

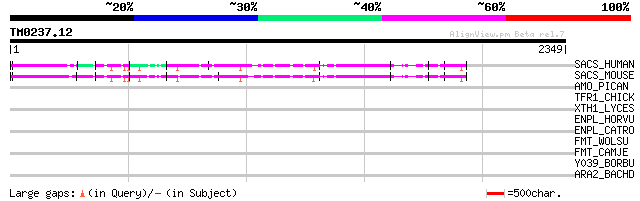
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0237.12
(2349 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SACS_HUMAN (Q9NZJ4) Sacsin 291 2e-77
SACS_MOUSE (Q9JLC8) Sacsin 282 7e-75
AMO_PICAN (P12807) Peroxisomal copper amine oxidase (EC 1.4.3.6)... 40 0.059
TFR1_CHICK (Q90997) Transferrin receptor protein 1 (TfR1) (TR) (... 38 0.29
XTH1_LYCES (Q40144) Probable xyloglucan endotransglucosylase/hyd... 37 0.65
ENPL_HORVU (P36183) Endoplasmin homolog precursor (GRP94 homolog) 36 1.4
ENPL_CATRO (P35016) Endoplasmin homolog precursor (GRP94 homolog) 35 1.9
FMT_WOLSU (Q7MA26) Methionyl-tRNA formyltransferase (EC 2.1.2.9) 34 5.5
FMT_CAMJE (Q9PJ28) Methionyl-tRNA formyltransferase (EC 2.1.2.9) 34 5.5
Y039_BORBU (O51068) Hypothetical protein BB0039 33 9.4
ARA2_BACHD (Q9K9D5) 3-phosphoshikimate 1-carboxyvinyltransferase... 33 9.4
>SACS_HUMAN (Q9NZJ4) Sacsin
Length = 3829
Score = 291 bits (744), Expect = 2e-77
Identities = 349/1460 (23%), Positives = 589/1460 (39%), Gaps = 212/1460 (14%)
Query: 13 DFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGDSLLSSSL 72
+FGQ LT RI+ +L YP +LKEL+QNADDA AT + D R H D +
Sbjct: 1771 EFGQKEKLTSRIKSILNAYPSEKEMLKELLQNADDAKATEICFVFDPRQHPVDRIFDDKW 1830
Query: 73 SQWQGPALLAYNDAVFTEDDFISISKIGGSSKHGQASKTGRFGVGFNSVYHLTDLPSFVS 132
+ QGPAL YN+ FTEDD I +G +K G KTG++G+GFNSVYH+TD PSF+S
Sbjct: 1831 APLQGPALCVYNNQPFTEDDVRGIQNLGKGTKEGNPYKTGQYGIGFNSVYHITDCPSFIS 1890
Query: 133 GKYVV-LFDPQGVYLPRVSAANPGKRIDFTGSSALSLYKDQFSPYCAFGCDMQSPFAGTL 191
G ++ +FDP Y P ++ +PG+ + + + D Y + + T+
Sbjct: 1891 GNDILCIFDPHARYAPGATSISPGRMFRDLDADFRTQFSDVLDLYLGTHFKLDNC---TM 1947
Query: 192 FRFPLRNADQASRSKLSRQAYTPEDISSMFVQLFEEGVLTLLFLKSVLCIEMYVWDAGEP 251
FRFPLRNA+ A S++S + + ++ +L +G L+FL + I + D
Sbjct: 1948 FRFPLRNAEMAKVSEISSVPASDRMVQNLLDKLRSDGAELLMFLNHMEKISICEIDKSTG 2007
Query: 252 KPKKIHSCSVSSVSDDTIWHRQALLRLSKSLNTTTEVDAFPLEFVTEAVRGVETVRQVDR 311
++S D + +Q + S+ ++ P++ +T + ++ +
Sbjct: 2008 ALNVLYSVKGKITDGDRLKRKQFHASVIDSVTKKRQLKDIPVQQITYTMDTEDSEGNLTT 2067
Query: 312 FYIVQTMA-SASSRIGSFAITASKEYDIQLLPWASIAACISDNSLNNDVLRTGQAFCFLP 370
+ I S+ ++ I+A K DI L P +AACI+ N + +AFCFLP
Sbjct: 2068 WLICNRSGFSSMEKVSKSVISAHKNQDITLFPRGGVAACITHN-----YKKPHRAFCFLP 2122
Query: 371 LPVRTGLSVQVNGFFEVSSNRRGIWYGDDMDRSGKVRSIWNRLLLEDLVAPAFVHM---- 426
L + TGL VNG F + S RR +W D+ VRS WN L+ L+APA+V +
Sbjct: 2123 LSLETGLPFHVNGHFALDSARRNLWRDDN---GVGVRSDWNNSLMTALIAPAYVELLIQL 2179
Query: 427 -------------------LHGVKELLGPTDIYYSLWPTGSFEEPWSI--LVQQIYINIC 465
+H VK+ L + S +P + + LV+ +Y I
Sbjct: 2180 KKRYFPGSDPTLSVLQNTPIHVVKDTLKK---FLSFFPVNRLDLQPDLYCLVKALYNCIH 2236
Query: 466 N-----APVIYS-NLGGG--------RWVSPSEA---------FLHDE-KFTKSKDLSLA 501
PV+ + N+ G W++ S + L DE + K+ D ++
Sbjct: 2237 EDMKRLLPVVRAPNIDGSDLHSAVIITWINMSTSNKTRPFFDNLLQDELQHLKNADYNIT 2296
Query: 502 ---------------LMQLGMPVVHLPN---SLFDMLLKYNSS-KVITPGTVRQFLRECE 542
L+++G +V+ + +L+ L+ + +TP +R FL
Sbjct: 2297 TRKTVAENVYRLKHLLLEIGFNLVYNCDETANLYHCLIDADIPVSYVTPADIRSFLMTFS 2356
Query: 543 S----CN-----------HLSRAHKL-LLLEYCLEDLVDDDVGKAAYNLPLLPLANGNFA 586
S C+ +L H L LL++YC +D ++++ LPLL +
Sbjct: 2357 SPDTNCHIGKLPCRLQQTNLKLFHSLKLLVDYCFKDAEENEI--EVEGLPLLITLDSVLQ 2414
Query: 587 SFLEASKGIPYFICDELEYKLLEPVSDRVIDQSIPPNILTRLSGIAMSSNTNIA-LFSIH 645
+F +A + P F+ ++L+ D ++ + + S I + N +A +F I
Sbjct: 2415 TF-DAKR--PKFLTTY--HELIPSRKDLFMN-----TLYLKYSNILL--NCKVAKVFDIS 2462
Query: 646 HFAHLFPVFMPDDWKYKCKVFWDPDSCQKPTSSWFVLFWQYLGK-------QSEILPLF- 697
FA L +P ++K K W + + SW W ++ + Q E P F
Sbjct: 2463 SFADLLSSVLPREYKTKSCTKWKDNFASE---SWLKNAWHFISESVSVKEDQEETKPTFD 2519
Query: 698 ------KDWPILPST------------SGHLLRPSRQLKM-INGSTLSDTVQDILVKIGC 738
KDW +LP T G +L P + + + + SD V L+K GC
Sbjct: 2520 IVVDTLKDWALLPGTKFTVSANQLVVPEGDVLLPLSLMHIAVFPNAQSDKVFHALMKAGC 2579
Query: 739 HILKPGYVVEHPDLFSYLCGGNAAGVLESIFNAFSSAENMQVSFSSLIAEE--RNELRRF 796
L + F L + A + + A + V S+ AE+ N+
Sbjct: 2580 IQLALNKICSKDSAFVPLLSCHTANIESP--TSILKALHYMVQTSTFRAEKLVENDFEAL 2637
Query: 797 LLDPQWYVGHSMDEFNIRFCRRLPIYQVYHREPTQDSQFSDLENPRKYLPPLDVPEFILV 856
L+ + H M + +I+ + LP Y+ +F K +P +V ++
Sbjct: 2638 LMYFNCNLNHLMSQDDIKILKSLPCYKSISGRYVSIGKFGTCYVLTKSIPSAEVEKWTQS 2697
Query: 857 GIE-FIVRSSNTEEDILSRYYGVERMGKAQFYKKHVFDRVGELQAEDRDSIMLSV---LQ 912
F+ + +E L G + + Y KH+ ++ L + + ++ + L
Sbjct: 2698 SSSAFLEEKIHLKE--LYEVIGCVPVDDLEVYLKHLLPKIENLSYDAKLEHLIYLKNRLS 2755
Query: 913 NLPLLSLEDASIRDLLRNLKFIPTVIGTLKCPSVLYDPRNEEIYALLEDSDSFPSGVFRE 972
+ LS + + L +L I LK YD +L + P+ F++
Sbjct: 2756 SAEELSEIKEQLFEKLESLLIIHDANSRLKQAKHFYDRTVRVFEVMLPEKLFIPNDFFKK 2815
