

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
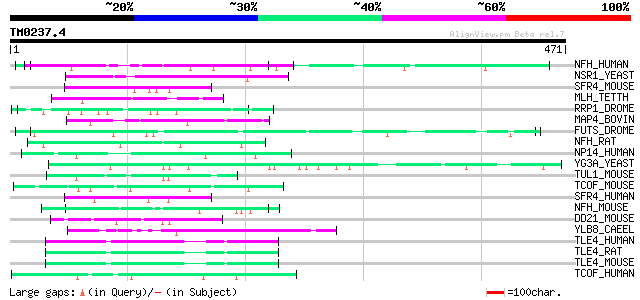
Query= TM0237.4
(471 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NFH_HUMAN (P12036) Neurofilament triplet H protein (200 kDa neur... 59 3e-08
NSR1_YEAST (P27476) Nuclear localization sequence binding protei... 53 2e-06
SFR4_MOUSE (Q8VE97) Splicing factor, arginine/serine-rich 4 50 2e-05
MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC ... 49 2e-05
RRP1_DROME (P27864) Recombination repair protein 1 (DNA-(apurini... 49 4e-05
MAP4_BOVIN (P36225) Microtubule-associated protein 4 (MAP 4) (Mi... 48 5e-05
FUTS_DROME (Q9W596) Microtubule-associated protein futsch 47 8e-05
NFH_RAT (P16884) Neurofilament triplet H protein (200 kDa neurof... 47 1e-04
NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130... 47 1e-04
YG3A_YEAST (P53278) Hypothetical 92.7 kDa protein in ASN2-PHB1 i... 46 2e-04
TUL1_MOUSE (Q9Z273) Tubby related protein 1 (Tubby-like protein 1) 46 2e-04
TCOF_MOUSE (O08784) Treacle protein (Treacher Collins syndrome p... 46 2e-04
SFR4_HUMAN (Q08170) Splicing factor, arginine/serine-rich 4 (Pre... 46 2e-04
NFH_MOUSE (P19246) Neurofilament triplet H protein (200 kDa neur... 46 2e-04
DD21_MOUSE (Q9JIK5) Nucleolar RNA helicase II (Nucleolar RNA hel... 46 2e-04
YLB8_CAEEL (P46582) Hypothetical protein C34E10.8 in chromosome III 45 3e-04
TLE4_HUMAN (Q04727) Transducin-like enhancer protein 4 45 3e-04
TLE4_RAT (Q07141) Transducin-like enhancer protein 4 (ESP2 protein) 45 4e-04
TLE4_MOUSE (Q62441) Transducin-like enhancer protein 4 (Groucho-... 45 4e-04
TCOF_HUMAN (Q13428) Treacle protein (Treacher Collins syndrome p... 45 4e-04
>NFH_HUMAN (P12036) Neurofilament triplet H protein (200 kDa
neurofilament protein) (Neurofilament heavy polypeptide)
(NF-H)
Length = 1026
Score = 58.9 bits (141), Expect = 3e-08
Identities = 56/214 (26%), Positives = 94/214 (43%), Gaps = 21/214 (9%)
Query: 13 SPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGV 72
S + I V S K+ +V E ++ + V+ ++ + ++ + K ++ A+G
Sbjct: 440 SEEKIKVVEKSEKETVIVEEQTEETQVTEEVTEEEEKEAKEEEGKEEEGGEEEEAEGGEE 499
Query: 73 STTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVE 132
T S ++ A A PE K + V E + S P+ SP AKS AE V+
Sbjct: 500 ETKSPPAEEA--ASPEKEAK---------SPVKEEAKS--PAEAKSPEKEEAKSPAE-VK 545
Query: 133 KPPKAASLAKKTSTSEGRRK------VRSKSGHKSPHRSKSSTNSSPSKESACQETQGAT 186
P KA S AK+ + S K +S + KSP ++KS + + + A
Sbjct: 546 SPEKAKSPAKEEAKSPPEAKSPEKEEAKSPAEVKSPEKAKSPAKEEAKSPAEAKSPEKAK 605
Query: 187 SPIREAEALPGKDDNPM-PPPKSTATVQPSPQAK 219
SP++E P + +P+ KS A V+ +AK
Sbjct: 606 SPVKEEAKSPAEAKSPVKEEAKSPAEVKSPEKAK 639
Score = 52.0 bits (123), Expect = 3e-06
Identities = 63/238 (26%), Positives = 100/238 (41%), Gaps = 31/238 (13%)
Query: 18 SPVAHSSKDKPVVIEDDDDDDEDD-NVSLADKLKG--RKRKPKPAASASQKRAKGAGVST 74
SP +K P + ++ + V +K K ++ PA + S ++AK S
Sbjct: 552 SPAKEEAKSPPEAKSPEKEEAKSPAEVKSPEKAKSPAKEEAKSPAEAKSPEKAK----SP 607
Query: 75 TSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKP 134
+K ++A+ V KE +A A+V + P++E + +P AKS P
Sbjct: 608 VKEEAKSPAEAKSPV--KE---EAKSPAEVKSPEKAKSPTKEEAKSPEKAKS-------P 655
Query: 135 PKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNS---------SPSKESACQETQGA 185
KA S K+ + S + K K+ KSP ++KS + SP KE A + + A
Sbjct: 656 EKAKSPEKEEAKSPEKAKSPVKAEAKSPEKAKSPVKAEAKSPEKAKSPVKEEA-KSPEKA 714
Query: 186 TSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSN--VSSEKLDTLFEEDPLA 241
SP++E P K +P+ T SP + S S EK TL + P A
Sbjct: 715 KSPVKEEAKSPEKAKSPVKEEAKTPEKAKSPVKEEAKSPEKAKSPEKAKTLDVKSPEA 772
Score = 50.4 bits (119), Expect = 1e-05
Identities = 102/471 (21%), Positives = 177/471 (36%), Gaps = 59/471 (12%)
Query: 6 AEVITQPSPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQK 65
AE P + SPV +K P + + ++ K + P+ A S +++
Sbjct: 506 AEEAASPEKEAKSPVKEEAKS-PAEAKSPEKEEA--------KSPAEVKSPEKAKSPAKE 556
Query: 66 RAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAK 125
AK + + + S A EV E + + + + + P + SP AK
Sbjct: 557 EAKSPPEAKSPEKEEAKSPA--EVKSPE-KAKSPAKEEAKSPAEAKSPEKAKSPVKEEAK 613
Query: 126 SKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNS-SPSKESACQETQG 184
S AE + P K + + S + K +K KSP ++KS + SP KE A + +
Sbjct: 614 SPAEA-KSPVKEEAKSPAEVKSPEKAKSPTKEEAKSPEKAKSPEKAKSPEKEEA-KSPEK 671
Query: 185 ATSPIREAEALPGKDDNPM-----PPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFEEDP 239
A SP++ P K +P+ P K+ + V+ ++ S V E + P
Sbjct: 672 AKSPVKAEAKSPEKAKSPVKAEAKSPEKAKSPVKEEAKSPEKAKSPVKEE------AKSP 725
Query: 240 LAALDGFLDGTLDWDSPPHQADATAETAQSSGVANLQQIEESMGRLKAIVFDIGFLDRIR 299
A SP + T E A+S + E++ KA D+
Sbjct: 726 EKA-----------KSPVKEEAKTPEKAKSPVKEEAKSPEKAKSPEKAKTLDV------- 767
Query: 300 VDPQASHAAKELLIFLLGQKLDSHQAAALANLQS-------FLVDALATFHQVKDVGQAV 352
P+A AKE + + ++ ++S DA A K++ +
Sbjct: 768 KSPEAKTPAKEEARSPADKFPEKAKSPVKEEVKSPEKAKSPLKEDAKA---PEKEIPKKE 824
Query: 353 SLKEAKVSSGKAQVAAMKEDFKKFTARKVLIADEISD-VDTKLEELHREIA---KLEKKR 408
+K K Q +KE KK K + + D+K EE ++ A K+E+K+
Sbjct: 825 EVKSPVKEEEKPQEVKVKEPPKKAEEEKAPATPKTEEKKDSKKEEAPKKEAPKPKVEEKK 884
Query: 409 ADLVEEGPAVKAQLDGLTKESKALI-ISRKDVIKELEVDQAVKKDLDAKIA 458
VE+ K + E K + K+ ++EV + K ++A
Sbjct: 885 EPAVEKPKESKVEAKKEEAEDKKKVPTPEKEAPAKVEVKEDAKPKEKTEVA 935
Score = 39.7 bits (91), Expect = 0.017
Identities = 39/203 (19%), Positives = 81/203 (39%), Gaps = 17/203 (8%)
Query: 52 RKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISN 111
++ P + S ++AK V + + +A+ + + ++ +V +
Sbjct: 747 KEEAKSPEKAKSPEKAKTLDVKSPEAKTPAKEEARSPADKFPEKAKSPVKEEVKSPEKAK 806
Query: 112 PPSRENSPAPN---PAKSKAEG------------VEKPPKAASLAKKTSTSEGRRKVRSK 156
P +E++ AP P K + + V++PPK A K +T + K SK
Sbjct: 807 SPLKEDAKAPEKEIPKKEEVKSPVKEEEKPQEVKVKEPPKKAEEEKAPATPKTEEKKDSK 866
Query: 157 SGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSP 216
++P + KE A ++ + + ++ EA K P P ++ A V+
Sbjct: 867 K-EEAPKKEAPKPKVEEKKEPAVEKPKESKVEAKKEEA-EDKKKVPTPEKEAPAKVEVKE 924
Query: 217 QAKGGFSSNVSSEKLDTLFEEDP 239
AK + V+ ++ D ++P
Sbjct: 925 DAKPKEKTEVAKKEPDDAKAKEP 947
Score = 37.0 bits (84), Expect = 0.11
Identities = 35/161 (21%), Positives = 65/161 (39%), Gaps = 16/161 (9%)
Query: 86 QPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTS 145
+P +E AT + + + S ++ +P P + K VEKP ++ AKK
Sbjct: 843 EPPKKAEEEKAPATPKTEEKKDSKKEEAPKKEAPKPKVEEKKEPAVEKPKESKVEAKK-E 901
Query: 146 TSEGRRKVRSKSGHKSPHRSKSSTNSSP-------SKESACQETQGATSPIREAEALPGK 198
+E ++KV + ++P + + ++ P KE + + + P + EA P K
Sbjct: 902 EAEDKKKVPTPE-KEAPAKVEVKEDAKPKEKTEVAKKEPDDAKAKEPSKPAEKKEAAPEK 960
Query: 199 DD-------NPMPPPKSTATVQPSPQAKGGFSSNVSSEKLD 232
D P PK+ A + + S +EK +
Sbjct: 961 KDTKEEKAKKPEEKPKTEAKAKEDDKTLSKEPSKPKAEKAE 1001
Score = 31.2 bits (69), Expect = 6.0
Identities = 35/178 (19%), Positives = 67/178 (36%), Gaps = 16/178 (8%)
Query: 15 QVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRK----------RKPKPAASASQ 64
+V SPV K + V +++ E++ K + +K PKP +
Sbjct: 825 EVKSPVKEEEKPQEVKVKEPPKKAEEEKAPATPKTEEKKDSKKEEAPKKEAPKPKVEEKK 884
Query: 65 KRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQ----ADVHEHSISNPPSRENSPA 120
+ A + A K ++ + +V E + A ++ A E + +++ A
Sbjct: 885 EPAVEKPKESKVEAKKEEAEDKKKVPTPEKEAPAKVEVKEDAKPKEKTEVAKKEPDDAKA 944
Query: 121 PNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESA 178
P+K + P K + +K E + K +K+ K ++ S S P E A
Sbjct: 945 KEPSKPAEKKEAAPEKKDTKEEKAKKPEEKPKTEAKA--KEDDKTLSKEPSKPKAEKA 1000
>NSR1_YEAST (P27476) Nuclear localization sequence binding protein
(P67)
Length = 414
Score = 52.8 bits (125), Expect = 2e-06
Identities = 44/192 (22%), Positives = 93/192 (47%), Gaps = 11/192 (5%)
Query: 48 KLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEH 107
K+KG K++ K + A +++AK S++ +S +S ++ E + E + +++
Sbjct: 6 KVKGNKKEVKASKQAKEEKAKAVSSSSSESSSSSSSSSESE-SESESESESS-------S 57
Query: 108 SISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKS 167
S S+ S +S + + ++S+AE ++ K +S + S+S+ + + K + S
