

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
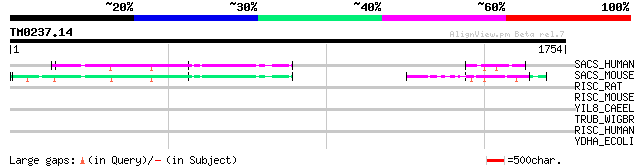
Query= TM0237.14
(1754 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SACS_HUMAN (Q9NZJ4) Sacsin 129 5e-29
SACS_MOUSE (Q9JLC8) Sacsin 128 2e-28
RISC_RAT (Q920A6) Retinoid-inducible serine carboxypeptidase pre... 41 0.026
RISC_MOUSE (Q920A5) Retinoid-inducible serine carboxypeptidase p... 37 0.37
YIL8_CAEEL (O01757) Hypothetical UPF0202 protein F55A12.8 in chr... 34 3.1
TRUB_WIGBR (Q8D2X8) tRNA pseudouridine synthase B (EC 4.2.1.70) ... 34 3.1
RISC_HUMAN (Q9HB40) Retinoid-inducible serine carboxypeptidase p... 33 5.3
YDHA_ECOLI (P28224) Hypothetical protein ydhA 33 7.0
>SACS_HUMAN (Q9NZJ4) Sacsin
Length = 3829
Score = 129 bits (325), Expect = 5e-29
Identities = 182/811 (22%), Positives = 330/811 (40%), Gaps = 94/811 (11%)
Query: 132 INDMVEEEFWSELKLITWCPVILDPAVRGLPWLKSS---KQVALPTVVRPKSQMWLVSSS 188
I D ++F ++ + I + P + PA L W +S + + T + +V
Sbjct: 1448 IRDPRAKDFAAKYQTIRFLPFLTKPAGFSLDWKGNSFKPETMFAATDLYTAEHQDIVCLL 1507
Query: 189 MLILDGE------CGTTYLQTK--LGWMDCPNIGVLSRQLIELSKSYQQLKTHSLLDPDF 240
IL+ CG+ L K LG + P + ++ QL E++KS +D
Sbjct: 1508 QPILNENSHSFRGCGSVSLAVKEFLGLLKKPTVDLVINQLKEVAKS---------VDDGI 1558
Query: 241 DVKLQKEIPCLYSKLQEYINTDDFNDLKA--RLDGVSWVWIGDDFVSPNALAFDSPVKFT 298
+ + Y L E + ++ + +L S++ + + +V ++F +
Sbjct: 1559 TLYQENITNACYKYLHEALMQNEITKMSIIDKLKPFSFILVENAYVDSEKVSFHLNFEAA 1618
Query: 299 PYLYVVSSEL-SEYKDLMIKLGVRLSFGILDYLHVLQRLQNDLNGVPLSTDQLNFVRC-- 355
PYLY + ++ + +++L +GVR S + D+ VL+ + + G T++ NF C
Sbjct: 1619 PYLYQLPNKYKNNFRELFETVGVRQSCTVEDFALVLESIDQE-RGTKQITEE-NFQLCRR 1676
Query: 356 -VHEAIAECCLEKP--LFEPFDSPLWIPDTFGVLMHAGDLVYNDAPWLENSSLIGRHFVH 412
+ E I EK E + +PDT +L+ A L YND PW++ + + H
Sbjct: 1677 IISEGIWSLIREKKQEFCEKNYGKILLPDTNLMLLPAKSLCYNDCPWIKVKDTTVK-YCH 1735
Query: 413 PSISNDLAERLGVQSVRCLSLVSEDMTKDLPCMGY---------NKVSELLALYGNSEFL 463
I ++A +LG R +L +G +++ +L Y + + +
Sbjct: 1736 ADIPREVAVKLGAVPKRHKALERYASNVCFTTLGTEFGQKEKLTSRIKSILNAYPSEKEM 1795
Query: 464 LFDLLELADCCKAKKLHLIYDKREHPRQSLLQHNLGEFQGPALVAIFEGACLSREEFSNF 523
L +LL+ AD KA ++ ++D R+HP + QGPAL ++ + ++
Sbjct: 1796 LKELLQNADDAKATEICFVFDPRQHPVDRIFDDKWAPLQGPAL-CVYNNQPFTEDDVRGI 1854
Query: 524 QLLPPWKLRGN---TLNYGLGLVGCYSICDLLSVVSGG-YFYMFDPRGLVLSAPSSNAPS 579
Q L GN T YG+G Y I D S +SG +FDP +S +P
Sbjct: 1855 QNLGKGTKEGNPYKTGQYGIGFNSVYHITDCPSFISGNDILCIFDPHARYAPGATSISP- 1913
Query: 580 GKMFSLIGTDLAQRFGDQFSPMLIDQNDLWSLSDSTIIRMPL-SSDCLKVGH----DVAS 634
