.... 28 6.0
GLGB_BACST (P30538) 1,4-alpha-glucan branching enzyme (EC 2.4.1.... 28 6.0
>YAQB_SCHPO (Q10110) Hypothetical protein C18G6.11c in chromosome I
Length = 599
Score = 51.2 bits (121), Expect = 9e-07
Identities = 34/139 (24%), Positives = 63/139 (44%), Gaps = 23/139 (16%)
Query: 1 AIMYVLCFRMRSLVDIPRLK------------LQLLNMPMEPIWKHKLSPLKVCLPTVVE 48
+I Y+ CFR R L + L++L+ + +L+PL+ C P +V
Sbjct: 457 SIFYIFCFRWRELCVSDESESMEPRPNEWIPGLEILHRSV----LSRLNPLRYCSPNIVL 512
Query: 49 EFLRQAKAAQLFMSSSETFVFNDLLESDDLSRSFGGIDRLDMFFPFDPCLSKKSESYIRP 108
+F + A +V++ ++E + G D +D +FPFDP KS ++P
Sbjct: 513 QFAKVANHLNFM------YVYS-IIEQNRKGIFREGFDTMDAYFPFDPYRLTKSSIIVQP 565
Query: 109 HFVRWSRVRTTYDSDDDED 127
+ W ++ D +++ED
Sbjct: 566 FYNEWQQIPGLDDDEEEED 584
>RRN3_YEAST (P36070) RNA polymerase I specific transcription
initiation factor RRN3
Length = 627
Score = 37.7 bits (86), Expect = 0.010
Identities = 31/149 (20%), Positives = 59/149 (38%), Gaps = 22/149 (14%)
Query: 1 AIMYVLCFRMRSLVDIPRLKLQLLNMPMEPIWKHKLSPLKVCLPTVVEEFLRQAKAAQL- 59
A+ Y+ CFR D L+ + + K +PLK C V+ F R A+ +
Sbjct: 479 ALCYIFCFRHNIFRDTDGNWECELDKFFQRMVISKFNPLKFCNENVMLMFARIAQQESVA 538
Query: 60 ----------------FMSSSETFVFNDLLESDDLSRSFGGIDR-----LDMFFPFDPCL 98
+ +++ + +++ S S+ R L +FP+DP
Sbjct: 539 YCFSIIENNNNERLRGIIGKADSDKKENSAQANTTSSSWSLATRQQFIDLQSYFPYDPLF 598
Query: 99 SKKSESYIRPHFVRWSRVRTTYDSDDDED 127
K + ++ +++ WS Y+SD +D
Sbjct: 599 LKNYKILMKEYYIEWSEASGEYESDGSDD 627
>URB1_MOUSE (Q7TMY8) E3 ubiquitin protein ligase URE-B1 (EC 6.3.2.-)
(Fragment)
Length = 2749
Score = 30.8 bits (68), Expect = 1.2
Identities = 15/25 (60%), Positives = 17/25 (68%), Gaps = 1/25 (4%)
Query: 121 DSDDDEDGSEMSHD-DFVDRNAKEL 144
D DDD+DGSEM D D+ D NA L
Sbjct: 827 DEDDDDDGSEMELDEDYPDMNASPL 851
>URB1_HUMAN (Q7Z6Z7) E3 ubiquitin protein ligase URE-B1 (EC 6.3.2.-)
(HSPC272)
Length = 3360
Score = 30.8 bits (68), Expect = 1.2
Identities = 15/25 (60%), Positives = 17/25 (68%), Gaps = 1/25 (4%)
Query: 121 DSDDDEDGSEMSHD-DFVDRNAKEL 144
D DDD+DGSEM D D+ D NA L
Sbjct: 1441 DEDDDDDGSEMELDEDYPDMNASPL 1465
>PYRD_SYNY3 (P74782) Dihydroorotate dehydrogenase (EC 1.3.3.1)
(Dihydroorotate oxidase) (DHOdehase) (DHODase) (DHOD)
Length = 381
Score = 28.9 bits (63), Expect = 4.6
Identities = 20/56 (35%), Positives = 26/56 (45%), Gaps = 3/56 (5%)
Query: 33 KHKLSPLKVCLPTVVEEFLRQAKAAQLFM---SSSETFVFNDLLESDDLSRSFGGI 85
K K++PL + V F A A F+ SS T L ESD+L R F G+
Sbjct: 161 KSKITPLDQAVEDYVFSFQTLAPVADYFVVNVSSPNTPGLRSLQESDELPRIFAGL 216
>GLGB_BACCL (P30537) 1,4-alpha-glucan branching enzyme (EC 2.4.1.18)
(Glycogen branching enzyme) (BE)
(1,4-alpha-D-glucan:1,4-alpha-D-glucan
6-glucosyl-transferase)
Length = 666
Score = 28.9 bits (63), Expect = 4.6
Identities = 19/65 (29%), Positives = 30/65 (45%), Gaps = 8/65 (12%)
Query: 94 FDPCLSKKSESYIRPHFVRWSRVRTTYDSDDDEDG--------SEMSHDDFVDRNAKELD 145
FD L +K + Y++ + R + Y+ D D G +E S F+ R KE D
Sbjct: 483 FDFELHRKMDEYVKQLIACYKRYKPFYELDHDPRGFEWIDVHNAEQSIFSFIRRGKKEGD 542
Query: 146 MMVSV 150
++V V
Sbjct: 543 VLVIV 547
>PK61_DROME (Q9W0V1) Serine/threonine-protein kinase Pk61C (EC
2.7.1.37) (dSTPK61)
Length = 819
Score = 28.5 bits (62), Expect = 6.0
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 2/34 (5%)
Query: 114 SRVRTTYDSDDDEDGSEMSH--DDFVDRNAKELD 145
S R + +DDED + + +DF DR+++ELD
Sbjct: 391 SEQRRSNSDEDDEDSDRLENEDEDFYDRDSEELD 424
>GLGB_BACST (P30538) 1,4-alpha-glucan branching enzyme (EC 2.4.1.18)
(Glycogen branching enzyme) (BE)
(1,4-alpha-D-glucan:1,4-alpha-D-glucan
6-glucosyl-transferase)
Length = 639
Score = 28.5 bits (62), Expect = 6.0
Identities = 20/75 (26%), Positives = 35/75 (46%), Gaps = 11/75 (14%)
Query: 85 IDRLD-MFFPFDPCLSKKSESYIRPHFVRWSRVRTTYDSDDDEDG--------SEMSHDD 135
+++LD M F FD + + Y++ + R + Y+ D DG +E S
Sbjct: 475 LEQLDWMLFDFD--MHRNMNMYVKELLKCYKRYKPLYELDHSPDGFEWIDVHNAEQSIFS 532
Query: 136 FVDRNAKELDMMVSV 150
F+ R KE D+++ V
Sbjct: 533 FIRRGKKEDDLLIVV 547
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.139 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,895,588
Number of Sequences: 164201
Number of extensions: 627966
Number of successful extensions: 2494
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 2484
Number of HSP's gapped (non-prelim): 11
length of query: 153
length of database: 59,974,054
effective HSP length: 101
effective length of query: 52
effective length of database: 43,389,753
effective search space: 2256267156
effective search space used: 2256267156
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0252a.4