Sbjct: 58 SSSSSDSESSSSSSSDSESEAETKKEESKDSSSSSSDSSSDEEEEEEKEETKKEESKESS 117
Query: 168 STNSSPSKESACQ-ETQGATSPIREAEALPGKDD--NPMPPPKSTATVQPSPQAKGGFSS 224
S++SS S S + E + + R++E ++D + K+ T +P+ G S
Sbjct: 118 SSDSSSSSSSDSESEKEESNDKKRKSEDAEEEEDEESSNKKQKNEETEEPATIFVGRLSW 177
Query: 225 NVSSEKLDTLFE 236
++ E L FE
Sbjct: 178 SIDDEWLKKEFE 189
>SFR4_MOUSE (Q8VE97) Splicing factor, arginine/serine-rich 4
Length = 489
Score = 49.7 bits (117), Expect = 2e-05
Identities = 40/139 (28%), Positives = 68/139 (48%), Gaps = 14/139 (10%)
Query: 47 DKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADV-- 104
DK KGRKR + S S+ +++ + ++ SK++S + K+ D + V
Sbjct: 349 DKRKGRKRSRDESRSRSRSKSERSRKHSSKRDSKVSSSSSSSKKKKDTDHSRSPSRSVSK 408
Query: 105 -HEHSISNPPSREN-----SPAPNP---AKSKAEGVEKP---PKAASLAKKTSTSEGRRK 152
EH+ + R + S APNP A+S++ KP ++ S +K S + R K
Sbjct: 409 EREHAKAESGQRGSRAEGESEAPNPEPRARSRSTSKSKPNVPAESRSRSKSASKTRSRSK 468
Query: 153 VRSKSGHKSPHRSKSSTNS 171
S+S +SP RS+S ++S
Sbjct: 469 SPSRSASRSPSRSRSRSHS 487
Score = 39.3 bits (90), Expect = 0.022
Identities = 50/210 (23%), Positives = 83/210 (38%), Gaps = 28/210 (13%)
Query: 47 DKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHE 106
+K + R R P + S S+ A+ + A K S + + K +A+
Sbjct: 254 NKSRSRSRSPDKSRSKSKDHAEDK-LQNNDSAGKAKSHSPSRHDSKSRSRSQERRAEEER 312
Query: 107 H-SISNPPSRENSPAPNPAKSKAEGVEKP-----PKAASLAKKTSTSEGRRKVRSKSGHK 160
S+S S+E S + + K+ + K +K S E R + RSKS
Sbjct: 313 RRSVSRARSQEKSRSQEKSLLKSRSRSRSRSRSRSKDKRKGRKRSRDESRSRSRSKSERS 372
Query: 161 SPHRSK--SSTNSSPSKESACQETQGATSPIR---------EAEA------LPGKDDNPM 203
H SK S +SS S ++T + SP R +AE+ G+ + P
Sbjct: 373 RKHSSKRDSKVSSSSSSSKKKKDTDHSRSPSRSVSKEREHAKAESGQRGSRAEGESEAPN 432
Query: 204 PPP----KSTATVQPSPQAKGGFSSNVSSE 229
P P +ST+ +P+ A+ S +S+
Sbjct: 433 PEPRARSRSTSKSKPNVPAESRSRSKSASK 462
Score = 37.4 bits (85), Expect = 0.084
Identities = 34/182 (18%), Positives = 75/182 (40%), Gaps = 11/182 (6%)
Query: 50 KGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSI 109
+ R +K K + + +++ S SK A+ ++ + G A
Sbjct: 239 RSRSKKEKSRSPSKDNKSRSRSRSPDKSRSKSKDHAEDKLQNNDSAGKAK---------- 288
Query: 110 SNPPSRENSPAPNPAKSK-AEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSS 168
S+ PSR +S + + ++ + AE + + + +++ S S+ + ++S+S +S RS+S
Sbjct: 289 SHSPSRHDSKSRSRSQERRAEEERRRSVSRARSQEKSRSQEKSLLKSRSRSRSRSRSRSK 348
Query: 169 TNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSS 228
K S + + S + K D+ + S++ + S +VS
Sbjct: 349 DKRKGRKRSRDESRSRSRSKSERSRKHSSKRDSKVSSSSSSSKKKKDTDHSRSPSRSVSK 408
Query: 229 EK 230
E+
Sbjct: 409 ER 410
Score = 31.2 bits (69), Expect = 6.0
Identities = 34/157 (21%), Positives = 60/157 (37%), Gaps = 13/157 (8%)
Query: 136 KAASLAKKTSTSEGRRKVRSKSGHKSPHRSKS---STNSSPSKESACQETQGATSPIREA 192
++ S ++ S S RK RS+SG SKS S + S S+ + +Q + +E
Sbjct: 187 RSRSHSRSRSRSRHSRKSRSRSGSSKSSHSKSRSRSRSGSHSRSKSRSRSQSRSRSKKEK 246
Query: 193 EALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFEEDPLAALDGFLDGTLD 252
P KD+ + + SP S + + +KL D D
Sbjct: 247 SRSPSKDN-------KSRSRSRSPDKSRSKSKDHAEDKLQ---NNDSAGKAKSHSPSRHD 296
Query: 253 WDSPPHQADATAETAQSSGVANLQQIEESMGRLKAIV 289
S + AE + V+ + E+S + K+++
Sbjct: 297 SKSRSRSQERRAEEERRRSVSRARSQEKSRSQEKSLL 333
>MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC LH)
[Contains: Micronuclear linker histone-alpha;
Micronuclear linker histone-beta; Micronuclear linker
histone-delta; Micronuclear linker histone-gamma]
Length = 633
Score = 49.3 bits (116), Expect = 2e-05
Identities = 42/151 (27%), Positives = 65/151 (42%), Gaps = 15/151 (9%)
Query: 36 DDDEDDNVSLADKLKGRKRKPKPAA-----SASQKRAKGAGVSTTSGASKLASDAQPEVN 90
D E+ + + + +GR+R+ K S S + K S T SK S ++ N
Sbjct: 462 DSSENASSTTQTRTRGRQREQKDMVNEKSNSKSSSKGKKNSKSNTRSKSKSKSASKSRKN 521
Query: 91 GKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGR 150
+ D T S S S+ S APN +K E +E+P K+ S+ R
Sbjct: 522 ASKSKKDTTNHGR-QTRSKSRSESKSKSEAPNKPSNKMEVIEQP-------KEESSDRKR 573
Query: 151 RKVRSKSGHKSPHRSKSSTNSSPSKESACQE 181
R+ RS+S K+ K S N S SK+ ++
Sbjct: 574 RESRSQSAKKT--SDKKSKNRSDSKKMTAED 602
Score = 42.4 bits (98), Expect = 0.003
Identities = 46/213 (21%), Positives = 82/213 (37%), Gaps = 26/213 (12%)
Query: 48 KLKGRKRKPKPAASASQKRAKGAGVSTTSGAS-------KLASDAQPEVNGKEGDGDATI 100
+ K K AS S ++KG S + G S K S ++P+ N + +
Sbjct: 400 RTKKANNKSASKASKSGSKSKGKSASKSKGKSSSKGKNSKSRSASKPKSNAAQNSNNTHQ 459
Query: 101 QADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRR--KVRSKSG 158
AD S EN+ + +++ E+ + S+S+G++ K ++S
Sbjct: 460 TAD----------SSENASSTTQTRTRGRQREQKDMVNEKSNSKSSSKGKKNSKSNTRSK 509
Query: 159 HKSPHRSKSSTNSSPSKESAC----QETQGATSPIREAEALPGKDDNPM---PPPKSTAT 211
KS SKS N+S SK+ Q + S + P K N M PK ++
Sbjct: 510 SKSKSASKSRKNASKSKKDTTNHGRQTRSKSRSESKSKSEAPNKPSNKMEVIEQPKEESS 569
Query: 212 VQPSPQAKGGFSSNVSSEKLDTLFEEDPLAALD 244
+ +++ + S +K + + A D
Sbjct: 570 DRKRRESRSQSAKKTSDKKSKNRSDSKKMTAED 602
Score = 40.8 bits (94), Expect = 0.008
Identities = 43/210 (20%), Positives = 83/210 (39%), Gaps = 12/210 (5%)
Query: 23 SSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLA 82
SS ++ D + A KGRK ++++++ + + ++S +K +
Sbjct: 222 SSSNRRKASSSKDQKGTRSSSRKASNSKGRKN-----STSNKRNSSSSSKRSSSSKNKKS 276
Query: 83 SDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAK 142
S ++ N K + S+ +R++S + + S ++G + + S +
Sbjct: 277 SSSK---NKKSSSSKGRKSSSSRGRKASSSKNRKSSKSKDRKSSSSKGRKSSSSSKSNKR 333
Query: 143 KTSTSEGRRKVRSK--SGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDD 200
K S+S GR+ SK KS R S ++S E Q+ + ++S + E+
Sbjct: 334 KASSSRGRKSSSSKGRKSSKSQERKNSHADTSKQMEDEGQKRRQSSSSAKRDESSKKSRR 393
Query: 201 NPMPP--PKSTATVQPSPQAKGGFSSNVSS 228
N M K S +K G S S
Sbjct: 394 NSMKEARTKKANNKSASKASKSGSKSKGKS 423
Score = 39.3 bits (90), Expect = 0.022
Identities = 34/144 (23%), Positives = 63/144 (43%), Gaps = 6/144 (4%)
Query: 50 KGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHS- 108
K R+ K +S S+ R K + +S +Q + + + Q S
Sbjct: 184 KSRRSSSKGKSSVSKGRTKSTSSKRRADSSASQGRSQSSSSNRRKASSSKDQKGTRSSSR 243
Query: 109 -ISNPPSRENSPAP---NPAKSKAEGVEKPPKAASLA-KKTSTSEGRRKVRSKSGHKSPH 163
SN R+NS + + + SK K K++S KK+S+S+GR+ S+ S
Sbjct: 244 KASNSKGRKNSTSNKRNSSSSSKRSSSSKNKKSSSSKNKKSSSSKGRKSSSSRGRKASSS 303
Query: 164 RSKSSTNSSPSKESACQETQGATS 187
+++ S+ S K S+ + + ++S
Sbjct: 304 KNRKSSKSKDRKSSSSKGRKSSSS 327
Score = 35.4 bits (80), Expect = 0.32
Identities = 40/192 (20%), Positives = 73/192 (37%), Gaps = 12/192 (6%)
Query: 48 KLKGRKRKPKPAASASQKRAKGAGVST-----------TSGASKLASDAQPEVNGKEGDG 96
+ K K + +SASQ R++ + + T +S+ AS+++ N
Sbjct: 200 RTKSTSSKRRADSSASQGRSQSSSSNRRKASSSKDQKGTRSSSRKASNSKGRKNSTSNKR 259
Query: 97 DATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSK 156
+++ + S + S + + +K + + KA+S + S+ RK S
Sbjct: 260 NSSSSSKRSSSSKNKKSSSSKNKKSSSSKGRKSSSSRGRKASSSKNRKSSKSKDRKSSSS 319
Query: 157 SGHKSPHRSKSS-TNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPS 215
G KS SKS+ +S S+ ++G S + D + + Q S
Sbjct: 320 KGRKSSSSSKSNKRKASSSRGRKSSSSKGRKSSKSQERKNSHADTSKQMEDEGQKRRQSS 379
Query: 216 PQAKGGFSSNVS 227
AK SS S
Sbjct: 380 SSAKRDESSKKS 391
Score = 33.9 bits (76), Expect = 0.93
Identities = 29/156 (18%), Positives = 63/156 (39%), Gaps = 7/156 (4%)
Query: 63 SQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPN 122
++K + A T + + + GK ++ + + S+ S + +
Sbjct: 164 NEKYGQAAQKKQTKRKNSTSKSRRSSSKGKSSVSKGRTKSTSSKRRADSSASQGRSQSSS 223
Query: 123 PAKSKAEGVEKPPKAASLAKKTSTSEGR-------RKVRSKSGHKSPHRSKSSTNSSPSK 175
+ KA + S ++K S S+GR R S S S ++K S++S K
Sbjct: 224 SNRRKASSSKDQKGTRSSSRKASNSKGRKNSTSNKRNSSSSSKRSSSSKNKKSSSSKNKK 283
Query: 176 ESACQETQGATSPIREAEALPGKDDNPMPPPKSTAT 211
S+ + + ++S R+A + + + KS+++
Sbjct: 284 SSSSKGRKSSSSRGRKASSSKNRKSSKSKDRKSSSS 319
Score = 31.6 bits (70), Expect = 4.6
Identities = 35/158 (22%), Positives = 60/158 (37%), Gaps = 26/158 (16%)
Query: 50 KGRK----RKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVH 105
KGRK R K ++S ++K +K ++S + +S + K +