Query: 973 SETL--------------DIMRGLGLKTSVSPDTVLESARCIE-HLMHEDQQKAYLKGKV 1017
E L + +R +GLK +S +L+ A+ I E+ K L+ V
Sbjct: 2816 LEQLIKPKNHVTFMTSWVEFLRNIGLKYILSQQQLLQFAKEISVRANTENWSKETLQNTV 2875
Query: 1018 --LFSYLEVNALKWLPDKFDDKKGAVNRILSQAATAFRSRNTKSDIEKFWNDLQLISWCP 1075
L ++ + L F + + + + A A R ++ L LI +
Sbjct: 2876 DILLHHIFQERMDLLSGNFLKELSLIPFLCPERAPAEFIR-FHPQYQEVNGTLPLIKFNG 2934
Query: 1076 VLVSPPFH-----SLPW---PVVSSMVAPPKVVRPPNDLWLVSAGMRILDGECSSTALLY 1127
V+P F L W P++ P + G + E L
Sbjct: 2935 AQVNPKFKQCDVLQLLWTSCPILPEKATPLSIKE--------QEGSDLGPQEQLEQVLNM 2986
Query: 1128 CLGWMCPPGGGVIAAQLLELGKNNEIVTDQVLRQELALAMPRIYSILTGMIGSDEIEIVK 1187
+ PP VI I L +E+ ++ + + +++ E +
Sbjct: 2987 LNVNLDPPLDKVI-------NNCRNICNITTLDEEMVKTRAKVLRSIYEFLSAEKREF-R 3038
Query: 1188 AVLEGCRWIWVGDGF--ATSDEVV--LDGPLHLAPYIRVIPVDLAVFKNLFLELGIREFL 1243
L G ++ V DG+ +EVV L+ PY+ +P++L F LF LG + +
Sbjct: 3039 FQLRGVAFVMVEDGWKLLKPEEVVINLEYESDFKPYLYKLPLELGTFHQLFKHLGTEDII 3098
Query: 1244 QPSDYVNILFRMANKKGSSPLDTQEIRAVMLIVHHLAEVYLHG-----------QKVQLY 1292
YV +L R+ LD E+R V +V L + + + LY
Sbjct: 3099 STKQYVEVLSRIFKNSEGKQLDPNEMRTVKRVVSGLFRSLQNDSVKVRSDLENVRDLALY 3158
Query: 1293 LPDVSGRLFLAGDLVYNDAP 1312
LP GRL + LV++DAP
Sbjct: 3159 LPSQDGRLVKSSILVFDDAP 3178
Score = 243 bits (620), Expect = 5e-63
Identities = 274/1033 (26%), Positives = 431/1033 (41%), Gaps = 166/1033 (16%)
Query: 843 KYLPPLDVPEFILVGIEFIVRSSNTEEDILSRYYGVERMGKAQFYKKHVFDRVGELQAED 902
K L +++P + +G E + ++ +L + + QF+ + F + E++AE
Sbjct: 1272 KNLCAVELPSSVKLGFE----EAGCKQILLENTFS-----EKQFFSEVFFPNIQEIEAEL 1322
Query: 903 RDSIMLSVLQNLPLLSLEDASIRDLLRNLKFIPTVIG--TLKCPSVLYDPRNEEIYALLE 960
RD +M+ VL +++ S +LR IP + L PS L P + L +
Sbjct: 1323 RDPLMIFVLNE----KVDEFS--GVLRVTPCIPCSLEGHPLVLPSRLIHPEGR-VAKLFD 1375
Query: 961 DSDS-FPSGVFRESETLDIMRGL---GL-KTSVSPDTVLESARCIEHLMHEDQQKAYLKG 1015
D FP G ++ I+ L G+ K + D +LE A + + D A L+
Sbjct: 1376 IKDGRFPYGSTQDYLNPIILIKLVQLGMAKDDILWDDMLERAVSVAEINKSDHVAACLRS 1435
Query: 1016 KVLFSYLEVNALKWLPDKFDDKKGAVNRILSQAATAFRSRNTKSDIEKFWNDLQLISWCP 1075
+L S + D+K R K K+ Q I + P
Sbjct: 1436 SILLSLI------------DEK------------LKIRDPRAKDFAAKY----QTIRFLP 1467
Query: 1076 VLVSPPFHSLPWPVVSSMVAPPKVVRPPNDLW------LVSAGMRILDGE------CSST 1123
L P SL W S P+ + DL+ +V IL+ C S
Sbjct: 1468 FLTKPAGFSLDWKGNSFK---PETMFAATDLYTAEHQDIVCLLQPILNENSHSFRGCGSV 1524
Query: 1124 ALLY--CLGWMCPPGGGVIAAQLLELGKNNEIVTDQVLRQELALAMPRIYSILTGMIGSD 1181
+L LG + P ++ QL E+ K+ V D + + + Y L + +
Sbjct: 1525 SLAVKEFLGLLKKPTVDLVINQLKEVAKS---VDDGITLYQENITNA-CYKYLHEALMQN 1580
Query: 1182 EIEIVKAV--LEGCRWIWVGDGFATSDEVVLDGPLHLAPYIRVIPVDLAV-FKNLFLELG 1238
EI + + L+ +I V + + S++V APY+ +P F+ LF +G
Sbjct: 1581 EITKMSIIDKLKPFSFILVENAYVDSEKVSFHLNFEAAPYLYQLPNKYKNNFRELFETVG 1640
Query: 1239 IREFLQPSDYVNILFRMANKKGSSPLDTQEIR--------AVMLIVHHLAEVYLHGQKVQ 1290
+R+ D+ +L + ++G+ + + + + ++ + + +
Sbjct: 1641 VRQSCTVEDFALVLESIDQERGTKQITEENFQLCRRIISEGIWSLIREKKQEFCEKNYGK 1700
Query: 1291 LYLPDVSGRLFLAGDLVYNDAPWLLGSEDPDGSFGNAPSVTWNAKRTVQKFVHGNISNDV 1350
+ LPD + L A L YND PW+ K T K+ H +I +V
Sbjct: 1701 ILLPDTNLMLLPAKSLCYNDCPWI------------------KVKDTTVKYCHADIPREV 1742
Query: 1351 AEKLGVRSLRRMLLAESADSMNFGLSGAAEAFGQHEALTTRLKHILEMYADGPGTLFELV 1410
A KLG R L A ++ F G FGQ E LT+R+K IL Y L EL+
Sbjct: 1743 AVKLGAVPKRHKALERYASNVCFTTLGTE--FGQKEKLTSRIKSILNAYPSEKEMLKELL 1800
Query: 1411 QNAEDAGASEVIFLLDKSQYGTSSVLSPEMADWQGPALYCFNDSVFTPQDLYAISRIGQE 1470
QNA+DA A+E+ F+ D Q+ + + A QGPAL +N+ FT D+ I +G+
Sbjct: 1801 QNADDAKATEICFVFDPRQHPVDRIFDDKWAPLQGPALCVYNNQPFTEDDVRGIQNLGKG 1860
Query: 1471 SKLEKAFAIGRFGLGFNCVYHFTDIPMFVSGENIV-MFDPHASNLPGISPSHPGLRIKFA 1529
+K + G++G+GFN VYH TD P F+SG +I+ +FDPHA PG + PG +
Sbjct: 1861 TKEGNPYKTGQYGIGFNSVYHITDCPSFISGNDILCIFDPHARYAPGATSISPGRMFRDL 1920
Query: 1530 GRKILEQFPDQFSSLL--HFGCDLQHPFPGTLFRFPLRTAGVASRSQIKKEVYTPEDVRS 1587
QF D L HF D T+FRFPLR A +A S+I + V++
Sbjct: 1921 DADFRTQFSDVLDLYLGTHFKLD-----NCTMFRFPLRNAEMAKVSEISSVPASDRMVQN 1975
Query: 1588 LFAAFSEVVSETLLFLRNVKSISIFLKEGTGHEMRLLHRVSRASLGESEIGSAEVQDVFN 1647
L +E L+FL +++ ISI + + + +L+ V ++ D
Sbjct: 1976 LLDKLRSDGAELLMFLNHMEKISICEIDKSTGALNVLYSV-----------KGKITD--- 2021
Query: 1648 FFKEDRLVGMNRAQFLKKLSLSIDRDLPYKCQKILITEQGTHGRNSHYWIMTECLGGGNV 1707
DRL R QF + ID + K + +Q T Y + TE
Sbjct: 2022 ---GDRL---KRKQFHASV---IDSVTKKRQLKDIPVQQIT------YTMDTE------- 2059
Query: 1708 LKGTSEASTSNSHNFVPW-ACVAAYLNSVKHGEDLVDSAEVEDDCLVSSDLFQFASLPMH 1766
+ N W C + +S++ V SA D LF +
Sbjct: 2060 ---------DSEGNLTTWLICNRSGFSSMEKVSKSVISAHKNQDI----TLFPRGGVAAC 2106
Query: 1767 PRENFE--GRAFCFLPLPISTGLPAHVNAYFELSSNRRDIWFGSDMTGGGRKRSDWNIYL 1824