G+MF + D +F D L + L + T+ R PL +++ KV +
Sbjct: 1914 GRMFRDLDADFRTQFSDVLDLYL---GTHFKLDNCTMFRFPLRNAEMAKVSEISSVPASD 1970
Query: 635 NRIKHITDVFMEHGSRTLLFLKSVLQVSISTWEE--GHSHPCQNFSISIDPSSSIMRNPF 692
++++ D G+ L+FL + ++SI ++ G + + I + R F
Sbjct: 1971 RMVQNLLDKLRSDGAELLMFLNHMEKISICEIDKSTGALNVLYSVKGKITDGDRLKRKQF 2030
Query: 693 SEKKWRKFQLSRLFSSSNTAIKMHVIDVSLYSEGTTFIDRWLLALSLGSGQ----TRNMA 748
R + +D SEG + WL+ G ++++
Sbjct: 2031 HASVIDSVTKKRQLKDIPVQQITYTMDTE-DSEGN--LTTWLICNRSGFSSMEKVSKSVI 2087
Query: 749 LDRRYLAYNLTPVAGIAALISRN---GHHADVYSTSSIMTPLPLSGCINLPVTVLGCFLV 805
+ L P G+AA I+ N H A + LPLS LP V G F +
Sbjct: 2088 SAHKNQDITLFPRGGVAACITHNYKKPHRAFCF--------LPLSLETGLPFHVNGHFAL 2139
Query: 806 CHNRGRYLFKYQDRRASAEGHFDAGNQLIELWNRELMSC-VCDSYVEMILEIQNL---RK 861
R R L++ D G + WN LM+ + +YVE++++++
Sbjct: 2140 DSAR-RNLWR-----------DDNGVGVRSDWNNSLMTALIAPAYVELLIQLKKRYFPGS 2187
Query: 862 DVSSSIIDSSACPGINLSLKAYGDNIYSFWP 892
D + S++ ++ + +LK + SF+P
Sbjct: 2188 DPTLSVLQNTPIHVVKDTLKKF----LSFFP 2214
Score = 50.8 bits (120), Expect = 3e-05
Identities = 105/468 (22%), Positives = 176/468 (37%), Gaps = 81/468 (17%)
Query: 144 LKLITWCPVILD--PAVRG-LPWLKSSKQVALPTVVRPKSQMWLVSSSMLILDGECGTTY 200
LK I W P + P G L W + P + L+ SS+ ++ E
Sbjct: 401 LKKIKWVPACKERPPNYPGSLVWKGDLCNLCAPPDMCDVGHAILIGSSLPLV--ESIHVN 458
Query: 201 LQTKLGWMDCPNIGVLSRQLIELSKSYQQLKTHSLLDPDFDVKLQKEIPCLYSKLQEYIN 260
L+ LG P++ + + ++ + KT S D D+ + Q + +Y + +++N
Sbjct: 459 LEKALGIFTKPSLSAVLKHF-KIVVDWYSSKTFS--DEDY-YQFQHILLEIYGFMHDHLN 514
Query: 261 T--DDFNDLKARLDGVSWVWIGDDFVSPNALAFDSPVK---FTPYLYVVSSELSEYKDLM 315
D F LK WVW G F P A A P+ PYL+ V ++++ L
Sbjct: 515 EGKDSFRALK-----FPWVWTGKKFC-PLAQAVIKPIHDLDLQPYLHNVPKTMAKFHQLF 568
Query: 316 ---------------------------------IKLGVRLSFGILDYLHVLQRLQNDLNG 342
K + L I+ +L+ Q +
Sbjct: 569 KVCGSIEELTSDHISMVIQKIYLKSDQDLSEQESKQNLHLMLNIIRWLYSNQIPASPNTP 628
Query: 343 VPLSTDQLNFVRCVHEAIAECCLEKPLFEPFDSPLWIPDTFGVLMHAGDLVYNDAPWLEN 402
VP+ + N + + + I ECC + + L +L+H D+ A WL+
Sbjct: 629 VPIHHSK-NPSKLIMKPIHECCYCDIKVDDLNDLLEDSVEPIILVHE-DIPMKTAEWLKV 686
Query: 403 SSLIGRHFVHPSISNDLAERLGVQSVRCLSLVSEDMTKDLPCMGYNKVSELLALYGNSEF 462
L R ++P E +G + S E +T ++ +L Y +
Sbjct: 687 PCLSTR-LINP-------ENMGFEQ----SGQREPLTV--------RIKNILEEYPSVSD 726
Query: 463 LLFDLLELADCCKAKKLHLIYDKREHP--RQSLLQHNLGEFQGPALVAIFEGACLSREEF 520
+ +LL+ AD A + + D R + R++LL + GPAL + F + S +F
Sbjct: 727 IFKELLQNADDANATECSFLIDMRRNMDIRENLLDPGMAACHGPALWS-FNNSQFSDSDF 785