Sbjct: 288 KGRKSSSSRGRKASSSKNRKSSKSKDRKSSSSKGRKSSSSSKSNKRKASSSRGRKSSSSK 347
Query: 106 EHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVR----------- 154
S R+NS A + + EG ++ ++S + S+ + RR
Sbjct: 348 GRKSSKSQERKNSHADTSKQMEDEGQKRRQSSSSAKRDESSKKSRRNSMKEARTKKANNK 407
Query: 155 -----SKSGHKSPHRS------KSSTNSSPSKESACQE 181
SKSG KS +S KSS+ SK + +
Sbjct: 408 SASKASKSGSKSKGKSASKSKGKSSSKGKNSKSRSASK 445
>RRP1_DROME (P27864) Recombination repair protein 1 (DNA-(apurinic
or apyrimidinic site) lyase) (EC 4.2.99.18)
Length = 679
Score = 48.5 bits (114), Expect = 4e-05
Identities = 54/218 (24%), Positives = 84/218 (37%), Gaps = 16/218 (7%)
Query: 7 EVITQPSPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKR 66
EV Q +P + VA K +P D DE+ + + K KGR +K A K
Sbjct: 66 EVEPQKAP---TAVARGKKKQP------KDTDENGQMEVVAKPKGRAKKATAEAEPEPKV 116
Query: 67 AKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKS 126
AG +T A K + A EV G +A + + + R+ P A
Sbjct: 117 DLPAGKATKPRAKKEPTPAPDEVTSSPPKG----RAKAEKPTNAQAKGRKRKELPAEANG 172
Query: 127 KAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGAT 186
AE +PPK + + T + + + + S K ++ + +K A E A
Sbjct: 173 GAEEAAEPPKQRARKEAVPTLKEQAEPGTISKEKVQKAETAAKRARGTKRLADSEIAAAL 232
Query: 187 SPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSS 224
E + +P K + K V+PSP+ G F S
Sbjct: 233 DE-PEVDEVPPKAASKR--AKKGKMVEPSPETVGDFQS 267
Score = 44.3 bits (103), Expect = 7e-04
Identities = 58/246 (23%), Positives = 91/246 (36%), Gaps = 45/246 (18%)
Query: 2 RIGQAEVITQPSPQVISPVAHSSKDK----PVVIEDDDDDDEDDNVSLADK-----LKGR 52
R +A +P P+V P ++K + P D+ + A+K KGR
Sbjct: 102 RAKKATAEAEPEPKVDLPAGKATKPRAKKEPTPAPDEVTSSPPKGRAKAEKPTNAQAKGR 161
Query: 53 KRKPKPA---------ASASQKRAKGAGVST--------TSGASKL--ASDAQPEVNGKE 93
KRK PA A ++RA+ V T T K+ A A G +
Sbjct: 162 KRKELPAEANGGAEEAAEPPKQRARKEAVPTLKEQAEPGTISKEKVQKAETAAKRARGTK 221
Query: 94 GDGDATIQADVHEHSISNPPSRENS----------PAPNPA---KSKAEGVEKPPKAASL 140
D+ I A + E + P + S P+P +S E VE PPK A+
Sbjct: 222 RLADSEIAAALDEPEVDEVPPKAASKRAKKGKMVEPSPETVGDFQSVQEEVESPPKTAAA 281
Query: 141 ----AKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALP 196
AKKT+ E ++ K+ K + P+ +++ + + E EA P
Sbjct: 282 PKKRAKKTTNGETAVELEPKTKAKPTKQRAKKEGKEPAPGKKQKKSADKENGVVEEEAKP 341
Query: 197 GKDDNP 202
+ P
Sbjct: 342 STETKP 347
Score = 38.1 bits (87), Expect = 0.049
Identities = 39/167 (23%), Positives = 60/167 (35%), Gaps = 12/167 (7%)
Query: 58 PAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSREN 117
P A +K+A+ T D P NG D +A A+ + P
Sbjct: 2 PRVKAVKKQAEALASEPT--------DPTPNANGNGVDENADSAAEELKVPAKGKPRARK 53
Query: 118 SPAPNPAKSKAEGVE--KPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSK 175
+ + +E VE K P A + KK + + + K R+K +T + +
Sbjct: 54 ATKTAVSAENSEEVEPQKAPTAVARGKKKQPKDTDENGQMEVVAKPKGRAKKATAEAEPE 113
Query: 176 ESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQ--PSPQAKG 220
AT P + E P D+ PPK A + + QAKG
Sbjct: 114 PKVDLPAGKATKPRAKKEPTPAPDEVTSSPPKGRAKAEKPTNAQAKG 160
Score = 31.6 bits (70), Expect = 4.6
Identities = 22/99 (22%), Positives = 38/99 (38%)
Query: 48 KLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEH 107
K + +K +PA QK++ +K +++ +P K+ A D+ E
Sbjct: 308 KQRAKKEGKEPAPGKKQKKSADKENGVVEEEAKPSTETKPAKGRKKAPVKAEDVEDIEEA 367
Query: 108 SISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTST 146
+ + P+R A A+ E K AKK T
Sbjct: 368 AEESKPARGRKKAAAKAEEPDVDEESGSKTTKKAKKAET 406
>MAP4_BOVIN (P36225) Microtubule-associated protein 4 (MAP 4)
(Microtubule-associated protein-U) (MAP-U)
Length = 1072
Score = 48.1 bits (113), Expect = 5e-05
Identities = 46/180 (25%), Positives = 75/180 (41%), Gaps = 21/180 (11%)
Query: 49 LKGRKRKPKPAASASQKRA--KGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHE 106
L G +KP AS S A K +T+ ++ + D +P+ +A + E
Sbjct: 680 LGGSNKKPMSLASGSVPAAPPKRPAAATSRPSTLPSKDTKPK---------PVAEAKIPE 730
Query: 107 HSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEG------RRKVRSKSGHK 160
+S P ++PA P + V K P A+LA STS +R K+ K
Sbjct: 731 KRVS-PSKPASAPAVKPGSKSTQAVPKAPATATLASPGSTSRNLSTPLPKRPTAIKTEGK 789
Query: 161 SPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKG 220
K +T S+P+ S + T TS ++++ +PG P ++ T P P+ G
Sbjct: 790 PAEIKKMATKSAPADLSRPKST--TTSSVKKSTTVPGTAPPAGAPSRARPTATP-PRPSG 846
Score = 37.7 bits (86), Expect = 0.064
Identities = 48/187 (25%), Positives = 79/187 (41%), Gaps = 25/187 (13%)
Query: 75 TSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKP 134
+S S+L S+ +EG ++ + I+ PP++E P+P K+K +P
Sbjct: 602 SSQTSELPSETSGVAKPEEGPPTGSVSGN----DITAPPNKELPPSPE-KKTKPLATTQP 656
Query: 135 PK-AASLAKKTSTSEGRRKVRSKSG--HKSP-HRSKSSTNSSPSKESACQETQGATSPIR 190
K + S AK TS ++ + G +K P + S ++P K A ++ +T P +
Sbjct: 657 AKTSTSKAKTQPTSLPKQTAPTTLGGSNKKPMSLASGSVPAAPPKRPAAATSRPSTLPSK 716
Query: 191 E------AEA-LPGKDDNPMPP---------PKSTATVQPSPQAKGGFSSNVSSEKLDTL 234
+ AEA +P K +P P KST V +P S +S L T
Sbjct: 717 DTKPKPVAEAKIPEKRVSPSKPASAPAVKPGSKSTQAVPKAPATATLASPGSTSRNLSTP 776
Query: 235 FEEDPLA 241
+ P A
Sbjct: 777 LPKRPTA 783
>FUTS_DROME (Q9W596) Microtubule-associated protein futsch
Length = 5412
Score = 47.4 bits (111), Expect = 8e-05
Identities = 91/449 (20%), Positives = 167/449 (36%), Gaps = 63/449 (14%)
Query: 6 AEVITQPSPQVISP--VAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASAS 63
AE PS + P VA S KD E + +E S+A+K +K AS +
Sbjct: 3715 AEKSPLPSKEASRPTSVAESVKD-----EAEKSKEESRRESVAEKSSLASKKASRPASVA 3769
Query: 64 QKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPS--RENSPAP 121
+ A S + ++ P + KE A++ V + + + RE+
Sbjct: 3770 ESVKDEAEKSKEESRRESVAEKSP-LASKEASRPASVAESVKDEAEKSKEESRRESVAEK 3828
Query: 122 NPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQE 181
+P SK +P A K + R KSP S ++ + ES E
Sbjct: 3829 SPLPSKE--ASRPTSVAESVKDEADKSKEESRRESGAEKSPLASMEASRPTSVAESVKDE 3886
Query: 182 TQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFEEDPLA 241
T+ + R + +P+P +++ + K + + +++ E+ PLA
Sbjct: 3887 TEKSKEESRRESV---TEKSPLPSKEASRPTSVAESVKDEAEKSKEESRRESVAEKSPLA 3943
Query: 242 ALDGFLDGTLDWDSPPHQADATAETAQSSGVANLQQIEESMGRLKAIVFDIGFLDRIRVD 301
+ + P A++ + A+ + + ++ G+ ++I D
Sbjct: 3944 SKES--------SRPASVAESIKDEAEGTKQESRRESMPESGKAESIKGD---------- 3985
Query: 302 PQASHAAKELLIFLLGQKLDSHQAAALANLQSFLVDALATFHQVKDVGQAVSLKEAKVSS 361
Q+S A+KE S + + +++ K G A+ +++V+S
Sbjct: 3986 -QSSLASKE----------TSRPDSVVESVKD---------ETEKPEGSAID--KSQVAS 4023
Query: 362 GKAQVAAMKEDFKK-FTARKVLIADEISDVDTKLEELHREIAKLEKKRADLVEEGPAVKA 420
VA +D K +R +AD+ D +E R ++ E + +EEGP A
Sbjct: 4024 RPESVAVSAKDEKSPLHSRPESVADKSPDAS---KEASRSLSVAETASSP-IEEGPRSIA 4079
Query: 421 QLD---GLTKESKALIISRKDVIKELEVD 446
L LT E+K + + I E D
Sbjct: 4080 DLSLPLNLTGEAKGKLPTLSSPIDVAEGD 4108
Score = 46.2 bits (108), Expect = 2e-04
Identities = 80/444 (18%), Positives = 161/444 (36%), Gaps = 53/444 (11%)
Query: 18 SPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSG 77
SP+A +P + + D+ + + + + ++ P P+ AS+ + + +
Sbjct: 1942 SPLASEEASRPASVAESVKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAE 2001
Query: 78 ASKLASDAQ-----PEVNGKEGDGDATIQADV-HEHSISNPPSRENSPA---PNPAKSKA 128
SK S + + KE A++ + E S SR S A P P+K
Sbjct: 2002 KSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKE-- 2059
Query: 129 EGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSP 188
+P A K + R KSP SK ++ + ES E + +
Sbjct: 2060 --ASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEE 2117
Query: 189 IREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFEEDPLAALDGFLD 248
R + +P+P +++ + K + + +++ E+ PL + +
Sbjct: 2118 SRRESV---AEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRP 2174
Query: 249 GTLDWDSPPHQADATAETAQSSGVANLQQIEESMGRLKAIVFDIGFLDRIRVDPQASHAA 308
++ +S +A+ + E ++ VA + A V + + +A +
Sbjct: 2175 ASVA-ESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAE-------SIKDEAEKSK 2226
Query: 309 KELLIFLLGQK--LDSHQAAALANLQSFLVDALATFHQVKDVGQAVSLKEAKVSSGKAQV 366
+E + +K L S +A+ A++ + D +++K S +