N++ RAFCFLPL + TGLP HVN +F L S RR++W + G RSDWN L
Sbjct: 2107 ITHNYKKPHRAFCFLPLSLETGLPFHVNGHFALDSARRNLWRDDNGVG---VRSDWNNSL 2163
Query: 1825 LENVVAPAYGRLL 1837
+ ++APAY LL
Sbjct: 2164 MTALIAPAYVELL 2176
Score = 213 bits (543), Expect = 4e-54
Identities = 260/1006 (25%), Positives = 434/1006 (42%), Gaps = 135/1006 (13%)
Query: 665 VFWDP--DSCQKPTSSWFVLFWQYLGKQ-SEILPLFKDWPILPST---SGHLLRPSRQLK 718
V W P ++ P+ SW + W+ L SE L LF + P++P T G +L+
Sbjct: 12 VQWYPFDENRNHPSVSWLKMVWKNLYIHFSEDLTLFDEMPLIPRTILEEGQTCVELIRLR 71
Query: 719 MINGSTLSDTVQ--------DILVKIGCHILKP-GYVVEHPDLFSYLCGGNAAGVLESIF 769
+ + L D + DI+ K+G +LK ++HP + Y+ + VL+ +
Sbjct: 72 IPSLVILDDESEAQLPEFLADIVQKLGGFVLKKLDASIQHPLIKKYIHSPLPSAVLQIME 131
Query: 770 NAFSSAENMQVSFSSLIAEERNELRRFLLDPQWYVGHSMDEFNIRFCRRLPIYQ-VYHRE 828
+ + +SL+ ++ LR+FL E R + L I++ + H
Sbjct: 132 KM--PLQKLCNQITSLLPTHKDALRKFLASLT-----DSSEKEKRIIQELAIFKRINHSS 184
Query: 829 PTQDSQFSDLENPRKYLPPLDVPEFILVGIEFIVRSSNTEEDILSRYYGVERMGKAQFYK 888
S ++ L+ + +P + + I ++ SS+ L+ +E++ K
Sbjct: 185 DQGISSYTKLKGCKVLHHTAKLPADLRLSIS-VIDSSDEATIRLANMLKIEQLKTTSCLK 243
Query: 889 KHVFDRVGELQA-EDRDSIMLSVLQNLPLLSLEDASIRDLLRNLKFIPTVIGTLKCPSVL 947
+ D + E+ +ML VL+NL L E+ ++ + L LKFI + L
Sbjct: 244 LVLKDIENAFYSHEEVTQLMLWVLENLSSLKNENPNVLEWLTPLKFIQISQEQMVSAGEL 303
Query: 948 YDPRNEEIYALL--EDSDSFPSGVFRESETLDIMRGLGLKT--SVSPDTVLESARCIEHL 1003
+DP E + L E+ FP VF + L +R +GLK S+ V++ A+ IE L
Sbjct: 304 FDPDIEVLKDLFCNEEGTYFPPSVFTSPDILHSLRQIGLKNEASLKEKDVVQVAKKIEAL 363
Query: 1004 MH---EDQQKAYLKGKVLFSYLEVNALKWLPDKFDDKKGAVNRILSQAATAFRSRNTKSD 1060
DQ K K L L N T +S K
Sbjct: 364 QVGACPDQDVLLKKAKTLLLVLNKNH-----------------------TLLQSSEGKMT 400
Query: 1061 IEKFWNDLQLISWCPVLVSPPFH---SLPWPV-VSSMVAPPKVVRPPNDLWLVSAGMRIL 1116
++K I W P P + SL W + ++ APP + + + L+ + + ++
Sbjct: 401 LKK-------IKWVPACKERPPNYPGSLVWKGDLCNLCAPPDMCDVGHAI-LIGSSLPLV 452
Query: 1117 DGECSSTALLYCLGWMCPPGGGVIAA--QLLELGKNNEIVTDQVLRQELALAMPRIYSIL 1174
E L LG P + +++ +++ +D+ Q + + IY +
Sbjct: 453 --ESIHVNLEKALGIFTKPSLSAVLKHFKIVVDWYSSKTFSDEDYYQFQHILL-EIYGFM 509
Query: 1175 TGMI--GSDEIEIVKAVLEGCRWIWVGDGFATSDEVVLDGPLH---LAPYIRVIPVDLAV 1229
+ G D +K W+W G F + V+ P+H L PY+ +P +A
Sbjct: 510 HDHLNEGKDSFRALKFP-----WVWTGKKFCPLAQAVIK-PIHDLDLQPYLHNVPKTMAK 563
Query: 1230 FKNLFLELGIREFLQPSDYVNILFRMANKKGSSPLDTQEIRAVMLIVHHLAEVYLHGQ-- 1287
F LF G E L SD+++++ + K L QE + + ++ ++ Q
Sbjct: 564 FHQLFKVCGSIEELT-SDHISMVIQKIYLKSDQDLSEQESKQNLHLMLNIIRWLYSNQIP 622
Query: 1288 -----KVQLYLPDVSGRLFLAG-------DLVYNDAPWLLGSEDPDGSFGNAPSVTWNAK 1335
V ++ +L + D+ +D LL ED P +
Sbjct: 623 ASPNTPVPIHHSKNPSKLIMKPIHECCYCDIKVDDLNDLL--ED-----SVEPII----- 670
Query: 1336 RTVQKFVHGNISNDVAEKLGVRSLRRMLLAESADSMNFGLSGAAEAFGQHEALTTRLKHI 1395
VH +I AE L V L L+ + ++M F E GQ E LT R+K+I
Sbjct: 671 -----LVHEDIPMKTAEWLKVPCLSTRLI--NPENMGF------EQSGQREPLTVRIKNI 717
Query: 1396 LEMYADGPGTLFELVQNAEDAGASEVIFLLD--KSQYGTSSVLSPEMADWQGPALYCFND 1453
LE Y EL+QNA+DA A+E FL+D ++ ++L P MA GPAL+ FN+
Sbjct: 718 LEEYPSVSDIFKELLQNADDANATECSFLIDMRRNMDIRENLLDPGMAACHGPALWSFNN 777
Query: 1454 SVFTPQDLYAISRIGQESKLEKAFAIGRFGLGFNCVYHFTDIPMFVSGENIVMFDPHASN 1513
S F+ D I+R+G+ K + +G+FGLGFN VYH TDIP+ +S E ++MFDP+ ++
Sbjct: 778 SQFSDSDFVNITRLGESLKRGEVDKVGKFGLGFNSVYHITDIPIIMSREFMIMFDPNINH 837
Query: 1514 LPG--ISPSHPGLRIKFA-GRKILEQFPDQFSSLLH-FGCDL------QHPFPGTLFRFP 1563
+ S+PG++I ++ +K L +FP+QF + FGC L + + GTLFR
Sbjct: 838 ISKHIKDKSNPGIKINWSKQQKRLRKFPNQFKPFIDVFGCQLPLTVEAPYSYNGTLFRLS 897
Query: 1564 LRTAGVASRSQIKKEVYTPEDVRSLFAAFSEVVSETLLFLRNVKSI 1609
RT A S++ Y D+ SL FS ++F ++VKS+
Sbjct: 898 FRTQQEAKVSEVSSTCYNTADIYSLVDEFSLCGHRLIIFTQSVKSM 943
Score = 162 bits (410), Expect = 1e-38
Identities = 101/292 (34%), Positives = 167/292 (56%), Gaps = 22/292 (7%)
Query: 5 SPESIFLEDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHAG 64
+PE++ E GQ LT RI+ +L YP + + KEL+QNADDA AT S +D R +
Sbjct: 695 NPENMGFEQSGQREPLTVRIKNILEEYPSVSDIFKELLQNADDANATECSFLIDMRRNMD 754
Query: 65 --DSLLSSSLSQWQGPALLAYNDAVFTEDDFISISKIGGSSKHGQASKTGRFGVGFNSVY 122
++LL ++ GPAL ++N++ F++ DF++I+++G S K G+ K G+FG+GFNSVY
Sbjct: 755 IRENLLDPGMAACHGPALWSFNNSQFSDSDFVNITRLGESLKRGEVDKVGKFGLGFNSVY 814
Query: 123 HLTDLPSFVSGKYVVLFDPQGVYLPR--VSAANPGKRIDFT-GSSALSLYKDQFSPYC-A 178
H+TD+P +S +++++FDP ++ + +NPG +I+++ L + +QF P+
Sbjct: 815 HITDIPIIMSREFMIMFDPNINHISKHIKDKSNPGIKINWSKQQKRLRKFPNQFKPFIDV 874
Query: 179 FGCDM----QSPFA--GTLFRFPLRNADQASRSKLSRQAYTPEDISSMFVQLFEEGVLTL 232
FGC + ++P++ GTLFR R +A S++S Y DI S+ + G +
Sbjct: 875 FGCQLPLTVEAPYSYNGTLFRLSFRTQQEAKVSEVSSTCYNTADIYSLVDEFSLCGHRLI 934