Query: 521 SNFQLLPPWKLRGNTLN---YGLGLVGCYSICDLLSVVSGGYFYMFDP 565
N L RG +GLG Y I D+ ++S + MFDP
Sbjct: 786 VNITRLGESLKRGEVDKVGKFGLGFNSVYHITDIPIIMSREFMIMFDP 833
Score = 48.9 bits (115), Expect = 1e-04
Identities = 50/219 (22%), Positives = 94/219 (42%), Gaps = 42/219 (19%)
Query: 1442 AQEEIEILRSLPIYKTVVGSYTKLKDQDQCMIPSNSFFKPHDEHCLSYATDSNGSSFL-- 1499
+Q++I+IL+SLP YK++ G Y + C + + S P E + T S+ S+FL
Sbjct: 2650 SQDDIKILKSLPCYKSISGRYVSIGKFGTCYVLTKSI--PSAE--VEKWTQSSSSAFLEE 2705
Query: 1500 --------RALGVLELNDQQILVRFGLPGYERKSMNEQEDILIY---------------- 1535
+G + ++D ++ ++ LP E S + + + LIY
Sbjct: 2706 KIHLKELYEVIGCVPVDDLEVYLKHLLPKIENLSYDAKLEHLIYLKNRLSSAEELSEIKE 2765
Query: 1536 -VFKNWHDLQSNQSVVEALKETKFVRNSDEFSTDLLKPKELFDPADAILISIFFGERKKF 1594
+F+ L LK+ K + +++ P++LF P D FF + ++
Sbjct: 2766 QLFEKLESLLIIHDANSRLKQAKHFYDRTVRVFEVMLPEKLFIPND------FFKKLEQL 2819
Query: 1595 PGER----FSTDGWLRILRKLGLRTATEVDVIIECAKRV 1629
+ F T W+ LR +GL+ +++ AK +
Sbjct: 2820 IKPKNHVTFMT-SWVEFLRNIGLKYILSQQQLLQFAKEI 2857
Score = 44.3 bits (103), Expect = 0.003
Identities = 38/168 (22%), Positives = 74/168 (43%), Gaps = 11/168 (6%)
Query: 241 DVKLQKEIPCLYSKLQEYINTDDFNDLKARLDGVSWVWIGDDF--VSPNALAFDSPVK-- 296
D ++ K + + E+++ + + + +L GV++V + D + + P + + +
Sbjct: 3012 DEEMVKTRAKVLRSIYEFLSAEK-REFRFQLRGVAFVMVEDGWKLLKPEEVVINLEYESD 3070
Query: 297 FTPYLYVVSSELSEYKDLMIKLGVRLSFGILDYLHVLQRLQNDLNGVPLSTDQLNFVRCV 356
F PYLY + EL + L LG Y+ VL R+ + G L +++ V+ V
Sbjct: 3071 FKPYLYKLPLELGTFHQLFKHLGTEDIISTKQYVEVLSRIFKNSEGKQLDPNEMRTVKRV 3130
Query: 357 HEAIAECC------LEKPLFEPFDSPLWIPDTFGVLMHAGDLVYNDAP 398
+ + L D L++P G L+ + LV++DAP
Sbjct: 3131 VSGLFRSLQNDSVKVRSDLENVRDLALYLPSQDGRLVKSSILVFDDAP 3178
Score = 35.4 bits (80), Expect = 1.4
Identities = 15/39 (38%), Positives = 25/39 (63%), Gaps = 1/39 (2%)
Query: 1253 PSSDSQGGPSSEWIKIFWKS-FRGSQEELSLFSDWPLIP 1290
P +++ PS W+K+ WK+ + E+L+LF + PLIP
Sbjct: 16 PFDENRNHPSVSWLKMVWKNLYIHFSEDLTLFDEMPLIP 54
>SACS_MOUSE (Q9JLC8) Sacsin
Length = 3830
Score = 128 bits (321), Expect = 2e-28
Identities = 206/963 (21%), Positives = 384/963 (39%), Gaps = 131/963 (13%)
Query: 10 KKMLHGDVFFPSDKFLDPEILDTLVCLGLRTTLG-FTGLLDCARSVS------------- 55
+K +VFFP+ + ++ E+ D L+ L L F+G+L V
Sbjct: 1303 EKQFFSEVFFPNIQEIEAELRDPLMNFVLNEKLDEFSGILRVTPCVPCSLEGHPLVLPSR 1362
Query: 56 LLHDSG------DTEASKHGKGLLV-FLDKLAC-KLSSNGESENDDQSLAVRSNITMDDA 107
L+H G DT+ + G +L+ + KL G +++D I DD
Sbjct: 1363 LIHPEGRVAKLFDTKDGRFPYGSTQDYLNPIILIKLVQLGMAKDD---------ILWDDM 1413