Sbjct: 2227 EETRRESVAEKSPLPSKEASRPASVAESIKD---------------EAEKSKEESRRESA 2271
Query: 367 AAMKEDFKKFTARKVLIADEISDVDTKLEELHREIAKLEKKRADLVEEGPAVKAQLDGLT 426
A K +R +A+ + D K +E E +R + E G A + D
Sbjct: 2272 AEKSPLPSKEASRPASVAESVKDEADKSKE--------ESRRESMAESGKAQSIKGD--- 2320
Query: 427 KESKALIISRKDVIKELEVDQAVK 450
+S +SR + + E D VK
Sbjct: 2321 -QSPLKEVSRPESVAESVKDDPVK 2343
Score = 43.1 bits (100), Expect = 0.002
Identities = 86/460 (18%), Positives = 173/460 (36%), Gaps = 53/460 (11%)
Query: 18 SPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSG 77
SP+A +P + + D+ + + + + ++ P P+ AS+ + V +
Sbjct: 3422 SPLASKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPTSVAESVKDEAE 3481
Query: 78 ASKLAS--DAQPE---VNGKEGDGDATIQADVHEHSISNPPS--RENSPAPNPAKSKAEG 130
SK S D+ E + KE A++ V + + + RE+ +P SK
Sbjct: 3482 KSKEESRRDSVAEKSPLASKEASRPASVAESVQDEAEKSKEESRRESVAEKSPLASKE-- 3539
Query: 131 VEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIR 190
+P A K + R KSP SK ++ + ES E + +
Sbjct: 3540 ASRPASVAESIKDEAEKSKEESRRESVAEKSPLASKEASRPTSVAESVKDEAEKSK---E 3596
Query: 191 EAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFEEDPLAALDGFLDGT 250
E+ + +P+ +++ + + + + +++ E+ PLA+ + +
Sbjct: 3597 ESSRDSVAEKSPLASKEASRPASVAESVQDEAEKSKEESRRESVAEKSPLASKEASRPAS 3656
Query: 251 LDWDSPPHQADATAETAQSSGVANLQQIEESMGRLKAIVFDIGFLDRIRVDPQASHAAKE 310
+ +S A+ + E ++ VA + A V + V +A + +E
Sbjct: 3657 VA-ESVKDDAEKSKEESRRESVAEKSPLASKEASRPASVAE-------SVKDEAEKSKEE 3708
Query: 311 LLIFLLGQK--LDSHQAAALANLQSFLVDALATFHQVKDVGQAVSLKEAKVSSGKAQVAA 368
+ +K L S +A+ ++ VKD +++K S + VA
Sbjct: 3709 SRRESVAEKSPLPSKEASRPTSVA----------ESVKD-----EAEKSKEESRRESVAE 3753
Query: 369 MKEDFKKFTARKVLIADEISD-VDTKLEELHRE-------IAKLEKKRADLVEEGPAVKA 420
K +R +A+ + D + EE RE +A E R V E +VK
Sbjct: 3754 KSSLASKKASRPASVAESVKDEAEKSKEESRRESVAEKSPLASKEASRPASVAE--SVKD 3811
Query: 421 QLDGLTKESKALIISRKDVIKELE------VDQAVKKDLD 454
+ + +ES+ ++ K + E V ++VK + D
Sbjct: 3812 EAEKSKEESRRESVAEKSPLPSKEASRPTSVAESVKDEAD 3851
Score = 40.8 bits (94), Expect = 0.008
Identities = 75/387 (19%), Positives = 150/387 (38%), Gaps = 39/387 (10%)
Query: 28 PVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQP 87
P V+E D ++S + + K S R S T ++K S
Sbjct: 2834 PSVVESTKADSTKGDISPSPESVLEGPKDDVEKSKESSRPPSVSASITGDSTKDVSRPAS 2893
Query: 88 EVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTS 147
V + + D +A+ SI+ S + + +KS ++ +K K+ +K+ S
Sbjct: 2894 VVESVKDEHD---KAESRRESIAKVESVIDEAGKSDSKSSSQDSQKDEKSTLASKEASRR 2950
Query: 148 EGRRKVRSKSGHKSPHRSKSSTNSS---PSKESACQETQGATSPIREAEALPGKDDNPMP 204
E + KS R +S S P + + +++ + P E++ +D+
Sbjct: 2951 ESVVESSKDDAEKSESRPESVIASGEPVPRESKSPLDSKDTSRPGSMVESVTAEDEKSEQ 3010
Query: 205 PPK--STATVQPSPQAKGGFSSNVSS-EKLDTLFEEDP------LAALDGFLDG--TLDW 253
+ S A + K G S S +D L ++D ++ G T+
Sbjct: 3011 QSRRESVAESVKADTKKDGKSQEASRPSSVDELLKDDDEKQESRRQSITGSHKAMSTMGD 3070
Query: 254 DSPPHQADATAETAQSSGVA------NLQQIEESMGRLK-----AIVFDIGFLDRIRVDP 302
+SP +AD + E ++ VA N + E +G + +I DI ++ +
Sbjct: 3071 ESPMDKADKSKEPSRPESVAESIKHENTKDEESPLGSRRDSVAESIKSDITKGEKSPLPS 3130
Query: 303 QASHAAKELLIFLLGQKLDSHQAAALANLQSFLVDALATFHQVKDVGQAVSLKEAKVSSG 362
+ + ++ + +K +S + + +++ KD A KE S
Sbjct: 3131 KEVSRPESVVGSIKDEKAESRRESVAESVKP---------ESSKDATSAPPSKEH--SRP 3179
Query: 363 KAQVAAMKEDFKKFTARKVLIADEISD 389
++ + ++K++ K T+R+V +AD I D
Sbjct: 3180 ESVLGSLKDEGDKTTSRRVSVADSIKD 3206
Score = 40.8 bits (94), Expect = 0.008
Identities = 80/441 (18%), Positives = 166/441 (37%), Gaps = 52/441 (11%)
Query: 21 AHSSKDKPVVIEDDDDDDEDDNVSLADKLKGR-----KRKPKPAASASQKRAKGAGVSTT 75
A S KD+ + D+ ++ S + + + K +PA+ A + +
Sbjct: 3288 AGSIKDEKSPLASKDEAEKSKEESRRESVAEQFPLVSKEVSRPASVAESVKDEAEKSKEE 3347
Query: 76 SGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPP 135
S + V G D + + S++ E SP P+ S+ P
Sbjct: 3348 SPLMSKEASRPASVAGSVKDEAEKSKEESRRESVA-----EKSPLPSKEASR-------P 3395
Query: 136 KAASLAKKTSTSEGRRKVRSKSG-HKSPHRSKSSTNSSPSKESACQETQGATSPIREAEA 194
+ + + K + + + R +SG KSP SK ++ + ES E + + R
Sbjct: 3396 ASVAESVKDEADKSKEESRRESGAEKSPLASKEASRPASVAESIKDEAEKSKEESRRESV 3455
Query: 195 LPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFEEDPLAALDGFLDGTLDWD 254
+ +P+P +++ + K + + D++ E+ PLA+ + ++ +
Sbjct: 3456 ---AEKSPLPSKEASRPTSVAESVKDEAEKSKEESRRDSVAEKSPLASKEASRPASVA-E 3511
Query: 255 SPPHQADATAETAQSSGVANLQQIEESMGRLKAIVFDIGFLDRIRVDPQASHAAKELLIF 314
S +A+ + E ++ VA + A V + + +A + +E
Sbjct: 3512 SVQDEAEKSKEESRRESVAEKSPLASKEASRPASVAE-------SIKDEAEKSKEE---- 3560
Query: 315 LLGQKLDSHQAAALANLQSFLVDALATFHQVKDVGQAVSLKEAKVSSGKAQVAAMKEDFK 374
++ + + LA+ ++ ++A VKD +++K S + VA
Sbjct: 3561 --SRRESVAEKSPLASKEASRPTSVA--ESVKD-----EAEKSKEESSRDSVAEKSPLAS 3611
Query: 375 KFTARKVLIADEISD-VDTKLEELHRE-------IAKLEKKRADLVEEGPAVKAQLDGLT 426
K +R +A+ + D + EE RE +A E R V E +VK +
Sbjct: 3612 KEASRPASVAESVQDEAEKSKEESRRESVAEKSPLASKEASRPASVAE--SVKDDAEKSK 3669
Query: 427 KESKALIISRKDVIKELEVDQ 447
+ES+ ++ K + E +
Sbjct: 3670 EESRRESVAEKSPLASKEASR 3690
Score = 38.9 bits (89), Expect = 0.029
Identities = 69/322 (21%), Positives = 120/322 (36%), Gaps = 33/322 (10%)
Query: 6 AEVITQPSPQVISP--VAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASAS 63
AE PS + P VA S KD E + +E S+A+K ++ AS +
Sbjct: 2198 AEKSPLPSKEASRPASVAESIKD-----EAEKSKEETRRESVAEKSPLPSKEASRPASVA 2252
Query: 64 QKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSI-SNPPSRENSPAPN 122
+ A S + A++ P + KE A++ V + + S SR S A +
Sbjct: 2253 ESIKDEAEKSKEESRRESAAEKSP-LPSKEASRPASVAESVKDEADKSKEESRRESMAES 2311
Query: 123 PAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQET 182
+G + P K S + + S V+SK + + S T S + + E+
Sbjct: 2312 GKAQSIKGDQSPLKEVSRPESVAESVKDDPVKSKEPSRRESVAGSVTADSARDDQSPLES 2371
Query: 183 QGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSN--VSSEKLDTLFEEDPL 240
+GA+ P +++ + + +S P+AK S + + ED
Sbjct: 2372 KGASRPESVVDSVKDEAEKQESRRESKTESVIPPKAKDDKSPKEVLQPVSMTETIRED-- 2429
Query: 241 AALDGFLDGTLDWDSPPHQADATAETAQSSGVANLQQIEESMGRLK----------AIVF 290
D P QA++ E+ S A+ + E+S K +I +
Sbjct: 2430 ----------ADQPMKPSQAESRRESIAESIKASSPRDEKSPLASKEASRPGSVAESIKY 2479
Query: 291 DIGFLDRIRVDPQASHAAKELL 312
D+ I+ D H+ +E L
Sbjct: 2480 DLDKPQIIKDDKSTEHSRRESL 2501
Score = 38.1 bits (87), Expect = 0.049
Identities = 88/461 (19%), Positives = 169/461 (36%), Gaps = 75/461 (16%)
Query: 18 SPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSG 77
SP+A +P + + D+ + S + + + P AS R S
Sbjct: 3496 SPLASKEASRPASVAESVQDEAEK--SKEESRRESVAEKSPLASKEASRPASVAESIKDE 3553
Query: 78 ASKLASDAQPE-------VNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEG 130
A K +++ E + KE ++ V + + S+E S + A+
Sbjct: 3554 AEKSKEESRRESVAEKSPLASKEASRPTSVAESVKDEA---EKSKEESSRDSVAEKSPLA 3610
Query: 131 VEKPPKAASLAKKTSTSEGRRKVRSKS---GHKSPHRSKSST-----------NSSPSKE 176
++ + AS+A+ + K S+ KSP SK ++ ++ SKE
Sbjct: 3611 SKEASRPASVAESVQDEAEKSKEESRRESVAEKSPLASKEASRPASVAESVKDDAEKSKE 3670
Query: 177 SACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFE 236
+ +E+ SP+ EA P S A K + + +++ E
Sbjct: 3671 ESRRESVAEKSPLASKEA---------SRPASVA-----ESVKDEAEKSKEESRRESVAE 3716
Query: 237 EDPLAALDGFLDGTLDWDSPPHQADATAETAQSSGVANLQQIEESMGRLKAIVFDIGFLD 296
+ PL + + ++ +S +A+ + E ++ VA + A V +
Sbjct: 3717 KSPLPSKEASRPTSVA-ESVKDEAEKSKEESRRESVAEKSSLASKKASRPASVAE----- 3770
Query: 297 RIRVDPQASHAAKELLIFLLGQK--LDSHQAAALANLQSFLVDALATFHQVKDVGQAVSL 354
V +A + +E + +K L S +A+ A++ VKD
Sbjct: 3771 --SVKDEAEKSKEESRRESVAEKSPLASKEASRPASVA----------ESVKD-----EA 3813
Query: 355 KEAKVSSGKAQVAAMKEDFKKFTARKVLIADEISD-VDTKLEELHRE-------IAKLEK 406
+++K S + VA K +R +A+ + D D EE RE +A +E
Sbjct: 3814 EKSKEESRRESVAEKSPLPSKEASRPTSVAESVKDEADKSKEESRRESGAEKSPLASMEA 3873
Query: 407 KRADLVEEGPAVKAQLDGLTKESKALIISRKDVIKELEVDQ 447
R V E +VK + + +ES+ ++ K + E +
Sbjct: 3874 SRPTSVAE--SVKDETEKSKEESRRESVTEKSPLPSKEASR 3912
Score = 37.7 bits (86), Expect = 0.064
Identities = 81/433 (18%), Positives = 164/433 (37%), Gaps = 50/433 (11%)