Query: 233 LFLKSVLCIEMYVWDAGEPKPKKIHSCSVSSVSDDTIWHRQALLRLSKSLNT 284
+F +SV MY+ K KI + S D I +++ SK+LNT
Sbjct: 935 IFTQSVK--SMYL------KYLKIEETNPSLAQDTVIIKKKSC--SSKALNT 976
Score = 74.7 bits (182), Expect = 3e-12
Identities = 43/153 (28%), Positives = 77/153 (50%), Gaps = 19/153 (12%)
Query: 363 GQAFCFLPLPVRTGLSVQVNGFFEVSSNRRGIWYGDDMDRSGKVRSIWNRLLLEDLVAPA 422
G+ FC+LPL ++TGL V +NG F V+SNR+ IW D R WN + ++ A
Sbjct: 1118 GEVFCYLPLRIKTGLPVHINGCFAVTSNRKEIWKTDTKGR-------WNTTFMRHVIVKA 1170
Query: 423 FVHMLHGVKELLGPTDI----YYSLWPTGSF-EEPWSILVQQIYINICNA-----PVIYS 472
++ +L +++L ++ YY++WP + +S++ Q Y +I + ++S
Sbjct: 1171 YLQVLSVLRDLATSGELMDYTYYAVWPDPDLVHDDFSVICQGFYEDIAHGKGKELTKVFS 1230
Query: 473 NLGGGRWVSPSEAFLHDEKFTKSKDLSLALMQL 505
+ G WVS D+ K +D+ A ++
Sbjct: 1231 D--GSTWVSMKNVRFLDDSILKRRDVGSAAFKI 1261
Score = 65.9 bits (159), Expect = 1e-09
Identities = 50/180 (27%), Positives = 85/180 (46%), Gaps = 26/180 (14%)
Query: 1773 GRAFCFLPLPISTGLPAHVNAYFELSSNRRDIWFGSDMTGGGRKRSDWNIYLLENVVAPA 1832
G FC+LPL I TGLP H+N F ++SNR++IW +D G WN + +V+ A
Sbjct: 1118 GEVFCYLPLRIKTGLPVHINGCFAVTSNRKEIW-KTDTKG------RWNTTFMRHVIVKA 1170
Query: 1833 YGRLLEKVALEIGPC-----YLFFSLWP-KTLGLEPWASVIRKLYQFVAEFNLRVL-YTE 1885
Y ++L V ++ Y ++++WP L + ++ + + Y+ +A + L
Sbjct: 1171 YLQVL-SVLRDLATSGELMDYTYYAVWPDPDLVHDDFSVICQGFYEDIAHGKGKELTKVF 1229
Query: 1886 ARGGQWISTKHAIFPDFSFPKADEL-----------IKALSGASLPVITLPQSLLERFME 1934
+ G W+S K+ F D S K ++ +K +L + LP S+ F E
Sbjct: 1230 SDGSTWVSMKNVRFLDDSILKRRDVGSAAFKIFLKYLKKTGSKNLCAVELPSSVKLGFEE 1289
Score = 46.2 bits (108), Expect = 0.001
Identities = 32/119 (26%), Positives = 64/119 (52%), Gaps = 15/119 (12%)
Query: 2077 LVSWTPGIHGQ--PSLEWLQLLWNYLKAN-CDDLLMFSKWPILP-----VGDDC--LIQL 2126
+V W P + PS+ WL+++W L + +DL +F + P++P G C LI+L
Sbjct: 11 IVQWYPFDENRNHPSVSWLKMVWKNLYIHFSEDLTLFDEMPLIPRTILEEGQTCVELIRL 70
Query: 2127 K-PNLNVIKNDGWS---EKMSSLLVKVGCLFLRP-DLQLDHPKLECFVQSPTARGVLNV 2180
+ P+L ++ ++ + E ++ ++ K+G L+ D + HP ++ ++ SP VL +
Sbjct: 71 RIPSLVILDDESEAQLPEFLADIVQKLGGFVLKKLDASIQHPLIKKYIHSPLPSAVLQI 129
>SACS_MOUSE (Q9JLC8) Sacsin
Length = 3830
Score = 282 bits (722), Expect = 7e-75
Identities = 353/1465 (24%), Positives = 591/1465 (40%), Gaps = 222/1465 (15%)
Query: 13 DFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGDSLLSSSL 72
+FGQ LT RI+ +L YP +LKEL+QNADDA AT + D R H D +
Sbjct: 1771 EFGQKEKLTSRIKSILNAYPSEKEMLKELLQNADDAKATEICFVFDPRQHPVDRIFDDKW 1830
Query: 73 SQWQGPALLAYNDAVFTEDDFISISKIGGSSKHGQASKTGRFGVGFNSVYHLTDLPSFVS 132
+ QGPAL YN+ FTEDD I +G +K G KTG +G+GFNSVYH+TD PSF+S
Sbjct: 1831 APLQGPALCVYNNQPFTEDDVRGIQNLGKGTKEGNPCKTGHYGIGFNSVYHITDCPSFIS 1890
Query: 133 GKYVV-LFDPQGVYLPRVSAANPGKRIDFTGSSALSLYKDQFSPYCAFGCDMQSPFAGTL 191
G ++ +FDP Y P ++ +PG+ + + + D Y + + T+
Sbjct: 1891 GNDILGIFDPHARYAPGATSVSPGRMFRDLDADFRTQFSDVLDLYLGNHFKLDNC---TM 1947
Query: 192 FRFPLRNADQASRSKLSRQAYTPEDISSMFVQLFEEGVLTLLFLKSVLCIEMYVWDAGEP 251
FRFPLRNA+ A S++S + + ++ +L +G L+FL + I + D
Sbjct: 1948 FRFPLRNAEMAQVSEISSVPSSDRMVQNLLDKLRSDGAELLMFLNHMEKISICEIDKATG 2007
Query: 252 KPKKIHSCSVSSVSDDTIWHRQALLRLSKSLNTTTEVDAFPLEFVTEAVRGVETVRQVDR 311
++S D + +Q + S+ ++ P++ +T + ++ +
Sbjct: 2008 GLNVLYSVKGKITDGDRLKRKQFHASVIDSVTKKRQLKDIPVQQITYTMDTEDSEGNLTT 2067
Query: 312 FYIVQTMA-SASSRIGSFAITASKEYDIQLLPWASIAACISDNSLNNDVLRTGQAFCFLP 370
+ I S+ ++ I+A K DI L P +AACI+ N + +AFCFLP
Sbjct: 2068 WLICNRSGFSSMEKVSKSVISAHKNQDITLFPRGGVAACITHN-----YKKPHRAFCFLP 2122
Query: 371 LPVRTGLSVQVNGFFEVSSNRRGIWYGDDMDRSGKVRSIWNRLLLEDLVAPAFVHM---- 426
L + TGL VNG F + S RR +W D+ VRS WN L+ L+APA+V +
Sbjct: 2123 LSLETGLPFHVNGHFALDSARRNLWRDDN---GVGVRSDWNNSLMTALIAPAYVELLIQL 2179
Query: 427 -------------------LHGVKELLGPTDIYYSLWPTGSFEEPWSI--LVQQIYINIC 465
+H VK+ L + S +P + + LV+ +Y I
Sbjct: 2180 KKRYFPGSDPTLSVLQNTPIHVVKDTLKK---FLSFFPVNRLDLQPDLYCLVKALYSCIH 2236
Query: 466 N-----APVIYS-NLGGG--------RWVSPSEA---------FLHDE-KFTKSKDLSLA 501
PV+ + N+ G W++ S + L DE + K+ D ++
Sbjct: 2237 EDMKRLLPVVRAPNIDGSDLHSAVIITWINMSTSNKTRPFFDNLLQDELQHLKNADYNIT 2296
Query: 502 ---------------LMQLGMPVVHLPN---SLFDMLLKYNSS-KVITPGTVRQFLRECE 542
L+++G +V+ + +L+ L+ + +TP VR FL
Sbjct: 2297 TRKTVAENVYRLKHLLLEIGFNLVYNCDETANLYHCLVDADIPVSYVTPADVRSFLMTFS 2356
Query: 543 S----CN-----------HLSRAHKL-LLLEYCLEDLVDDDVGKAAYNLPLLPLANGNFA 586
S C+ +L H L LL++YC +D + + LPLL +
Sbjct: 2357 SPDTNCHIGKLPCRLQQTNLKLFHSLKLLVDYCFKDAEESEF--EVEGLPLLITLD---- 2410
Query: 587 SFLEASKGI-PYFICDELEYKLLEPVSDRVIDQSIPPNILTRLSGIAMSSNTNIA-LFSI 644
S L+ G P F+ ++L+ D ++ + + S + + N +A +F I
Sbjct: 2411 SVLQIFDGKRPKFLTTY--HELIPSRKDLFMN-----TLYLKYSSVLL--NCKVAKVFDI 2461