Query: 108 VVYDGFPKDENSLIDDVDLFMSSLINDMVEE----------EFWSELKLITWCPVILDPA 157
+ + N SS++ +++E +F ++ + I + P + PA
Sbjct: 1414 LERAESVAEINKSDHAAACLRSSILLSLIDEKLKIKDPRAKDFAAKYQTIPFLPFLTKPA 1473
Query: 158 VRGLPWLKSS---KQVALPTVVRPKSQMWLVSSSMLILDGE------CGTTYLQTK--LG 206
L W +S + + T + +V IL+ CG+ L K LG
Sbjct: 1474 GFSLEWKGNSFKPETMFAATDIYTAEYQDIVCLLQPILNENSHSFRGCGSVSLAVKEFLG 1533
Query: 207 WMDCPNIGVLSRQLIELSKSYQQLKTHSLLDPDFDVKLQKEIPCLYSKLQEYINTDDFND 266
+ P + ++ QL +++KS +D + + Y L E + ++
Sbjct: 1534 LLKKPTVDLVINQLKQVAKS---------VDDGITLYQENITNACYKYLHEAVLQNEMAK 1584
Query: 267 --LKARLDGVSWVWIGDDFVSPNALAFDSPVKFTPYLYVVSSEL-SEYKDLMIKLGVRLS 323
+ +L ++ + + +V ++F + PYLY + ++ + +++L +GVR S
Sbjct: 1585 ATIIEKLKPFCFILVENVYVESEKVSFHLNFEAAPYLYQLPNKYKNNFRELFESVGVRQS 1644
Query: 324 FGILDYLHVLQRLQNDLNGVPLSTDQLNFVR-CVHEAIAECCLEK--PLFEPFDSPLWIP 380
F + D+ VL+ + + ++ + R + E I EK E + +P
Sbjct: 1645 FTVEDFALVLESIDQERGKKQITEENFQLCRRIISEGIWSLIREKRQEFCEKNYGKILLP 1704
Query: 381 DTFGVLMHAGDLVYNDAPWLENSSLIGRHFVHPSISNDLAERLGVQSVRCLSLVSEDMTK 440
DT +L+ A L YND PW++ + + H I ++A +LG R +L
Sbjct: 1705 DTNLLLLPAKSLCYNDCPWIKVKDSTVK-YCHADIPREVAVKLGAIPKRHKALERYASNI 1763
Query: 441 DLPCMGY---------NKVSELLALYGNSEFLLFDLLELADCCKAKKLHLIYDKREHPRQ 491
+G +++ +L Y + + +L +LL+ AD KA ++ ++D R+HP
Sbjct: 1764 CFTALGTEFGQKEKLTSRIKSILNAYPSEKEMLKELLQNADDAKATEICFVFDPRQHPVD 1823
Query: 492 SLLQHNLGEFQGPALVAIFEGACLSREEFSNFQLLPPWKLRGN---TLNYGLGLVGCYSI 548
+ QGPAL ++ + ++ Q L GN T +YG+G Y I
Sbjct: 1824 RIFDDKWAPLQGPAL-CVYNNQPFTEDDVRGIQNLGKGTKEGNPCKTGHYGIGFNSVYHI 1882
Query: 549 CDLLSVVSGG-YFYMFDPRGLVLSAPSSNAPSGKMFSLIGTDLAQRFGDQFSPMLIDQND 607
D S +SG +FDP +S +P G+MF + D +F D L +
Sbjct: 1883 TDCPSFISGNDILGIFDPHARYAPGATSVSP-GRMFRDLDADFRTQFSDVLDLYL---GN 1938
Query: 608 LWSLSDSTIIRMPLSSDCLKVGHDVAS-----NRIKHITDVFMEHGSRTLLFLKSVLQVS 662
+ L + T+ R PL + + +++S ++++ D G+ L+FL + ++S
Sbjct: 1939 HFKLDNCTMFRFPLRNAEMAQVSEISSVPSSDRMVQNLLDKLRSDGAELLMFLNHMEKIS 1998
Query: 663 ISTWEE--GHSHPCQNFSISIDPSSSIMRNPFSEKKWRKFQLSRLFSSSNTAIKMHVIDV 720
I ++ G + + I + R F R + +D
Sbjct: 1999 ICEIDKATGGLNVLYSVKGKITDGDRLKRKQFHASVIDSVTKKRQLKDIPVQQITYTMDT 2058
Query: 721 SLYSEGTTFIDRWLLALSLGSGQ----TRNMALDRRYLAYNLTPVAGIAALISRN---GH 773
SEG + WL+ G ++++ + L P G+AA I+ N H
Sbjct: 2059 E-DSEGN--LTTWLICNRSGFSSMEKVSKSVISAHKNQDITLFPRGGVAACITHNYKKPH 2115
Query: 774 HADVYSTSSIMTPLPLSGCINLPVTVLGCFLVCHNRGRYLFKYQDRRASAEGHFDAGNQL 833
A + LPLS LP V G F + R R L++ D G +