Query: 18 SPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSG 77
SP +P + + DD S+ D P AS R AG + +S
Sbjct: 2666 SPQVLKESSRPAWVAESKDDAAQLKSSVEDLRS-------PVASTEISRPASAGETASSP 2718
Query: 78 ASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKA 137
+ D ++ TI+ + ++S+P + P + + +
Sbjct: 2719 IEEAPKDFAEFEQAEKAVLPLTIELKGNLPTLSSPVDVAHGDFPQTSTPTSSPTVASVQP 2778
Query: 138 ASLAK----KTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAE 193
A L+K KT++S +S G + R +S S+ + ++++ A+ P E
Sbjct: 2779 AELSKVDIEKTASSPIDEAPKSLIGCPAEERPESPAESAKDAAESVEKSKDASRPPSVVE 2838
Query: 194 ALPGKDDNPMPPPKSTATVQPSPQAK-GGFSSNVSSEKLDTLFEEDPLAALDGFLDGTLD 252
+ K D+ + + PSP++ G +V K ++ A++ G D T D
Sbjct: 2839 ST--KADS------TKGDISPSPESVLEGPKDDVEKSK-ESSRPPSVSASITG--DSTKD 2887
Query: 253 WDSPPHQADATAETAQSSGVANLQQIEESMGRLKAIVFDIGFLDRIRVDPQASHAAKELL 312
P ++ + + + ES+ ++++++ + G D + K L
Sbjct: 2888 VSRPASVVESVKDEHDKA-----ESRRESIAKVESVIDEAGKSDSKSSSQDSQKDEKSTL 2942
Query: 313 IFLLGQKLDSHQAAALANLQSFLVDALATFHQVKDVGQAVSLKEAKVSSGKAQVAAMKED 372
A+ A+ + +V++ D ++ S E+ ++SG+ K
Sbjct: 2943 ------------ASKEASRRESVVES-----SKDDAEKSESRPESVIASGEPVPRESKSP 2985
Query: 373 F-KKFTARKVLIADEISDVDTKLEELHREIAKLEKKRADLVEEGPAVKA----QLDGLTK 427
K T+R + + ++ D K E+ R + E +AD ++G + +A +D L K
Sbjct: 2986 LDSKDTSRPGSMVESVTAEDEKSEQQSRRESVAESVKADTKKDGKSQEASRPSSVDELLK 3045
Query: 428 ESKALIISRKDVI 440
+ SR+ I
Sbjct: 3046 DDDEKQESRRQSI 3058
>NFH_RAT (P16884) Neurofilament triplet H protein (200 kDa
neurofilament protein) (Neurofilament heavy polypeptide)
(NF-H) (Fragment)
Length = 831
Score = 47.0 bits (110), Expect = 1e-04
Identities = 52/224 (23%), Positives = 84/224 (37%), Gaps = 23/224 (10%)
Query: 16 VISPVAHSSKDK---PVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGV 72
V SPV S + PV ++ + V +K PA + S AK V
Sbjct: 411 VKSPVEAKSPAEVKSPVTVKSPAEAKSPVEVKSPASVKSPSEAKSPAGAKSPAEAKSPVV 470
Query: 73 STTSGASKLASDAQPEVNGK-----------EGDGDATIQADVHEHSISNPPSRENSPAP 121
+ + +K + A+P K + +A A+ P + SP
Sbjct: 471 AKSPAEAKSPAGAKPPAEAKSPAEAKSPAEAKSPAEAKSPAEAKSPVEVKSPEKAKSPVK 530
Query: 122 NPAKSKAEGVEKPPKAASLAKK------TSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSK 175
AKS AE + P KA S K+ S + K + KSP ++K+ SP
Sbjct: 531 EGAKSLAE-AKSPEKAKSPVKEEIKPPAEVKSPEKAKSPMRKEAKSPEKAKTLDVKSPEA 589
Query: 176 ESACQETQGATSPIREAEAL--PGKDDNPMPPPKSTATVQPSPQ 217
+ +E + IR E + P K++ P + T T + +P+
Sbjct: 590 KPPAKEEAKRPADIRSPEQVKSPAKEEAKSPEKEETRTEKVAPK 633
Score = 40.4 bits (93), Expect = 0.010
Identities = 53/204 (25%), Positives = 81/204 (38%), Gaps = 33/204 (16%)
Query: 48 KLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQ----AD 103
K G + P A S ++ ++ A V + A A P + + ++ A
Sbjct: 388 KSPGEAKSPAEAKSPAEVKSP-ATVKSPVEAKSPAEVKSPVTVKSPAEAKSPVEVKSPAS 446
Query: 104 VHEHSISNPPSRENSPA----PNPAKSKAE-----GVEKPPKAASLAKKTSTSEGRRKVR 154
V S + P+ SPA P AKS AE G + P +A S A+ S +E +
Sbjct: 447 VKSPSEAKSPAGAKSPAEAKSPVVAKSPAEAKSPAGAKPPAEAKSPAEAKSPAEAKSPAE 506
Query: 155 SKSGH--------KSPHRSKSSTNSSPSKESACQETQGATSPIRE-----AEA-LPGKDD 200
+KS KSP ++KS + + + A SP++E AE P K
Sbjct: 507 AKSPAEAKSPVEVKSPEKAKSPVKEGAKSLAEAKSPEKAKSPVKEEIKPPAEVKSPEKAK 566
Query: 201 NPM-----PPPKSTATVQPSPQAK 219
+PM P K+ SP+AK
Sbjct: 567 SPMRKEAKSPEKAKTLDVKSPEAK 590
Score = 34.7 bits (78), Expect = 0.54
Identities = 36/142 (25%), Positives = 63/142 (44%), Gaps = 14/142 (9%)
Query: 88 EVNGKEGDGDATIQADV--HEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTS 145
E KE G+ +A+ E + ++PP+ E A +P K V++ K+ + AK +
Sbjct: 239 EEEDKEAQGEEEEEAEEGGEEAATTSPPAEE---AASPEKETKSPVKEEAKSPAEAKSPA 295
Query: 146 TSEGRRKVRSKSGHKSPHRSKSSTN-SSPSKESACQETQGATSPIREAEA-------LPG 197
++ + +S + KSP +KS SP++ + E + AE PG
Sbjct: 296 EAKSPAEAKSPAEVKSPAVAKSPAEVKSPAEVKSPAEAKSPAEAKSPAEVKSPATVKSPG 355
Query: 198 KDDNPMPPPKSTATVQPSPQAK 219
+ +P KS A V+ +AK
Sbjct: 356 EAKSP-AEAKSPAEVKSPAEAK 376
Score = 32.3 bits (72), Expect = 2.7
Identities = 35/189 (18%), Positives = 65/189 (33%), Gaps = 22/189 (11%)
Query: 48 KLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEH 107
K ++ +PA S ++ K K + + KE +++ V E
Sbjct: 590 KPPAKEEAKRPADIRSPEQVKSPAKEEAKSPEKEETRTEKVAPKKE-----EVKSPVEEV 644
Query: 108 SISNPPSR---ENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHR 164
PP + E +PA + K ++ PK A K K + G
Sbjct: 645 KAKEPPKKVEEEKTPATPKTEVKESKKDEAPKEAQKPKAEEKEPLTEKPKDSPGEAKKEE 704
Query: 165 SKSSTNSSPSKESACQ-------------ETQGATSPIREAE-ALPGKDDNPMPPPKSTA 210
+K ++P +E+ + E A P + +E P K++ P P K
Sbjct: 705 AKEKKAAAPEEETPAKLGVKEEAKPKEKAEDAKAKEPSKPSEKEKPKKEEVPAAPEKKDT 764
Query: 211 TVQPSPQAK 219
+ + ++K
Sbjct: 765 KEEKTTESK 773
>NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130
kDa protein) (140 kDa nucleolar phosphoprotein)
(Nopp140) (Nucleolar and coiled-body phosphoprotein 1)
Length = 699
Score = 46.6 bits (109), Expect = 1e-04
Identities = 55/254 (21%), Positives = 96/254 (37%), Gaps = 45/254 (17%)
Query: 11 QPSPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRK-------------PK 57
QP + + P A ++K P + D D D+ S + K +K K PK
Sbjct: 144 QPVQKGVKPQAKAAKAPPKKAKSSDSDS--DSSSEDEPPKNQKPKITPVTVKAQTKAPPK 201
Query: 58 PAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSREN 117
PA +A + A S++S +S +SD D E + P +
Sbjct: 202 PARAAPKIANGKAASSSSSSSSSSSSD------------------DSEEEKAAATPKKTV 243
Query: 118 SPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHR---------SKSS 168
AK+ + P + +S ++ +S+ E + + P+ K S
Sbjct: 244 PKKQVVAKAPVKAATTPTRKSSSSEDSSSDEEEEQKKPMKNKPGPYSYAPPPSAPPPKKS 303
Query: 169 TNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPK---STATVQPSPQAKGGFSSN 225
+ P K++ ++ +S E+ ++ PP K S AT +P P K SS+
Sbjct: 304 LGTQPPKKAVEKQQPVESSEDSSDESDSSSEEEKKPPTKAVVSKATTKPPPAKKAAESSS 363
Query: 226 VSSEKLDTLFEEDP 239
SS+ + +E P
Sbjct: 364 DSSDSDSSEDDEAP 377
Score = 42.0 bits (97), Expect = 0.003
Identities = 67/318 (21%), Positives = 120/318 (37%), Gaps = 48/318 (15%)
Query: 10 TQPSPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKG 69
TQP + + K +PV +D D+ D + ++KP A S+ K
Sbjct: 306 TQPPKKAVE------KQQPVESSEDSSDESDSSSE-------EEKKPPTKAVVSKATTKP 352
Query: 70 AGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAE 129
+ +S +SD+ + E D + A ++S + P SPA PA + +
Sbjct: 353 PPAKKAAESSSDSSDS----DSSEDDEAPSKPAGTTKNSSNKPAVTTKSPAVKPAAAPKQ 408
Query: 130 GVEKPPK-------AASLAKKTSTSEGRR----------KVRSKSGHKSPHRS--KSSTN 170
V K ++S +++S+SE + K +K+ P + + S +
Sbjct: 409 PVGGGQKLLTRKADSSSSEEESSSSEEEKTKKMVATTKPKATAKAALSLPAKQAPQGSRD 468
Query: 171 SSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEK 230
SS +S+ E + + + P K P K + + ++ SS+ SSE+
Sbjct: 469 SSSDSDSSSSEEEEEKTSKSAVKKKPQKVAGGAAPSKPASAKKGKAESSNSSSSDDSSEE 528
Query: 231 LDTLFEEDPLAALDGFLDGTLDWDSPPHQADATAETAQSSGVA-NLQQIEESMGRLKAIV 289
EE+ L P +A TAQ+ A N ++ EE + +V
Sbjct: 529 -----EEEKLKGKGS------PRPQAPKANGTSALTAQNGKAAKNSEEEEEEKKKAAVVV 577
Query: 290 FDIGFLDRIRVDPQASHA 307
G L + + + A A
Sbjct: 578 SKSGSLKKRKQNEAAKEA 595
Score = 32.0 bits (71), Expect = 3.5
Identities = 37/166 (22%), Positives = 55/166 (32%), Gaps = 28/166 (16%)
Query: 10 TQPSPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKG 69
T P QV++ + P +D D ++ K K KP P + A A
Sbjct: 242 TVPKKQVVAKAPVKAATTPTRKSSSSEDSSSDEEE--EQKKPMKNKPGPYSYAPPPSAPP 299
Query: 70 AGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAE 129
S + K A + Q V E D + S +E
Sbjct: 300 PKKSLGTQPPKKAVEKQQPVESSEDSSDES-------------------------DSSSE 334
Query: 130 GVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSK 175
+KPP A ++K T+ +K S S S S + +PSK
Sbjct: 335 EEKKPPTKAVVSKATTKPPPAKKAAESSSDSSDSDS-SEDDEAPSK 379
>YG3A_YEAST (P53278) Hypothetical 92.7 kDa protein in ASN2-PHB1
intergenic region
Length = 816
Score = 46.2 bits (108), Expect = 2e-04
Identities = 104/519 (20%), Positives = 192/519 (36%), Gaps = 112/519 (21%)
Query: 34 DDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASD--------- 84
D+ DD D S + ++S+S +TTS A++ +D
Sbjct: 154 DEKDDTDTTTSSSTSTSSSSSSSSSSSSSSSSDEGDVTSTTTSEATEATADTATTTTTTT 213