Query: 645 HHFAHLFPVFMPDDWKYKCKVFWDPDSCQKPTSSWFVLFWQYLGK-------QSEILPLF 697
FA L +P ++K K W + + SW W ++ + Q E P F
Sbjct: 2462 SSFADLLSSVLPREYKTKNCAKWKDNFASE---SWLKNAWHFISESVSVTDDQEEPKPAF 2518
Query: 698 -------KDWPILPSTS------------GHLLRPSRQLKM-INGSTLSDTVQDILVKIG 737
KDW +LP T G +L P + + + + SD V L+K G
Sbjct: 2519 DVIVDILKDWALLPGTKFTVSTSQLVVPEGDVLIPLSLMHIAVFPNAQSDKVFHALMKAG 2578
Query: 738 CHILKPGYVVEHPDLFSYLCGGNAAGVLESIFNAFSSAENMQVSFSSLIAEE--RNELRR 795
C L + L + A + + A + V S+ E+ N+
Sbjct: 2579 CIQLALNKICSKDSALVPLLSCHTANIDSPA--SILKAVHYMVQTSTFRTEKLMENDFEA 2636
Query: 796 FLLDPQWYVGHSMDEFNIRFCRRLPIYQVYHREPTQDSQFSDLENPRKYLPPLDVPEFIL 855
L+ + H M + +I+ + LP Y+ ++F K +P +V ++
Sbjct: 2637 LLMYFNCNLSHLMSQDDIKILKSLPCYKSISGRYMSIAKFGTCYVLTKSIPSAEVEKWTQ 2696
Query: 856 VGIE-FIVRSSNTEEDILSRYYGVERMGKAQFYKKHVFDRVGELQAEDRDSIMLSVLQNL 914
F+ + +E L G + + Y KH+ ++ L + D L L+N
Sbjct: 2697 SSSSAFLEEKVHLKE--LYEVLGCVPVDDLEVYLKHLLPKIENL-SYDAKLEHLIYLKNR 2753
Query: 915 PLLSLEDAS-----IRDLLRNLKFIPTVIGTLKCPSVLYDPRNEEIYALLEDSDSFPSGV 969
L S+E+ S + + L +L I LK YD +L + P
Sbjct: 2754 -LASIEEPSEIKEQLFEKLESLLIIHDANNRLKQAKHFYDRTVRVFEVMLPEKLFIPKEF 2812
Query: 970 FRESETL--------------DIMRGLGLKTSVSPDTVLESARCIEHLMH-EDQQKAYLK 1014
F++ E + + +R +GLK ++S +L+ A+ I + E+ K L+
Sbjct: 2813 FKKLEQVIKPKNQAAFMTSWVEFLRNIGLKYALSQQQLLQFAKEISVRANTENWSKETLQ 2872
Query: 1015 GKV--LFSYLEVNALKWLPDKFDDKKGAVNRILSQAATAFRSRNTKSDIEKFWNDLQLIS 1072
V L ++ + L F + + + + A A R ++ L LI
Sbjct: 2873 STVDILLHHIFQERMDLLSGNFLKELSLIPFLCPERAPAEYIR-FHPQYQEVNGTLPLIK 2931
Query: 1073 WCPVLVSPPFHS-----LPW---PVVSSMVAPPKVVRPPNDLWLVSAGMRILDGECSSTA 1124
+ V+P F L W P++ P + G + E
Sbjct: 2932 FNGAQVNPKFKQCDVLQLLWTSCPILPEKATPLSIKE--------QEGSDLAPQEQLEQV 2983
Query: 1125 LLYCLGWMCPPGGGVIAAQLLELGKNN--EIVTDQVLRQELALAMPRIYSILTGMIGSDE 1182
L + PP VI NN I L +E+ ++ + + +++
Sbjct: 2984 LNMLNVNLDPPLDKVI---------NNCRNICNITTLDEEMVKTRAKVLRSIYEFLSAEK 3034
Query: 1183 IEIVKAVLEGCRWIWVGDGFAT--SDEVV--LDGPLHLAPYIRVIPVDLAVFKNLFLELG 1238
E + L G ++ V DG+ +EVV L+ PY+ +P++L F LF LG
Sbjct: 3035 REF-RFQLRGVAFVMVEDGWKLLKPEEVVINLEYEADFKPYLYKLPLELGTFHQLFKHLG 3093
Query: 1239 IREFLQPSDYVNILFRMANKKGSSPLDTQEIRAVMLIVHHLAEVYL-----------HGQ 1287
+ + YV +L R+ LD E+R V +V L + + +
Sbjct: 3094 TEDIISTKQYVEVLSRIFKSSEGKQLDPNEMRTVKRVVSGLFKSLQNDSVKVRSDLENAR 3153
Query: 1288 KVQLYLPDVSGRLFLAGDLVYNDAP 1312
+ LYLP G+L + LV++DAP
Sbjct: 3154 DLALYLPSQDGKLVKSSILVFDDAP 3178
Score = 234 bits (597), Expect = 2e-60
Identities = 265/992 (26%), Positives = 412/992 (40%), Gaps = 159/992 (16%)
Query: 885 QFYKKHVFDRVGELQAEDRDSIMLSVLQNLPLLSLEDASIRDLLRNLKFIPTVIG--TLK 942
QF+ + F + E++AE RD +M VL L++ S +LR +P + L
Sbjct: 1305 QFFSEVFFPNIQEIEAELRDPLMNFVLNE----KLDEFS--GILRVTPCVPCSLEGHPLV 1358
Query: 943 CPSVLYDPRNEEIYALLEDSDS-FPSGVFRESETLDIMRGL---GL-KTSVSPDTVLESA 997
PS L P + L + D FP G ++ I+ L G+ K + D +LE A
Sbjct: 1359 LPSRLIHPEGR-VAKLFDTKDGRFPYGSTQDYLNPIILIKLVQLGMAKDDILWDDMLERA 1417
Query: 998 RCIEHLMHEDQQKAYLKGKVLFSYLEVNALKWLPDKFDDKKGAVNRILSQAATAFRSRNT 1057
+ + D A L+ +L S + D+K +I A F ++
Sbjct: 1418 ESVAEINKSDHAAACLRSSILLSLI------------DEKL----KIKDPRAKDFAAK-- 1459
Query: 1058 KSDIEKFWNDLQLISWCPVLVSPPFHSLPWPVVSSMVAPPKVVRPPNDLW------LVSA 1111
Q I + P L P SL W S P+ + D++ +V
Sbjct: 1460 ----------YQTIPFLPFLTKPAGFSLEWKGNSFK---PETMFAATDIYTAEYQDIVCL 1506
Query: 1112 GMRILDGE------CSSTALLY--CLGWMCPPGGGVIAAQLLELGKNNEIVTDQVLRQEL 1163
IL+ C S +L LG + P ++ QL ++ K+ V D + +
Sbjct: 1507 LQPILNENSHSFRGCGSVSLAVKEFLGLLKKPTVDLVINQLKQVAKS---VDDGITLYQE 1563
Query: 1164 ALAMPRIYSILTGMIGSDEIE---IVKAVLEGCRWIWVGDGFATSDEVVLDGPLHLAPYI 1220
+ Y L + +E+ I++ + C +I V + + S++V APY+
Sbjct: 1564 NITNA-CYKYLHEAVLQNEMAKATIIEKLKPFC-FILVENVYVESEKVSFHLNFEAAPYL 1621
Query: 1221 RVIPVDLAV-FKNLFLELGIREFLQPSDYVNILFRMANKKGSSPLDTQEIR--------A 1271
+P F+ LF +G+R+ D+ +L + ++G + + +
Sbjct: 1622 YQLPNKYKNNFRELFESVGVRQSFTVEDFALVLESIDQERGKKQITEENFQLCRRIISEG 1681
Query: 1272 VMLIVHHLAEVYLHGQKVQLYLPDVSGRLFLAGDLVYNDAPWLLGSEDPDGSFGNAPSVT 1331
+ ++ + + ++ LPD + L A L YND PW+
Sbjct: 1682 IWSLIREKRQEFCEKNYGKILLPDTNLLLLPAKSLCYNDCPWI----------------- 1724
Query: 1332 WNAKRTVQKFVHGNISNDVAEKLGVRSLRRMLLAESADSMNFGLSGAAEAFGQHEALTTR 1391
K + K+ H +I +VA KLG R L A ++ F G FGQ E LT+R
Sbjct: 1725 -KVKDSTVKYCHADIPREVAVKLGAIPKRHKALERYASNICFTALGTE--FGQKEKLTSR 1781
Query: 1392 LKHILEMYADGPGTLFELVQNAEDAGASEVIFLLDKSQYGTSSVLSPEMADWQGPALYCF 1451
+K IL Y L EL+QNA+DA A+E+ F+ D Q+ + + A QGPAL +
Sbjct: 1782 IKSILNAYPSEKEMLKELLQNADDAKATEICFVFDPRQHPVDRIFDDKWAPLQGPALCVY 1841