Sbjct: 2116 RAFCF--------LPLSLETGLPFHVNGHFALDSAR-RNLWR-----------DDNGVGV 2155
Query: 834 IELWNRELMSC-VCDSYVEMILEIQNL---RKDVSSSIIDSSACPGINLSLKAYGDNIYS 889
WN LM+ + +YVE++++++ D + S++ ++ + +LK + S
Sbjct: 2156 RSDWNNSLMTALIAPAYVELLIQLKKRYFPGSDPTLSVLQNTPIHVVKDTLKKF----LS 2211
Query: 890 FWP 892
F+P
Sbjct: 2212 FFP 2214
Score = 65.9 bits (159), Expect = 1e-09
Identities = 91/411 (22%), Positives = 173/411 (41%), Gaps = 74/411 (18%)
Query: 1253 PSSDSQGGPSSEWIKIFWKS-FRGSQEELSLFSDWPLIP-AFLGRPVLC----RVRERHL 1306
P S+ + PS W+K+ WK+ + E+L+LF + PLIP L C R+R +
Sbjct: 16 PFSEDKRHPSLSWLKMVWKNLYIHFSEDLTLFDEMPLIPRTLLNEDQTCVELIRLRIPSV 75
Query: 1307 VFIPPPLVQRTLQSGILDRESTENHVGGVSVSRDDTSVAEAYTSAFDRLKISYPWLLSLL 1366
V + + L + D +GG+ + R DTS+ + + S L
Sbjct: 76 VILDDE-TEAQLPEFLAD---IVQKLGGIVLKRLDTSIQHPLVKKY---------IHSPL 122
Query: 1367 NQCNVPIFDEAFIDCATSSNCFSMPGGSLGQVIASKLVAAKQAGYFTEPNNLSASSCDAL 1426
+ I ++ +P L IAS L K A AS D
Sbjct: 123 PSAILQIMEK-------------IPLQKLCNQIASLLPTHKDA-----LRKFLASLTDT- 163
Query: 1427 FSLFCDELFSNGFHYAQEEIEILRSLPIYKTV-------VGSYTKLKDQDQCMIPSNSFF 1479
+++E I++ L I+K + + SYTKLK C + ++
Sbjct: 164 ---------------SEKEKRIIQELTIFKRINHSSDQGISSYTKLKG---CKVLDHTAK 205
Query: 1480 KPHDEHCLSYATDSNGSSFLRALGVLELNDQQIL--VRFGLPGYERKSMNEQE--DILIY 1535
P D DS+ + +R +L++ + ++F L ++E ++++
Sbjct: 206 LPTDLRLSVSVIDSSDEATIRLANMLKIEKLKTTSCLKFVLKDIGNAFYTQEEVTQLMLW 265
Query: 1536 VFKNWHDLQS-NQSVVEALKETKFVRNSDEFSTDLLKPKELFDPADAILISIFFGERKK- 1593
+ +N L++ N +V++ L KF+ S ++ +LFDP +L +F+ E +
Sbjct: 266 ILENLSSLKNENSNVLDWLMPLKFIHMSQGH---VVAAGDLFDPDIEVLRDLFYNEEEAC 322
Query: 1594 FPGERFSTDGWLRILRKLGLRTATEVDV--IIECAKRVEFLGIECMKSGDL 1642
FP F++ L LR++GL+ + + +++ A+++E L + ++ D+
Sbjct: 323 FPPTIFTSPDILHSLRQIGLKNESSLKEKDVVQVARKIEALQVSSCQNQDV 373
Score = 53.5 bits (127), Expect = 5e-06
Identities = 59/279 (21%), Positives = 109/279 (38%), Gaps = 45/279 (16%)
Query: 1442 AQEEIEILRSLPIYKTVVGSYTKLKDQDQCMIPSNSFFKPHDEHCLSYATDSNGSSFL-- 1499
+Q++I+IL+SLP YK++ G Y + C + + S P E + T S+ S+FL
Sbjct: 2650 SQDDIKILKSLPCYKSISGRYMSIAKFGTCYVLTKSI--PSAE--VEKWTQSSSSAFLEE 2705
Query: 1500 --------RALGVLELNDQQILVRFGLPGYERKSMNEQEDILIYVFKNWHDLQSNQSVVE 1551
LG + ++D ++ ++ LP E S + + + LIY+ ++ + E
Sbjct: 2706 KVHLKELYEVLGCVPVDDLEVYLKHLLPKIENLSYDAKLEHLIYLKNRLASIEEPSEIKE 2765
Query: 1552 ALKETKFVRNSDEFSTDLLKPKELFDPADAILISIFFGERKKFPGERFS----------- 1600
L E + + LK + F + + E+ P E F
Sbjct: 2766 QLFEKLESLLIIHDANNRLKQAKHFYDRTVRVFEVMLPEKLFIPKEFFKKLEQVIKPKNQ 2825