Query: 85 -----AQPEVNGKEGDGD-ATIQADVHEHSISNPPSRENSPAPNPAKSKAE------GVE 132
+ N E D AT + HE +S S A + + AE GV
Sbjct: 214 STSTTSTSTTNAVENSADEATSVEEEHEDKVSESTSIGKGTADSAQINVAEPISSENGVL 273
Query: 133 KP----PKAASLAKKTSTSEGR------RKVRSKSGHKSPHRSKSSTNSSPSKESACQET 182
+P S + T E + +KV SG + +++S+ +E+
Sbjct: 274 EPRTTDQSGGSKSGVVPTDEQKEEKSDVKKVNPPSGEEKKEVEAEGDAEEETEQSSAEES 333
Query: 183 QGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAK---GGFS-----SNVSSEKLDTL 234
TS +E +D++P+ P K+ P K G F+ ++ S++K T
Sbjct: 334 AERTSTPETSEPESEEDESPIDPSKAPKVPFQEPSRKERTGIFALWKSPTSSSTQKSKTA 393
Query: 235 FEEDPLAALD-----------GFLDGT----LDWDSPPHQA---DATAETAQSSGVANLQ 276
+P+A + G+L +++D HQA D A+ N +
Sbjct: 394 APSNPVATPENPELIVKTKEHGYLSKAVYDKINYDEKIHQAWLADLRAKEKDKYDAKNKE 453
Query: 277 ----------QIEESMGRLKA--------IVFDIGFLDRIRVDPQASHAAKELLIFLLGQ 318
QI+E +KA I L + +D A H K+L+I
Sbjct: 454 YKEKLQDLQNQIDEIENSMKAMREETSEKIEVSKNRLVKKIIDVNAEHNNKKLMI----- 508
Query: 319 KLDSHQAAALANLQSFLVDALATFHQVKDVGQAVSLKEAKVSSGKAQVAAMKEDFKKFTA 378
L + ++ L ++V D V + +++ K V +++F +T
Sbjct: 509 ---------LKDTENMKNQKLQEKNEVLDKQTNVKSEIDDLNNEKTNV---QKEFNDWTT 556
Query: 379 RKVLIADE-------ISDVDTKLEELHREIAKLEKKRADLVEEGPAVKAQLDGLTKESKA 431
++ + I+ ++ K ++ EI LEKK+ DLV + T+E+K
Sbjct: 557 NLSNLSQQLDAQIFKINQINLKQGKVQNEIDNLEKKKEDLVTQ-----------TEENKK 605
Query: 432 LIISRKDVIKELEVDQAVKK--DLDAKIATFADRLNSFK 468
L V++ +E + + + D+D +I++ + + K
Sbjct: 606 LHEKNVQVLESVENKEYLPQINDIDNQISSLLNEVTIIK 644
>TUL1_MOUSE (Q9Z273) Tubby related protein 1 (Tubby-like protein 1)
Length = 543
Score = 45.8 bits (107), Expect = 2e-04
Identities = 45/184 (24%), Positives = 73/184 (39%), Gaps = 31/184 (16%)
Query: 32 EDDDDDDEDDNVSLADKLKGRKRK----PKPAASASQKRAKGAGVSTTSGASKLASDAQP 87
ED DDDD DD+ +K +G+K K PK A +K+AK G G+ + +P
Sbjct: 117 EDSDDDDNDDDEEEEEKKEGKKEKSSLPPKKAPKEREKKAKALGPRGDVGSPD--APRKP 174
Query: 88 EVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAE----------------GV 131
K+ G+ T + + P + PA +PA + E GV
Sbjct: 175 LRTKKKEVGEGT---KLRKAKKKGPGETDKDPAGSPAALRKEFPAAMFLVGEGGAAEKGV 231
Query: 132 EK--PPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPI 189
+K PPK + KK E +V S S + ++ + +E + ++ P+
Sbjct: 232 KKKGPPKGSEEEKKEEEEEVEEEVASAVMKNSNQKGRAKGKG----KKKVKEERASSPPV 287
Query: 190 REAE 193
E
Sbjct: 288 EVGE 291
>TCOF_MOUSE (O08784) Treacle protein (Treacher Collins syndrome
protein homolog)
Length = 1320
Score = 45.8 bits (107), Expect = 2e-04
Identities = 65/253 (25%), Positives = 97/253 (37%), Gaps = 30/253 (11%)
Query: 4 GQAEVITQPSPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPK------ 57
G A V TQ + + +A +D + ED ++ED+ + A L GR + K
Sbjct: 641 GVAPVHTQKTGPSVKAMAQ--EDSESLEEDSSSEEEDETPAQATPL-GRLPQAKANPPPT 697
Query: 58 --PAASASQKRA----KGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQ----ADVHEH 107
P ASAS K KG S S + P GK G Q A E
Sbjct: 698 KTPPASASGKAVAAPTKGKPPVPNSTVSARGQRSVPAA-GKAGAPATQAQKGPVAGTGED 756
Query: 108 SISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRR-----KVRSKSGHKSP 162
S S+ +S PA+ K G +AAS K S +G K S + P
Sbjct: 757 SESSSKEESDSEEETPAQIKPVGKTSQVRAASAPAKESPKKGAHPGTPGKTGSSATQAQP 816
Query: 163 HRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDN---PMPPPKSTATVQPSPQAK 219
+++ S +S S+ES + I+ ++ + P P P+ V + +A
Sbjct: 817 GKTEDSDSS--SEESDSDTEMPSAQAIKSPPVSVNRNSSPAVPAPTPEGVQAVNTTKKAS 874
Query: 220 GGFSSNVSSEKLD 232
G + + SSE D
Sbjct: 875 GTTAQSSSSESED 887
Score = 40.4 bits (93), Expect = 0.010
Identities = 58/263 (22%), Positives = 89/263 (33%), Gaps = 29/263 (11%)
Query: 6 AEVITQPSPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPA-ASASQ 64
+E + + Q + P A S V ED ++S + K KPA A AS
Sbjct: 169 SETEEEGNAQALGPTAKSGT---VSAGQGSSSSEDSSISSDETDVEVKSPAKPAQAKASA 225
Query: 65 KRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQA---------DVHEHSISNPPSR 115
AK T G +KL + A A A + E S + S
Sbjct: 226 APAKDPPARTAPGPTKLGNVAPTPAKPARAAAAAAAAAVAAAAAAAAEESESSEEDSDSE 285
Query: 116 ENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEG----------------RRKVRSKSGH 159
+ +PA P++ KA G +A S++ K + +G + + K
Sbjct: 286 DEAPAGLPSQVKASGKGPHVRADSVSAKGISGKGPILATPGKTGPAATQAKAERPEKDSE 345
Query: 160 KSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAK 219
S S + P + Q SP + P K+ + P T A+
Sbjct: 346 TSSEDDSDSEDEMPVTVNTPQARTSGKSPRARGTSAPAKESSQKGAPAVTPGKARPVAAQ 405
Query: 220 GGFSSNVSSEKLDTLFEEDPLAA 242
G SSE+ ++ E P AA
Sbjct: 406 AGKPEAKSSEESESDSGETPAAA 428
Score = 36.6 bits (83), Expect = 0.14
Identities = 39/167 (23%), Positives = 64/167 (37%), Gaps = 14/167 (8%)
Query: 12 PSPQVISP----VAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRA 67
P Q ++P SS+D D +DE+D + P P SQK
Sbjct: 974 PEKQQLAPGYPKAPRSSEDS----SDTSSEDEEDAKRPQMPKSAHRLDPDP----SQKET 1025
Query: 68 KGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSK 127
T S ++ + +Q ++G G T+ + P S + + K
Sbjct: 1026 VVEETPTESSEDEMVAPSQSLLSGYMTPG-LTVANSQASKATPRPDSNSLASSAPATKDN 1084
Query: 128 AEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPS 174
+G +K K+ A GR++ S S + P + K ST+SSP+
Sbjct: 1085 PDGKQKS-KSQHAADTALPKTGRKEASSGSTPQKPKKLKKSTSSSPA 1130
Score = 34.3 bits (77), Expect = 0.71
Identities = 47/199 (23%), Positives = 75/199 (37%), Gaps = 32/199 (16%)
Query: 57 KPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRE 116
KPA +A+ A A V+ + A+ S++ E + E + A + + V + S P R
Sbjct: 251 KPARAAAA--AAAAAVAAAAAAAAEESESSEEDSDSEDEAPAGLPSQV-KASGKGPHVRA 307
Query: 117 NSPA------------------PNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSG 158
+S + P ++KAE EK + +S S E V +
Sbjct: 308 DSVSAKGISGKGPILATPGKTGPAATQAKAERPEKDSETSSEDDSDSEDEMPVTVNTPQA 367
Query: 159 HKSPHRSKSSTNSSPSKES----ACQETQGATSPIREAEALP-------GKDDNPMPPPK 207
S ++ S+P+KES A T G P+ P + D+ P
Sbjct: 368 RTSGKSPRARGTSAPAKESSQKGAPAVTPGKARPVAAQAGKPEAKSSEESESDSGETPAA 427
Query: 208 STATVQPSPQAKGGFSSNV 226
+T T P+ G SS V
Sbjct: 428 ATLTTSPAKVKPLGKSSQV 446
Score = 31.2 bits (69), Expect = 6.0
Identities = 45/190 (23%), Positives = 80/190 (41%), Gaps = 17/190 (8%)
Query: 13 SPQVISPVAHSSKDKPVVIEDDDDDDEDDNV-------SLADKLKGRKRKPKPAASASQK 65
SP + P+ SS+ +PV + N+ S A +++ K++ +S+++
Sbjct: 433 SPAKVKPLGKSSQVRPVSTVTPGSSGKGANLPCPGKVGSAALRVQMVKKEDVSESSSAEL 492
Query: 66 RAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAK 125
+ G G S A AS A P+ T + H S E AP +
Sbjct: 493 DSDGPG----SPAKAKASLALPQKVRPVATQVKTDRGKGHSGSSEESSDSEEEAAPAASA 548
Query: 126 SKAE-GVEKPPKAASLAKKTSTSEGRRKVRSKSGH-KSPHRSKSSTNSSPSKESACQETQ 183
++A+ +EK KA+S +++ G S S H K+ + S++ SSP+ Q +
Sbjct: 549 AQAKPALEKQMKASSRKGTPASATGA----STSSHCKAGAVTSSASLSSPALAKGTQRSD 604
Query: 184 GATSPIREAE 193
+S E+E
Sbjct: 605 VDSSSESESE 614
>SFR4_HUMAN (Q08170) Splicing factor, arginine/serine-rich 4
(Pre-mRNA splicing factor SRP75) (SRP001LB)
Length = 494
Score = 45.8 bits (107), Expect = 2e-04
Identities = 37/141 (26%), Positives = 61/141 (43%), Gaps = 16/141 (11%)
Query: 47 DKLKGRKRKPKPAASASQKRAKGA-----GVSTTSGASKLASDAQPEVNGKEGDGDATIQ 101
DK KGRKR + + S S+ R+K G S A + + + + +
Sbjct: 352 DKRKGRKRSREESRSRSRSRSKSERSRKRGSKRDSKAGSSKKKKKEDTDRSQSRSPSRSV 411
Query: 102 ADVHEHSISNPPSRE------NSPAPNPAKSKAEGVEK-----PPKAASLAKKTSTSEGR 150
+ EH+ S RE N+ +S++ K P ++ S +K S + R
Sbjct: 412 SKEREHAKSESSQREGRGESENAGTNQETRSRSRSNSKSKPNLPSESRSRSKSASKTRSR 471
Query: 151 RKVRSKSGHKSPHRSKSSTNS 171
K RS+S +SP RS+S ++S
Sbjct: 472 SKSRSRSASRSPSRSRSRSHS 492
Score = 41.2 bits (95), Expect = 0.006
Identities = 46/191 (24%), Positives = 72/191 (37%), Gaps = 21/191 (10%)
Query: 14 PQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVS 73
P+ SP H SK K + +E+ S++ +GR ++ S S+ R+KG S
Sbjct: 286 PKSRSPSRHKSKSKSRSRSQERRVEEEKRGSVS---RGRSQEKSLRQSRSRSRSKGGSRS 342
Query: 74 TTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEK 133
+ SK ++ + G++ E S S SR S +S+ G ++
Sbjct: 343 RSRSRSK----SKDKRKGRKRS---------REESRSRSRSRSKS-----ERSRKRGSKR 384