Query: 1452 NDSVFTPQDLYAISRIGQESKLEKAFAIGRFGLGFNCVYHFTDIPMFVSGENIV-MFDPH 1510
N+ FT D+ I +G+ +K G +G+GFN VYH TD P F+SG +I+ +FDPH
Sbjct: 1842 NNQPFTEDDVRGIQNLGKGTKEGNPCKTGHYGIGFNSVYHITDCPSFISGNDILGIFDPH 1901
Query: 1511 ASNLPGISPSHPGLRIKFAGRKILEQFPDQFSSLL--HFGCDLQHPFPGTLFRFPLRTAG 1568
A PG + PG + QF D L HF D T+FRFPLR A
Sbjct: 1902 ARYAPGATSVSPGRMFRDLDADFRTQFSDVLDLYLGNHFKLD-----NCTMFRFPLRNAE 1956
Query: 1569 VASRSQIKKEVYTPEDVRSLFAAFSEVVSETLLFLRNVKSISIFLKEGTGHEMRLLHRVS 1628
+A S+I + V++L +E L+FL +++ ISI + + +L+ V
Sbjct: 1957 MAQVSEISSVPSSDRMVQNLLDKLRSDGAELLMFLNHMEKISICEIDKATGGLNVLYSV- 2015
Query: 1629 RASLGESEIGSAEVQDVFNFFKEDRLVGMNRAQFLKKLSLSIDRDLPYKCQKILITEQGT 1688
++ D DRL R QF + ID + K + +Q T
Sbjct: 2016 ----------KGKITD------GDRL---KRKQFHASV---IDSVTKKRQLKDIPVQQIT 2053
Query: 1689 HGRNSHYWIMTECLGGGNVLKGTSEASTSNSHNFVPW-ACVAAYLNSVKHGEDLVDSAEV 1747
Y + TE + N W C + +S++ V SA
Sbjct: 2054 ------YTMDTE----------------DSEGNLTTWLICNRSGFSSMEKVSKSVISAHK 2091
Query: 1748 EDDCLVSSDLFQFASLPMHPRENFE--GRAFCFLPLPISTGLPAHVNAYFELSSNRRDIW 1805
D LF + N++ RAFCFLPL + TGLP HVN +F L S RR++W
Sbjct: 2092 NQDI----TLFPRGGVAACITHNYKKPHRAFCFLPLSLETGLPFHVNGHFALDSARRNLW 2147
Query: 1806 FGSDMTGGGRKRSDWNIYLLENVVAPAYGRLL 1837
+ G RSDWN L+ ++APAY LL
Sbjct: 2148 RDDNGVG---VRSDWNNSLMTALIAPAYVELL 2176
Score = 214 bits (546), Expect = 2e-54
Identities = 263/1008 (26%), Positives = 435/1008 (43%), Gaps = 139/1008 (13%)
Query: 665 VFWDPDSCQK--PTSSWFVLFWQYLGKQ-SEILPLFKDWPILP-------STSGHLLR-- 712
V W P S K P+ SW + W+ L SE L LF + P++P T L+R
Sbjct: 12 VQWYPFSEDKRHPSLSWLKMVWKNLYIHFSEDLTLFDEMPLIPRTLLNEDQTCVELIRLR 71
Query: 713 -PSRQLKMINGST---LSDTVQDILVKIGCHILKP-GYVVEHPDLFSYLCGGNAAGVLES 767
PS + +++ T L + + DI+ K+G +LK ++HP + Y+ + +L+
Sbjct: 72 IPS--VVILDDETEAQLPEFLADIVQKLGGIVLKRLDTSIQHPLVKKYIHSPLPSAILQI 129
Query: 768 IFNAFSSAENMQVSFSSLIAEERNELRRFLLDPQWYVGHSMDEFNIRFCRRLPIYQ-VYH 826
+ + + +SL+ ++ LR+FL E R + L I++ + H
Sbjct: 130 MEKI--PLQKLCNQIASLLPTHKDALRKFLASLT-----DTSEKEKRIIQELTIFKRINH 182
Query: 827 REPTQDSQFSDLENPRKYLPPLDVPEFILVGIEFIVRSSNTEEDILSRYYGVERMGKAQF 886
S ++ L+ + +P + + + ++ SS+ L+ +E++ K
Sbjct: 183 SSDQGISSYTKLKGCKVLDHTAKLPTDLRLSVS-VIDSSDEATIRLANMLKIEKL-KTTS 240
Query: 887 YKKHVFDRVGEL--QAEDRDSIMLSVLQNLPLLSLEDASIRDLLRNLKFIPTVIGTLKCP 944
K V +G E+ +ML +L+NL L E++++ D L LKFI G +
Sbjct: 241 CLKFVLKDIGNAFYTQEEVTQLMLWILENLSSLKNENSNVLDWLMPLKFIHMSQGHVVAA 300
Query: 945 SVLYDPRNEEIYALL--EDSDSFPSGVFRESETLDIMRGLGLK--TSVSPDTVLESARCI 1000
L+DP E + L E+ FP +F + L +R +GLK +S+ V++ AR I
Sbjct: 301 GDLFDPDIEVLRDLFYNEEEACFPPTIFTSPDILHSLRQIGLKNESSLKEKDVVQVARKI 360
Query: 1001 EHLMHEDQQKAYLKGKVLFSYLEVNALKWLPDKFDDKKGAVNRILSQAATAFRSRNTKSD 1060
E L Q D K + +L++ T +S K
Sbjct: 361 EALQVSSCQNQ--------------------DVLMKKAKTLLLVLNKNQTLLQSSEGKMA 400
Query: 1061 IEKFWNDLQLISWCPVLVSPPFH---SLPWPV-VSSMVAPPKVVRPPNDLWLVSAGMRIL 1116
++K I W P P + SL W + ++ APP + + + LV + + ++
Sbjct: 401 LKK-------IKWVPACKERPPNYPGSLVWKGDLCNLCAPPDMCDAAHAV-LVGSSLPLV 452
Query: 1117 DGECSSTALLYCLGWMCPPGGGVIAAQLLELGKNNEIVTDQVLRQELALAMPRIYSILTG 1176
E L L P I A L + T + E I + G
Sbjct: 453 --ESVHVNLEQALSIFTKP---TINAVLKHFKTVVDWYTSKTFSDEDYYQFQHILLEIYG 507
Query: 1177 MI------GSDEIEIVKAVLEGCRWIWVGDGFATSDEVVLDGPLH---LAPYIRVIPVDL 1227
+ G D + +K W+W G F + V+ P H L PY+ +P +
Sbjct: 508 FMHDHLSEGKDSFKALKFP-----WVWTGKNFCPLAQAVIK-PTHDLDLQPYLYNVPKTM 561
Query: 1228 AVFKNLFLELGIREFLQPSDYVNILFRMANKKGSSPLDTQEIRAVMLIVHHLAEVYLHGQ 1287
A F LF G E L SD+++++ + K L +E + + ++ ++ Q
Sbjct: 562 AKFHQLFKACGSIEELT-SDHISMVIQKVYLKSDQELSEEESKQNLHLMLNIMRWLYSNQ 620
Query: 1288 -------KVQLYLPDVSGRLFLAG-------DLVYNDAPWLLGSEDPDGSFGNAPSVTWN 1333
V +Y +L + D+ +D LL ED P +
Sbjct: 621 IPASPNTPVPIYHSRNPSKLVMKPIHECCYCDIKVDDLNDLL--ED-----SVEPII--- 670
Query: 1334 AKRTVQKFVHGNISNDVAEKLGVRSLRRMLLAESADSMNFGLSGAAEAFGQHEALTTRLK 1393
VH +I AE L V L L+ + ++M F E GQ E LT R+K
Sbjct: 671 -------LVHEDIPMKTAEWLKVPCLSTRLI--NPENMGF------EQSGQREPLTVRIK 715
Query: 1394 HILEMYADGPGTLFELVQNAEDAGASEVIFLLD--KSQYGTSSVLSPEMADWQGPALYCF 1451
+ILE Y EL+QNA+DA A+E F++D ++ ++L P MA GPAL+ F
Sbjct: 716 NILEEYPSVSDIFKELLQNADDANATECSFMIDMRRNMDIRENLLDPGMAACHGPALWSF 775
Query: 1452 NDSVFTPQDLYAISRIGQESKLEKAFAIGRFGLGFNCVYHFTDIPMFVSGENIVMFDPHA 1511
N+S F+ D I+R+G+ K + +G+FGLGFN VYH TDIP+ +S E ++MFDP+
Sbjct: 776 NNSEFSDSDFLNITRLGESLKRGEVDKVGKFGLGFNSVYHITDIPIIMSREFMIMFDPNI 835
Query: 1512 SNLPG--ISPSHPGLRIKFA-GRKILEQFPDQFSSLLH-FGCDL------QHPFPGTLFR 1561
+++ S+PG++I ++ +K L +FP+QF + FGC L + + GTLFR
Sbjct: 836 NHISKHIKDRSNPGIKINWSKQQKRLRKFPNQFKPFIDVFGCQLPLAVEAPYSYNGTLFR 895
Query: 1562 FPLRTAGVASRSQIKKEVYTPEDVRSLFAAFSEVVSETLLFLRNVKSI 1609