Query: 1601 ---TDGWLRILRKLGLRTATEVDVIIECAKRVEFLGIECMKSGDLDDFEADTTNSRAEVS 1657
W+ LR +GL+ A +++ AK + + +++ +T S ++
Sbjct: 2826 AAFMTSWVEFLRNIGLKYALSQQQLLQFAKEISV-------RANTENWSKETLQSTVDI- 2877
Query: 1658 PEVWALGGSVVEFIFSNFALFFSNNFCDLLGKIAFVPSE 1696
++ IF S NF L I F+ E
Sbjct: 2878 ---------LLHHIFQERMDLLSGNFLKELSLIPFLCPE 2907
Score = 49.7 bits (117), Expect = 7e-05
Identities = 136/618 (22%), Positives = 222/618 (35%), Gaps = 140/618 (22%)
Query: 1 LYDPRVPELKKMLHGD--VFFPSDKFLDPEILDTLVCLGLR--TTLGFTGLLDCARSVSL 56
L+DP + L+ + + + FP F P+IL +L +GL+ ++L ++ AR +
Sbjct: 303 LFDPDIEVLRDLFYNEEEACFPPTIFTSPDILHSLRQIGLKNESSLKEKDVVQVARKIEA 362
Query: 57 LHDSGDTEAS---KHGKGLLVFLDKLACKLSSNGESENDDQSLAVRSNITMDDAVVYDGF 113
L S K K LL+ L+K +Q+L S M
Sbjct: 363 LQVSSCQNQDVLMKKAKTLLLVLNK--------------NQTLLQSSEGKM--------- 399
Query: 114 PKDENSLIDDVDLFMSSLINDMVEEEFWSELKLITWCPVILD--PAVRG-LPWLKSSKQV 170
LK I W P + P G L W +
Sbjct: 400 -----------------------------ALKKIKWVPACKERPPNYPGSLVWKGDLCNL 430
Query: 171 ALPTVVRPKSQMWLVSSSMLILDGECGTTYLQTKLGWMDCPNIGVLSRQLIELSKSYQQL 230
P + + LV SS+ ++ E L+ L P I + + + Y
Sbjct: 431 CAPPDMCDAAHAVLVGSSLPLV--ESVHVNLEQALSIFTKPTINAVLKHFKTVVDWYTS- 487
Query: 231 KTHSLLDPDFDVKLQKEIPCLYSKLQEYINT--DDFNDLKARLDGVSWVWIGDDFVSPNA 288
KT S D D+ + Q + +Y + ++++ D F LK WVW G +F P A
Sbjct: 488 KTFS--DEDY-YQFQHILLEIYGFMHDHLSEGKDSFKALK-----FPWVWTGKNFC-PLA 538
Query: 289 LAFDSP---VKFTPYLYVVSSELSEYKDLMIKLG-------------------------- 319
A P + PYLY V ++++ L G
Sbjct: 539 QAVIKPTHDLDLQPYLYNVPKTMAKFHQLFKACGSIEELTSDHISMVIQKVYLKSDQELS 598
Query: 320 -------VRLSFGILDYLHVLQRLQNDLNGVPLSTDQLNFVRCVHEAIAECCLEKPLFEP 372
+ L I+ +L+ Q + VP+ + N + V + I ECC +
Sbjct: 599 EEESKQNLHLMLNIMRWLYSNQIPASPNTPVPIYHSR-NPSKLVMKPIHECCYCDIKVDD 657
Query: 373 FDSPLWIPDTFGVLMHAGDLVYNDAPWLENSSLIGRHFVHPSISNDLAERLGVQSVRCLS 432
+ L +L+H D+ A WL+ L R ++P E +G + S
Sbjct: 658 LNDLLEDSVEPIILVHE-DIPMKTAEWLKVPCLSTR-LINP-------ENMGFEQ----S 704
Query: 433 LVSEDMTKDLPCMGYNKVSELLALYGNSEFLLFDLLELADCCKAKKLHLIYDKREHP--R 490
E +T ++ +L Y + + +LL+ AD A + + D R + R
Sbjct: 705 GQREPLTV--------RIKNILEEYPSVSDIFKELLQNADDANATECSFMIDMRRNMDIR 756
Query: 491 QSLLQHNLGEFQGPALVAIFEGACLSREEFSNFQLLPPWKLRGNTLN---YGLGLVGCYS 547
++LL + GPAL + F + S +F N L RG +GLG Y
Sbjct: 757 ENLLDPGMAACHGPALWS-FNNSEFSDSDFLNITRLGESLKRGEVDKVGKFGLGFNSVYH 815
Query: 548 ICDLLSVVSGGYFYMFDP 565
I D+ ++S + MFDP
Sbjct: 816 ITDIPIIMSREFMIMFDP 833
Score = 45.1 bits (105), Expect = 0.002
Identities = 38/168 (22%), Positives = 74/168 (43%), Gaps = 11/168 (6%)