Query: 134 PPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAE 193
KA S KK R + RS S S R + + SS + E G R
Sbjct: 385 DSKAGSSKKKKKEDTDRSQSRSPSRSVSKEREHAKSESSQREGRGESENAGTNQETRSRS 444
Query: 194 ALPGKDDNPMP 204
K +P
Sbjct: 445 RSNSKSKPNLP 455
Score = 36.6 bits (83), Expect = 0.14
Identities = 31/102 (30%), Positives = 49/102 (47%), Gaps = 7/102 (6%)
Query: 88 EVNGKE----GDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKK 143
EVNG++ D + + + S S+ SR S ++S++ G K + S ++
Sbjct: 164 EVNGRKIRLVEDKPGSRRRRSYSRSRSHSRSRSRSRHSRKSRSRS-GSSKSSHSKSRSRS 222
Query: 144 TSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGA 185
S S R K RS+S +S RSK + SPSKE + + A
Sbjct: 223 RSGSRSRSKSRSRSQSRS--RSKKEKSRSPSKEKSRSRSHSA 262
Score = 33.9 bits (76), Expect = 0.93
Identities = 31/139 (22%), Positives = 57/139 (40%), Gaps = 15/139 (10%)
Query: 50 KGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSI 109
K R R +S S+ R++ S + S+ S ++ ++ S
Sbjct: 203 KSRSRSGSSKSSHSKSRSRSRSGSRSRSKSRSRSQSRSRSKKEKS----------RSPSK 252
Query: 110 SNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHR----- 164
SR +S + +KSK + EK ++ K S S R K +SKS +S R
Sbjct: 253 EKSRSRSHSAGKSRSKSKDQAEEKIQNNDNVGKPKSRSPSRHKSKSKSRSRSQERRVEEE 312
Query: 165 SKSSTNSSPSKESACQETQ 183
+ S + S+E + ++++
Sbjct: 313 KRGSVSRGRSQEKSLRQSR 331
Score = 33.1 bits (74), Expect = 1.6
Identities = 36/157 (22%), Positives = 65/157 (40%), Gaps = 24/157 (15%)
Query: 50 KGRKRKPK---PAASASQKRAKGAGVSTTSGASKLASDAQPEVN-------------GKE 93
+ R +K K P+ S+ R+ AG S + + Q N K
Sbjct: 239 RSRSKKEKSRSPSKEKSRSRSHSAGKSRSKSKDQAEEKIQNNDNVGKPKSRSPSRHKSKS 298
Query: 94 GDGDATIQADVHEH---SISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGR 150
+ + V E S+S S+E S + ++S+++G + S ++ S S+ +
Sbjct: 299 KSRSRSQERRVEEEKRGSVSRGRSQEKSLRQSRSRSRSKGGSR-----SRSRSRSKSKDK 353
Query: 151 RKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATS 187
RK R +S +S RS+S + S S++ + A S
Sbjct: 354 RKGRKRSREESRSRSRSRSKSERSRKRGSKRDSKAGS 390
>NFH_MOUSE (P19246) Neurofilament triplet H protein (200 kDa
neurofilament protein) (Neurofilament heavy polypeptide)
(NF-H)
Length = 1087
Score = 45.8 bits (107), Expect = 2e-04
Identities = 44/184 (23%), Positives = 68/184 (36%), Gaps = 4/184 (2%)
Query: 48 KLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEH 107
++K PAA S AK + G +K +A+ K +A +V
Sbjct: 709 EVKSPGEAKSPAAVKSPAEAKSPAAVKSPGEAKSPGEAKSPAEAKS-PAEAKSPIEVKSP 767
Query: 108 SISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKS 167
+ P +E + +P AKS E + P K + S + K K G K P ++K
Sbjct: 768 EKAKTPVKEGAKSPAEAKSP-EKAKSPVKEDIKPPAEAKSPEKAKSPVKEGAKPPEKAKP 826
Query: 168 STNSSPSKESACQETQGATSPIREAEAL--PGKDDNPMPPPKSTATVQPSPQAKGGFSSN 225
SP ++ QE + IR E + P K+ P + T + K S
Sbjct: 827 LDVKSPEAQTPVQEEATVPTDIRPPEQVKSPAKEKAKSPEKEEAKTSEKVAPKKEEVKSP 886
Query: 226 VSSE 229
V E
Sbjct: 887 VKEE 890
Score = 45.1 bits (105), Expect = 4e-04
Identities = 49/205 (23%), Positives = 82/205 (39%), Gaps = 17/205 (8%)
Query: 28 PVVIEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQP 87
P ++ + + K PA + S K G + + K ++A+
Sbjct: 635 PATVKSPGEAKSPSEAKSPAEAKSPAEAKSPAEAKSPAEVKSPGEAKSPAEPKSPAEAKS 694
Query: 88 EVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTS 147
K +A A+V + P+ SPA AKS A V+ P +A S + S +
Sbjct: 695 PAEVKS-PAEAKSPAEVKSPGEAKSPAAVKSPAE--AKSPA-AVKSPGEAKSPGEAKSPA 750
Query: 148 EGRRKVRSKSGH--KSPHRSKSSTNSSPSKESACQETQGATSPIRE-----AEA-LPGKD 199
E + +KS KSP ++K+ + + + A SP++E AEA P K
Sbjct: 751 EAKSPAEAKSPIEVKSPEKAKTPVKEGAKSPAEAKSPEKAKSPVKEDIKPPAEAKSPEKA 810
Query: 200 DNPM-----PPPKSTATVQPSPQAK 219
+P+ PP K+ SP+A+
Sbjct: 811 KSPVKEGAKPPEKAKPLDVKSPEAQ 835
Score = 39.7 bits (91), Expect = 0.017
Identities = 43/190 (22%), Positives = 80/190 (41%), Gaps = 11/190 (5%)
Query: 32 EDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNG 91
E ++ ++E+ + + + K S ++ AK G + + G +K ++A+
Sbjct: 483 EGEEKEEEELAAATSPPAEEAASPEKETKSRVKEEAKSPGEAKSPGEAKSPAEAKSPGEA 542
Query: 92 KEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRR 151
K G+A + + P+ SPA AKS AE PK+ + K ++
Sbjct: 543 KS-PGEAKSPGEAKSPAEPKSPAEPKSPAE--AKSPAE-----PKSPATVKSPGEAKSPS 594
Query: 152 KVRSKSGHKSPHRSKSSTNS-SPSKESACQETQGATSPIREAEA-LPGKDDNPMPPPKST 209
+ +S + KSP +KS + SP++ + E + A PG+ +P KS
Sbjct: 595 EAKSPAEAKSPAEAKSPAEAKSPAEAKSPAEAKSPAEAKSPATVKSPGEAKSP-SEAKSP 653
Query: 210 ATVQPSPQAK 219
A + +AK
Sbjct: 654 AEAKSPAEAK 663
Score = 38.1 bits (87), Expect = 0.049
Identities = 37/162 (22%), Positives = 63/162 (38%), Gaps = 12/162 (7%)
Query: 81 LASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASL 140
+ + E G+EG+ +A + E ++ S A +P K V++ K+
Sbjct: 465 VTEEEDKEAQGQEGE-EAEEGEEKEEEELAAATSPPAEEAASPEKETKSRVKEEAKSPGE 523
Query: 141 AKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDD 200
AK ++ + +S KSP +K SP + + E + P AEA +
Sbjct: 524 AKSPGEAKSPAEAKSPGEAKSPGEAK-----SPGEAKSPAEPKSPAEPKSPAEAKSPAE- 577
Query: 201 NPMPPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFEEDPLAA 242
PKS ATV+ +AK + +E + P A
Sbjct: 578 -----PKSPATVKSPGEAKSPSEAKSPAEAKSPAEAKSPAEA 614
Score = 36.6 bits (83), Expect = 0.14
Identities = 40/175 (22%), Positives = 73/175 (40%), Gaps = 13/175 (7%)
Query: 58 PAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSREN 117
PA + S ++AK S +K A+P ++ K + +Q + + PP +
Sbjct: 801 PAEAKSPEKAK----SPVKEGAKPPEKAKP-LDVKSPEAQTPVQEEATVPTDIRPPEQVK 855
Query: 118 SPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKES 177
SPA K KA+ EK S E + V+ + K P + + P+ ++
Sbjct: 856 SPA----KEKAKSPEKEEAKTSEKVAPKKEEVKSPVKEEVKAKEPPKKVEEEKTLPTPKT 911
Query: 178 ACQETQGATSPIREA--EALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEK 230
+E++ +P +EA + K + P PK +T + + G V+SE+
Sbjct: 912 EAKESKKDEAP-KEAPKPKVEEKKETPTEKPKD-STAEAKKEEAGEKKKAVASEE 964
>DD21_MOUSE (Q9JIK5) Nucleolar RNA helicase II (Nucleolar RNA
helicase Gu) (RH II/Gu) (DEAD-box protein 21)
Length = 851
Score = 45.8 bits (107), Expect = 2e-04
Identities = 41/173 (23%), Positives = 72/173 (40%), Gaps = 33/173 (19%)
Query: 35 DDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEV--NGK 92
+++ +DD S + K K+K +P + K + T + + A +P+ GK
Sbjct: 80 EEEPQDDTASTS---KTSKKKKEPLEKQADSETKE--IITEEPSEEEADMPKPKKMKKGK 134
Query: 93 EGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKA----------------EGVEKP-- 134
E +GDA ++ ++ +S P S E + P P K K G+ +P
Sbjct: 135 EANGDAGEKSPKLKNGLSQP-SEEEADIPKPKKMKKGKEANGDAGEKSPKLKNGLSQPSE 193
Query: 135 -------PKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQ 180
PK K+ S G + R K G P KS+++ +P +ES+ +
Sbjct: 194 EEVDIPKPKKMKKGKEASGDAGEKSPRLKDGLSQPSEPKSNSSDAPGEESSSE 246
>YLB8_CAEEL (P46582) Hypothetical protein C34E10.8 in chromosome III
Length = 1024
Score = 45.4 bits (106), Expect = 3e-04
Identities = 57/231 (24%), Positives = 95/231 (40%), Gaps = 27/231 (11%)
Query: 50 KGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSI 109
KG ++ P A S K A GA T++ AS + PE K + Q+DV S
Sbjct: 779 KGGRKAGAPGAKESSKAAGGAQKGTSA-----ASTSVPEPT-KSSESSVDPQSDV---SF 829
Query: 110 SNPPSRENSPAPNPAKSKAEGVEKPPKAASL--AKKTSTSEGRRKVRSKSGHKSPHRSKS 167
SNP SPAP E +EK AA + K E + S H + S
Sbjct: 830 SNP-----SPAP-------EVIEKVAPAAMTITSNKRKIDETLASESTSSEATLIHDTTS 877
Query: 168 STNSSP-SKESACQETQGATSPIREAEALPGKDDNPMPPPKSTATVQPSPQAKGGFSSNV 226
S+++ S E +++ ++P+ E K D P P ST P+P + ++ +
Sbjct: 878 SSSAETVSGEPPAKKSSDVSAPVPSPEKEKEKIDRPKTPKSSTKRTTPTPSGRTPRAAAI 937
Query: 227 SSEKLDTLFEEDPLAALDGFLDGTLDWDSPPHQADATAETAQSSGVANLQQ 277
++ + + + + +A + D ++P D AE + ++ QQ
Sbjct: 938 AANQAISHSKPNVPSASTSSSAASTDQENP---LDLLAELSVAAAAEEQQQ 985
>TLE4_HUMAN (Q04727) Transducin-like enhancer protein 4
Length = 766
Score = 45.4 bits (106), Expect = 3e-04
Identities = 46/199 (23%), Positives = 80/199 (40%), Gaps = 24/199 (12%)
Query: 31 IEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVN 90
I+D+ ++D+ D +K P + ++K A S+ S K + E+
Sbjct: 177 IKDEKKHHDNDHQRDRDSIKSSSVSPSASFRGAEKHRNSADYSSESKKQKTE---EKEIA 233