RT A S++ Y D+ SL FS ++F ++V S+
Sbjct: 896 LSFRTQQEAKVSEVSSTCYNTADIYSLVDEFSLCGHRLIIFTQSVNSM 943
Score = 159 bits (403), Expect = 7e-38
Identities = 98/291 (33%), Positives = 166/291 (56%), Gaps = 22/291 (7%)
Query: 5 SPESIFLEDFGQTVDLTRRIREVLLNYPEGTTVLKELIQNADDAGATTVSLCLDRRSHAG 64
+PE++ E GQ LT RI+ +L YP + + KEL+QNADDA AT S +D R +
Sbjct: 695 NPENMGFEQSGQREPLTVRIKNILEEYPSVSDIFKELLQNADDANATECSFMIDMRRNMD 754
Query: 65 --DSLLSSSLSQWQGPALLAYNDAVFTEDDFISISKIGGSSKHGQASKTGRFGVGFNSVY 122
++LL ++ GPAL ++N++ F++ DF++I+++G S K G+ K G+FG+GFNSVY
Sbjct: 755 IRENLLDPGMAACHGPALWSFNNSEFSDSDFLNITRLGESLKRGEVDKVGKFGLGFNSVY 814
Query: 123 HLTDLPSFVSGKYVVLFDPQGVYLPR--VSAANPGKRIDFT-GSSALSLYKDQFSPYC-A 178
H+TD+P +S +++++FDP ++ + +NPG +I+++ L + +QF P+
Sbjct: 815 HITDIPIIMSREFMIMFDPNINHISKHIKDRSNPGIKINWSKQQKRLRKFPNQFKPFIDV 874
Query: 179 FGCDM----QSPFA--GTLFRFPLRNADQASRSKLSRQAYTPEDISSMFVQLFEEGVLTL 232
FGC + ++P++ GTLFR R +A S++S Y DI S+ + G +
Sbjct: 875 FGCQLPLAVEAPYSYNGTLFRLSFRTQQEAKVSEVSSTCYNTADIYSLVDEFSLCGHRLI 934
Query: 233 LFLKSVLCIEMYVWDAGEPKPKKIHSCSVSSVSDDTIWHRQALLRLSKSLN 283
+F +SV + + E P S++ DTI ++ + SK+LN
Sbjct: 935 IFTQSVNSMYLKYLKIEETNP---------SLAQDTIIIKKKVCP-SKALN 975
Score = 70.1 bits (170), Expect = 7e-11
Identities = 41/153 (26%), Positives = 76/153 (48%), Gaps = 19/153 (12%)
Query: 363 GQAFCFLPLPVRTGLSVQVNGFFEVSSNRRGIWYGDDMDRSGKVRSIWNRLLLEDLVAPA 422
G+ FC+LPL ++TGL + +NG F V+SNR+ IW D R WN + ++ A
Sbjct: 1118 GEVFCYLPLRIKTGLPIHINGCFAVTSNRKEIWKTDTKGR-------WNTTFMRHVIVKA 1170
Query: 423 FVHMLHGVKELLGPTDI----YYSLWPTGSF-EEPWSILVQQIYINICNA-----PVIYS 472
++ L +++L ++ YY++WP + +S++ + Y +I + ++S
Sbjct: 1171 YLQALSVLRDLAIGGELTDYTYYAVWPDPDLVHDDFSVICKGFYEDIAHGKGKELTRVFS 1230
Query: 473 NLGGGRWVSPSEAFLHDEKFTKSKDLSLALMQL 505
+ G WVS D+ + KD+ A ++
Sbjct: 1231 D--GSMWVSMKNVRFLDDSILQRKDVGSAAFKI 1261
Score = 67.4 bits (163), Expect = 4e-10
Identities = 51/179 (28%), Positives = 85/179 (46%), Gaps = 24/179 (13%)
Query: 1773 GRAFCFLPLPISTGLPAHVNAYFELSSNRRDIWFGSDMTGGGRKRSDWNIYLLENVVAPA 1832
G FC+LPL I TGLP H+N F ++SNR++IW +D G WN + +V+ A
Sbjct: 1118 GEVFCYLPLRIKTGLPIHINGCFAVTSNRKEIW-KTDTKG------RWNTTFMRHVIVKA 1170
Query: 1833 YGRLLEKVA-LEIG---PCYLFFSLWP-KTLGLEPWASVIRKLYQFVAEFNLRVL-YTEA 1886
Y + L + L IG Y ++++WP L + ++ + + Y+ +A + L +
Sbjct: 1171 YLQALSVLRDLAIGGELTDYTYYAVWPDPDLVHDDFSVICKGFYEDIAHGKGKELTRVFS 1230
Query: 1887 RGGQWISTKHAIFPDFSFPKADEL-----------IKALSGASLPVITLPQSLLERFME 1934
G W+S K+ F D S + ++ +K +L + LP S+ F E
Sbjct: 1231 DGSMWVSMKNVRFLDDSILQRKDVGSAAFKIFLKYLKKTGSKNLCAVELPSSVKAGFEE 1289
Score = 44.7 bits (104), Expect = 0.003
Identities = 32/119 (26%), Positives = 59/119 (48%), Gaps = 15/119 (12%)
Query: 2077 LVSWTPGIHGQ--PSLEWLQLLWNYLKAN-CDDLLMFSKWPILP---VGDD--C--LIQL 2126
+V W P + PSL WL+++W L + +DL +F + P++P + +D C LI+L
Sbjct: 11 VVQWYPFSEDKRHPSLSWLKMVWKNLYIHFSEDLTLFDEMPLIPRTLLNEDQTCVELIRL 70
Query: 2127 KPNLNVIKNDGWSEKMSSLLVKV-----GCLFLRPDLQLDHPKLECFVQSPTARGVLNV 2180
+ VI +D ++ L + G + R D + HP ++ ++ SP +L +
Sbjct: 71 RIPSVVILDDETEAQLPEFLADIVQKLGGIVLKRLDTSIQHPLVKKYIHSPLPSAILQI 129
>AMO_PICAN (P12807) Peroxisomal copper amine oxidase (EC 1.4.3.6)
(Methylamine oxidase)
Length = 692
Score = 40.4 bits (93), Expect = 0.059
Identities = 34/111 (30%), Positives = 53/111 (47%), Gaps = 18/111 (16%)
Query: 791 NELRRFLLDPQWYVGHSMDEFNIRFCRRLPIYQVYHREPTQDSQFSDLENPRKYLPPLDV 850
NE+ + DP W +G+ DE +RL VY+R DSQ+S +P + P +D
Sbjct: 146 NEMHKVYCDP-WTIGY--DE-RWGTGKRLQQALVYYRSDEDDSQYS---HPLDFCPIVDT 198
Query: 851 PEFILVGIEFIVRSSNTEEDILSRYYGVERMGKAQFYKKHVFDRVGELQAE 901
E ++ I DI +R V + A FY KH+ ++VG ++ E
Sbjct: 199 EEKKVIFI-----------DIPNRRRKVSKHKHANFYPKHMIEKVGAMRPE 238
>TFR1_CHICK (Q90997) Transferrin receptor protein 1 (TfR1) (TR) (TfR)
(Trfr)
Length = 776
Score = 38.1 bits (87), Expect = 0.29
Identities = 40/173 (23%), Positives = 66/173 (38%), Gaps = 21/173 (12%)
Query: 1324 FGNAPSVTWNAKRTVQKFVHGNISNDVAEKLG----VRSLRRMLLAESADS--------M 1371
F N WN + ++ V G+ N V+ + + ++A S +
Sbjct: 183 FRNFLDKVWNDEHYIKLQVRGSTKNQVSISINGKEEILETPDAIVAYSESGSVSGKPVYV 242
Query: 1372 NFGLSGAAEAFGQHEALTTRLKHILEMYADGPGTLFELVQNAEDAGASEVIFLLDKSQYG 1431
N+GL E + + L + + G TL E V NA++AGA+ V+ +D YG
Sbjct: 243 NYGLKKDFEII---QKVVASLNGTIVIVRAGKITLAEKVANAKEAGAAGVLMYVDSLMYG 299
Query: 1432 TSSVLSP------EMADWQGPALYCFNDSVFTPQDLYAISRIGQESKLEKAFA 1478
+ L P D P FN + F P + + I ++ A A
Sbjct: 300 ITDTLIPFGHAHLGTGDPYTPGFPSFNHTQFPPVESSGLPHIAVQTISSSAAA 352
>XTH1_LYCES (Q40144) Probable xyloglucan
endotransglucosylase/hydrolase 1 precursor (EC
2.4.1.207) (LeXTH1)