Query: 241 DVKLQKEIPCLYSKLQEYINTDDFNDLKARLDGVSWVWIGDDF--VSPNALAFDSPVK-- 296
D ++ K + + E+++ + + + +L GV++V + D + + P + + +
Sbjct: 3012 DEEMVKTRAKVLRSIYEFLSAEK-REFRFQLRGVAFVMVEDGWKLLKPEEVVINLEYEAD 3070
Query: 297 FTPYLYVVSSELSEYKDLMIKLGVRLSFGILDYLHVLQRLQNDLNGVPLSTDQLNFVRCV 356
F PYLY + EL + L LG Y+ VL R+ G L +++ V+ V
Sbjct: 3071 FKPYLYKLPLELGTFHQLFKHLGTEDIISTKQYVEVLSRIFKSSEGKQLDPNEMRTVKRV 3130
Query: 357 HEAIAECC------LEKPLFEPFDSPLWIPDTFGVLMHAGDLVYNDAP 398
+ + + L D L++P G L+ + LV++DAP
Sbjct: 3131 VSGLFKSLQNDSVKVRSDLENARDLALYLPSQDGKLVKSSILVFDDAP 3178
>RISC_RAT (Q920A6) Retinoid-inducible serine carboxypeptidase
precursor (EC 3.4.16.-) (Serine carboxypeptidase 1)
Length = 452
Score = 41.2 bits (95), Expect = 0.026
Identities = 52/218 (23%), Positives = 96/218 (43%), Gaps = 30/218 (13%)
Query: 172 LPTVVRPKSQMWLVSSSMLILDGECGT--TYLQTKLGWMD-----CPNIGVLSRQLIELS 224
L T ++P++ WL +S+L +D GT +Y+ T + ++ VL + +
Sbjct: 94 LDTRLKPRNTTWLQWASLLFVDNPVGTGFSYVNTTDAYAKDLDTVASDMMVLLKSFFDCH 153
Query: 225 KSYQQLKTHSLLDPDFDVKLQKEIPC-LYSKLQEYINTDDFNDLKARLDGVSWVWIGDDF 283
K +Q + + + + K+ I L+ +Q+ +K GV+ +GD +
Sbjct: 154 KEFQTVPFY-IFSESYGGKMAAGISLELHKAIQQ-------GTIKCNFSGVA---LGDSW 202
Query: 284 VSPNALAFDSPVKFTPYLYVVS----SELSEYKDLMIKLGVRLSFGILDYLHVLQRLQND 339
+SP DS + + PYLY VS L+E D+ ++ ++ G Y Q
Sbjct: 203 ISP----VDSVLSWGPYLYSVSLLDNKGLAEVSDIAEQVLNAVNKGF--YKEATQLWGKA 256
Query: 340 LNGVPLSTDQLNFVRCVHEAIAECCLEKPLFEPFDSPL 377
+ +TD +NF + ++ + +E L E F SPL
Sbjct: 257 EMIIEKNTDGVNFYNILTKSTPDTSMESSL-EFFRSPL 293
>RISC_MOUSE (Q920A5) Retinoid-inducible serine carboxypeptidase
precursor (EC 3.4.16.-) (Serine carboxypeptidase 1)
Length = 452
Score = 37.4 bits (85), Expect = 0.37
Identities = 35/142 (24%), Positives = 65/142 (45%), Gaps = 23/142 (16%)
Query: 172 LPTVVRPKSQMWLVSSSMLILDGECGT--TYLQTKLGWMD-----CPNIGVLSRQLIELS 224
L T ++P++ WL +S+L +D GT +Y+ T + ++ VL + +
Sbjct: 94 LDTQLKPRNTTWLQWASLLFVDNPVGTGFSYVNTTDAYAKDLDTVASDMMVLLKSFFDCH 153
Query: 225 KSYQQLKTHSLLDPDFDVKLQKEIPC-LYSKLQEYINTDDFNDLKARLDGVSWVWIGDDF 283
K +Q + + + + K+ I LY +Q+ +K GV+ +GD +
Sbjct: 154 KEFQTVPFY-IFSESYGGKMAAGISVELYKAVQQ-------GTIKCNFSGVA---LGDSW 202
Query: 284 VSPNALAFDSPVKFTPYLYVVS 305
+SP DS + + PYLY +S
Sbjct: 203 ISP----VDSVLSWGPYLYSMS 220
>YIL8_CAEEL (O01757) Hypothetical UPF0202 protein F55A12.8 in
chromosome I
Length = 1043
Score = 34.3 bits (77), Expect = 3.1
Identities = 23/104 (22%), Positives = 47/104 (45%), Gaps = 10/104 (9%)