Query: 91 GK-EGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEG 149
+ + DG+ + V + S +P S SPA +P ++ + K A ++ + S
Sbjct: 234 ARYDSDGEKSDDNLVVDVSNEDPSSPRGSPAHSPRENGLDKTRLLKKDAPISPASIASS- 292
Query: 150 RRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKST 209
SST SS SKE + E +T+P+ ++ + D P P ST
Sbjct: 293 -----------------SSTPSSKSKELSLNEK--STTPVSKSNTPTPRTDAPTPGSNST 333
Query: 210 ATVQPSPQAKGGFSSNVSS 228
++P P G SS
Sbjct: 334 PGLRPVPGKPPGVDPLASS 352
>TLE4_RAT (Q07141) Transducin-like enhancer protein 4 (ESP2 protein)
Length = 741
Score = 45.1 bits (105), Expect = 4e-04
Identities = 46/199 (23%), Positives = 79/199 (39%), Gaps = 24/199 (12%)
Query: 31 IEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVN 90
I+D+ ++D+ D +K P + S+K S+ S K + E+
Sbjct: 152 IKDEKKHHDNDHQRDRDSIKSSSVSPSASFRGSEKHRNSTDYSSESKKQKTE---EKEIA 208
Query: 91 GK-EGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEG 149
+ + DG+ + V + S +P S SPA +P ++ + K A ++ + S
Sbjct: 209 ARYDSDGEKSDDNLVVDVSNEDPSSPRGSPAHSPRENGLDKTRLLKKDAPISPASVASS- 267
Query: 150 RRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKST 209
SST SS SKE + E +T+P+ ++ + D P P ST
Sbjct: 268 -----------------SSTPSSKSKELSLNEK--STTPVSKSNTPTPRTDAPTPGSNST 308
Query: 210 ATVQPSPQAKGGFSSNVSS 228
++P P G SS
Sbjct: 309 PGLRPVPGKPPGVDPLASS 327
>TLE4_MOUSE (Q62441) Transducin-like enhancer protein 4
(Groucho-related protein 4) (Grg-4)
Length = 766
Score = 45.1 bits (105), Expect = 4e-04
Identities = 46/199 (23%), Positives = 79/199 (39%), Gaps = 24/199 (12%)
Query: 31 IEDDDDDDEDDNVSLADKLKGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVN 90
I+D+ ++D+ D +K P + S+K S+ S K + E+
Sbjct: 177 IKDEKKHHDNDHQRDRDSIKSSSVSPSASFRGSEKHRNSTDYSSESKKQKTE---EKEIA 233
Query: 91 GK-EGDGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEG 149
+ + DG+ + V + S +P S SPA +P ++ + K A ++ + S
Sbjct: 234 ARYDSDGEKSDDNLVVDVSNEDPSSPRGSPAHSPRENGLDKTRLLKKDAPISPASVASS- 292
Query: 150 RRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKDDNPMPPPKST 209
SST SS SKE + E +T+P+ ++ + D P P ST
Sbjct: 293 -----------------SSTPSSKSKELSLNEK--STTPVSKSNTPTPRTDAPTPGSNST 333
Query: 210 ATVQPSPQAKGGFSSNVSS 228
++P P G SS
Sbjct: 334 PGLRPVPGKPPGVDPLASS 352
>TCOF_HUMAN (Q13428) Treacle protein (Treacher Collins syndrome
protein)
Length = 1411
Score = 45.1 bits (105), Expect = 4e-04
Identities = 65/263 (24%), Positives = 96/263 (35%), Gaps = 26/263 (9%)
Query: 2 RIGQAEVITQPSPQVISPVAHSSKDKPVVIEDDDDDDEDDNVSLADKLKGRKRKP---KP 58
++G A + SP+ + A K P V + E+D+ S +++ + P KP
Sbjct: 293 QVGAASAPAKESPRKGAAPAPPGKTGPAVAKAQAGKREEDSQSSSEESDSEEEAPAQAKP 352
Query: 59 AASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQA-----DVHEHSISNPP 113
+ A Q RA A K A+ A P K G A +Q D S +
Sbjct: 353 SGKAPQVRA--ASAPAKESPRKGAAPAPPR---KTGPAAAQVQVGKQEEDSRSSSEESDS 407
Query: 114 SRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKV-RSKSGHKSPH------RSK 166
RE A N A+ K G K AS +G V K G +P
Sbjct: 408 DREALAAMNAAQVKPLGKSPQVKPASTMGMGPLGKGAGPVPPGKVGPATPSAQVGKWEED 467
Query: 167 SSTNSSPSKESACQETQGATSPIRE------AEALPGKDDNPMPPPKSTATVQPSPQAKG 220
S ++S S +S+ E A +P +E +A P PP K+ K
Sbjct: 468 SESSSEESSDSSDGEVPTAVAPAQEKSLGNILQAKPTSSPAKGPPQKAGPVAVQVKAEKP 527
Query: 221 GFSSNVSSEKLDTLFEEDPLAAL 243
+S S E D+ E+ AA+
Sbjct: 528 MDNSESSEESSDSADSEEAPAAM 550
Score = 42.4 bits (98), Expect = 0.003
Identities = 50/216 (23%), Positives = 78/216 (35%), Gaps = 24/216 (11%)
Query: 39 EDDNVSLADKLKGRKRKPKPAASASQKRAKG----AGVSTTSGASKLASDAQPEVNGKEG 94
E+D+ S +++ + + A + Q +A G G ++ A P GK G
Sbjct: 259 EEDSESSSEESSDSEEETPAAKALLQAKASGKTSQVGAASAPAKESPRKGAAPAPPGKTG 318
Query: 95 DGDATIQADVHEHSISNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGR---- 150
A QA E + +S PA++K G +AAS K S +G
Sbjct: 319 PAVAKAQAGKREEDSQSSSEESDSEEEAPAQAKPSGKAPQVRAASAPAKESPRKGAAPAP 378
Query: 151 -RKVRSKSGH----KSPHRSKSSTNSSPSKESACQ-------ETQGATSPIREAEALP-- 196
RK + K S+SS+ S S A + G + ++ A +
Sbjct: 379 PRKTGPAAAQVQVGKQEEDSRSSSEESDSDREALAAMNAAQVKPLGKSPQVKPASTMGMG 438
Query: 197 --GKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEK 230
GK P+PP K + K S SSE+
Sbjct: 439 PLGKGAGPVPPGKVGPATPSAQVGKWEEDSESSSEE 474
Score = 40.4 bits (93), Expect = 0.010
Identities = 50/220 (22%), Positives = 77/220 (34%), Gaps = 29/220 (13%)
Query: 51 GRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEV----------NGKEGDGDATI 100
GRKRK + A+ K+ + + +TS +S+ +A+ E N D
Sbjct: 60 GRKRKAEEDAALQAKKTRVSDPISTSESSEEEEEAEAETAKATPRLASTNSSVLGADLPS 119
Query: 101 QADVHEHSISNPPSRENSPAPNPAKSKA-----EGVEKPPKAASLAKKTSTSEGRRK--- 152
+ + + + P+PA K G A A T SE +
Sbjct: 120 SMKEKAKAETEKAGKTGNSMPHPATGKTVANLLSGKSPRKSAEPSANTTLVSETEEEGSV 179
Query: 153 ----VRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIRE-------AEALPGKDDN 201
+K G S ++ SS+ + S A+ I + A+ PGK
Sbjct: 180 PAFGAAAKPGMVSAGQADSSSEDTSSSSDETDVEVKASEKILQVRAASAPAKGTPGKGAT 239
Query: 202 PMPPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFEEDPLA 241
P PP K+ A + K S SSE+ EE P A
Sbjct: 240 PAPPGKAGAVASQTKAGKPEEDSESSSEESSDSEEETPAA 279
Score = 37.0 bits (84), Expect = 0.11
Identities = 50/191 (26%), Positives = 76/191 (39%), Gaps = 26/191 (13%)
Query: 58 PAASASQKRAK-GAGVSTTSGASKLASDAQPEVNGKEGDGDATIQADVHEHSISNPPSRE 116
PA A+ K AG + +S +S + +V K + +Q P +
Sbjct: 180 PAFGAAAKPGMVSAGQADSSSEDTSSSSDETDVEVKASE--KILQVRAASAPAKGTPGKG 237
Query: 117 NSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKE 176
+PAP P KA ++A +T G+ + S+S S S S +P+ +
Sbjct: 238 ATPAP------------PGKAGAVASQTKA--GKPEEDSES---SSEESSDSEEETPAAK 280
Query: 177 SACQ-ETQGATSPIREAEA----LPGKDDNPMPPPKSTATVQPSPQAKGGFSSNVSSEKL 231
+ Q + G TS + A A P K P PP K+ V + K S SSE+
Sbjct: 281 ALLQAKASGKTSQVGAASAPAKESPRKGAAPAPPGKTGPAVAKAQAGKREEDSQSSSEES 340
Query: 232 DTLFEEDPLAA 242
D+ EE P A
Sbjct: 341 DS-EEEAPAQA 350
Score = 35.8 bits (81), Expect = 0.24
Identities = 51/220 (23%), Positives = 76/220 (34%), Gaps = 45/220 (20%)
Query: 32 EDDDDDDEDDNVSLADKLKGRKRKPKPA-ASASQKRAKGAGVS----TTSGASKLASDAQ 86
E++ D +E+ K G+ + + A A A + KGA + T A++
Sbjct: 795 EEESDSEEEAETLAQAKPSGKTHQIRAALAPAKESPRKGAAPTPPGKTGPSAAQAGKQDD 854
Query: 87 PEVNGKEGDGDATIQADVHEHSISNPP-------SRENSPAPNPAKSKAEGVEKPPKAAS 139
+ +E D D A V + PP PA PA++ +AAS
Sbjct: 855 SGSSSEESDSDGEAPAAVTSAQVIKPPLIFVDPNRSPAGPAATPAQA---------QAAS 905
Query: 140 LAKKTSTSEGRRKVRSKSGHKSPHRSKSSTNSSPSKESACQETQGATSPIREAEALPGKD 199
+K SE ST S S ES ++ AT + PG
Sbjct: 906 TPRKARASE-------------------STARSSSSESEDEDVIPATQCL-----TPGIR 941
Query: 200 DNPMPPPKSTATVQPSPQAKGGFSSNVSSEKLDTLFEEDP 239
N + P + + P G SS SS D +E P
Sbjct: 942 TNVVTMPTAHPRIAPKASMAGASSSKESSRISDGKKQEGP 981
Score = 34.7 bits (78), Expect = 0.54
Identities = 30/121 (24%), Positives = 44/121 (35%), Gaps = 14/121 (11%)
Query: 50 KGRKRKPKPAASASQKRAKGAGVSTTSGASKLASDAQPEVNGKEGDGDATIQA--DVHEH 107
+ +K+K A + STTS A +V K+G G Q D E
Sbjct: 1283 RSKKKKKLGAGEGGEASVSPEKTSTTSKGKAKRDKASGDVKEKKGKGSLGSQGAKDEPEE 1342
Query: 108 SI------------SNPPSRENSPAPNPAKSKAEGVEKPPKAASLAKKTSTSEGRRKVRS 155
+ SNP S++ + K E EK KA + K S S ++K +
Sbjct: 1343 ELQKGMGTVEGGDQSNPKSKKEKKKSDKRKKDKEKKEKKKKAKKASTKDSESPSQKKKKK 1402
Query: 156 K 156
K
Sbjct: 1403 K 1403
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.307 0.125 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,873,927
Number of Sequences: 164201
Number of extensions: 2305555
Number of successful extensions: 18057
Number of sequences better than 10.0: 929
Number of HSP's better than 10.0 without gapping: 164
Number of HSP's successfully gapped in prelim test: 801
Number of HSP's that attempted gapping in prelim test: 15123
Number of HSP's gapped (non-prelim): 2822
length of query: 471
length of database: 59,974,054
effective HSP length: 114
effective length of query: 357
effective length of database: 41,255,140
effective search space: 14728084980
effective search space used: 14728084980
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0237.4