Length = 296
Score = 37.0 bits (84), Expect = 0.65
Identities = 21/72 (29%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query: 714 SRQLKMINGSTLSDTVQDILVKIGCHILKPGYVVEHPDLFSYLCGGNAAGVLESIFNAFS 773
S +K +NG T +D + D G K Y+ H + L GG++AGV+ + + + +
Sbjct: 43 SHHIKFLNGGTTTDLILDRSSGAGFQS-KKSYLFGHFSMKMRLVGGDSAGVVTAFYLSSN 101
Query: 774 SAENMQVSFSSL 785
+AE+ ++ F L
Sbjct: 102 NAEHDEIDFEFL 113
>ENPL_HORVU (P36183) Endoplasmin homolog precursor (GRP94 homolog)
Length = 809
Score = 35.8 bits (81), Expect = 1.4
Identities = 37/127 (29%), Positives = 58/127 (45%), Gaps = 17/127 (13%)
Query: 23 RIREVLLN--YPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGD---SLLSSSLSQWQG 77
R+ ++++N Y L+ELI NA DA L L + G+ + L + +
Sbjct: 88 RLMDIIINSLYSNKDIFLRELISNASDALDKIRFLALTDKEVMGEGDTAKLEIQIKLDKE 147
Query: 78 PALLAYND--AVFTEDDFI----SISKIGGSS------KHGQASKTGRFGVGFNSVYHLT 125
+L+ D T++D I +I+K G S+ G + G+FGVGF SVY +
Sbjct: 148 NKILSIRDRGVGMTKEDLIKNLGTIAKSGTSAFVEKMQTGGDLNLIGQFGVGFYSVYLVA 207
Query: 126 DLPSFVS 132
D VS
Sbjct: 208 DYVEVVS 214
Score = 33.9 bits (76), Expect = 5.5
Identities = 50/176 (28%), Positives = 74/176 (41%), Gaps = 23/176 (13%)
Query: 1350 VAEKLG-----VRSLRRMLLAESADSMNFGLSGAAEAFGQHEALTTRLKHIL--EMYADG 1402
V EKLG + + ++ ES L +AE F + +A +RL I+ +Y++
Sbjct: 43 VEEKLGAVPHGLSTDSEVVQRESESISRKTLRNSAEKF-EFQAEVSRLMDIIINSLYSNK 101
Query: 1403 PGTLFELVQNAEDA-GASEVIFLLDKSQYGTSSVLSPEMA---DWQGPALYCFNDSV-FT 1457
L EL+ NA DA + L DK G E+ D + L + V T
Sbjct: 102 DIFLRELISNASDALDKIRFLALTDKEVMGEGDTAKLEIQIKLDKENKILSIRDRGVGMT 161
Query: 1458 PQDLY----AISRIGQESKLEKAFA------IGRFGLGFNCVYHFTDIPMFVSGEN 1503
+DL I++ G + +EK IG+FG+GF VY D VS N
Sbjct: 162 KEDLIKNLGTIAKSGTSAFVEKMQTGGDLNLIGQFGVGFYSVYLVADYVEVVSKHN 217
>ENPL_CATRO (P35016) Endoplasmin homolog precursor (GRP94 homolog)
Length = 817
Score = 35.4 bits (80), Expect = 1.9
Identities = 36/127 (28%), Positives = 58/127 (45%), Gaps = 17/127 (13%)
Query: 23 RIREVLLN--YPEGTTVLKELIQNADDAGATTVSLCLDRRSHAGD---SLLSSSLSQWQG 77
R+ ++++N Y L+ELI NA DA L L + G+ + L + +
Sbjct: 88 RLMDIIINSLYSNKDIFLRELISNASDALDKIRFLALTDKEILGEGDTAKLEIQIKLDKE 147
Query: 78 PALLAYNDAVF--TEDDFI----SISKIGGSS------KHGQASKTGRFGVGFNSVYHLT 125
+L+ D T++D I +I+K G S+ G + G+FGVGF SVY +
Sbjct: 148 KKILSIRDRGIGMTKEDLIKNLGTIAKSGTSAFVEKMQTSGDLNLIGQFGVGFYSVYLVP 207
Query: 126 DLPSFVS 132
D +S
Sbjct: 208 DYVEVIS 214
>FMT_WOLSU (Q7MA26) Methionyl-tRNA formyltransferase (EC 2.1.2.9)
Length = 305
Score = 33.9 bits (76), Expect = 5.5
Identities = 19/56 (33%), Positives = 28/56 (49%), Gaps = 1/56 (1%)
Query: 1815 RKRSDWNIYLLENVVAPAYGRLLEKVALEIGPCY-LFFSLWPKTLGLEPWASVIRK 1869
R +W + +V AYG++L KV L+I PC L S+ P G P +R+
Sbjct: 73 RWAKEWRALEPDFIVVAAYGKILPKVILDIAPCINLHASILPLYRGASPIHESLRR 128
>FMT_CAMJE (Q9PJ28) Methionyl-tRNA formyltransferase (EC 2.1.2.9)
Length = 305
Score = 33.9 bits (76), Expect = 5.5
Identities = 18/41 (43%), Positives = 23/41 (55%), Gaps = 1/41 (2%)
Query: 1828 VVAPAYGRLLEKVALEIGPCY-LFFSLWPKTLGLEPWASVI 1867
+V AYG++L K L++ PC L SL PK G P S I
Sbjct: 86 IVVAAYGKILPKAILDLAPCVNLHASLLPKYRGASPIQSAI 126
>Y039_BORBU (O51068) Hypothetical protein BB0039
Length = 500
Score = 33.1 bits (74), Expect = 9.4
Identities = 20/58 (34%), Positives = 35/58 (59%), Gaps = 5/58 (8%)
Query: 2272 SDMERVIMRRYLEIKEPTRVEFYKDHIFNRMSEFL--LKPEVVSSILNDVQLLIKEDI 2327
SD + ++ Y+E EP+ +E Y + N++S+FL +K +S +L QL +KE+I
Sbjct: 335 SDRDISSVKNYIETYEPSVIEGYLE---NKLSDFLGDVKFSSLSQVLEKYQLSLKEEI 389
>ARA2_BACHD (Q9K9D5) 3-phosphoshikimate 1-carboxyvinyltransferase 2
(EC 2.5.1.19) (5-enolpyruvylshikimate-3-phosphate
synthase 2) (EPSP synthase 2) (EPSPS 2)
Length = 447
Score = 33.1 bits (74), Expect = 9.4
Identities = 23/75 (30%), Positives = 34/75 (44%), Gaps = 9/75 (12%)
Query: 265 SDDTIWHRQALLRLSKSLNTTTEVDAFPLEFVTEAVRGVETVRQVDRFYIVQTMASASSR 324
SDDT W QAL RL +N E T ++RG+ + YI A
Sbjct: 65 SDDTYWCIQALKRLGVQINVQGE---------TTSIRGIGGQWKSSSLYIGAAGTLARFL 115
Query: 325 IGSFAITASKEYDIQ 339
+G+ AI+ S ++I+
Sbjct: 116 LGALAISRSGNWEIE 130
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 279,765,737
Number of Sequences: 164201
Number of extensions: 12361307
Number of successful extensions: 25864
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 25799
Number of HSP's gapped (non-prelim): 38
length of query: 2349
length of database: 59,974,054
effective HSP length: 127
effective length of query: 2222
effective length of database: 39,120,527
effective search space: 86925810994
effective search space used: 86925810994
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0237.12