Query: 1236 NHVMGSNMAPWLSWKKLPSSDSQGGPSSEWIKIFWKSFRGSQEELSLFSDWPLIPAFLGR 1295
N + G + L + SDS PS+ W+ ++W+ FR L F D+ PA +
Sbjct: 736 NDITGEHTCIILKGLEHGGSDSDEEPSATWLPVYWREFRRRIVNLLSF-DFSSFPAQMAL 794
Query: 1296 PVLCRVRERHLVFIPPPLVQRTLQSGILDRESTENHVGGVSVSR 1339
+L +++ +H V++ ++ +++R H+ + R
Sbjct: 795 SLL-QLKNKH--------VEKQMKRLVIERSELAIHLSNTDLRR 829
>TRUB_WIGBR (Q8D2X8) tRNA pseudouridine synthase B (EC 4.2.1.70) (tRNA
pseudouridine 55 synthase) (Psi55 synthase)
(Pseudouridylate synthase) (Uracil hydrolyase)
Length = 327
Score = 34.3 bits (77), Expect = 3.1
Identities = 16/41 (39%), Positives = 24/41 (58%)
Query: 1577 DPADAILISIFFGERKKFPGERFSTDGWLRILRKLGLRTAT 1617
DP + ++ IFFG+ KF +T+ W +I KLG +T T
Sbjct: 69 DPIASGMLPIFFGDATKFSDYLLNTNKWYKIKAKLGEKTNT 109
>RISC_HUMAN (Q9HB40) Retinoid-inducible serine carboxypeptidase
precursor (EC 3.4.16.-) (Serine carboxypeptidase 1)
(MSTP034) (UNQ265/PRO302)
Length = 452
Score = 33.5 bits (75), Expect = 5.3
Identities = 31/141 (21%), Positives = 61/141 (42%), Gaps = 21/141 (14%)
Query: 172 LPTVVRPKSQMWLVSSSMLILDGECGTTYLQTKLGWMDCPNIGVLSRQLIELSKSY---- 227
L + ++P+ WL ++S+L +D GT + ++ +++ ++ L K++
Sbjct: 94 LDSDLKPRKTTWLQAASLLFVDNPVGTGFSYVNGSGAYAKDLAMVASDMMVLLKTFFSCH 153
Query: 228 --QQLKTHSLLDPDFDVKLQKEIPC-LYSKLQEYINTDDFNDLKARLDGVSWVWIGDDFV 284
Q + + K+ I LY +Q +K GV+ +GD ++
Sbjct: 154 KEFQTVPFYIFSESYGGKMAAGIGLELYKAIQR-------GTIKCNFAGVA---LGDSWI 203
Query: 285 SPNALAFDSPVKFTPYLYVVS 305
SP DS + + PYLY +S
Sbjct: 204 SP----VDSVLSWGPYLYSMS 220
>YDHA_ECOLI (P28224) Hypothetical protein ydhA
Length = 109
Score = 33.1 bits (74), Expect = 7.0
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 5/74 (6%)
Query: 829 AGNQLIELWNRELMSCVCDSYVEMILEIQNLRKDVSSSIIDSSAC---PGINLSLKAYGD 885
A NQL+E + + CD + +++ N R++VS + D+ GI+ S Y D
Sbjct: 20 AFNQLVERMQTDTLEYQCDEK-PLTVKLNNPRQEVSF-VYDNQLLHLKQGISASGARYTD 77
Query: 886 NIYSFWPRSSEGHV 899
IY FW + E V
Sbjct: 78 GIYVFWSKGDEATV 91
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 212,945,288
Number of Sequences: 164201
Number of extensions: 9474716
Number of successful extensions: 20118
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 20080
Number of HSP's gapped (non-prelim): 32
length of query: 1754
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1630
effective length of database: 39,613,130
effective search space: 64569401900
effective search space used: 64569401900
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0237.14


